Abstract
Production of disease symptoms represents the final phase of infectious diseases and is a main cause of crop loss and/or marketability. However, little is known about the molecular basis of disease symptom development. In this study, a genetic screening was conducted to identify Arabidopsis (Arabidopsis thaliana) mutants that are impaired specifically in the development of disease symptoms (leaf chlorosis and/or necrosis) after infection with the bacterial pathogen Pseudomonas syringae pv tomato (Pst) DC3000. An ethyl methanesulfonate-induced Arabidopsis mutant (no chlorosis1 [noc1]) was identified. In wild-type plants, the abundance of chlorophylls decreased markedly after Pst DC3000 infection, whereas the total amount of chlorophylls remained relatively unchanged in the noc1 mutant. Interestingly, noc1 mutant plants also exhibited reduced disease symptoms in response to the fungal pathogen Alternaria brassicicola. Genetic and molecular analyses showed that the nuclear gene STAYGREEN (SGR; or Mendel’s I locus) is mutated (resulting in the aspartic acid to tyrosine substitution at amino acid position 88) in noc1 plants. Transforming wild-type SGR cDNA into the noc1 mutant rescued the chlorosis phenotype in response to Pst DC3000 infection. The SGR transcript was highly induced by Pst DC3000, A. brassicicola, or coronatine (COR), a bacterial phytotoxin that promotes chlorosis. The induction of SGR expression by COR is dependent on COI1, a principal component of the jasmonate receptor complex. These results suggest that pathogen/COR-induced expression of SGR is a critical step underlying the development of plant disease chlorosis.
Tissue chlorosis and necrosis are among the most frequently observed disease symptoms associated with infection by diverse plant pathogens. These disease symptoms generally occur at the late stages of disease, but are highly relevant to agriculture because they cause actual damage to plant tissues, resulting in yield loss and/or poor marketability of crops. Symptomless infection is often associated with benign or symbiotic plant-microbe associations. Despite its importance in disease, the molecular basis of disease chlorosis and necrosis remain poorly understood.
Pseudomonas syringae pv tomato (Pst) DC3000, the bacterial pathogen used in this study, is well known for causing localized necrosis and diffuse chlorosis in its hosts tomato (Solanum lycopersicum) and Arabidopsis (Arabidopsis thaliana; Ma and Cuppels, 1991; Whalen et al., 1991; Katagiri et al., 2002). Many virulence genes are needed for Pst DC3000 to cause disease, among which two virulence systems have been studied extensively: the type III secretion system and the phytotoxin coronatine (COR). The type III secretion system delivers dozens of effector proteins into plant cells (Alfano and Collmer, 2004; He et al., 2004; Büttner and He, 2009). Many of these effectors suppress host immune responses (Boller and He, 2009; Cui et al., 2009; Lewis et al., 2009) and some are also linked to production of disease necroses (Badel et al., 2003; DebRoy et al., 2004; Cohn and Martin, 2005). COR, a molecular mimic of the plant hormone jasmonate, is not only involved in suppression of host immune responses, but also is important for the development of chlorosis symptoms (Feys et al., 1994; Mittal and Davis, 1995; Bender et al., 1999; Kloek et al., 2001; Brooks et al., 2004, 2005; Block et al., 2005; Melotto et al., 2008b). However, because effector- and COR-deficient bacterial mutants are generally affected in multiple steps of pathogenesis, it is often difficult to determine whether these virulence factors contribute to symptom development directly or indirectly through promoting bacterial colonization and/or multiplication.
An alternative approach to elucidate the molecular control of disease symptom production would be to isolate plant mutants that exhibit reduced disease necroses and/or chlorosis in response to pathogen infection. Indeed, numerous Arabidopsis constitutive defense mutants that show no or reduced disease symptoms to P. syringae infection have been isolated since the early 1990s (Bowling et al., 1994). However, similar to the situation with effector- or COR-defective Pst DC3000 mutants, bacterial populations in such plant mutants are often reduced compared to those in susceptible plants, making it difficult to conclude whether corresponding plant genes have a direct role in mediating symptom development or indirectly through affecting bacterial multiplication. There are a few exceptions: The Arabidopsis mutant ethylene-insensitive2 and the Arabidopsis mutant suppressor of G2 allele of skp1 (affected in jasmonate signaling) exhibit reduced disease symptoms to Pst DC3000 infection without significantly affecting bacterial growth (Bent et al., 1992; Uppalapati et al., 2011), implicating the involvement of ethylene and jasmonate signaling in the production of Pst DC3000-elicited disease symptoms. How ethylene or jasmonate signaling lead to downstream disease symptom is not understood.
To increase our understanding of the molecular basis of disease symptom development during Pst DC3000 infection of Arabidopsis, we conducted a genetic screen for Arabidopsis mutants that are reduced in disease symptom development, but not Pst DC3000 multiplication. In this article, we report the identification and characterization of such an Arabidopsis mutant and cloning of the corresponding gene. Our results suggest that pathogen-responsive STAYGREEN (SGR)/NON-YELLOWING/Mendel’s I locus plays a critical role in controlling disease chlorosis induced by Pst DC3000 and, interestingly, also by a fungal pathogen, Alternaria brassicicola.
