Abstract
Selective inhibition of gene expression by antisense oligodeoxynucleotides (ODNs) is widely applied in gene function analyses; however, experiments with ODNs in plants are scarce. In this work, we extend the use of ODNs in different plant species, optimizing the uptake, stability, and efficiency of ODNs with a combination of molecular biological and biophysical techniques to transiently inhibit the gene expression of different chloroplast proteins. We targeted the nucleus-encoded phytoene desaturase (pds) gene, encoding a key enzyme in carotenoid biosynthesis, the chlorophyll a/b-binding (cab) protein genes, and the chloroplast-encoded psbA gene, encoding the D1 protein. For pds and psbA, the in vivo stability of ODNs was increased by phosphorothioate modifications. After infiltration of ODNs into juvenile tobacco (Nicotiana benthamiana) leaves, we detected a 25% to 35% reduction in mRNA level and an approximately 5% decrease in both carotenoid content and the variable fluorescence of photosystem II. In detached etiolated wheat (Triticum aestivum) leaves, after 8 h of greening, the mRNA level, carotenoid content, and variable fluorescence were inhibited up to 75%, 25%, and 20%, respectively. Regarding cab, ODN treatments of etiolated wheat leaves resulted in an up to 59% decrease in the amount of chlorophyll b, a 41% decrease of the maximum chlorophyll fluorescence intensity, the cab mRNA level was reduced to 66%, and the protein level was suppressed up to 85% compared with the control. The psbA mRNA and protein levels in Arabidopsis (Arabidopsis thaliana) leaves were inhibited by up to 85% and 72%, respectively. To exploit the potential of ODNs for photosynthetic genes, we propose molecular design combined with fast, noninvasive techniques to test their functional effects.
Antisense oligodeoxynucleotides (ODNs) are short synthetic strands of DNA or analogs that consist of 15 to 20 nucleotides. They specifically target their complementary stretches of RNA by duplex formation and inhibit protein biosynthesis. In principle, they are able to interfere with each step of nucleic acid metabolism, preferentially with transcription, splicing, and translation (Crooke, 2004; Ravichandran et al., 2004; Gleave and Monia, 2005).
The inhibition of translation from mRNA into protein by ODNs occurs via two major mechanisms. Antisense ODNs are able to block the ribosome translocation sterically by hybridization, which is called hybridizational arrest. A second inhibitory mechanism operates in cells and is mediated by the cellular enzyme RNase H. The RNA:DNA heteroduplex is recognized by this enzyme and results in subsequent RNA cleavage (Gewirtz et al., 1998; Kurreck, 2003; Shi and Hoekstra, 2004; Chan et al., 2006). In this case, the antisense ODN remains intact and can trigger the cleavage of other RNA molecules like a catalyst.
As shown by studies on mammalian cells, antisense ODNs possess a number of desired features compared with alternative technologies such as short hairpin RNA (shRNA) and artificial microRNA. ODNs can be introduced directly into cells, whereas shRNA and artificial microRNA have to be inserted into a plasmid before they can be introduced into the cell (Scherer and Rossi, 2003; Sandy et al., 2005; Behlke, 2008). This means as well that the inhibition level of gene expression depends on the expression level of the plasmid inside the cell. Antisense ODNs, in contrast, are ready to use in a dose-dependent way (Kurreck, 2003). Their properties and functions can be tuned by chemical modifications. Another technology, small interfering RNAs (siRNAs), can also be introduced directly into the cell; however, compared with ODNs, they have two disadvantages: (1) their design and synthesis is more complicated; (2) ODNs can tolerate a 1 to 2 nucleotide mismatch and still be (at least partially) effective, whereas this would make siRNAs completely ineffective (Vickers et al., 2003). Moreover, nonspecific off-target effects form a bigger risk when using siRNAs. To our knowledge, methodologies based on synthetic shRNA, siRNA, and artificial microRNA molecules have not been tested in plants.
Antisense ODNs have a broad applicability, allow direct utilization of sequence information, are low cost, and have a high probability of success (Scherer and Rossi, 2003). Finally, homologous sequences can be targeted with a single antisense ODN; this means that a single antisense ODN can inhibit more than one gene from the same gene family (Bennett and Cowsert, 1999).
Sequence-selective inhibition of gene expression is applied extensively for the elucidation of complex gene expression patterns or the validation of results gained from high-throughput genomic experiments such as DNA arrays (Gleave and Monia, 2005; Rayburn and Zhang, 2008). Antisense ODNs have gained great interest, not only as genomic tools but also as possible therapeutic entities. These synthetic molecules are capable of interfering with the expression of the targeted gene, thus enabling the selective and rational design of genomic or therapeutic agents (Gleave and Monia, 2005; Rayburn and Zhang, 2008). Potentially, antisense ODNs could be used to cure any disease that is caused by the expression of a deleterious gene (e.g. viral infection, cancer growth, or inflammatory disease). It was first demonstrated by Zamecnik and Stephenson (1978) that antisense ODNs can be successfully used to inhibit viral replication in cell cultures, and since then, a number of applications have been reported (Scherer and Rossi, 2003).
One of the major challenges in the application of antisense ODNs is stabilization, since the natural molecular structure of ODNs is exposed to quick endonucleolytic and exonucleolytic degradation. In order to extend the biological life span and improve accessibility, a number of chemical modifications were introduced successfully into antisense ODNs (Chan et al., 2006; Lebleu et al., 2008). Most commonly, the stability of ODNs is increased by phosphorothioate (PS) modification (Eckstein, 1985). Phosphorothioated ODNs are the major representatives of first-generation DNA analogs that are the best known and most widely used antisense agents to date. In this class of analogs, one of the nonbridging oxygen atoms in the phophodiester bond is replaced by sulfur (Matsukura et al., 1987; Eckstein, 2000; Kurreck, 2003). The PS modification is particularly popular because it provides sufficient stabilization against nucleolytic degradation, whereas the duplex of PS-ODN and RNA is still recognized by cellular RNase H. This is an advantage compared with most other modifications. The cleavage of target mRNA by RNase H is considered an important factor for the activity of antisense ODNs (Stein et al., 1993; Stein, 1995). Equally important is the cellular delivery. A further challenge is to find the optimal target sites inside the RNA, which provide maximum efficacy and minimize off-target effects at the same time.
