Abstract
Nonphotosynthetic plastids are important sites for the biosynthesis of starch, fatty acids, and amino acids. The uptake and subsequent use of cytosolic ATP to fuel these and other anabolic processes would lead to the accumulation of inorganic phosphate (Pi) if not balanced by a Pi export activity. However, the identity of the transporter(s) responsible for Pi export is unclear. The plastid-localized Pi transporter PHT4;2 of Arabidopsis (Arabidopsis thaliana) is expressed in multiple sink organs but is nearly restricted to roots during vegetative growth. We identified and used pht4;2 null mutants to confirm that PHT4;2 contributes to Pi transport in isolated root plastids. Starch accumulation was limited in pht4;2 roots, which is consistent with the inhibition of starch synthesis by excess Pi as a result of a defect in Pi export. Reduced starch accumulation in leaves and altered expression patterns for starch synthesis genes and other plastid transporter genes suggest metabolic adaptation to the defect in roots. Moreover, pht4;2 rosettes, but not roots, were significantly larger than those of the wild type, with 40% greater leaf area and twice the biomass when plants were grown with a short (8-h) photoperiod. Increased cell proliferation accounted for the larger leaf size and biomass, as no changes were detected in mature cell size, specific leaf area, or relative photosynthetic electron transport activity. These data suggest novel signaling between roots and leaves that contributes to the regulation of leaf size.
Unlike chloroplasts, nonphotosynthetic plastids cannot synthesize the ATP needed to fuel anabolic processes within these organelles, which include the biosynthesis of starch, fatty acids, and amino acids (Neuhaus and Emes, 2000). Metabolism in such plastids, therefore, is dependent on the import of ATP from the cytosol. Nucleotide transporters (NTTs) located in the plastid inner envelope fulfill this energy requirement by catalyzing the stoichiometric exchange of cytosolic ATP for stromal ADP (Reiser et al., 2004; Reinhold et al., 2007). A consequence of the unbalanced phosphate moieties associated with this exchange is that inorganic phosphate (Pi) would accumulate to deleterious levels within the stroma if not countered by Pi export. Despite the fundamental importance of Pi homeostasis to plastid functions and, in turn, to plant growth and development, the identity of the transporter(s) responsible for this activity has been elusive.
In addition to ATP/ADP exchange, NTTs can accept Pi as a third substrate to transport Pi-Pi homoexchange or to cotransport Pi plus ADP in exchange for ATP (Trentmann et al., 2008). Although the latter activity would preclude the necessity for a distinct Pi transporter, studies with cauliflower (Brassica oleracea) bud amyloplasts revealed Pi export activity in the absence of potential counterexchange substrates (i.e. unidirectional transport), indicating that transporters other than NTTs contribute to net Pi export (Neuhaus and Maass, 1996).
Plastid-localized Pi transport systems identified to date include members of the plastidic Pi translocator (pPT), PHT2 and PHT4 families. pPTs catalyze the exchange of Pi with phosphorylated compounds and therefore would not support net Pi export (Flügge, 1999; Eicks et al., 2002). In contrast, members of the PHT2 and PHT4 families mediate H+- and/or Na+-dependent Pi transport (Daram et al., 1999; Versaw and Harrison, 2002; Zhao et al., 2003; Guo et al., 2008b; Ruiz Pavón et al., 2008). The role of these transporters in nonphotosynthetic plastids is unclear. Gene expression studies suggest that all of these transporters are present in limited types of heterotrophic tissues, but most are expressed predominantly in autotrophic tissues (Rausch et al., 2004; Guo et al., 2008a). PHT4;2 (At2g38060) is the sole exception to this expression pattern, as its transcripts are nearly restricted to roots (Guo et al., 2008a).
In this study, we used Arabidopsis (Arabidopsis thaliana) mutants that lack a functional copy of the PHT4;2 gene to gain insight into the roles of the encoded transporter in plastid and whole plant physiology. We confirmed that PHT4;2 contributes to Pi transport in isolated root plastids and found that starch accumulation is reduced in both roots and leaves of the mutant plants. We also discovered a surprising conditional growth phenotype in which rosette leaves of pht4;2 mutants were larger than those of the wild type when plants were grown with a short (8-h) photoperiod, and increased cell proliferation accounted for the greater leaf area and biomass.
Mature leaf size is an intrinsic trait that is a function of both the size and number of cells in the organ. The mechanisms that control final leaf size are poorly understood. In some cases, a reduction in cell number through mutation or transgenes is offset by a compensatory increase in cell size (Hemerly et al., 1995; De Veylder et al., 2001; Horiguchi et al., 2006). In contrast, a number of modifiers of leaf size have been identified that primarily affect cell number, but these represent diverse functional classes, including transcriptional regulation, auxin signaling, protein synthesis, and DNA replication (for review, see Gonzalez et al., 2009; Krizek, 2009). A recent comparative analysis of a subset of these modifiers confirmed that multiple independent pathways contribute to the control of cell proliferation in leaves (Gonzalez et al., 2010). Because none of the previously described modifiers of leaf size appear to be involved in plastid functions, and because PHT4;2 expression during vegetative growth is restricted to roots, the effect of pht4;2 mutations on cell proliferation suggests novel signaling between roots and developing leaves that also contributes to the regulation of leaf size.
RESULTS
Localization of PHT4;2 Expression in Arabidopsis
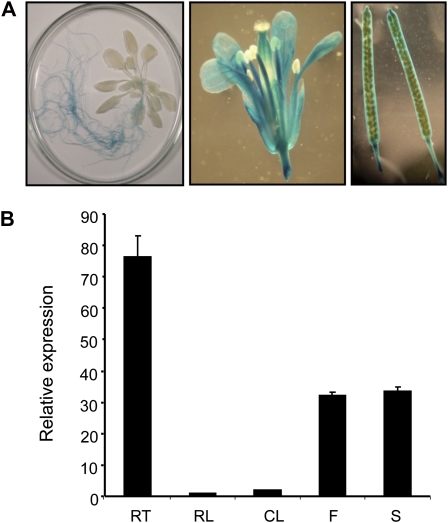
We reported previously that PHT4;2 expression was restricted to roots and exhibited a diurnal but not circadian pattern in which transcript abundance was greater in the dark than in the light (Guo et al., 2008a). Expression data in the Genevestigator microarray database (Hruz et al., 2008) support these patterns and suggest that PHT4;2 is also expressed in floral tissues. To investigate spatial expression in greater detail, a PHT4;2 promoter-GUS transcriptional fusion construct was introduced into Arabidopsis and GUS activity was assessed in progeny of the transgenic plants. To maximize expression, plants were grown with a short (8-h) photoperiod.
As expected, GUS activity was readily detected in roots but not in leaves (Fig. 1A). GUS activity was also detected in sepals, stamens, and carpels as well as silique valves and septum (Fig. 1A), but not in the mature seeds. Quantitative reverse transcription (RT)-PCR analysis of PHT4;2 transcripts in wild-type plants corroborated these findings and indicated that expression was greatest in roots, followed by intermediate levels in flowers and siliques and barely detectable levels in rosette and cauline leaves (Fig. 1B). These results extend our previous studies to suggest that, in addition to roots, PHT4;2 is broadly expressed in carbon import-dependent sink organs. However, even in roots, PHT4;2 transcript levels were low, suggesting that the abundance of the encoded protein would also be limited. Indeed, PHT4;2 was detected by western-blot analysis in small amounts in total protein extracts from roots but not in extracts from other organs, and the detection was further complicated by the presence of several nonspecific bands (data not shown). Next, we isolated microsomes from different organs to enrich the cellular membrane protein fractions and detected PHT4;2 as a 35-kD band in samples prepared from roots, but the abundance of this protein in flowers and siliques still remained below our limit of detection (Supplemental Fig. S1). The identity of the 35-kD band as PHT4;2 was confirmed with blots prepared from mutant and wild-type root plastids, and this enrichment also eliminated nonspecific signals (see below). The significance of expression in other carbon import-dependent sink organs such as flowers and siliques is unclear, because the encoded protein could be detected by western-blot analysis only in roots (Supplemental Fig. S1).
Figure 1.
Localization of PHT4;2 expression in Arabidopsis. A, Representative PHT4;2-GUS staining patterns for a 5-week-old plant (left), flower (center), and mature siliques (right). B, Quantitative RT-PCR analysis of PHT4;2 expression in wild-type plants (RT, root; RL, rosette leaf; CL, cauline leaf; F, flower; S, silique). Expression levels were normalized to EIF-4A2. Values shown are means ± se for three independent analyses.