RESULTS
Identification of the no chlorosis1 Mutant
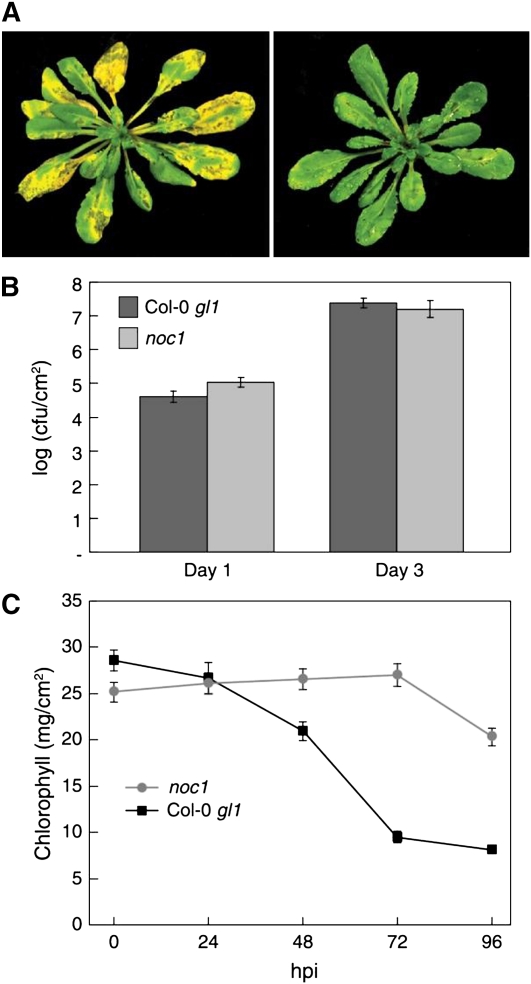
Approximately 10,000 ethyl methanesulfonate-mutagenized Arabidopsis ecotype Columbia-0 (Col-0) glabrous1 (gl1) plants were screened for altered symptom development after the plants were dipped in a suspension containing 1 × 108 colony forming units (CFU)/mL Pst DC3000 bacteria. One mutant isolated from this screen, no chlorosis1 (noc1), was found to be defective in symptom development. In all experiments, noc1 leaves remained green whereas wild-type leaves began to show chlorosis between 48 and 72 h after inoculation (Fig. 1A). In most experiments reduced severity of necrosis symptom was also observed in noc1 plants, although this phenotype was not as obvious as the lack-of-chlorosis phenotype. There were no noticeable differences between wild-type and noc1 plants in size, morphology, growth, or development in the absence of pathogen inoculation. Most importantly, the reduction in disease symptoms in noc1 plants was not caused by reduced bacterial multiplication because Pst DC3000 populations in Col-0 gl1 and noc1 plants were similar at 1 and 3 d postinfection (dpi; Fig. 1B). Thus, noc1 is a bona fide disease symptom mutant.
Figure 1.
The phenotypes of the noc1 mutant following Pst DC3000 infection. A, Arabidopsis Col-0 gl1 plant exhibiting typical chlorosis and water-soaking phenotype associated with Pst DC300 infection at 72 hpi (left); noc1 mutants display water soaking but little chlorosis following Pst DC3000 infection (right). B, Col-0 gl1 and noc1 plants were dip inoculated with Pst DC3000 at 108 CFU/mL. Bacterial populations were determined at 1 and 3 dpi. C, Chlorophyll amounts in the noc1 mutant and wild-type Col-0 gl1 subsequent to Pst DC3000 infection at 0, 24, 48, 72, and 96 hpi.
In addition to the noc1 mutant, we also isolated several other mutants that exhibited reduced disease symptom development during this study. However, these other mutants were dwarf and/or necrotic, suggestive of constitutive activation of nonspecific disease resistance (Bowling et al., 1994). We did not conduct further characterization of such mutants.
Maintenance of the Chlorophyll Level in noc1 Plants after Infection with Pst DC3000
To quantify the chlorotic response to Pst DC3000, we conducted a chlorophyll abundance assay using leaf tissue infiltrated with 2 × 106 CFU/mL of Pst DC3000 and collected at 0, 24, 48, 72, and 96 h postinoculation (hpi). The results from one representative experiment are shown in Figure 1C. Prior to inoculation with Pst DC3000, noc1 and wild-type plants had approximately equal amounts of total chlorophyll (25.2 and 28.6 mg/cm2, respectively). Wild-type plants began to lose chlorophyll by 24 hpi, with levels decreasing through 96 hpi. At 72 hpi, noc1 plants contained almost 3 times more chlorophyll than wild-type plants (27.0 mg/cm2 in noc1 plants versus 9.5 mg/cm2 in wild-type plants). This experiment demonstrates that wild-type plants lose chlorophyll much faster than noc1 plants after Pst DC3000 infection.
The noc1 Phenotype Results from a Single Nucleotide Change in AtSGR1 (At4g22920)
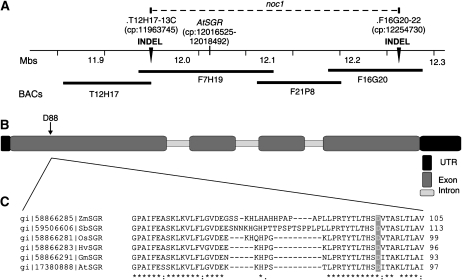
To identify the NOC1 gene, noc1 plants were crossed with Landsberg erecta (Ler) plants and the F1 progeny were selfed to create an F2 population for mapping. The noc1 mutation shows normal Mendelian genetics and is recessive. We initially used bulk segregant analysis to analyze a pool of approximately 100 F2 individuals that exhibited the mutant phenotype (homozygous for the noc1 mutation). One marker, NGA107, located on the long arm of chromosome 4, showed linkage to the mutation. We tested a larger population of F2 individuals using additional chromosome-4-specific insertion and deletion (INDEL) markers identified from the Monsanto Arabidopsis Polymorphism, Ler Sequence Collection (St. Louis, MO). This delimited the region of the noc1 mutation to the area between two INDEL markers (T12H17-13C and F16G20-22) located on chromosome 4 at 11.96 Mb and 12.25 Mb, respectively (Fig. 2A).
Figure 2.