The application of sequence-selective gene-silencing ODNs has been described for several organisms, including mammalian cells, and it is an important emerging therapeutic tool in clinical medicine (Gewirtz et al., 1998; Dagle and Weeks, 2001; Hu et al., 2002; Shi and Hoekstra, 2004; Yang et al., 2004). In spite of the general applicability, the antisense ODN technology has not yet been truly exploited in plant tissue, although it was observed almost two decades ago that plant cell suspension cultures are capable of taking up single-stranded ODNs (Tsutsumi et al., 1992). Similarly, it has been suggested that pollen tubes can take up antisense ODNs (Moutinho et al., 2001a, 2001b). One advantage of applying ODNs in plants would be that the functions of vital genes can be studied and pleiotropic effects minimized, which represent common problems when creating mutants by genetic transformation. In itself, it is also an advantage that ODN-treated plants would not be genetically modified organisms; therefore, no special measures would be required during cultivation and transportation. Sun et al. (2005, 2007) were the first to apply ODNs in green leaves: antisense ODNs with their natural structure were used to inhibit the expression of SUSIBA2, a transcription factor involved in starch synthesis. The direct application of nucleic acids in plant tissues was then suggested to open the way for high-throughput screening for gene function (Roberts, 2005), but the use of this technique in plant science fell short of expectations.
The major aim of our work was to extend the applicability of antisense ODNs in leaves of dicotyledonous and monocotyledonous plant species, namely in tobacco (Nicotiana benthamiana), Arabidopsis (Arabidopsis thaliana), and wheat (Triticum aestivum). To this end, we combined techniques of the molecular design of ODNs and chlorophyll (Chl) fluorescence induction, a fast, noninvasive technique, which is widely used for screening, to identify mutants with modified photosynthetic performance (Niyogi et al., 1998; Eberhard et al., 2008). As model genes, we chose phytoene desaturase (pds), encoding a key enzyme in carotenoid biosynthesis, the chlorophyll a/b-binding protein gene (cab), encoding the light-harvesting proteins, and psbA, encoding the D1 protein. In order to maximize the inhibitory effects, we applied a complex optimization process in choosing accessible spots on the targeted mRNA. Our data show that ODNs are efficiently transported within the leaf and reach the nucleus and the chloroplast. Antisense ODNs efficiently knock down the gene expression of the pds gene, resulting in decreased carotenoid contents and diminished PSII activity; these inhibitory effects were enhanced by PS modification of the antisense ODNs. Regarding the cab gene, we show with western-blot analysis that a single antisense ODN is capable of inhibiting more than one gene from the same gene family with high efficiency. Using ODNs to inhibit the psbA gene, encoding for the D1 protein, we show, to our knowledge for the first time, that it is possible to inhibit a chloroplast-encoded gene with antisense ODN technology.
RESULTS
Uptake and Transport of ODNs in Plants as Investigated by Fluorescence Microscopy
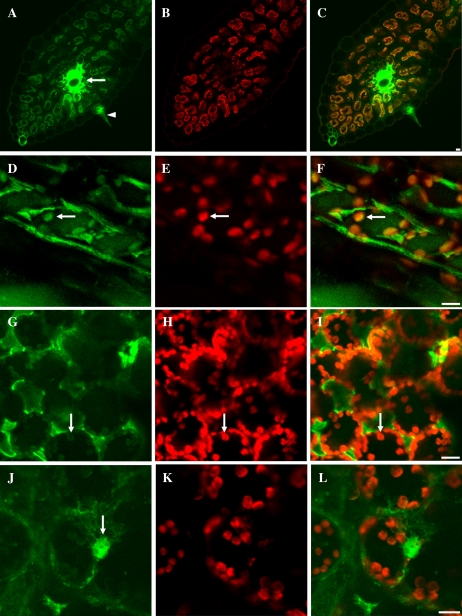
ODNs were introduced into intact tobacco leaves by infiltration with a syringe, as described for plasmid uptake by Sparkes et al. (2006), and into detached wheat leaves through their vascular system. To monitor the presence of ODN uptake in wheat and tobacco leaf cells, 17-mer ODNs representing random nonsense sequences were synthesized with 5′6-FAM fluorescent dye covalently attached. The localization of ODNs was analyzed by confocal laser scanning microscopy. In cross sections of leaves, after 24 h of feeding, the fluorescence intensity was high in the veins of wheat leaves (Fig. 1A). ODNs accumulated both in the epidermal and parenchymal cells, indicating the transport of ODNs inside the leaves. High-resolution confocal laser scanning microscopy showed that the ODNs accumulated both in the cytoplasm and the chloroplasts of parenchyma cells of wheat (Fig. 1, D and F) and tobacco (Fig. 1, G and I). Chl a fluorescence emission was used to localize the position of the chloroplasts (Fig. 1, B, E, H, and K). ODNs accumulated also in the nucleus (Fig. 1J). These data indicate that ODNs efficiently penetrate through cell walls and plasma membranes.
Figure 1.
Cellular distribution of fluorescein-labeled antisense ODNs (green) and Chl autofluorescence (red) in wheat (A–F) and tobacco (G–L) leaves. Low-magnification imaging of leaf blade cross sections is shown in A to C, where the arrow indicates a strong accumulation of antisense ODNs in the veins of a wheat leaf. The arrowhead in A points to a trichome with fluorescent ODNs. Merged images are shown in the right column. Arrows in D to I show chloroplast accumulation of antisense ODNs. The arrow in J shows a nucleus filled with fluorescent antisense ODNs. Bars = 10 μm.
pds as a Model Gene to Test the Efficacy of Antisense ODNs and the Selection of Target Sequences
To demonstrate the applicability of the antisense ODN technology in plants, we chose the nucleus-encoded pds and cab genes as model genes. The pds gene has been used as a marker in several studies on gene silencing in plants (Chamovitz et al., 1993; Kumagai et al., 1995; Lindgren et al., 2003; Tao and Zhou, 2004; Wang et al., 2005, 2009). pds encodes a key enzyme in carotenoid biosynthesis, which converts phytoene to the colored ξ-carotene in a two-step desaturation reaction (Bartley and Scolnik, 1995). Low expression of the pds gene results in a general suppression of carotenoid biosynthesis (Wetzel and Rodermel, 1998).
The mRNA molecules form complex secondary and tertiary structures; therefore, in antisense technology, the identification of mRNA sites that can be targeted efficiently is the key to success (Stein, 2001). The percentage of active antisense ODNs is known to vary from one target to the next. A variety of strategies have been developed to successfully design ODNs (Sohail and Southern, 2000). In this work, ODN sequences were designed based on a search for the presence of accessible single-stranded loops in the mRNA secondary structure (Zuker, 2003).