Identification of pht4;2 T-DNA Insertion Mutants
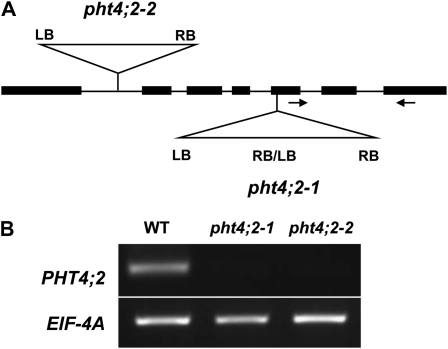
To obtain insight into the role of PHT4;2 in plastid function and overall plant metabolism, we required mutants that lacked a functional PHT4;2 allele. We obtained two independent T-DNA insertion lines from the SALK collection (Alonso et al., 2003), SALK_019289 (pht4;2-1) and SALK_070992 (pht4;2-2). Insertions were confirmed by PCR and sequencing of amplicons that spanned the insertion junctions. The pht4;2-1 line has a tandem T-DNA insertion 2,170 bp downstream of the translation start site and a 7-bp deletion at the insertion site. The pht4;2-2 line has a single T-DNA inserted within the first intron, 738 bp downstream of the translation start site, and also has a deletion of 4 bp at the insertion site. The structures of the mutant loci are represented in Figure 2A.
Figure 2.
Molecular characterization of pht4;2 T-DNA insertion mutants. A, Scheme indicating the positions of T-DNA insertions in pht4;2-1 and pht4;2-2 lines. Insertions were confirmed by PCR and DNA sequence. Arrows represent primers used for RT-PCR. LB, Left border; RB, right border. B, RT-PCR analysis of total RNA isolated from roots of wild-type (WT) and pht4;2 plants.
Homozygous mutant lines were identified by PCR and then screened by DNA gel blot for the presence of additional insertions. At least one backcross to the wild type was required to segregate ectopic insertions from the genetic backgrounds. Only homozygous mutants that lacked ectopic insertions were used in subsequent studies.
RT-PCR analysis of root RNA using primers that anneal downstream of the insertion sites revealed no PHT4;2 transcripts in the homozygous mutants, suggesting that both mutant alleles are null (Fig. 2B).
Localization of PHT4;2 to Root Plastids
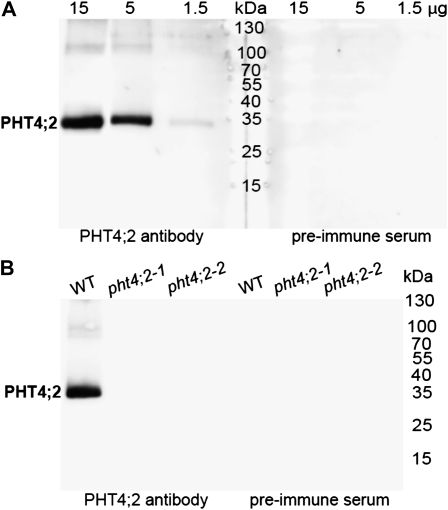
To confirm the localization of the endogenous PHT4;2 protein to root plastids, intact plastids were isolated from roots of hydroponically grown wild-type Arabidopsis, and the proteins were subjected to western-blot analysis. As shown in Figure 3A, a single 35-kD band reacted with the antibody. The signal intensity varied in proportion to the amount of total protein loaded in the well, and no reaction was detected when the corresponding preimmune serum was used, indicating that the antibody is highly specific. No reacting proteins were detected in leaf chloroplasts (data not shown), in accord with transcript localization results (Fig. 1).
Figure 3.
Localization of PHT4;2 to root plastids. A, Western blot of proteins isolated from wild-type root plastids using a peptide-specific antibody and preimmune serum. B, Western blot of wild-type (WT) and pht4;2 root plastid proteins (15 μg protein lane−1) with antibody and preimmune serum. Molecular mass markers are indicated.
Western-blot analysis was also carried out with plastids isolated from roots of pht4;2 mutants. Equal amounts of total plastid protein (15 μg of protein per lane) from the wild type and each mutant were evaluated. The 35-kD band was detected in the wild-type preparation but in neither of the mutants (Fig. 3B), which further verified that the mutations are null.
PHT4;2 encodes a 512-amino acid protein (UniProt Q7XJR2) that is predicted by TargetP (Emanuelsson et al., 2007) to include an N-terminal 44-amino acid transit peptide. The theoretical molecular mass of the processed protein is 50.5 kD, which differs significantly from the 35 kD estimated from SDS-PAGE. Two other members of the PHT4 family, PHT4;1 and PHT4;4, also exhibit anomalous migration by SDS-PAGE (Roth et al., 2004; Ruiz Pavón et al., 2008). The faster gel migrations of PHT4;2 and these other PHT4 proteins may be a function of overproportional binding of SDS, which is common for highly hydrophobic integral membrane proteins (Rath et al., 2009).
Pi Transport in Isolated Root Plastids
Previous studies indicated that PHT4;2, as well as the other five members of the Arabidopsis PHT4 family, mediate Pi transport that is dependent on the presence of a H+ electrochemical gradient when expressed in yeast (Guo et al., 2008b). Interestingly, one of these same proteins, PHT4;1, was found to catalyze Na+-dependent Pi transport when expressed in Escherichia coli (Ruiz Pavón et al., 2008). Thus, different heterologous systems, perhaps due to differences in membrane lipid composition and cell physiology, impart or reveal unique qualities of the Pi transport process. Here, we have taken advantage of the availability of pht4;2 knockout lines to study PHT4;2 in its native context of root plastids.
Plastids isolated from roots of hydroponically grown wild-type and pht4;2 plants had equivalent intactness (70%–75%) as measured by phase-contrast microscopy. To assess the purity and yield of the plastid preparations, the activities of organelle-specific marker enzymes were measured in crude extracts (filtered root homogenates) and in the enriched plastid preparations (Table I). Specific activities of the marker enzymes could be determined only for the enriched plastid preparations due to the presence of bovine serum albumin (BSA) in the isolation buffer. Approximately 7% of the total activity of the plastid marker nitrite reductase was recovered in each plastid preparation, indicating that plastid enrichment was equivalent for each plant genotype. Contamination of these preparations by other organelles was also equivalent for each plant genotype, with 0.33% to 0.44% of the total activity of NADH-malate dehydrogenase (MDH), a marker for combined mitochondrial, cytosolic, and peroxisomal contamination, and 0.55% to 0.86% of the total activity of the vacuolar marker α-mannosidase. Microbial contamination of plastid preparations, which was assessed by growth on Luria-Bertani agar medium, was also similar, with 200 to 250 colony-forming units mg−1 protein, which corresponds to a low number of microbial cells (six to eight) in samples used for Pi transport measurements. Pi uptake by root plastids was linear for 60 s regardless of genotype, but the rate of transport measured with wild-type plastids was twice that of plastids isolated from pht4;2 mutants (Supplemental Fig. S2). Because the purity and enrichment of root plastids were equivalent, we attribute the reduced transport rates with mutant plastids solely to the absence of PHT4;2.
Table I. Distribution of organelle marker enzyme activities in isolated root plastid preparations.
Yields are defined as the percentage of total activity in crude extracts (filtered root homogenates). Specific activities are expressed as nkat mg−1 protein. Values are means ± se of three independent determinations. No significant differences from the wild type were found in the pht4;2-1 and pht4;2-2 mutants (P < 0.05, Student’s t test).