Map-based identification of the noc1 mutation. A, A portion of the long arm of Arabidopsis chromosome 4 where the noc1 mutation is located between the two INDEL markers (T12H17-13C and F16G20-22; indicated by black arrowheads) at chromosomal positions (cp) 11963745 and 12254730, respectively. B, Gene structure of AtSGR (At4g22920; cp: 12016525–12018492), showing the location of the noc1 mutation, an Asp to Tyr substitution at position 88 resulting from a guanine to thymidine nucleotide mutation. C, The residue D88 is conserved among SGR orthologs in diverse plants, including Zea mays (gi 58866285), Sorghum bicolor (gi 59506606), Oryza sativa (gi 58866281), Hordeum vulgare (gi 58866283), and Glycine max (gi 58866291). Orthologous sequences were arranged using CLUSTALW software.
Based on the impaired chlorosis phenotype of the noc1 mutant, we reasoned that the mutation might lie in a gene coding for a chloroplast-targeted protein. The mapped region for the noc1 mutation includes six genes encoding predicted chloroplast localization signals. The cDNA clones of candidate genes from noc1 and Col-0 gl1 plants were subjected to sequencing and revealed a single guanine to thymidine nucleotide mutation, resulting in an Asp (D) to Tyr (Y) amino acid substitution at position 88 of At4g22920/AtSGR/NON-YELLOWING/Mendel’s I locus (AtSGR hereafter; Fig. 2B). Orthologs of AtSGR (Ren et al., 2007) in monocot and dicot species are tightly conserved overall with no variation occurring at this particular residue (Fig. 2C).
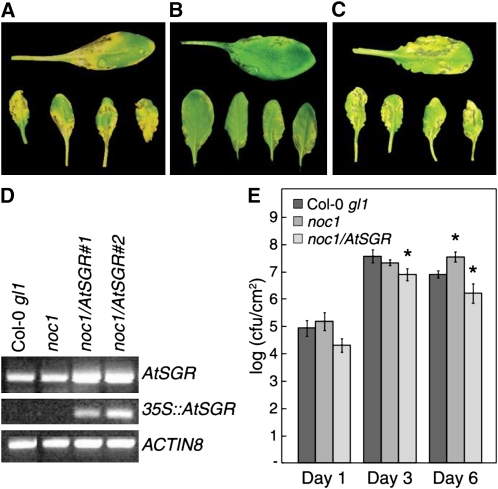
To determine whether the D88Y mutation in AtSGR is responsible for the noc1 phenotypes, we transformed noc1 plants with the full-length AtSGR cDNA from Col-0 gl1 cloned in pBAR-35S, which contains the cauliflower mosaic virus 35S promoter and Basta (glufosinate)-resistance gene. Ten independent T2 lines were confirmed to exhibit Basta resistance and to harbor the AtSGR transgene. Three homozygous T3 lines were chosen for disease symptom observations after infection by Pst DC3000. In preliminary tests, all three were restored in chlorosis symptom development. We then focused on line 1 for further disease symptom and bacterial growth analyses. Results from this line are shown in Figure 3. By 72 to 96 hpi, Pst DC3000-infected noc1/35S:AtSGR plants showed disease symptoms, including chlorosis, that were more pronounced than Pst DC3000-infected Col-0 gl1 plants (Fig. 3, A–C). Interestingly, in these experiments we noticed that noc1/35S:AtSGR plants did not support bacterial multiplication to the level observed in either noc1 or Col-0 gl1 plants at day 3 (Fig. 3E). This observation raised the possibility that accelerated disease symptoms may negatively affect Pst DC3000 growth and, accordingly, the noc1 mutant may allow better bacterial growth. However, we did not observe an enhanced Pst DC3000 growth at day 3 in noc1 plants (Figs. 1B and 4E). We commonly use a 3-d period for assessing Pst DC3000 multiplication in the laboratory; however, bacterial infection in the field involves longer durations. We therefore extended our multiplication assay to 6 d, which led to an interesting finding. Pst DC3000 populations declined in Col-0 gl1 and noc1/35S:AtSGR plants, as infected tissues senesced, whereas Pst DC3000 maintained a high population in the noc1 plants (Fig. 3E). These results suggest that disease symptom development restricts Pst DC3000 persistence in infected tissues.
Figure 3.
Complementation of the noc1 mutation by 35S:AtSGR. Disease symptoms on Col-0 gl1 (A), noc1 (B), and noc1/35S:AtSGR line 1 (C) leaves 4 d after dip inoculation with Pst DC3000 at 1 × 108 CFU/mL. D, RT-PCR using AtSGR-specific primers showing native expression of endogenous AtSGR in Col-0 gl1 and noc1 plants and increased expression in noc1 lines complemented with 35S:AtSGR transgene (top). RT-PCR using primers utilizing transgene-specific primers (see “Materials and Methods”) shows expression of 35S:AtSGR only in complemented noc1 lines (middle). RT-PCR of ACTIN8 was used as a loading reference (bottom). E, Pst DC3000 multiplication assay. Col-0 gl1, noc1, and noc1/AtSGR plants (line 1) were dip inoculated with Pst DC3000 at 108 CFU/mL. Bacterial populations were determined at 1, 3, and 6 dpi. t tests were performed in Microsoft Excel using a two-sample, equal variance formula. Comparisons are between Col-0 gl1 and noc1 and between Col-0 gl1 and noc1/AtSGR at each time point. P values of <0.05 are indicated with a single asterisk (*).