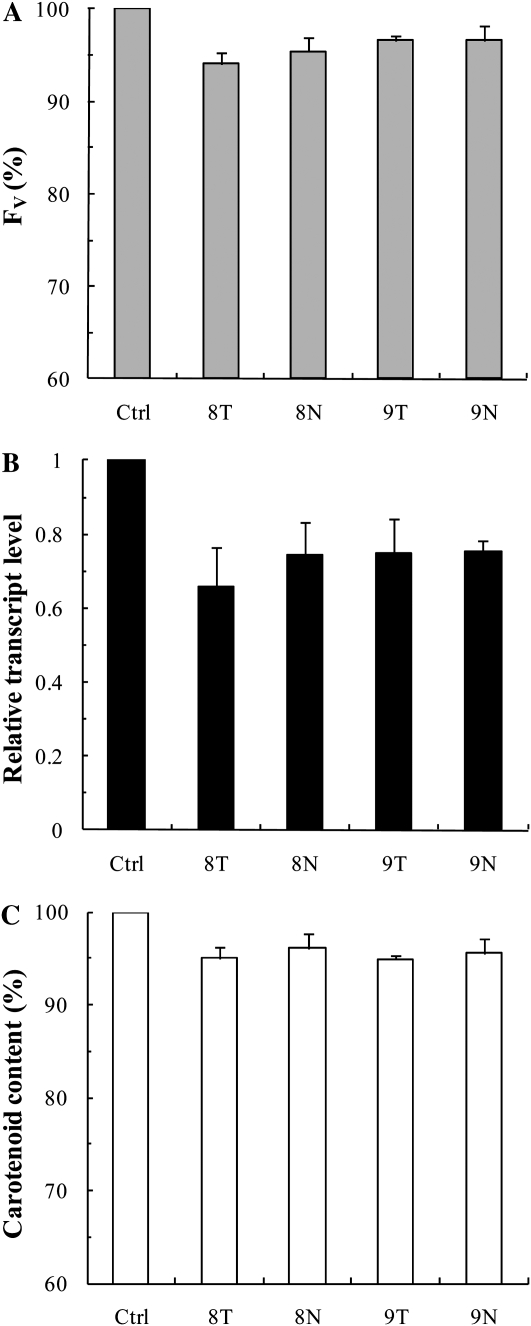
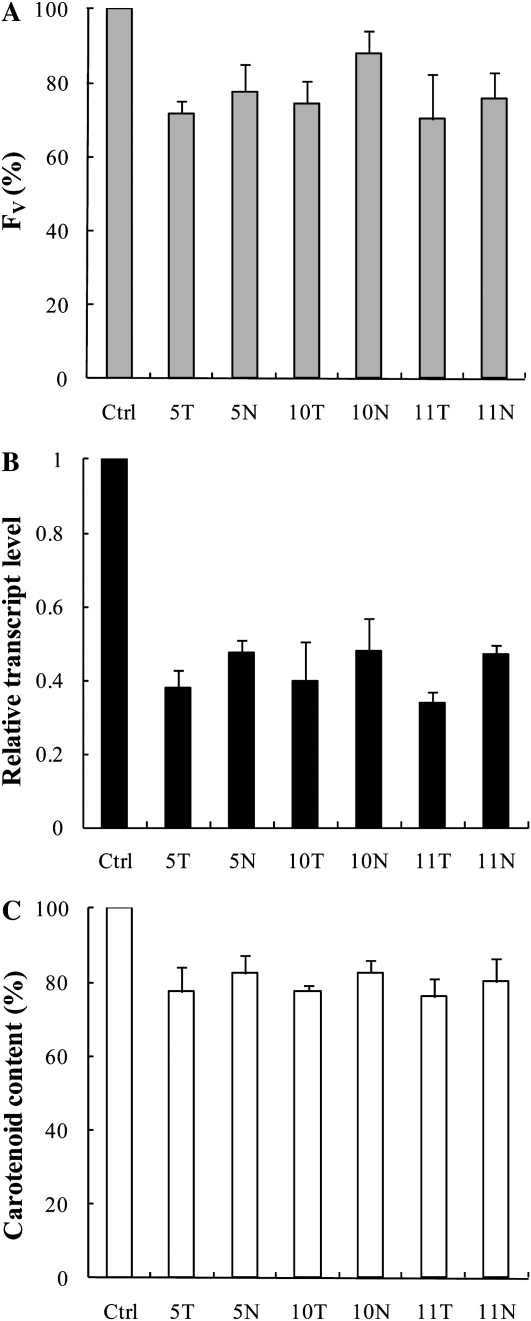
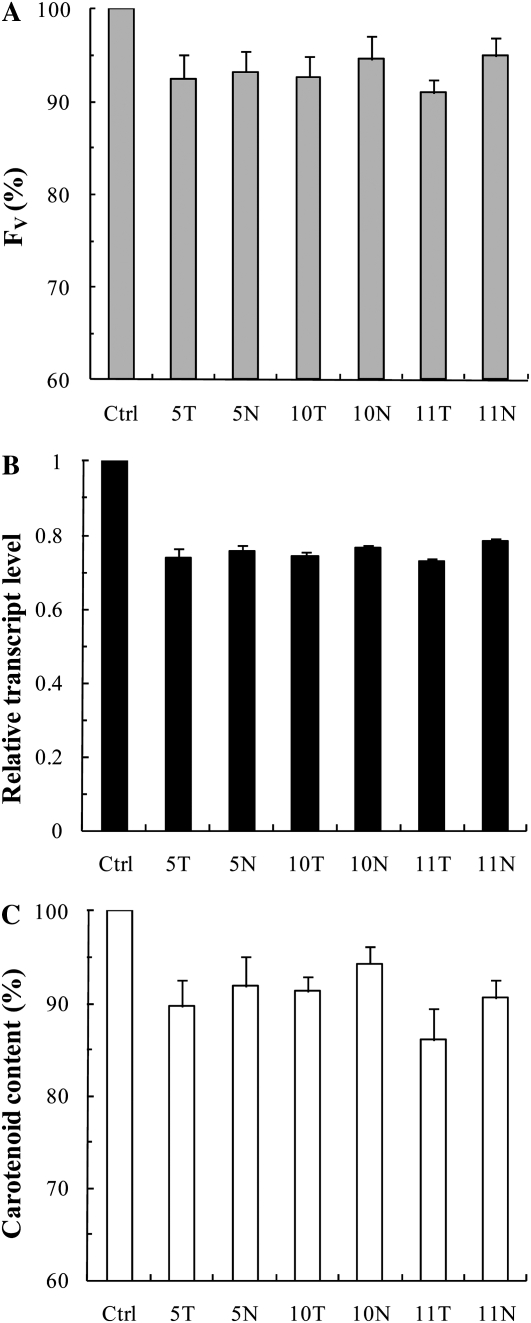
For the targeted pds mRNA, 10 different antisense ODNs were designed and synthesized both for wheat and tobacco, with their natural phosphodiester and PS structures. It has been shown that a decrease in the carotenoid content due to the suppression of the pds gene results in a decrease of the Chl content and a decrease of the photosynthetic efficiency as well (Wang et al., 2010), which is probably due to the instability of the synthesized Chl-binding protein complexes in the absence of carotenoids (Plumley and Schmidt, 1987). Therefore, to select the most efficient ODNs and also to determine the time points when the largest differences are found between the control and the ODN-treated plants, we used the fast Chl a fluorescence (OJIP) transient technique, a noninvasive and very sensitive method for measuring photosynthetic efficiency (Govindjee, 2004; Schansker et al., 2005, Lazár and Schansker, 2009). For monitoring the effects of antisense ODNs, we used the variable fluorescence parameter Fv (maximum fluorescence minus minimum fluorescence [Fm − F0]). Based on the Fv parameter, two pairs of antisense ODNs were selected in preliminary experiments for tobacco, which we called 8T, 8N, 9T, and 9N (where T stands for the PS structure and N indicates the natural, phosphodiester structure). For wheat, three pairs of antisense ODNs were selected, called 5T, 5N, 10T, 10N, 11T, and 11N. The preliminary experiments also showed that the largest differences between the Fv values of the antisense ODN-treated and control samples were observed after 8 h of ODN treatment; besides the 8-h illumination time, we also chose the 24-h time point in order to investigate the effects of ODNs at a later stage of greening.