| Marker Enzyme | Wild Type |
pht4;2-1 |
pht4;2-2 |
|||
| Yield | Specific Activity | Yield | Specific Activity | Yield | Specific Activity | |
| % | % | % | ||||
| Nitrite reductase (plastid) | 6.91 ± 0.52 | 42.51 ± 14.34 | 7.73 ± 1.01 | 40.17 ± 16.17 | 7.46 ± 0.36 | 35.51 ± 0.17 |
| NADH-MDH (mitochondrion, cytosol, peroxisome) | 0.33 ± 0.21 | 3.83 ± 1.17 | 0.39 ± 0.32 | 8.17 ± 1.67 | 0.44 ± 0.05 | 3.83 ± 2.67 |
| α-Mannosidase (vacuole) | 0.56 ± 0.03 | 3.00 ± 0.67 | 0.86 ± 0.61 | 3.17 ± 2.33 | 0.55 ± 0.07 | 2.33 ± 0.17 |
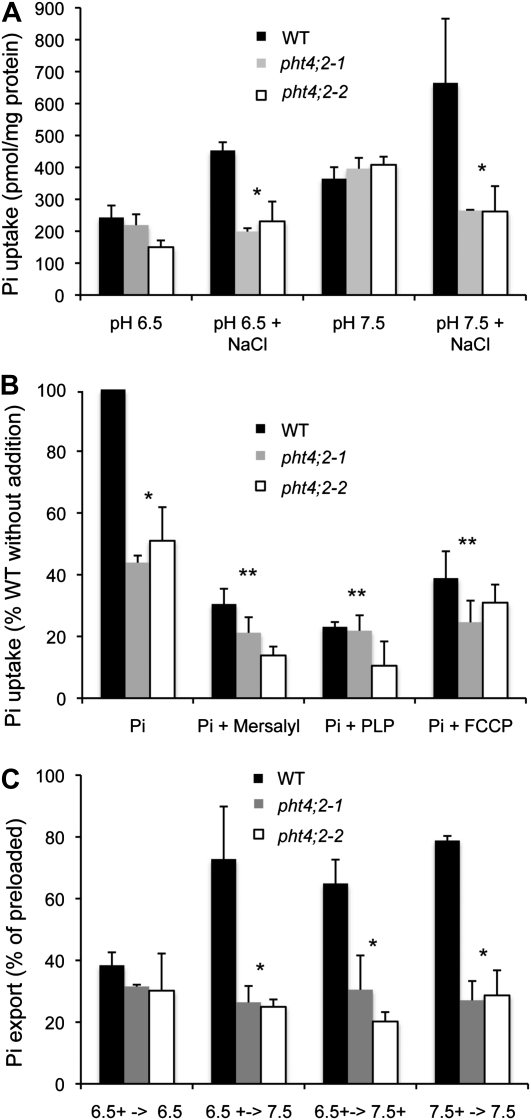
Pi uptake by root plastids isolated from wild-type plants was significantly greater than those isolated from pht4;2 mutants when 5 mm NaCl was included in the assay, but not when this salt was omitted (Fig. 4A) or when all sources of Na+ were substituted with K+ (data not shown). Pi transport activities for all plastids were greater at pH 7.5 than at pH 6.5, but significant differences between mutants and the wild type were observed only in the presence of NaCl. These results suggest that PHT4;2 catalyzes Na+-dependent Pi transport in root plastids.
Figure 4.
Characterization of Pi transport in isolated root plastids. A, Effect of pH and NaCl on Pi uptake in wild-type (WT) and pht4;2 plastids. B, Effect of inhibitors on Pi uptake. Plotted values are relative to untreated wild-type plastids. All assays were conducted at pH 6.5 and in the presence of 5 mm NaCl. C, Pi export from preloaded plastids. Plotted values are expressed as the percentage of preloaded Pi. Preloading and export buffer conditions indicated on the x axis are separated by arrows, and plus signs indicate the presence of 5 mm NaCl. The data shown represent means ± sd for three independent experiments. * Significantly different from the wild type (P < 0.05, Student’s t test); ** significantly different from untreated plastids of the same genotype (P < 0.05).
To better understand the mechanism of the observed Pi uptake activity, we investigated the effects of select pharmacological agents. Transport assays were conducted in the presence of NaCl and at pH 6.5, and Pi uptake was expressed as a percentage of the amount of Pi accumulated in untreated wild-type plastids. All three of the compounds we tested, mersalyl, pyridoxal 5′-phosphate (PLP), and carbonyl cyanide p-(trifluoromethoxy)-phenyl-hydrazone (FCCP), had a pronounced inhibitory effect on Pi transport in wild-type plastids and a lower but consistent effect in mutant plastids (Fig. 4B), suggesting that PHT4;2 is the predominant but not sole source of Pi transport activity under these assay conditions. Addition of mersalyl, a thiol-reactive agent (Amores et al., 1994), reduced wild-type transport to 35%, indicating the importance of sulfhydryl groups for PHT4;2 activity. Similarly, treatment with PLP, a Lys-reactive agent known to inhibit pPTs (Gross et al., 1990; Flügge, 1992), reduced wild-type activity to 23%. Addition of the protonophore FCCP reduced transport in wild-type plastids to 40%, indicating that the concentration and/or membrane potential components of the H+ electrochemical gradient also affect PHT4;2 activity. Dissipation of membrane potential by FCCP would also diminish the magnitude of a Na+ electrochemical gradient.
The Pi uptake assays described above verified that root plastids could be preloaded with Pi to levels sufficient to assess its export. Plastids were preloaded in the presence of 5 mm NaCl at pH 6.5 or 7.5, washed, and resuspended in buffer with or without NaCl. After incubation, the amounts of Pi exported and retained in the plastid were both measured, and export was calculated as a percentage of total Pi to account for differences in preloading. As shown in Figure 4C, Pi export was significantly reduced in root plastids isolated from pht4;2 mutants under most of the conditions tested, confirming that PHT4;2 contributes to this activity in wild-type plastids. Export activities for wild-type plastids measured at pH 7.5 were equivalent when pH, NaCl, or both were varied after preloading (Fig. 4C), suggesting an apparent lack of specificity for either H+ or Na+. In contrast, similarly low export activities were detected in mutant and wild-type plastids when the compositions of the preloading and export solutions were identical (data not shown).
Starch Accumulation
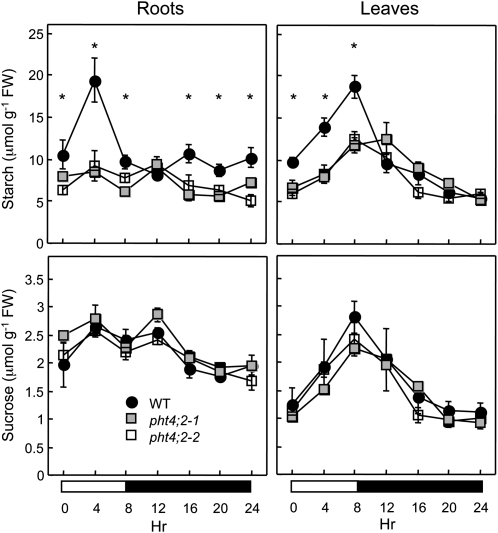
Pi is an allosteric inhibitor of ADP-Glc pyrophosphorylase (AGPase), which catalyzes the first committed step in starch biosynthesis within plastids (Preiss, 1982; Ballicora et al., 2004). Therefore, we reasoned that starch accumulation in roots could serve as an in vivo indicator for defects in plastidic Pi homeostasis associated with the absence of PHT4;2. To test this hypothesis, we measured the starch contents of roots that were harvested from hydroponically grown, 6-week-old wild-type and pht4;2 plants at multiple times throughout an 8-h-light/16-h-dark photocycle. As reported previously for hydroponically grown Arabidopsis (Malinova et al., 2011), starch was distributed throughout the entire root rather than restricted to the root tip (data not shown). Starch levels in wild-type roots doubled during the first half of the photoperiod, returned to near-starting levels by the end of the photoperiod, and remained relatively constant throughout the dark portion of the photocycle (Fig. 5). In contrast, starch levels in pht4;2 roots were nearly constant throughout the entire photocycle and were significantly lower than those in wild-type roots at nearly all time points measured. This reduced capacity for starch accumulation is consistent with Pi inhibition of starch synthesis due to a defect in Pi export.
Figure 5.
Starch and Suc contents. Roots and rosette leaves of 6-week-old wild-type (WT) and pht4;2 plants were collected throughout an 8-h-light/16-h-dark photocycle. Values shown are means ± se for five independent samples. * Significantly different from the wild type (P < 0.05, Student’s t test). FW, Fresh weight.