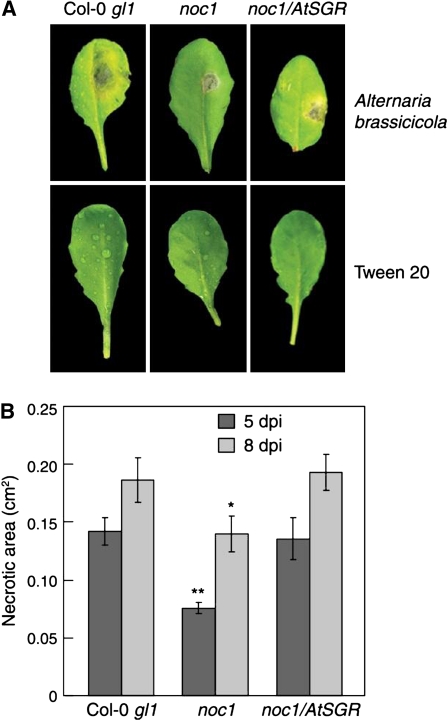
Figure 4.
Effects of the noc1 mutation on A. brassicicola-induced disease symptoms. A, Top sections: Chlorosis and/or necrosis caused by A. brassicicola infection in Col-0 gl1 (left), the noc1 mutant (center), and noc1/35S:AtSGR (right) leaves 8 dpi. Bottom sections: Mock inoculations with 0.1% Tween 20 are shown. B, Necrotic lesion areas were measured at 5 and 8 dpi in A. brassicicola-infected Col-0 gl1, noc1, and noc1/AtSGR plants using ImageJ software. Student’s t tests were performed in Microsoft Excel using a two-sample, equal variance formula. Comparisons are between Col-0 gl1 and the noc1 mutant and between Col-0 gl1 and noc1/35S:SGR at each time point. P values of <0.05 are indicated with a single asterisk (*); P values < 0.005 are indicated by **.
Effect of the noc1 Mutation on A. brassicicola-Induced Chlorosis
To determine whether noc1 plants are also affected in disease symptoms caused by a fungal pathogen, Col-0 gl1 and noc1 plants were inoculated with spores of the necrotrophic fungus A. brassicicola. A necrotic lesion developed at the site of inoculation in Col-0 gl1, noc1, and complemented noc1/35S:AtSGR plants at 5 to 10 dpi. In some experiments, a chlorotic halo, surrounding the necrotic lesion, may also develop within 5 to 10 dpi in only Col-0 gl1 and noc1/35S:AtSGR plants (Fig. 4A). However, the chlorosis phenotype induced by A. brassicicola was variable between experiments and could not be quantified reproducibly. To quantify disease symptoms, we therefore measured necrotic lesion areas using ImageJ software. The area of necrotic lesion development was smaller in noc1 plants infected with A. brassicicola than that in Col-0 gl1 plants infected in the same manner (Fig. 4). Fungus-induced disease necroses were restored in noc1/35S:AtSGR plants infected with A. brassicicola (Fig. 4). Plants inoculated with buffer control (0.1% Tween 20) alone showed no signs of chlorosis or necroses (Fig. 4A). These results demonstrate that AtSGR1 is required for the development of disease symptoms caused by a necrotrophic pathogen.
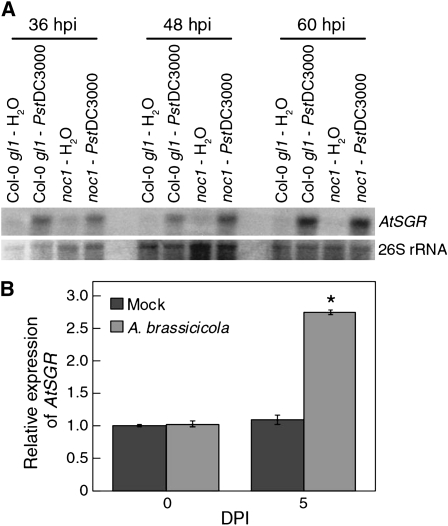
AtSGR Is Highly Induced during Pst DC3000 and A. brassicicola Infection
Previous studies have shown that SGR expression is critical for initiation of developmentally regulated chlorophyll degradation in a number of plant species (Armstead et al., 2006, 2007; Park et al., 2007; Ren et al., 2007). The requirement of AtSGR for disease chlorosis suggests that the expression of AtSGR might be induced during pathogen infection. To examine this possibility, we collected total RNA from water- and Pst DC3000-inoculated noc1 and Col-0 gl1 plants at 36, 48, and 60 hpi and performed northern-blot analyses using an AtSGR-specific probe. We found that the AtSGR expression is strongly induced by Pst DC3000, but not water, in both Col-0 gl1 and noc1 plants at all sampled time points (Fig. 5A). Thus, AtSGR expression is regulated during Pst DC3000 infection and the noc1 mutation does not significantly affect this expression. Quantitative reverse transcription (RT)-PCR analysis was also performed with RNA from A. brassicicola-infected leaves, showing that AtSGR expression was induced by fungal infection (Fig. 5B; Supplemental Fig. S1).
Figure 5.
Induction of AtSGR expression during infection. A, Northern-blot analysis of AtSGR expression in Pst DC3000-infected Col-0 gl1 and noc1 mutant plants. 26S rRNA visualized by ethidium bromide staining was used as a loading reference. B, Quantitative RT-PCR analysis of AtSGR expression in mock- and Alternaria-infected Col-0 gl1 plants at 0 and 5 dpi. Error bars represent sds from three biological replicates. The experiment was repeated once and similar results were obtained. Asterisks indicate significant difference (P < 0.05) between mock- and Alternaria-infected Col-0 gl1 plants using the Student’s t test.