The Effects of Antisense ODNs on pds in Young Tobacco Leaves
To investigate the effects of pds antisense ODNs on a dicotyledonous species, young leaves of 2-month-old tobacco plants were used. A 10 μm pds antisense ODN solution was infiltrated into intact leaf segments with the help of a syringe (Sparkes et al., 2006), and samples were taken 24 h after the infiltration.
The 24-h antisense ODN treatment resulted in 5% to 10% lower Fv values compared with the random nonsense controls (Fig. 2A). This was likely the consequence of the decrease in the transcript level of the pds gene, which was suppressed by about 25% to 35% compared with the random nonsense control (Fig. 2B). This inhibition led to a 5% to 10% decrease in the carotenoid content (Fig. 2C), a decrease similar to the one observed for Fv. The data also show that the antisense ODN 8 with PS modification (8T) was somewhat more efficient than its natural structure (8N). These relatively small effects of the antisense ODNs on the mRNA level, the carotenoid content, and the Fv value can be explained by the slow turnover of the PDS enzyme and the high background levels of the pigments and photosynthetic complexes. Therefore, we carried out experiments on greening leaves (i.e. during the synthesis of the photosynthetic machinery).
Figure 2.
Effects of pds antisense ODNs on the Chl a Fv (A), the relative transcript level (B), and the carotenoid content (C), expressed in percentage of the control in young tobacco leaves 24 h after infiltration with ODNs. Ctrl, Control, random nonsense sequence; T, PS structure; N, phosphorodiester (natural) structure. The values are averages ± sd of at least eight measurements derived from four independent experiments.
The Effects of pds Antisense ODNs during Greening of Wheat Leaves
It is well known that etiolated leaves contain no Chl, whereas lutein and violaxanthin can be found at low levels when compared with light-grown plants, and they contain only traces of any other carotenoid (Park et al., 2002). When etiolated leaves are exposed to light, Chl biosynthesis starts and the carotenoid content increases (von Lintig et al., 1997). Based on these observations, we expected that pds antisense ODNs could efficiently slow down or inhibit the deetiolation process. Etiolated leaves were detached and their basal part was submerged for 12 h in a 10 μm pds antisense ODN solution in the dark. Subsequently, they were illuminated with white light of 100 μmol photons m−2 s−1 for 24 h. The carotenoid content of etiolated leaves was low compared with green leaves and remained constant during the 12-h incubation in the presence of antisense ODNs in the dark (data not shown).
After 8 h of illumination of wheat leaves, the Fv value was 12% to 30% lower in the antisense ODN-treated plants (Fig. 3A) compared with controls (i.e. random nonsense ODNs). At the same time, the expression level of the pds gene was suppressed dramatically, by about 50% to 70% (Fig. 3B). This inhibition led to a significant decrease in the carotenoid content: 18% to 24% depending on the antisense ODN applied (Fig. 3C; the total carotenoid content of the control was about 0.2 μg mg−1 fresh weight). It is important to note that the differences in all three parameters were more pronounced in plants treated with PS-modified ODNs compared with ODNs with a natural structure; the largest difference was found between ODNs 11T and 11N.
Figure 3.
Effects of pds antisense ODNs on the Chl a Fv (A), the relative transcript level (B), and the carotenoid content (C) expressed in percentage of the control in etiolated wheat leaves after 8 h of incubation. Abbreviations are as in Figure 2. The values are averages ± sd of eight measurements derived from two independent experiments.
These data show that pds ODNs slow photomorphogenesis down. Indeed, the Chl content was also lower in the antisense ODN-treated plants (by about 25%–42%, depending on the ODN applied; data not shown). This is consistent with the well-established fact that the stability of pigment-protein complexes depends on the presence of carotenoids. It is interesting that following an 8 h deetiolation, the Chl a/b ratio was higher (typically between 10 and 16) in ODN-treated plants than in the control plants (Chl a/b ratio around 8), suggesting a much slower synthesis of the outer (Chl a- and b-containing) antennae in the ODN-treated leaves. This difference vanished after 24 h of illumination, which is in agreement with the results of Wang et al. (2009), who showed that in mature transgenic plants, the silencing of the pds gene does not change the Chl a/b ratio.
Seeing the interference of ODN treatments with early greening events, we looked at wheat leaves in a more advanced stage of their photomorphogenesis as well. After 24 h of deetiolation, the effect of ODNs on the Fv value was less than 10% (Fig. 4A). Concerning the transcript level of the pds gene, the decrease induced by the antisense ODNs was found to be about 25% during this later phase of deetiolation (Fig. 4B), while these values varied between 50% and 70% after 8 h (Fig. 3B). Also, the reduction in the carotenoid content became considerably smaller (Fig. 4C; the total carotenoid content was about 0.27 μg mg−1 fresh weight in the control leaves) compared with leaves that had been measured at an earlier stage of greening. This result is in accordance with the idea that ODNs can only inhibit de novo synthesis.
Figure 4.
Effects of pds antisense ODNs on the Chl a Fv (A), the relative transcript level (B), and the carotenoid content (C) expressed in percentage of the control in etiolated wheat leaves after 24 h of incubation. Abbreviations are as in Figure 2. The values are averages ± sd of eight measurements derived from two independent experiments.
The Effect of cab Gene Antisense ODNs on Antenna Development during the Greening of Wheat Leaves
Other model genes were the genes coding for the Chl a/b-binding proteins (cab genes). The CAB proteins (also called light-harvesting complexes [LHCs]) form a family of nucleus-encoded thylakoid proteins. At least 10 distinct types of CAB proteins have been recognized in higher plants. Some are found in the antenna of PSI and some in the antenna of PSII. Lhca genes encode the polypeptides of LHCI, while Lhcb genes encode PSII antenna complexes; in particular, Lhcb1, Lhcb2, and Lhcb3 genes encode the polypeptides of trimeric LHCII. The Lhcb4, Lhcb5, and Lhcb6 proteins (often called CP29, CP26, and CP24, respectively) are monomeric proteins linking the LHCII trimers to the reaction center core (Jansson, 1999).