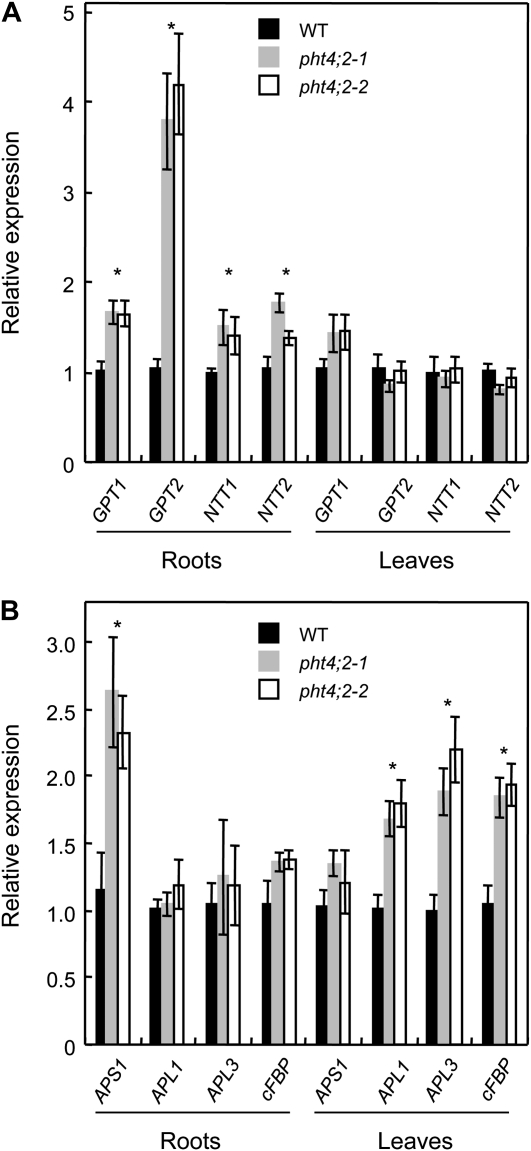
Starch contents of leaves were also lower in the mutants than in the wild type, but only during the light portion of the photocycle (Fig. 5). Because PHT4;2 transcripts and protein in wild-type plants were restricted to roots during vegetative growth, the diminished accumulation of starch in mutant leaves presumably reflects a secondary consequence of metabolic changes in the roots. Consistent with this idea, we detected root- and leaf-specific changes in the expression of a number of genes that are either directly or indirectly involved in starch synthesis (Fig. 6). Specifically, transcript levels for GPT1 and GPT2, which encode Glc-6-P/Pi translocators (GPTs), and NTT1 and NTT2, which encode ATP/ADP nucleotide translocators (NTTs), were induced in roots but not in leaves of pht4;2 plants (Fig. 6A). The proposed functions of GPT and NTT are to provide nonphotosynthetic plastids with the carbon and energy, respectively, needed for starch synthesis and the oxidative pentose phosphate pathway, although mutant analyses indicate that these proteins also have roles in photosynthetic tissues (Niewiadomski et al., 2005; Reinhold et al., 2007). In addition, expression of the AGPase small subunit gene, APS1, was induced only in roots of pht4;2 plants, whereas AGPase large subunit genes, APL1 and APL3, were induced only in leaves (Fig. 6B). Because transcript levels for these genes were inversely correlated with starch levels, the altered expression patterns may reflect compensatory responses to the effects of pht4;2 mutations. Similarly, the cytosolic fructose-1,6-bisphosphatase gene (cFBP), which encodes a key enzyme in Suc biosynthesis (Stitt et al., 2010), was induced only in pht4;2 leaves (Fig. 6B), but we detected no significant changes in the accumulation of Suc in leaves or roots of pht4;2 plants (Fig. 5).
Figure 6.
PHT4;2 modifies the expression of multiple genes involved in plastid transport and carbon metabolism. Expression levels were determined by quantitative RT-PCR of total RNA from plants grown in hydroponics and expressed relative to wild-type (WT) levels. A, Genes involved in plastid transport, including Glc-6-P/Pi exchange (GPT1 and GPT2) and ATP/ADP exchange (NTT1 and NTT2). B, Genes involved in starch synthesis (APS1, APL1, and APL3) and Suc synthesis (cFBP). Values shown are means ± se for three independent analyses. * Significantly different from the wild type (P < 0.05, Student’s t test).
Plant Size and Biomass
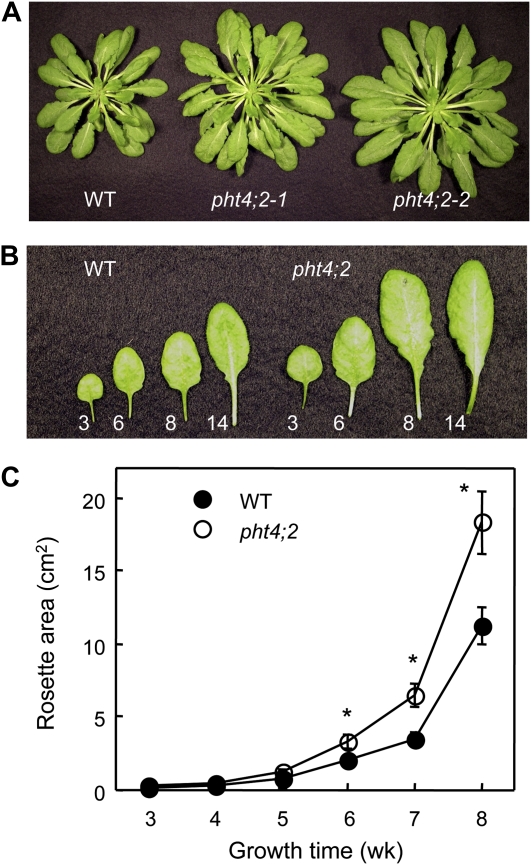
The pht4;2 mutants have no obvious morphological or developmental phenotype when grown with a 14-h photoperiod. However, when plants were grown with a shorter photoperiod (8 h of light), fully expanded pht4;2 rosettes were notably larger than those of the wild type (Fig. 7A). Both mutant and wild-type plants had 43 ± 1 rosette leaves when flowering initiated, indicating that the increase in rosette size was not due to additional leaves but rather an increase in leaf size (Fig. 7B). Because individual leaves emerge and complete the expansion phase of organ growth at different chronological times, differences in rosette sizes were not detected until plants had grown for at least 5 weeks but then increased to a maximum when flowering initiated at 8 to 9 weeks (Fig. 7C).
Figure 7.
Increased size of pht4;2 rosette leaves. A, Rosettes at the end of vegetative growth (8 weeks). B, Rosette leaves 3, 6, 8, and 14 of representative 8-week-old wild-type (WT) and pht4;2 plants. C, Increase in rosette area during vegetative growth. Values shown are means ± se (n = 8). * Significantly different from the wild type (P < 0.05, Student’s t test). As shown in A, both the allelic loss-of-function mutants exhibited the same growth pattern, so for clarity, only the results for pht4;2-1 are shown in B and C.
The large-leaf phenotype was also observed when plants were grown hydroponically, and the magnitude of the size difference was typically greater than that seen with plants grown in soil. The rosette area of fully expanded pht4;2 mutants was 45% to 65% greater than in the wild type, and biomass was at least twice that of the wild type (Table II). Roots at the time of harvest (8 weeks) were tangled, so it was not possible to accurately assess the growth of individual root systems. Therefore, we used the combined weight of root systems to derive an average fresh weight per plant. Results from four independent growth experiments consisting of 74 to 113 plants indicated no significant differences in root growth (0.86 ± 0.17, 0.88 ± 0.18, and 0.73 ± 0.10 g plant−1 for the wild type, pht4;2-1, and pht4;2-2, respectively). In addition, when plants were allowed to complete their life cycle, no differences between pht4;2 mutants and the wild type were detected in flowering time, flower size, or seed size (data not shown).
Table II. Rosette area and biomass at the end of the vegetative growth stage.
Plants were grown in hydroponics for 8 weeks. Values are means ± se (n = 10). * Significantly different from the wild type (P < 0.05, Student’s t test).
| Plant | Area | Fresh Weight | Dry Weight |
| cm2 | g | g | |
| Wild type | 37.06 ± 2.82 | 0.78 ± 0.08 | 0.06 ± 0.01 |
| pht4;2-1 | 53.76 ± 3.52* | 1.47 ± 0.13* | 0.12 ± 0.01* |
| pht4;2-2 | 61.20 ± 3.87* | 1.78 ± 0.13* | 0.16 ± 0.01* |
It was possible that, in addition to increased area, leaf thickness also contributed to the greater biomass of pht4;2 leaves. To estimate the thickness of leaves throughout a rosette, we took advantage of the relationship between leaf thickness and the product of specific leaf area (leaf area per unit of dry mass) and leaf dry matter content (ratio of dry mass to fresh mass; Vile et al., 2005). Leaf punches from the centers of 10 fully expanded rosette leaves were collected from mutant and wild-type plants. Both specific leaf area (0.51 ± 0.02 cm2 mg−1) and leaf dry matter content (0.10 ± 0.01 mg mg−1) were identical for pht4;2 and wild-type leaves. From these values and a leaf density of 1 g cm−3 (Vile et al., 2005), we calculated an average thickness of 196 ± 2 μm for both mutant and wild-type leaves, which is similar to values obtained from direct measurements of leaf sections and three-dimensional imaging of mature leaves (Wuyts et al., 2010). Consequently, the increased area of pht4;2 leaves fully accounts for the increased biomass.