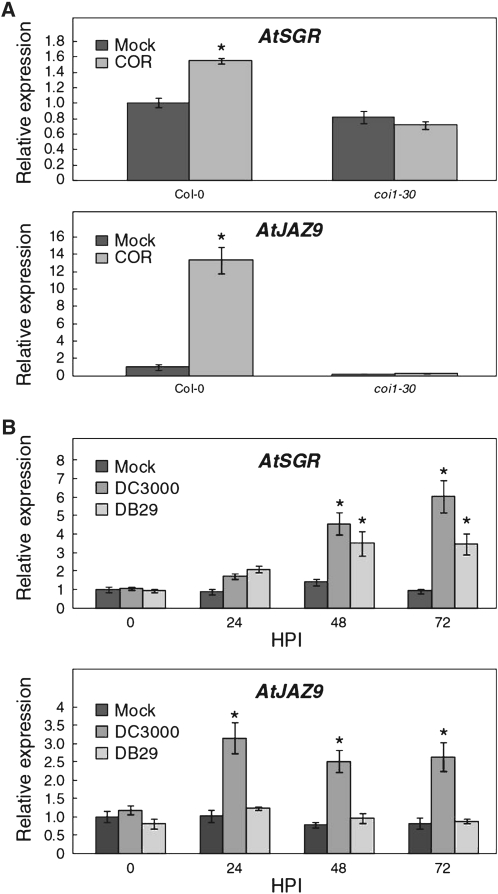
We next investigated whether specific virulence factors of Pst DC3000 could induce the expression of AtSGR. COR is a well-known bacterial virulence factor that promotes the development of disease chlorosis in plants (Uppalapati et al., 2005, 2007; Ishiga et al., 2009). Recent studies have demonstrated that COR mimics the active form of the plant hormone jasmonate and directly targets the jasmonate receptor complex in which the CORONATINE INSENSITIVE1 (COI1) F-box protein is a principal component (Katsir et al., 2008; Melotto et al., 2008a; Fonseca et al., 2009; Sheard et al., 2010). To determine whether COR could induce expression of AtSGR, we treated 8-d-old, Col-0 gl1 seedlings with buffer or 10 μm COR. Seedlings were collected at 3 h posttreatment and total RNA was isolated for each group. Quantitative RT-PCR analysis showed that, like JAZ9 (a known COR/jasmonate-responsive gene; Thines et al., 2007), AtSGR expression was induced by COR (Fig. 6A; Supplemental Fig. S2A). We next examined whether COR-induced expression of AtSGR requires COI1-dependent jasmonate signaling. As shown in Figure 6A, COR could not induce AtSGR expression in the coi1 mutant (Feys et al., 1994). These results suggest that COR induction of AtSGR expression requires an intact jasmonate signaling pathway.
Figure 6.
Role of COR in the induction AtSGR expression. A, Quantitative RT-PCR showing AtSGR and AtJAZ9 expression in mock- and COR-treated wild-type and coi1 background plants. Error bars represent sd from three biological replicates. B, AtSGR expression is induced by 48 hpi in Col-0 gl1 plants infected with Pst DC3000 (COR+) or DB29 (COR−). Quantitative RT-PCR shows AtSGR and AtJAZ9 expression in mock- and P. syringae-infected Col-gl1 plants. Error bars represent sds from three biological replicates. Experiments were repeated once and similar results were obtained. Asterisks indicate significant difference (P < 0.05) when comparing bacterial infections to mock inoculations using the Student’s t test.
Finally, we examined whether COR production is necessary for Pst DC3000 to induce the expression of AtSGR. Col-0 gl1 plants were infected with Pst DC3000 or a COR-deficient mutant strain, DB29 (Uppalapati et al., 2007). Quantitative RT-PCR analysis revealed that expression of AtSGR is strongly induced by 48 hpi, continuing through 72 hpi, in Col-0 gl1 plants infected with either DB29 or Pst DC3000 (Fig. 6B; Supplemental Fig. S2B), although expression of AtSGR was significantly higher in plants infected with DC3000 as compared to those infected with DB29. These data indicate that COR is sufficient but not necessary for Pst DC3000 to induce AtSGR expression during infection.
DISCUSSION
Development of disease symptoms represents the final stage of a pathogenic infection and is particularly destructive to the plant because it damages plant tissues, affecting not only crop productivity, but also crop marketability. Reduced disease symptoms, in the form of disease-tolerant cultivars, could be exploited as a method of disease management. Despite its importance in disease, the molecular control of disease symptom production remains one of the least-understood aspects of plant diseases.
Although ethylene and jasmonate signaling pathways are known to be important for Pst DC3000-induced disease chlorosis (Bent et al., 1992; Uppalapati et al., 2011), it is not understood how these pathways ultimately impact chlorophyll homeostasis in infected tissues. Our isolation of the noc1 mutant and subsequent identification of AtSGR gene begin to provide molecular insight into how Pst DC3000 infection perturbs chlorophyll homeostasis and causes tissue chlorosis. Previous research has suggested that SGR-family proteins, once produced during senescence, enter the chloroplast and destabilize photosystem complexes, which seems to be a prerequisite for regulated chlorophyll degradation (Park et al., 2007). We found that Pst DC3000 infection induces the expression of AtSGR and transgenic constitutive expression of AtSGR accelerates disease symptom development caused by Pst DC3000 infection. These results indicate that pathogen-induced disease chlorosis results from activation of a key regulator of an endogenous, senescence-associated chlorophyll degradation program.
The identification and characterization of the noc1 mutant also raises several important issues regarding the relationship between disease symptom development and bacterial pathogenesis. First, our data show that, in the case of Pst DC3000 infection at the inoculums of 1 × 106 CFU/mL, the bacterial population reaches a maximum level at 3 dpi in both wild-type and noc1 plants. However, whereas the Pst DC3000 population declined in wild-type Col-0 gl1 plants, it persisted at a high level for a longer time in noc1 mutant plants (Fig. 3). This finding has significant ramifications in terms of the role of disease symptom in bacterial pathogenesis. Disease symptoms are commonly believed to be a consequence of collateral damages caused by infection. However, our result suggests that this may not be the case: Disease symptom development can play an active role in negatively impacting the persistence of hemibiotrophic pathogens like Pst DC3000 at late stages of pathogenesis. Interestingly, a similar result was observed in the study of XopD, a type III effector of Xanthomonas campestris pv vesicatoria, which delays symptom development and tissue degeneration in tomato via modulation of host transcriptional programming, resulting in increased pathogen multiplication (Kim et al., 2008).