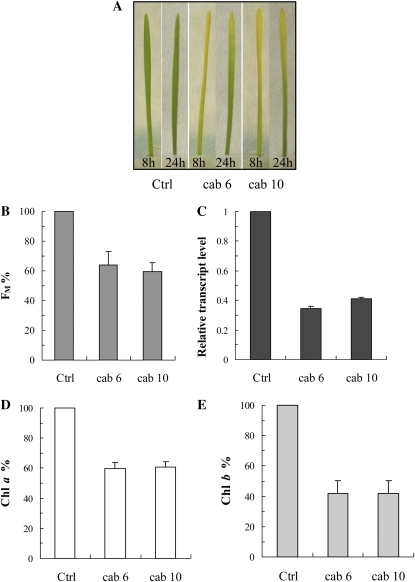
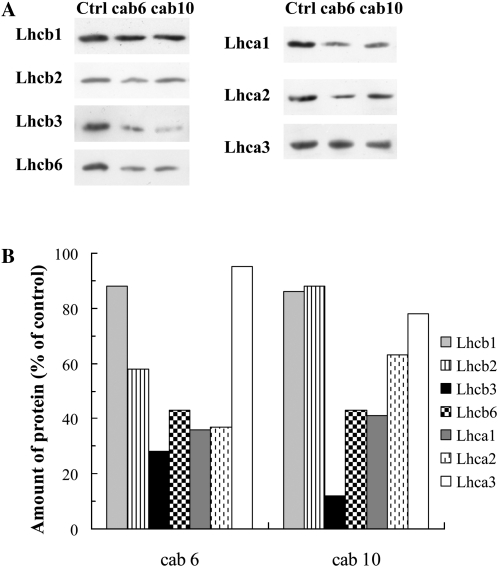
For the inhibition of the cab gene mRNAs of wheat, 10 different antisense ODNs were designed and synthesized using the natural (phosphodiester) structure based on the single mRNA sequence found in the GenBank database for wheat. Although all ODNs were able to inhibit the cab genes to some extent, the two most effective ODNs were selected using the fast Chl a fluorescence transients as a selection criterion. The fluorescence data also indicated that the maximum effect of the ODN treatments was obtained after 8 h of greening. Figure 5A, which shows photographs of etiolated wheat seedlings treated with CAB ODNs, also demonstrates very clear effects after 8 h of greening. After 8 h of illumination, the Fm values were 36% to 41% lower in the antisense ODN-treated plants than in the controls (Fig. 5B). At the same time, the expression levels of the cab genes were suppressed by about 59% to 66% (Fig. 5C). This inhibition led to a significant decrease in the Chl contents of the leaves (Chl a, 41%; Chl b, 59%; Fig. 5, D and E). We performed western-blot analyses for the components of the trimeric LHCII components (Lhcb1, Lhcb2, and Lhcb3 proteins), for one of the monomeric LHCs of PSII, the Lhcb6 protein (CP24), as well as for three components of the PSI LHCs (Lhca1, Lhca2, and Lhca3). In the case of the cab6 antisense ODN, of these complexes, the most affected proteins were Lhcb3, Lhcb6, Lhca1, and Lhca2 (66%–72% inhibition; Fig. 6). The cab10 antisense ODN inhibited the synthesis of Lhcb3 with a very high efficiency (approximately 88% compared with the control), whereas the other proteins analyzed were less affected. The least affected proteins were Lhcb1, Lhcb2, and Lhca3 for both ODNs tested (Fig. 6).
Figure 5.
Effects of two different cab antisense ODNs, cab6 and cab10, on wheat seedlings upon deetiolation of the leaves. Photographs of 8- and 24-h deetiolated leaves (A) and effects on the Chl a Fm (B), the relative transcript level (C), the Chl a content (D), and the Chl b content (E) after 8 h of greening are shown. Values are expressed in percentage of the control. The values are averages ± sd of eight measurements from two independent experiments.
Figure 6.
Amounts of Lhcb1, -2, -3, and -6 and Lhca1, -2, and -3 proteins in control and ODN-treated (cab6 and cab10) wheat leaves after 8 h of deetiolation, determined by western-blot analyses. Typical blots (A) and band intensities determined by densitometry (B) are presented.
Effects of Antisense ODNs on psbA in Arabidopsis Leaves
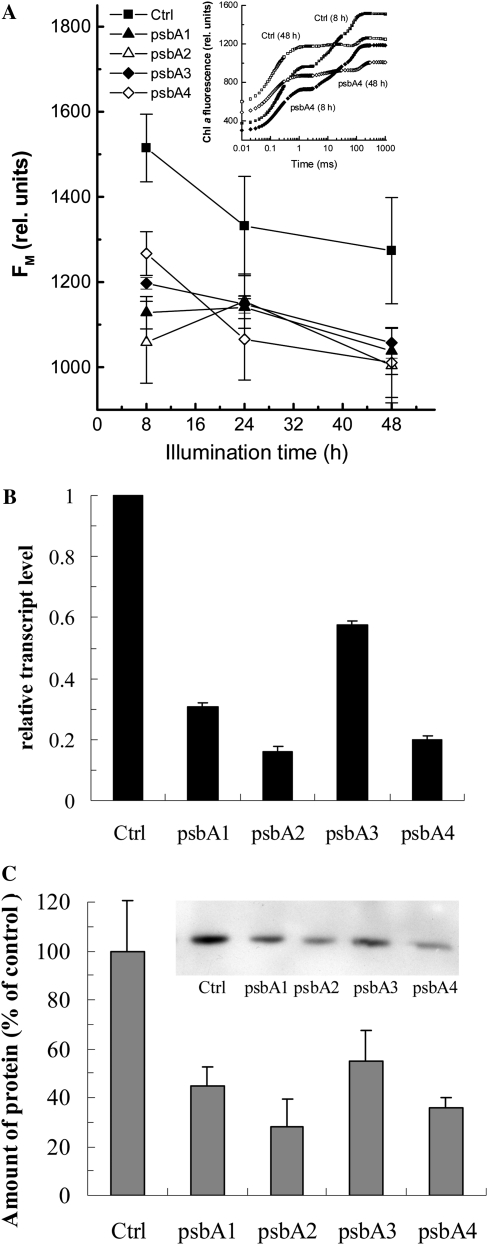
As a chloroplast-encoded model gene, we choose the psbA gene, which encodes the D1 protein of the PSII reaction center. For the inhibition of the psbA gene of Arabidopsis, four different antisense ODNs were designed and synthesized using the PS structure, based on the single psbA mRNA sequence of Arabidopsis. Fluorescence measurements were made after 8, 24, and 48 h of illumination, and of each fluorescence transient, the Fm value was determined (Fig. 7A). The inset in Figure 7A shows fluorescence transients of control and psbA4-treated leaves after 8 and 48 h of illumination. Both for the control and the ODN-treated leaves, the transients had a more reduced plastoquinone pool after 48 h of illumination (compare with Schansker et al., 2005). Since the Fm value is not affected by the redox state of the photosynthetic electron transport chain, it was used to assess the effect of the ODNs in Figure 7A. The main difference between the control and the ODN-treated leaves was already observed after 8 h of illumination. After 48 h of illumination, the four ODNs were inhibiting the psbA gene by 42% for the least effective ODN and up to 85% for the most effective ODN (Fig. 7B). We performed western-blot analyses for the D1 protein. The psbA2 and psbA4 antisense ODNs gave the strongest inhibition of the D1 protein: 72% and 64% inhibition, respectively (Fig. 7C).