In the course of our investigation of leaf size, we noted that PHT4;2 is located adjacent to DEETIOLATED2 (DET2) on chromosome 2 (loci At2g38060 and At2g38050, respectively), with 550 bp between the PHT4;2 stop codon and the DET2 translation start site. DET2 encodes a steroid 5α-reductase that catalyzes a key step in BR biosynthesis (Noguchi et al., 1999). This close proximity raised the possibility that the large-leaf phenotype we attributed to a disruption of PHT4;2 was instead caused by an overexpression of DET2 via T-DNA activation (Weigel et al., 2000; Ren et al., 2004). However, this possibility is inconsistent with our finding that the large-leaf phenotype (assessed by rosette fresh weight) segregated as a recessive trait in F2 progeny of a cross between the wild type and the homozygous mutant (14 wild type [1.69 ± 0.06 g]:45 hemizygous mutant [1.56 ± 0.10 g]:15 homozygous mutant [2.17 ± 0.12 g]; χ2 = 3.48, P = 0.17). Furthermore, quantitative RT-PCR indicated that DET2 transcript level was unaffected by the pht4;2 mutation (data not shown).
Leaf Cell Size, Number, and Ploidy
Leaf size is dependent on both the number and the size of cells in the organ. To determine whether the enlargement of pht4;2 leaves was associated with increased cell number, cell size, or a combination of these parameters, we compared the ninth rosette leaves of pht4;2 and wild-type plants at different stages of leaf expansion (Table III). When plants were grown for 7 weeks, the ninth rosette leaves were fully expanded and the leaf-size phenotype was apparent, with 23% greater leaf area for pht4;2 mutants. The size of epidermal cells, however, was not significantly different. Consequently, pht4;2 leaves had 30% to 35% more cells than the wild type. Expansion of the ninth rosette leaf was incomplete when plants were grown for 5 weeks, and the sizes of pht4;2 and wild-type leaves were indistinguishable. Nevertheless, the mutant leaves had 34% to 44% more epidermal cells than the wild type, but the cells were also 26% smaller. Together, these results indicate that the large-leaf phenotype of pht4;2 plants is the result of increased cell proliferation, with subsequent expansion of individual cells to normal size at maturity. This phenotype appears to be conditional for growth with a short photoperiod, because when the same analysis was conducted with plants grown with a longer (14-h) photoperiod, all genotypes exhibited equivalent leaf areas, epidermal cell sizes, and epidermal cell numbers (2.64 ± 0.16 cm2, 2,120 ± 120 μm2, and 124,530 ± 10,330 cells, respectively).
Table III. Leaf cell sizes and numbers.
The ninth rosette leaf was harvested from plants at growth times corresponding to incomplete and complete leaf expansion. Values are means ± se for three leaves. Cell areas are calculated from three regions within each leaf. * Significantly different from the wild type (P < 0.05, Student’s t test).
| Plant and Sample | Leaf 9 Area | Epidermal Cell Area | Epidermal Cell No. |
| cm2 | μm2 | ||
| Incomplete expansion (5 weeks) | |||
| Wild type | 1.6 ± 0.1 | 2,540 ± 140 | 63,000 ± 3,940 |
| pht4;2-1 | 1.6 ± 0.1 | 1,900 ± 60* | 84,410 ± 5,280* |
| pht4;2-2 | 1.7 ± 0.1 | 1,870 ± 250* | 90,980 ± 5,350* |
| Complete expansion (7 weeks) | |||
| Wild type | 2.2 ± 0.1 | 2,790 ± 450 | 78,870 ± 3,590 |
| pht4;2-1 | 2.7 ± 0.2* | 2,640 ± 200 | 102,250 ± 7,570* |
| pht4;2-2 | 2.7 ± 0.1* | 2,520 ± 210 | 107,030 ± 3,960* |
Repeated cycles of endoreduplication give rise to somatic polyploidy, which is positively correlated with cell and leaf size (Melaragno et al., 1993; Vlieghe et al., 2005). However, endoreduplication appears to be unaffected by the loss of PHT4;2, because the distribution of ploidy levels in cells of mature pht4;2 and wild-type leaves was nearly identical (Supplemental Fig. S3). Endoreduplication occurs after cell division ceases and continues until leaf expansion is complete (Beemster et al., 2005). Consequently, the similar distribution of ploidy levels also suggests that additional cell proliferation in pht4;2 leaves occurs predominantly, if not exclusively, at early stages in leaf development.
Photosynthetic Activity
Leaf enlargement and altered starch accumulation in pht4;2 mutants suggested the possibility of a change in photosynthetic activity. The two mutant lines presented similar fluorescence characteristics to wild-type plants, as reflected in the dark-adapted state fluorescence yield (F0) and maximum fluorescence yield (Fm) values (data not shown). Therefore, the maximum quantum yield for PSII photochemistry (Fv/Fm) was not significantly different in wild-type, pht4;2-1, and pht4;2-2 plants, 0.812 ± 0.011, 0.809 ± 0.005, and 0.801 ± 0.003, respectively.
To assess and compare the light saturation of photosynthetic performance in mutant and wild-type plants, we recorded a light response curve of chlorophyll fluorescence and calculated the relative electron transport rate (ETR). This analysis revealed that all plants display similar ETR values and reach saturation at approximately 500 μmol photons m−2 s−1 (Supplemental Fig. S4). Together, these data indicate that the lack of PHT4;2 does not modify photosynthetic electron transport in leaves. As expected from these data, total chlorophyll contents were not significantly different in wild-type, pht4;2-1, and pht4;2-2 leaves (1.14 ± 0.1, 1.15 ± 0.1, and 1.15 ± 0.1 mg g−1 fresh weight, respectively).
DISCUSSION
Although ATP/ADP exchange fulfills the energy requirement of plastids with limited or no photosynthetic activity, the unbalanced phosphate moieties would lead to detrimental accumulation of Pi and negative charge within the stroma if not countered by Pi export. Earlier biochemical data suggested that an unknown transporter in cauliflower bud amyloplasts catalyzes Pi export in the absence of exogenous counterexchange substrates (Neuhaus and Maass, 1996), but the effect of Na+ and H+ levels on this activity was not investigated. Our data presented here suggest that PHT4;2 mediates Na+-dependent Pi transport in Arabidopsis root plastids and, given its spatial expression pattern, is likely to carry out the same function in other sink organs. Our analysis of pht4;2 null mutants also revealed pleiotropic effects in starch accumulation throughout the plant and cell proliferation in leaves, which underscore the importance of plastidic Pi homeostasis to plant growth and physiology.
We evaluated Pi transport in root plastids isolated from pht4;2 and wild-type plants and found that export and import activities were both reduced in mutant plastids, indicating that PHT4;2 is capable of catalyzing reversible Pi transport under our assay conditions (Fig. 4). Given that ATP consumption would generate considerable amounts of Pi within the stroma, it is possible that Pi import does not occur under physiological conditions. Moreover, because root plastids and chloroplasts are developmentally related and share many of the same biosynthetic and transport processes (Bräutigam and Weber, 2009; Daher et al., 2010), stromal pH and inner envelope membrane potential of root plastids are likely to be similar to those of dark-adapted chloroplasts (pH approximately 7; 20–100 mV, interior negative; Demmig and Gimmler, 1983; Wu et al., 1991), and these conditions would favor Pi export.
Pi uniport, such as the activity described by Neuhaus and Maass (1996), is an attractive possible mechanism for PHT4;2, as this would rectify both the Pi concentration and charge imbalances derived from ATP/ADP exchange. Nevertheless, coupled transport (e.g. symport), as indicated from the effects of Na+ and H+ we observed with isolated plastids, is also possible, provided that the associated charge imbalance is rectified by an independent activity. Indeed, the addition of Na+, but not K+, to plastids clearly stimulated the Pi import activity of PHT4;2. Pi import was not stimulated by external acidic conditions (pH 6.5) in the absence of Na+ (Fig. 4A), indicating that the presence of a H+ gradient alone is insufficient to activate Pi transport. However, Na+-dependent Pi import was largely inhibited when the imposed H+ gradient was dissipated by a protonophore (FCCP; Fig. 4B), suggesting that other aspects of the H+ gradient are important for Pi transport. For example, a change in stromal pH would affect the fraction of Pi ionic forms, H2PO4−HPO42−. Thus, if PHT4;2 preferentially transports monoanionic H2PO4−then its inward electrochemical gradient would be considerably less when the stroma and external solution have the same pH than when the stroma is more alkaline than the external solution. Alkalinization of the stroma may occur via Na+/H+ exchange, as this transport process has been reported to play an important role in the homeostasis of cytosolic and stromal pH (Song et al., 2004).