Second, previous reports show that bacterial phytotoxin COR induces chlorosis in plants and that COR-deficient mutants have reduced ability to cause chlorosis (Ma and Cuppels, 1991; Whalen et al., 1991; Uppalapati et al., 2005, 2007). Consistent with these observations, we found that COR was sufficient to activate AtSGR expression, providing a molecular basis for the involvement of COR in disease chlorosis. Additionally, COR-treated coi1 mutants showed no induction of AtSGR expression (Fig. 6), suggesting that the jasmonate receptor complex is required for COR-induced expression of AtSGR. Interestingly, we found a putative MYC2-binding motif (ACGTG; Boter et al., 2004; Yadav et al., 2005) in the promoter region of AtSGR at nucleotide position −23 (data not shown). MYC2 is a major transcription factor involved in jasmonate/COR-induced gene expression (Lorenzo et al., 2004; Laurie-Berry et al., 2006), further suggesting an involvement of jasmonate signaling in COR-mediated induction of AtSGR expression. It should be pointed out, however, that in Arabidopsis, purified COR was shown to induce purpling, instead of chlorosis (Bent et al., 1992). The exact reason for this phenomenon is not known. Because COR structurally and functionally mimics jasmonate, which is known to induce anthocyanin production in Arabidopsis (Ellis and Turner, 2001), it is possible that the chlorosis induced by COR in Arabidopsis is masked by purpling associated with anthocyanin production. In any case, although COR is sufficient for induction of AtSGR, we found that it is not required for AtSGR induction during infection with Pst DC3000 (Fig. 6). Thus, it is likely that COR is not the only virulence factor in Pst DC3000 that is involved in the development of disease chlorosis, but that the action of additional virulence factors (possibly type III effectors) may also be involved in the induction of AtSGR and contribute to the production of disease chlorosis. Indeed, disease chlorosis is often observed in Arabidopsis mutants that allow high multiplication of COR-deficient Pst DC3000 (Melotto et al., 2006; Zeng and He, 2010).
Third, in addition to affecting disease chlorosis, the noc1 mutation also reduced disease necrosis caused by Pst DC3000 and A. brassicicola infection, although this effect is less obvious compared to that of disease chlorosis (Figs. 1 and 4). This is an interesting finding in light of a recent study that investigated the effect of AtSGR overexpression and RNAi-mediated suppression on the hypersensitive response (HR) elicited by Pst DC3000 (avrRpm1) in Arabidopsis (Mur et al., 2010). It was found that increased and decreased AtSGR expression, respectively, accelerated and suppressed the kinetics of HR-associated cell death in resistant Arabidopsis plants. Mur and colleagues postulate that some phototoxic chlorophyll catabolites contribute to HR cell death in resistant plants (Mur et al., 2010). If so, we speculate that such chlorophyll catabolites could also contribute to the formation of disease necrosis in susceptible Arabidopsis plants infected by Pst DC3000 or A. brassicicola, as observed in our study (Figs. 1 and 4).
The stay-green phenotype was first described in 1866 by Gregor Mendel in differentiating yellow and green cotyledon color in segregating populations of pea (Pisum sativum; Mendel, 1866). SGR orthologs have been cloned from a wide range of dicot and monocot species (Armstead et al., 2006, 2007; Jiang et al., 2007; Park et al., 2007; Sato et al., 2007; Aubry et al., 2008; Barry et al., 2008). Consequently, we hypothesize that pathogen induction of SGR genes may be a common mechanism underlying disease chlorosis across a wide spectrum of plant-pathogen interactions. As such, further study of SGR genes and their regulation could lead to transgenic plants with not only controlled senescence, but also disease symptom expression, thereby benefiting agriculture.
MATERIALS AND METHODS
Plant Material, Mutagenesis, and Growth Conditions
Approximately 1 g of Arabidopsis (Arabidopsis thaliana) ecotype Col-0 gl1 seeds was mixed with 100 mL of distilled water and 250 μL of ethyl methanesulfonate. The mixture was incubated overnight at room temperature in the dark with gentle agitation. The seeds were washed six times with 500 mL of distilled water, resuspended in 300 mL of 0.1% agarose, and sown onto a soil mixture (equal portions of Baccto high-porosity professional plant mix, perlite, and vermiculite, covered with a thin layer of fine vermiculite). The flats were covered with lids and incubated in the dark at 4°C for 3 d. The flats were then transferred to a growth chamber (20°C with 12 h of fluorescent light [100 μE m−2 s−1] and 12 h of darkness). The plants were self fertilized to create a population of M2 plants.
Screening and Isolation of Arabidopsis Mutants
Four- to 6-week-old M2 plants were dipped in a 1 × 108 CFU/mL suspension of Pst DC3000 and 0.05% Silwet l-77 (Lehle Seeds, www.arabidopsis.com) for 2 to 3 s. The inoculated plants were incubated in high (80%–90%) humidity conditions for 96 h and screened for a lack of symptom development.
Bacteria Enumeration in Inoculated Leaves of noc1 Mutants and Wild-Type Col-0 gl1 Plants
Four- to 5-week-old plants were used for bacteria enumeration. Pst DC3000 was grown in low-salt Luria-Bertani broth to the mid-to-late logarithmic phase at 28°C. Bacterial cultures were pelleted and resuspended in sterile water to a final OD600 of 0.2 (equivalent to 1 × 108 CFU/mL) for dip inoculation. Fully expanded leaves were dip inoculated with bacterial suspensions. Plants were placed in trays with standing water and covered with plastic wrap to maintain high humidity. During the experimental period there was no obvious tissue desiccation in inoculated plants. Bacteria enumeration followed the protocol described by Katagiri et al. (2002). P values were derived from multiplication data utilizing Microsoft Excel software for t test statistical analysis.