Figure 7.
Effects of psbA antisense ODNs on the Chl a Fm (A), the relative transcript level (B), and the amount of D1 protein in ODN-treated Arabidopsis leaves after 48 h of illumination (C). Control leaves were treated with random nonsense ODNs. The inset in A shows OJIP transients of control and psbA4-treated leaves. A representative western blot is shown in the inset in C.
DISCUSSION
Key to identifying the functions of genes and for target validation is the ability to perturb the function of a particular gene in a given biological system. In plant science, several techniques have been applied for assigning the function to genes and proteins, including the creation of mutants and the knockout or knockdown of certain genes (e.g. by antisense cloning). The use of expression libraries, small molecule inhibitors, and peptide or antibody inhibitors are also common techniques for assigning the functions of genes.
Here, we have demonstrated that a sequence-specific transient gene inhibition induced by antisense ODNs in tobacco, Arabidopsis, and wheat leaves can be used to study gene function. We used three experimental approaches: infiltration of ODNs into undetached leaves of tobacco plants, vacuum infiltration of detached Arabidopsis leaves, and the uptake of ODNs by detached etiolated wheat leaves. Three model genes were chosen: the nucleus-encoded pds and cab genes and the chloroplast-encoded psbA gene.
In undetached tobacco leaves, a clear effect of the pds ODNs could be observed (Fig. 2), although it was not as pronounced as during the greening of etiolated wheat leaves. In tobacco leaves, carotenoids were already present at the start of the treatment; therefore, a smaller effect was expected than in the case of deetiolation. It should be noted that the uptake of the ODNs was not continuous when they were introduced by infiltration into the leaves. Since antisense ODNs can inhibit only the de novo biosynthesis, a larger effect is to be expected in leaves in which the pigment content is still increasing, preferably from a low level.
In the case of wheat leaves, the inhibitory effect of pds ODNs on the carotenoid content after 8 h of deetiolation was striking (Fig. 3C). After 24 h of greening, the effects were less pronounced but still noticeable (Fig. 4C), similar to the effects in juvenile tobacco leaves (Fig. 2C). The fact that the effects of antisense ODNs were less pronounced after 24 h of greening compared with 8 h of greening is a consequence of the process studied. During greening, the LHCs of PSII and PSI and their associated Chls and carotenoids accumulate. The ODNs targeting the pds and cab genes slow this accumulation process down but do not prevent it completely. Therefore, the difference between the control and the ODN-treated leaves will gradually disappear. Indeed, in an unreported experiment, in which the greening process was followed for 72 h, the difference disappeared completely.
The inhibitory effect of antisense ODNs on the cab proteins was strong despite the use of only the natural (phosphodiester) structure (Figs. 5 and 6). For example, in the case of the cab10 antisense ODN, the protein level of Lhcb3 was inhibited by about 88%. Our results also show a strong differential effect on the different members of the CAB protein family (Fig. 6). This illustrates that it is possible to design ODNs targeting a gene family, which is a clear advantage over creating mutants. For wheat, at present, only one cab sequence is available in the database; therefore, it was not possible to design antisense ODNs specific for a particular cab gene. With information on the whole wheat genome, it would be possible to align the cab genes, which can then be used as a starting point for the design of ODNs that target one gene or several genes that show high sequence homology. Nevertheless, our data show that already on the basis of a single gene sequence, ODNs could be designed that suppress the expression of several light-harvesting proteins during the deetiolation of detached leaves. The third target gene was psbA, which is a chloroplast-encoded gene. The inhibition of psbA by ODNs was as strong as the inhibition of the cab genes, suggesting that ODNs are as effective in chloroplasts as in the cytoplasm/nucleus. This experiment also shows that ODNs work as well in Arabidopsis as in wheat or tobacco.
ODN Application and Delivery
In this study, it is shown that ODNs dissolved in distilled water are efficiently taken up by and transported in plant leaves (Fig. 1). When testing the effect of different sugars, we found no evidence for enhanced uptake (data not shown). This result contrasts with the observation of Sun et al. (2005, 2007) that the uptake by barley (Hordeum vulgare) leaves of the SUSIBA2 transcriptional factor antisense ODNs was promoted by high concentrations of Suc (200 mm). During our preliminary experiments, we found that wheat, the spring cv CY-45 in particular, was sensitive to osmotic stress, which could already be observed in the presence of 200 mm Suc. This prompted us to make an attempt on sugar-independent uptake of ODNs, which appeared to be an efficient method under our experimental conditions, using the simple techniques described in “Materials and Methods.”
The application method was adapted to the type of plant used. Tobacco leaves are too large for an efficient passive uptake of ODNs from a solution. In the literature, it has been shown that fluorescent fusion proteins (Sparkes et al., 2006) and fluorescent sensors of reactive oxygen species (Hideg et al., 2002) can be injected into the leaf. We show here that this method also works for ODNs (Fig. 1, G–L). Arabidopsis leaves are too small for passive transport or injection; thus, we used vacuum infiltration to treat detached leaves with ODNs.
A special feature of the application of ODNs is the timing. The expression of both the pds and the cab genes is under circadian control (http://diurnal.cgrb.oregonstate.edu). Although the circadian rhythm does not play a role yet in etiolated wheat leaves, except for the experiments with juvenile tobacco leaves, the same daily schedule has been followed to avoid the effects of the circadian rhythm.
PS Modification of Antisense ODNs
One way to increase the efficiency of the inhibition of gene expression by antisense ODNs is to increase the stability of ODNs by chemical modification. PS modification (Eckstein, 1985; Campbell et al., 1990; Agrawal et al., 1991; Ghosh et al., 1993; Uhlmann et al., 2000) was used in several studies to protect ODNs against exonucleases and endonucleases, of which the most active enzyme is the 3′ exonuclease (Uhlmann and Peyman, 1990; Hoke et al., 1991; Shaw et al., 1991). Capping of the 3′ end, or both the 3′ and 5′ ends, by PS linkages protects the ODN against exonuclease degradation (Shaw et al., 1991; Gillardon et al., 1995), but these PS end-capped ODNs are still subject to endonuclease degradation. The combination of the end-capping technique and protection at internal pyrimidine residues, which are the major sites of endonuclease degradation, has been shown to be more efficient (Uhlmann et al., 2000; Samani et al., 2001).
PS-modified antisense ODNs are highly water soluble (Levin, 1999) and can efficiently recruit RNase H to cleave the target RNA (Zamaratski et al., 2001). All these properties make PS ODNs superior to the natural (phosphodiester) ODNs in antisense activity (Ravichandran et al., 2004).