Pi export mediated by PHT4;2 was also found to be Na+ dependent (Fig. 4C). We propose that this export activity is the in vivo function of PHT4;2 in nonphotosynthetic plastids, as this would enable homeostasis with respect to Pi concentration. Although Pi export under conditions where Na+ was present both during loading and export would suggest that transport can occur in the absence of a Na+ gradient, a Na+ gradient may in fact exist under these conditions if Na+/H+ exchangers use the imposed H+ gradient to import Na+ into the stroma. Additional studies using mutants with limited plastidic Na+/H+ exchange activities are needed to resolve this possibility. However, it is important to note that the lack of significant Pi export when the composition of the import and export solutions were the same (no pH or Na+ gradient) strongly supports the idea that PHT4;2 requires a Na+ gradient. Furthermore, this requirement may be common to PHT4 transporters, because mutational analysis identified a Ser that is critical for the Na+ dependency of Pi transport by PHT4;1, and this amino acid is fully conserved in the PHT4 family (Ruiz Pavón et al., 2010).
pht4;2 mutants exhibited abnormal starch accumulation as well as increased leaf cell proliferation with a concomitant increase in biomass. These pleiotropic effects could initiate with altered homeostasis of Pi levels in the stroma of root plastids. Specifically, we expected a defect in Pi export to result in elevated stromal Pi levels. Confirmation of this effect through direct measurement in isolated plastids was compromised by ongoing metabolism during the isolation procedure. However, reduced starch accumulation in pht4;2 roots was consistent with an increase in stromal Pi concentrations, as Pi is an inhibitor of starch biosynthesis (Preiss, 1982; Ballicora et al., 2004).
Starch levels were also reduced in pht4;2 leaves. The lack of PHT4;2 transcripts and protein in wild-type leaves suggests that the starch phenotype in mutant leaves is a secondary consequence of the metabolic defect in roots. For example, although the altered expression of other plastid transporter genes and APS1 in roots but not leaves (Fig. 6A) suggests a local response to the defect in root plastids, altered expression of a subset of starch and Suc metabolism genes in leaves but not roots (Fig. 6B) implicates a defect in the long-distance signaling of metabolic status. The strong induction of GPT2 in pht4;2 roots further substantiates the relationship between PHT4;2 and starch accumulation, because GPT2 is also induced in a number of starch-deficient mutants (Kunz et al., 2010).
The partitioning of carbon between starch and Suc biosynthetic pathways is tightly regulated to coordinate carbon demands throughout the day and night. Thus, it might be expected that reduced starch accumulation in pht4;2 plants would be accompanied by an increase in Suc levels. However, we detected no significant changes in Suc levels in roots or leaves of pht4;2 mutants. This finding does not preclude the possibility that the partitioning of carbon to other sugars, amino acids, or lipids is altered and that one or more of these metabolites are directly involved in stimulating cell proliferation in pht4;2 leaves. Additional studies that include defining the full metabolite profiles of roots and leaves of pht4;2 and wild-type plants are needed to resolve these possibilities.
An increase in the number of cells in pht4;2 leaves fully accounted for the greater rosette area and biomass, as epidermal cell sizes and leaf thickness were equivalent to the wild type when leaves were fully expanded. When leaves were examined prior to full expansion, the increase in cell number was already evident, indicating that cell proliferation was augmented early in leaf development, which is in accord with the timing of the proliferative phase of leaf growth (Beemster et al., 2005). However, there was a corresponding reduction in cell size at this time point, such that leaf sizes were equivalent to the wild type. Thus, the leaf-size phenotype was apparent only after cells expanded to normal sizes. Although relative photosynthetic ETR (normalized to leaf area) was unaffected by pht4;2 mutations, the greater total leaf area of the mutants would provide a proportional increase in the photosynthetic activity per plant and thereby support a concomitant increase in biomass.
The mechanisms controlling the sizes of leaves and other organs are poorly understood, largely because this trait is regulated through multiple pathways (Gonzalez et al., 2009; Krizek, 2009). Moreover, Gonzalez et al. (2010) recently demonstrated that several of these pathways, including those associated with auxin, jasmonate, brassinosteroid, and GA synthesis or signaling, as well as transcriptional regulation by GROWTH-REGULATING FACTOR5 (GRF5), which is not associated with changes in phytohormones, independently control cell proliferation and leaf size. Indeed, the phenotypes of lines evaluated in this study differ from each other and from pht4;2, primarily with respect to leaf positions. For example, overexpression of AVP1, a vacuolar pyrophosphate-dependent H+ pump that is involved in auxin transport (Li et al., 2005), yields rosettes with a greater number of leaves, all of which are enlarged. Overexpression of JAW1, which is involved in the regulation of jasmonate biosynthesis (Palatnik et al., 2003; Schommer et al., 2008), leads to the enlargement of only the first rosette leaves, and these have a characteristic uneven shape. Leaf enlargement is also restricted to the first few rosette leaves in lines overexpressing GRF5 and BRI1, a brassinosteroid receptor (Wang et al., 2001; Horiguchi et al., 2005), whereas overexpression of GA20OX1, an enzyme involved in GA synthesis (Huang et al., 1998), yields enlarged young but not older leaves.
Cell cycle progression is responsive to carbon status (Riou-Khamlichi et al., 2000), and it is possible that the influence of pht4;2 mutations on starch accumulation constitutes one of the multiple pathways that converge on the control of cell proliferation and organ size. Starch-deficient mutants do not have abnormally large leaves (Ventriglia et al., 2008), suggesting that reduced starch accumulation is unlikely to be causal for the increase in cell proliferation underlying the pht4;2 leaf-size phenotype. However, starch accumulation in such mutants is affected throughout the entire plant, so comparisons may be misleading if the leaf-size phenotype is related to carbon balance between source and sink organs. In addition, our results indicate that the mechanism responsible for increased cell proliferation is nonautonomous, suggesting that PHT4;2 plays a key role in coordinating metabolic signals throughout the entire plant.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines SALK_019289 (pht4;2-1) and SALK_070992 (pht4;2-2) were obtained from the Arabidopsis Biological Resource Center. Insertion sites and zygosity were confirmed by PCR using combinations of PHT4;2-specific primers and primers that anneal to the T-DNA right and left borders. The sequence of amplicons that spanned insertion junctions was confirmed. Homozygous pht4;2-1 and pht4;2-2 plants were screened by DNA gel-blot analysis using the nptII segment of the binary plasmid pROK2 as a probe to identify individuals with insertions at a single locus.
Plants were grown in chambers at 21°C with 70% relative humidity and an 8-h photoperiod (150 μmol m−2 s−1) in soil (SunGro Redi-earth) or hydroponically as described previously (Noren et al., 2004; Guo et al., 2008b).
Analysis of PHT4;2 Promoter-GUS Expression
A PHT4;2 promoter-GUS transcriptional fusion (Guo et al., 2008a) was introduced into Agrobacterium tumefaciens strain GV3101 and used to transform Arabidopsis (Clough and Bent, 1998). Transgenic seedlings were selected on half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) containing 25 μg mL−1 kanamycin. At least 12 independent lines were examined for GUS activity by histochemical detection. Seedlings, flowers, and siliques were fixed in 50 mm sodium phosphate, pH 7.0, and 4% (v/v) formaldehyde, infiltrated with 50 mm sodium phosphate, pH 7.0, 0.1% (v/v) Triton X-100, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 10 mm EDTA, and 0.05% (w/v) 5-bromo-4-chloro-3-indolyl glucuronide, and then incubated in the dark at 37°C overnight. Green tissues were destained with 70% (v/v) ethanol prior to observation. Images were captured with a Nikon Coolpix 4300 camera mounted on an Olympus LMS 225R dissection microscope.
Quantitative RT-PCR Analysis
Total RNA was isolated from roots or leaves using TRI reagent (Sigma-Aldrich), and traces of contaminating DNA were eliminated with TURBO DNA-free (Ambion). Three biological replicates were used for each analysis. First-strand cDNA was synthesized from 1 μg of total RNA using the SuperScript first-strand cDNA synthesis kit (Invitrogen). Real-time PCR was conducted with Power SYBR Green Master Mix and the ABI Prism 7500 sequence detection system (Applied Biosystems). Expression levels were normalized to EIF4A-2 (Guo et al., 2008b).