Alternaria brassicicola Infection
We inoculated 10 leaves of each plant with 10 μL of Alternaria brassicicola spores at a concentration of 6.4 × 105 spores/mL suspended in 0.1% Tween 20. For control inoculations we used 0.1% Tween 20. Plants were covered with humidity domes and kept at high humidity throughout the infection process.
For quantification of disease symptoms caused by fungal infection, the areas of necrotic lesions caused by Alternaria inoculation were measured on 30 infected plant leaves. The average lesion area was calculated from 30 to 50 lesions at 5 and 8 dpi using ImageJ software. Older leaves were excluded from the sample set to avoid senescence-associated chlorosis and necrosis.
AtSGR expression was measured in A. brassicicola-infected tissue at 0 and 5 dpi. RNA was isolated from leaf tissue using an RNAeasy kit (Qiagen, www.qiagen.com). Semiquantitative RT-PCR was performed as described by the manufacturer (Takara, www.takara-bio.com).
Chlorophyll Extraction and Quantification
For chlorophyll abundance assays we infiltrated leaf tissue with 2 × 106 CFU/mL Pst DC3000 and collected samples at 0, 24, 48, 72, and 96 hpi. All chlorophyll extraction steps were conducted in near darkness. Leaf disks from four separate leaves at each time point were frozen in liquid nitrogen and stored at 80°C. The frozen tissue was homogenized in 600 μL of 80% acetone, the homogenates were centrifuged at 500g for three minutes at 4°C, and the supernatant was transferred to a new tube and kept on ice. The absorbance of four dilutions (1:10, 1:5, 1:3, and 1:2.5) of each sample was determined using a spectrophotometer. The amount of chlorophyll was calculated as previously described (Arnon, 1949).
Gene Mapping
Mapping of the noc1 mutation was conducted as described by Lukowitz and colleagues (Lukowitz et al., 2000). Initial genome-wide screening was conducted using an array of primers (Invitrogen, www.invitrogen.com) to detect simple sequence length polymorphisms from each chromosome. Additional INDELs, identified in the Monsanto Arabidopsis polymorphism and Ler sequence collection (www.arabidopsis.org/browse/Cereon/index.jsp), were used to further define the region containing the noc1 mutation (Jander et al., 2002).
Amino acid sequences of AtSGR and its orthologs were aligned using ClustalW (www.ddbj.nig.ac.jp/search/clustalw-j.html) and were then adjusted manually. Complementation of the noc1 phenotype was accomplished by transforming noc1 plants with 35S:AtSGR (At4g22920) in binary vector pBAR-35S (provided by J. Dangl, University of North Carolina, Chapel Hill, NC). The cDNA was amplified from a pool of total Col-0 gl1 RNA using LA Taq polymerase (Takara) and AtSGR-specific primers carrying XhoI (5′) and SpeI (3′) restriction sites (F: ggctcgagatgtgtagtttgtcggc and R: ggactagtctagagtttctccggatt) to facilitate eventual cloning into pBAR-35S. The PCR product was first cloned into pTOPO 2.1 (Invitrogen) and the AtSGR insert sequenced. The sequence-verified AtSGR insert was then transferred into the XhoI and SpeI sites of the binary vector pBAR-35S. Plant transformation was conducted as described (Clough and Bent, 1998). Transformants were screened in the T1 and T2 generations for Basta herbicide resistance and in T2 and T3 generations for the presence of the AtSGR transgene using RT-PCR with primers specific to the 5′ and 3′ untranslated regions (pBAR1 vector sequence) of the transgene: F-ggattgatgtgatatctccac and R-caagaccggcaacaggattc.
Northern Blotting
For northern-blotting analysis, total RNA was isolated from Arabidopsis leaves using the RNAgents total RNA isolation system (Promega, www.promega.com). The RNA concentration was determined based on A260, and RNA was separated on 2% formaldehyde agarose gels. The RNA was blotted onto Hybond N+ nylon membranes (Amersham, www.gelifesciences.com) using 10× saline sodium citrate buffer and UV cross-linked using a Stratalinker (Stratagene, www.genomics.agilent.com) with the auto-cross-link setting. Approximately 100 ng of a 300-bp exon-coding fragment, from AtSGR cDNA, was amplified by PCR using the following primers: F: atgtgtagtttgtcggcgat and R: cccggtatagcctatttgcc. The resulting fragment was purified using the Qiagen gel extraction kit (Qiagen) and labeled with 32P-CTP using random hexamers. Blotted membranes were hybridized with labeled probes using PerfectHyb plus hybridization buffer (Sigma-Aldrich, www.sigmaaldrich.com), following the manufacturer’s protocol, and then washed in 0.5× saline sodium citrate buffer and exposed to x-ray film for approximately 16 h.
Quantitative RT-PCR
Total RNAs were isolated from Arabidopsis leaves using the RNAgents total RNA isolation system (Promega, www.promega.com). Poly d(T) cDNA was prepared from 500 ng total RNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and quantified on an ABI7500 fast RT-PCR system (Applied Biosystems) with the fast SYBR green master kit (Applied Biosystems) according to the manufacturer’s protocols. Expression levels were normalized to AtPP2AA3 (PROTEIN PHOSPHATASE 2A SUBUNIT A, At1g13320; Czechowski et al., 2005). The primers used for RT-PCR were: AtSGR, 5′-ACTACCTGTGGTGTTGAAGG-3′ and 5′-CGACTTTGTTGAACTCATTGAC-3′, AtPP2AA3, 5′-GGTTACAAGACAAGGTTCACTC-3′ and 5′-CATTCAGGACCAAACTCTTCAG-3′, and AtJAZ9 (At1g70700), 5′-ATGAGGTTAACGATGATGCTG-3′ and 5′-CTTAGCCTCCTGGAAATCTG-3′.