In our study, we used a combination of end-capping and internal pyrimidine protection by PS linkages that indeed resulted in a significantly higher inhibition in the transcript level compared with the natural structure in the case of the pds gene (Figs. 2B, 3B and 4B). For tobacco leaves, the influence of the PS modification could be detected in the case of the 8T-8N ODNs on the mRNA levels. A similar but smaller effect existed for the 9T-9N ODN pair (Fig. 2B). Studying the wheat pds antisense ODNs with PS modification, the inhibition was about 15% higher compared with the natural structures (Fig. 3B). These data suggest that nucleases are active in plant cells and that PS modification can efficiently protect ODNs. Moutinho et al. (2001a, 2001b) have already used ODNs having PS modifications in the three bases adjacent to each terminus in a pollen tube model system, but to our knowledge, our study is the first that demonstrates that PS modification increases the efficacy of ODNs in leaves of higher plants. New generations of modified ODN structures, bearing, for example, a 3′-O substitution or locked nucleic acids, provide higher hybridization efficiency and cellular stability. Therefore, we anticipate a further increase in the inhibitory efficiency in future studies (Kurreck, 2003).
Mechanism of the Knockdown of Gene Expression by Antisense ODNs
The first proven mechanism of antisense ODN inhibition was the so-called hybridizational arrest mechanism, in which the ODN sterically blocks the ribosome translocation (Crooke, 1999; Kurreck, 2003). However, the predominant mechanism of inhibition of antisense ODNs involves the formation of an RNA-antisense ODN duplex through complementary Watson-Crick base pairing, leading to RNase H-mediated cleavage of the target mRNA (Crooke, 1999; Wu et al., 2004; Galarneau et al., 2005). RNase H is a ubiquitous enzyme that hydrolyzes the RNA strand of an RNA/DNA duplex. Oligonucleotide-assisted RNase H-dependent reduction of targeted RNA expression can be quite efficient, reaching 80% to 95% down-regulation of protein and mRNA expression. This fact can be understood when we consider the catalysis-like nature of RNase H cleavage.
The importance of RNase H-induced cleavage of mRNA has been demonstrated in several systems (Minshull and Hunt, 1986; Cazenave et al., 1993; Giles et al., 1995). The RNase H mechanism works in the case of ODNs that have a natural structure (phosphodiester) and PS modifications as well (Wilds and Damha, 2000; Damha et al., 2001).
For the three studied genes, we observed in all cases that inhibition at the mRNA level is higher than at the protein level. The smallest difference is observed for the psbA gene, but this is probably due to the high turnover rate of the D1 protein. However, the complexity of the system that we studied does not allow us to discriminate on this basis between the working mechanisms of ODNs that have been proposed in the literature (see above). A strategy that may be used to address the action mechanism of ODNs in plants is to use ODNs that have been modified in such a way that they are no longer a target for RNase H like 2′-O-methyl RNA, locked nucleic acid, or morpholino phosphoroamidate-based ODNs (Kurreck, 2003).
CONCLUSION AND PERSPECTIVE
In this paper, we have shown that ODNs can be efficiently transported within leaves of vascular plants and that they reach the chloroplasts in monocotyledonous and dicotyledonous plant species. Antisense ODNs were used to knock down the expression of the nucleus-encoded pds and cab genes and the chloroplast-encoded psbA gene. We have also shown that the efficiency of the inhibition of a target gene, the pds gene, can be increased by PS modification of the antisense ODNs. We also show, to our knowledge for the first time, that ODNs are efficiently transported into the chloroplasts and that chloroplast-encoded genes can be targeted by antisense ODNs. Antisense ODNs can be designed and synthesized in a very short period of time, no selection markers are required, and handling does not require special care, since plants treated with antisense ODNs are not genetically modified. With this approach, the functions of essential genes can also be readily studied and fewer pleiotropic effects are expected to occur, since the effects of the antisense ODNs develop on the time scale of hours to tens of hours. Although the effect of antisense ODNs is transient and full inhibition might not be possible to achieve, a disadvantage in many applications, these features can also be regarded as essential in certain important applications. For instance, in order to enhance the hydrogen evolution in green algae, Melis et al. (2000) used sulfur deprivation to transiently inhibit the de novo synthesis of the PSII reaction center complex protein D1. An additional advantage is that ODNs can be designed to target multiple genes at the same time. We propose that antisense ODN technology can be used as a high-throughput screening method for studies on the roles of genes and proteins with yet unknown functions. It is also conceivable that the interaction of genes can be studied with the help of antisense ODNs, since ODNs can be applied in combination. Photosynthesis is one of the research areas to which this method can be successfully applied. A special advantage of the study of photosynthetic genes is the availability of a range of sensitive, noninvasive techniques, such as time-resolved Chl a fluorescence and fluorescence lifetime measurements (Krumova et al., 2010), from which information can be derived on the operation of PSII and variations in the light-harvesting antenna system, as well as different time-resolved absorption measurements at various wavelengths, providing information, for example, on the operation of the PSI reaction center, the cytochrome b6/f complex, or the ability of the thylakoid membranes to generate and maintain a transmembrane electric field (Junge, 1977; Kramer et al., 2004). These techniques can be applied on small leaf areas and for some of these techniques even on individual cyanobacterial or algal cells and chloroplast elements as small as micrometer-sized voxels (volumetric pixels).
MATERIALS AND METHODS
Design and Synthesis of Antisense ODNs
ODN sequences were selected based on a search for freely available single-stranded loops in the mRNA secondary structure (M. Zuker, Mfold Web server for nucleic acid folding and hybridization prediction [http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form]; Zuker, 2003). The synthesis of ODNs was performed using an Expedite 8909 DNA synthesizer (Applied Biosystems) by standard cyanoethyl phosphoramidite chemistry at a nominal scale of 3 μmol. All of the reagents for the automated ODN synthesis were also from Applied Biosystems. The ODNs were purified on Poly-PAK cartridges (Glen Research), yielding more than 97% full-sized ODNs, as shown by analytical ion-exchange HPLC. PSs were built in with more than 98% efficiency by using EDITH sulfurizing reagent (Link Technologies) according to the manufacturer’s instructions.
For uptake experiments, random nonsense ODNs were labeled with fluorescein cyanoethyl phosphoramidite (6-FAM; Link Technologies; 494-nm excitation, 525-nm green emission) at the 5′ position and purified by HPLC.
ODNs Used
In the experiments, a random nonsense control was used. A nucleotide BLAST search of the whole GenBank database with this sequence did not yield a single hit of a sequenced angiosperm gene.