Root Plastid Isolation and Pi Transport Assays
Plastids were isolated from the entire root system of 6-week-old plants as described (Emes and England, 1986). Briefly, roots (10–15 g) of hydroponically grown plants were washed in distilled water, chilled at 4°C for 30 min, and then ground in four 5-s bursts in a blender with 40 mL of cold isolation buffer: 50 mm potassium-Tricine (pH 8.0), 0.3 m sorbitol, 1 mm EDTA, 2 mm MgCl2, and 0.1% (w/v) freshly added BSA. The homogenate was filtered through two layers of Miracloth to yield a crude extract fraction. The crude extract was centrifuged at 1,500g for 3 min at 4°C. The pellet was suspended in 0.5 mL of isolation buffer and then transferred to a tube containing 40 mL of 10% (v/v) Percoll in isolation buffer. The mixture was centrifuged at 3,000g for 10 min at 4°C. Intact plastids were pelleted, while broken plastids and cell debris remained at the top of the Percoll solution. The intact plastids were suspended in 20 mL of isolation buffer without BSA and then pelleted again by centrifugation at 1,500g for 3 min. The pellet was finally suspended in 0.5 mL of isolation buffer without BSA and maintained at 4°C. Root plastids were typically 70% to 75% intact as estimated by phase-contrast microscopy. The protein content of the plastid suspension was determined using the Bradford assay with BSA as a standard (Bradford, 1976). For plastid enrichment, the activity of nitrite reductase was assayed as described previously (Takahashi et al., 2001). For plastid purity, vacuolar contamination was determined by α-mannosidase activity (Stitt et al., 1989), and combined mitochondrial, cytosolic, and peroxisomal contamination was determined by NADH-MDH activity (Schneider and Keller, 2009). Yields of marker enzyme activities in plastid preparations were calculated as a percentage of the activities in crude extracts.
Pi uptake by freshly isolated root plastids was assayed in 30 μL of assay buffer at a final protein concentration of 1 mg mL−1 for up to 60 s at 22°C. The assay buffer contained 50 mm Mg-HEPES adjusted to either pH 6.5 or 7.5 as indicated, 0.3 m sorbitol, 1 mm MgCl2, and 0 or 5 mm NaCl as indicated. The final concentration of [32P]orthophosphate (60 mCi mmol−1; 1 mCi = 37 MBq; Perkin-Elmer) was 50 μm. Transport inhibition studies were carried out with 150 μm mersalyl, 6 mm PLP, or 50 μm FCCP, added 1 min before the addition of Pi. Transport was terminated by the addition of 170 μL of cold assay buffer followed by centrifugation at 16,000g for 1 min. The pellet was washed with 200 μL of cold assay buffer, suspended in 30 μL of 2 mm dodecyl maltoside, and incubated for 5 min on ice. Accumulated Pi was measured by liquid scintillation spectrometry in 1.5 mL of distilled water. Control experiments performed in the presence of 10 mm potassium phosphate buffer (pH 6.5 and 7.5) indicated 10% to 15% nonspecific binding, which was subtracted from the corresponding measured activity. Root plastids isolated from the wild type and mutants were assayed in parallel for each experiment. Results are averages of three independent experiments performed in triplicate ± sd.
Pi export studies were conducted with the same buffers as for uptake. Plastids were first preloaded with Pi for 60 s in the indicated buffer and then washed twice with the same buffer to remove excess Pi. Preloaded plastids were then incubated for 60 s in the indicated assay buffer at the same temperature. The assay was terminated by centrifugation, and the amounts of Pi remaining in the plastid pellet and exported to the assay buffer were determined by scintillation counting as described above. Results are averages of two independent experiments performed in triplicate ± sd.
SDS-PAGE and Western Blotting
Microsomes were prepared as described (Bush, 1989) from wild-type organs to enrich for cellular membrane proteins. The resulting membrane protein fractions and root plastid proteins were separated by electrophoresis using 14% (w/v) acrylamide SDS gels. Following electroblotting to polyvinylidene fluoride membranes (Millipore), the PHT4;2 protein was immunodetected using a specific antibody against the N-terminal peptide RYSSESDGKRRNA produced in rabbit by Innovagen. The blots were further reacted with secondary donkey anti-rabbit antibody conjugated with horseradish peroxidase and a chemiluminescent substrate kit (GE Healthcare).
Extraction and Measurement of Suc and Starch
Leaf and root samples were harvested at the indicated times in the photoperiod and then immediately frozen in liquid nitrogen. Suc was extracted with 80% (v/v) ethanol and 5% (v/v) formic acid at 80°C for 20 min, then the extraction was repeated with 80% ethanol. The supernatants were pooled, lyophilized, suspended in 10 mm Na acetate (pH 5.5), and then assayed for Suc. Pellets were dried and starch was gelatinized and hydrolyzed as described (Smith and Zeeman, 2006). Suc and starch concentrations were quantified using coupled assays in which the formation of NADH was monitored by A340 using a Synergy HT microplate reader. All assays were performed in duplicate, and values were determined from standard curves. At least four independent samples were collected for each time point, and results are reported as means ± se.
Leaf and Cell Size Analysis
The area of rosettes and individual rosette leaves was determined from digital images that included reference objects using the histogram function of Adobe Photoshop. To determine epidermal cell area, nail polish impressions of the abaxial epidermis were prepared from the ninth rosette leaf of at least three wild-type and pht4;2 plants. Images were captured with constant magnification (400×) for three different locations in each leaf between 25% and 75% of the distance between the tip and the base of the blade, halfway between the midrib and the leaf margin. Cells in each field of view were counted, and cell areas (means ± se) were determined from a total of at least 500 cells.
Ploidy Analysis
Nuclei were isolated from fully expanded leaves of 8-week-old plants as described previously (Galbraith et al., 1983). Briefly, leaves were finely chopped with a razor blade in 2 mL of ice-cold buffer containing 45 mm MgCl2, 30 mm sodium citrate, 20 mm MOPS, pH 7, and 1 mg mL−1 Triton X-100. Tissue fragments were removed by filtration through 36-μm nylon mesh, and DNase-free RNase (10 μg mL−1 final) and propidium iodide (100 μg mL−1 final) were added. Nuclei were incubated for 30 min in the dark at room temperature before analyzing the distribution of DNA content using a FACScan flow cytometer. Results are from three independent plants of each genotype and at least 4000 nuclei for each analysis.
Photosynthetic Activity in Detached Leaves
Chlorophyll fluorescence was measured using a pulse-amplitude fluorometer (model PAM-210; Walz) in leaves detached from 16-h dark-adapted plants. The F0 was recorded by using a weak measuring light, whereas the Fm was measured after application of a 1-s pulse of saturating visible light (Schreiber et al., 1986). The Fv/Fm was calculated using the equation Fv/Fm = (Fm − F0)/Fm. The photosynthetic ETR was determined in detached leaves using PAM-210 by measuring the quantum yield of PSII photochemistry (Y′) after every 20 s of illumination with photosynthetically active radiation (PAR) of 0 to 1,850 μmol photons m−2 s−1, increased stepwise. The relative ETR was calculated by the equation ETR = 0.84 × 0.5 × PAR × Y′ (Genty et al., 1989). It was assumed that 84% of the incident light was absorbed (factor 0.84) and that an equal fraction of the absorbed quanta is distributed to PSII and PSI (factor 0.5). Results are means of three independent measurements ± sd.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PHT4;2 (NM_129362), EIF4A-2 (NM_104305), GPT1 (NM_124861), GPT2 (NM_104862), NTT1 (NM_106679), NTT2 (NM_101419), APS1 (NM_124205), APL1 (NM_121927), APL3 (NM_120081), cFBP (NM_103492), and DET2 (NM_129361).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Localization of the PHT4;2 protein in Arabidopsis organs by western-blot analysis.
Supplemental Figure S2. Rate of Pi uptake by isolated root plastids.
Supplemental Figure S3. Distribution of ploidy in rosette leaves.
Supplemental Figure S4. Plot of relative ETR as a function of the intensity of PAR.