Induction of AtSGR Expression by COR
Arabidopsis seeds (Col-0 gl1 and coi1) were germinated and grown on Murashige and Skoog agar plates for 4 d with coi1 seedlings being selected on 10 μm methyl jasmonate (Sigma-Aldrich). Seedlings were transferred to fresh Murashige and Skoog plates and grown for another 4 d and were treated with 10 μm COR (Sigma-Aldrich) or 0.1% ethanol by spraying. Seedlings were harvested at 3 h posttreatment and total RNA was isolated using the RNeasy plant mini kit (Qiagen) per manufacturer’s instructions. cDNA synthesis of AtSGR, JAZ9, and UBC was completed using avian reverse transcriptase and oligo-DT primers followed by RT-PCR using gene-specific primers (AtSGR: F: 5′atgtgtagtttgtcggcgat3′ and R: 5′ctagagtttctccggatttg3′; JAZ9: F: 5′atggaaagagattttctgggtttg3′ and R: 5′ttatgtaggagaagtagaagagta3′; UBC21: F: 5′tcaaatggaccgctcttatc3′ and R: 5′cacagactgaagcgtccaag3′).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Semiquantitative RT-PCR analysis of AtSGR expression in Alternaria-infected Col-0 gl1, the noc1 mutant, and noc1/AtSGR plants at 0 and 5 dpi.
Supplemental Figure S2. A, Semiquantitative RT-PCR showing levels of expression for AtSGR and JAZ9 in Col-0 seedlings 3 h after treatment with 10 μm COR (+) or buffer (−).
Acknowledgments
We thank Wanessa Wight and the laboratory of Jonathan Walton for providing A. brassicicola spores and Natasha Raikhel for the initial mapping primer set from Invitrogen.
References
- Alfano JR, Collmer A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42: 385–414 [DOI] [PubMed] [Google Scholar]
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al. (2006) From crop to model to crop: identifying the genetic basis of the staygreen mutation in the Lolium/Festuca forage and amenity grasses. New Phytol 172: 592–597 [DOI] [PubMed] [Google Scholar]
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al. (2007) Cross-species identification of Mendel’s I locus. Science 315: 73. [DOI] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Mani J, Hörtensteiner S. (2008) Stay-green protein, defective in Mendel’s green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol Biol 67: 243–256 [DOI] [PubMed] [Google Scholar]
- Badel JL, Nomura K, Bandyopadhyay S, Shimizu R, Collmer A, He SY. (2003) Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol Microbiol 49: 1239–1251 [DOI] [PubMed] [Google Scholar]
- Barry CS, McQuinn RP, Chung MY, Besuden A, Giovannoni JJ. (2008) Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol 147: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CL, Alarcón-Chaidez F, Gross DC. (1999) Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63: 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ. (1992) Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact 5: 372–378 [DOI] [PubMed] [Google Scholar]
- Block A, Schmelz E, Jones JB, Klee HJ. (2005) Coronatine and salicylic acid: the battle between Arabidopsis and Pseudomonas for phytohormone control. Mol Plant Pathol 6: 79–83 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S. (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DM, Bender CL, Kunkel BN. (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol Plant Pathol 6: 629–639 [DOI] [PubMed] [Google Scholar]
- Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza-Vázquez A, Bender CL, Kunkel BN. (2004) Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 17: 162–174 [DOI] [PubMed] [Google Scholar]
- Büttner D, He SY. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol 150: 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohn JR, Martin GB. (2005) Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J 44: 139–154 [DOI] [PubMed] [Google Scholar]
- Cui H, Xiang T, Zhou JM. (2009) Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol 11: 1453–1461 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-RD. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101: 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- He P, Chintamanani S, Chen Z, Zhu L, Kunkel BN, Alfano JR, Tang X, Zhou JM. (2004) Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J 37: 589–602 [DOI] [PubMed] [Google Scholar]
- Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, Bender CL. (2009) The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol 181: 147–160 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G. (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52: 197–209 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY. (2002) Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book. 1: e0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Hotson A, Keegan M, Schmelz EA, Mudgett MB. (2008) XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in xanthomonas-infected tomato leaves. Plant Cell 20: 1915–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN. (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Laurie-Berry N, Joardar V, Street IH, Kunkel BN. (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact 19: 789–800 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Guttman DS, Desveaux D. (2009) The targeting of plant cellular systems by injected type III effector proteins. Semin Cell Dev Biol 20: 1055–1063 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR. (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SW, Cuppels DA. (1991) Characterization of a DNA region required for production of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 4: 69–77 [Google Scholar]
- Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, et al. (2008a) A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, He SY. (2008b) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol 46: 101–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mendel G. (1866) Versuche über pflanzen-hybriden. Verh Naturforsch Ver Brünn 4: 3–47 [Google Scholar]
- Mittal S, Davis KR. (1995) Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Mur LA, Aubry S, Mondhe M, Kingston-Smith A, Gallagher J, Timms-Taravella E, James C, Papp I, Hörtensteiner S, Thomas H, et al. (2010) Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol 188: 161–174 [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B. (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Morita R, Nishimura M, Yamaguchi H, Kusaba M. (2007) Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc Natl Acad Sci USA 104: 14169–14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ1 is a target of the SCFCOI1 ubiquitin ligase during jasmonate signaling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL. (2005) The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J 42: 201–217 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Ryu CM, Ishiga T, Wang K, Noël LD, Parker JE, Mysore KS. (2011) SGT1 contributes to coronatine signaling and Pseudomonas syringae pv. tomato disease symptom development in tomato and Arabidopsis. New Phytol 189: 83–93 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, Mysore KS, Bender CL. (2007) The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 20: 955–965 [DOI] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ. (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacteriallocus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, He SY. (2010) A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol 153: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]