Acknowledgments
We thank David Galbraith, Georgina Lambert, and Katy Kao for assistance with flow cytometry and Pallavi Mukherjee and Tom McKnight for critical reading of the manuscript.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amores MV, Hortelano P, García-Salguero L, Lupiáñez JA. (1994) Metabolic adaptation of renal carbohydrate metabolism. V. In vivo response of rat renal-tubule gluconeogenesis to different diuretics. Mol Cell Biochem 137: 117–125 [DOI] [PubMed] [Google Scholar]
- Ballicora MA, Iglesias AA, Preiss J. (2004) ADP-glucose pyrophosphorylase: a regulatory enzyme for plant starch synthesis. Photosynth Res 79: 1–24 [DOI] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Weber AP. (2009) Proteomic analysis of the proplastid envelope membrane provides novel insights into small molecule and protein transport across proplastid membranes. Mol Plant 2: 1247–1261 [DOI] [PubMed] [Google Scholar]
- Bush DR. (1989) Proton-coupled sucrose transport in plasmalemma vesicles isolated from sugar beet (Beta vulgaris L. cv Great Western) leaves. Plant Physiol 89: 1318–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Daher Z, Recorbet G, Valot B, Robert F, Balliau T, Potin S, Schoefs B, Dumas-Gaudot E. (2010) Proteomic analysis of Medicago truncatula root plastids. Proteomics 10: 2123–2137 [DOI] [PubMed] [Google Scholar]
- Daram P, Brunner S, Rausch C, Steiner C, Amrhein N, Bucher M. (1999) Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 11: 2153–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig B, Gimmler H. (1983) Properties of the isolated intact chloroplast at cytoplasmic K concentrations. I. Light-induced cation uptake into intact chloroplasts is driven by an electrical potential difference. Plant Physiol 73: 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicks M, Maurino V, Knappe S, Flügge UI, Fischer K. (2002) The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol 128: 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Emes MJ, England S. (1986) Purification of plastids from higher-plant roots. Planta 168: 161–166 [DOI] [PubMed] [Google Scholar]
- Flügge UI. (1992) Reaction mechanism and asymmetric orientation of the reconstituted chloroplast phosphate translocator. Biochim Biophys Acta 1110: 112–118 [DOI] [PubMed] [Google Scholar]
- Flügge UI. (1999) Phosphate translocators in plastids. Annu Rev Plant Physiol Plant Mol Biol 50: 27–45 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR. (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Gonzalez N, Beemster GT, Inzé D. (2009) David and Goliath: what can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr Opin Plant Biol 12: 157–164 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A, Brückner G, Heldt HW, Flügge UI. (1990) Comparison of the kinetic properties, inhibition and labelling of the phosphate translocators from maize and spinach mesophyll chloroplasts. Planta 180: 262–271 [DOI] [PubMed] [Google Scholar]
- Guo B, Irigoyen S, Fowler TB, Versaw WK. (2008a) Differential expression and phylogenetic analysis suggest specialization of plastid-localized members of the PHT4 phosphate transporter family for photosynthetic and heterotrophic tissues. Plant Signal Behav 3: 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Jin Y, Wussler C, Blancaflor EB, Motes CM, Versaw WK. (2008b) Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol 177: 889–898 [DOI] [PubMed] [Google Scholar]
- Hemerly A, Engler JdeA, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. (2006) Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J Plant Res 119: 37–42 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. (1998) Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol 118: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA. (2009) Making bigger plants: key regulators of final organ size. Curr Opin Plant Biol 12: 17–22 [DOI] [PubMed] [Google Scholar]
- Kunz HH, Häusler RE, Fettke J, Herbst K, Niewiadomski P, Gierth M, Bell K, Steup M, Flügge UI, Schneider A. (2010) The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol (Stuttg) (Suppl 1) 12: 115–128 [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Malinova I, Steup M, Fettke J. (2011) Starch-related cytosolic heteroglycans in roots from Arabidopsis thaliana. J Plant Physiol 168: 1406–1414 [DOI] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Neuhaus HE, Emes MJ. (2000) Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51: 111–140 [DOI] [PubMed] [Google Scholar]
- Neuhaus HE, Maass U. (1996) Unidirectional transport of orthophosphate across the envelope of isolated cauliflower-bud amyloplasts. Planta 198: 542–548 [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A. (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J. (1999) Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol 120: 833–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren H, Svennsson P, Andersson B. (2004) A convenient and versatile hydroponic cultivation system for Arabidopsis thaliana. Physiol Plant 121: 343–348 [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Preiss J. (1982) Regulation of the biosynthesis and degradation of starch. Annu Rev Plant Biol 33: 431–454 [Google Scholar]
- Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci USA 106: 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C, Zimmermann P, Amrhein N, Bucher M. (2004) Expression analysis suggests novel roles for the plastidic phosphate transporter Pht2;1 in auto- and heterotrophic tissues in potato and Arabidopsis. Plant J 39: 13–28 [DOI] [PubMed] [Google Scholar]
- Reinhold T, Alawady A, Grimm B, Beran KC, Jahns P, Conrath U, Bauer J, Reiser J, Melzer M, Jeblick W, et al. (2007) Limitation of nocturnal import of ATP into Arabidopsis chloroplasts leads to photooxidative damage. Plant J 50: 293–304 [DOI] [PubMed] [Google Scholar]
- Reiser J, Linka N, Lemke L, Jeblick W, Neuhaus HE. (2004) Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol 136: 3524–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Johnston JS, Shippen DE, McKnight TD. (2004) TELOMERASE ACTIVATOR1 induces telomerase activity and potentiates responses to auxin in Arabidopsis. Plant Cell 16: 2910–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JM, Murray JA. (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Menzel G, Petétot JM, Rochat-Hacker S, Poirier Y. (2004) Characterization of a protein of the plastid inner envelope having homology to animal inorganic phosphate, chloride and organic-anion transporters. Planta 218: 406–416 [DOI] [PubMed] [Google Scholar]
- Ruiz-Pavón L, Karlsson PM, Carlsson J, Samyn D, Persson B, Persson BL, Spetea C. (2010) Functionally important amino acids in the Arabidopsis thylakoid phosphate transporter: homology modeling and site-directed mutagenesis. Biochemistry 49: 6430–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavón LR, Lundh F, Lundin B, Mishra A, Persson BL, Spetea C. (2008) Arabidopsis ANTR1 is a thylakoid Na+-dependent phosphate transporter: functional characterization in Escherichia coli. J Biol Chem 283: 13520–13527 [DOI] [PubMed] [Google Scholar]
- Schneider T, Keller F. (2009) Raffinose in chloroplasts is synthesized in the cytosol and transported across the chloroplast envelope. Plant Cell Physiol 50: 2174–2182 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U, Schliwa U, Bilger W. (1986) Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth Res 10: 51–62 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC. (2006) Quantification of starch in plant tissues. Nat Protoc 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Song CP, Guo Y, Qiu Q, Lambert G, Galbraith DW, Jagendorf A, Zhu JK. (2004) A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 10211–10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Lunn J, Usadel B. (2010) Arabidopsis and primary photosynthetic metabolism: more than the icing on the cake. Plant J 61: 1067–1091 [DOI] [PubMed] [Google Scholar]
- Stitt M, McLilley R, Gerhardt R, Heldt HW. (1989) Metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol 174: 518–552 [Google Scholar]
- Takahashi M, Sasaki Y, Ida S, Morikawa H. (2001) Nitrite reductase gene enrichment improves assimilation of NO2 in Arabidopsis. Plant Physiol 126: 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentmann O, Jung B, Neuhaus HE, Haferkamp I. (2008) Nonmitochondrial ATP/ADP transporters accept phosphate as third substrate. J Biol Chem 283: 36486–36493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventriglia T, Kuhn ML, Ruiz MT, Ribeiro-Pedro M, Valverde F, Ballicora MA, Preiss J, Romero JM. (2008) Two Arabidopsis ADP-glucose pyrophosphorylase large subunits (APL1 and APL2) are catalytic. Plant Physiol 148: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ. (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vile D, Garnier E, Shipley B, Laurent G, Navas ML, Roumet C, Lavorel S, Díaz S, Hodgson JG, Lloret F, et al. (2005) Specific leaf area and dry matter content estimate thickness in laminar leaves. Ann Bot (Lond) 96: 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GT, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L. (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Peters J, Berkowitz GA. (1991) Surface charge-mediated effects of Mg2+ on K+ flux across the chloroplast envelope are associated with regulation of stromal pH and photosynthesis. Plant Physiol 97: 580–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts N, Palauqui JC, Conejero G, Verdeil JL, Granier C, Massonnet C. (2010) High-contrast three-dimensional imaging of the Arabidopsis leaf enables the analysis of cell dimensions in the epidermis and mesophyll. Plant Methods 6: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Versaw WK, Liu J, Harrison MJ. (2003) A phosphate transporter from Medicago truncatula is expressed in the photosynthetic tissues of the plant and is located in the chloroplast envelope. New Phytol 157: 291–302 [DOI] [PubMed] [Google Scholar]