In Tobacco
pds antisense ODN(8) 5′-TTCTGAGTCACTACGATTT-3′ (19-mer) and ODN(9) 5′-GCTTCGCTAGTTCCTTC-3′ (17-mer) and their PS analogs 5′-TSTSCSTGAGTSCACSTACSGASTSTST-3′ and 5′-GSCSTSTCSGCSTAGTSTSCCSTSTSC-3′, respectively, were used. The ODNs were complementary to the tobacco (Nicotiana benthamiana) pds mRNA sequence (GenBank accession no. EU165355.1) at positions 1,643 to 1,624 and 1,677 to 1,660, respectively. The label “S” in the sequence string denotes the positions of PS linkages.
In Wheat
pds antisense ODN(5) 5′-GGCCAAGTAAGCATTTCA-3′, ODN(10) 5′-ATCAACTGGTGCTGCAA-3′ (18-mers), and ODN(11) 5′-CAAGGTTCGCAGTTCGGG-3′ (17-mer) and their PS analogs having the structures 5′-GSGSCSCAAGTSAAGSCASTTSTSCSA-3′, 5′-ASTSCSAACSTGGTSGCSTGSCSASA-3′, and 5′-CSASASGGTSTCSGCSAGTSTCSGSGSG-3′ were applied. Sequences were directed against the wheat (Triticum aestivum) pds mRNA sequence (GenBank accession no. BT009315) at nucleotide positions 179 to 161, 624 to 607, and 1,040 to 1,022, respectively. As controls, a 17-mer random nonsense 5′-GGCGGCTAACGCTTCGA-3′ ODN and its respective PS 5′-GSGSCSGGCSTAACSGCSTTSCSGSA-3′ ODN were used.
In wheat, cab antisense ODN(6) 5′-AGAGCACACGGTCAGAG-3′ (17-mer) and cab ODN(10) 5′-GCGAGGCGGCCATTCTTG-3′ (18-mer) were used. ODNs were complementary to the wheat cab mRNA sequence (GenBank accession no. M10144.1) at positions 227 to 210 and 716 to 698. As a control, a 17-mer random nonsense 5′-GGCGGCTAACGCTTCGA-3′ ODN was used.
In Arabidopsis
psbA antisense ODN(1) 5′-CSASTSAGGCSTTSTSCGSCTSTSTSC-3′ (17-mer), ODN(2) 5′-TSCSCSCSTGATSCAAACSTASGSASA-3′ (18-mer), ODN(3) 5′-GSCSCSCSGAASTCSTGTSAASCCSTSTSC-3′ (19-mer), and ODN(4) 5′-GSTSTSGTSGAGCSATSTACGSTSTSC-3′ (18-mer) were used. ODNs were complementary to the Arabidopsis (Arabidopsis thaliana) psbA mRNA sequence (GenBank accession no. NC000932) at positions 41 to 25, 677 to 660, 721 to 703, and 1,014 to 997. As a control, a 17-mer random nonsense PS 5′-GSGSCSGGCSTAACSGCSTTSCSGSA-3′ ODN was used.
Antisense ODN Treatment of Tobacco Leaves
Tobacco plants were grown in a greenhouse at 20°C to 25°C. Supplemental light was provided by metal halide lamps for 12 h per day, and the light intensity at the plant surface was about 200 μmol photons m−2 s−1. Expanding leaves of 2-month-old plants were used for the experiments. Antisense ODNs and random nonsense ODNs were dissolved in sterile distilled water at a concentration of 10 μm and infiltrated in intact leaves with the help of a syringe, as described for transient plasmid uptake (Sparkes et al., 2006). The ODN treatments were carried out between 8 and 10 am. After 24 h, fast Chl a fluorescence transients were measured and treated leaf discs were cut and stored at −80°C until further use. Besides the random nonsense ODNs, we always used water controls as well (i.e. treating the leaves with distilled water).
Antisense ODN Treatment of Wheat Leaves
Wheat seeds (genotype CY-45) were sown in pots filled with one part sand and two parts soil. The pots were kept in a growth chamber in total darkness for 11 d at 19°C and 70% humidity. Ten-centimeter-long leaf segments of etiolated wheat seedlings were prepared. The leaves were cut under water to avoid the formation of air bubbles, and the lower 1- to 2-cm part was submerged in 10 μm water solution of pds and cab antisense ODNs, random nonsense ODNs, and pure water in a 1.5-mL Eppendorf tube. The tube was sealed with Parafilm in order to avoid the evaporation of solution. After 12 h of incubation in the dark, leaves were illuminated at 100 μmol photons m−2 s−1 for 24 h. Based on preliminary experiments using fast Chl a fluorescence transients (see below), we chose the 8- and 24-h illumination times, after which fluorescence measurements were performed and leaf segments were harvested and stored at −80°C until further use. Four independent repetitions were made for each ODN.
Antisense ODN Treatment of Arabidopsis Leaves
Arabidopsis plants (genotype Columbia-0) were grown in a greenhouse under short-day conditions (8 h of light, 16 h of dark) at approximately 100 μmol photons m−2 s−1 during the light period. The temperature was kept between 20°C and 24°C. Six-week-old plants were used for the experiments. Antisense ODNs and random nonsense ODNs were dissolved in sterile distilled water at a concentration of 10 μm and vacuum infiltrated into detached Arabidopsis leaves. After 12 h of incubation in the dark, leaves were illuminated at 250 μmol photons m−2 s−1 for 48 h. After 8, 24, and 48 h, fast Chl a fluorescence transients were measured and leaf discs were cut and stored at −80°C until further use. Eight independent repetitions were made for each ODN.
Confocal Laser Scanning Microscopy
Microscopy was performed using an Olympus Fluoview FV1000 confocal laser scanning microscope (Olympus Life Science Europa). The microscope configuration was as follows: objective lenses, LUMPLFL 40× (water; numerical aperture 0.8) and LUMPLFL 60× (water; numerical aperture 0.9); sampling speed, 8 μs pixel−1; line averaging, 2×; zoom, 1× and 3×; scanning mode, sequential unidirectional; excitation, 488 nm; laser transmissivity, 5%; main dichroic beam splitter, DM405/488; intermediate dichroic beam splitter, SDM560; FAM was detected between 500 and 555 nm, and Chl was detected between 650 and 750 nm. Unlabeled leaves were used to determine the level of green autofluorescence, and the laser intensity/detector voltage parameters were set accordingly, so that no green autofluorescence was visible in unlabeled leaves.
Wheat leaf photographs were taken with an Olympus Camedia C7070 wide zoom digital camera.
OJIP Measurements
Fluorescence measurements were carried out at room temperature with a Handy-PEA instrument (Hansatech Instruments). Leaf samples were illuminated with continuous red light emitted by three light-emitting diodes (3,500 μmol photons m−2 s−1, 650-nm peak wavelength; the spectral half-width was 22 nm; the emitted light was cut off at 700 nm by a near-infrared short-pass filter). The first reliably measured point of the fluorescence transient was at 20 μs, which was taken as F0. The length of the measurements was 1 s. To study the effects of antisense ODNs on the photosynthetic machinery, Fm and Fv = Fm − F0 were used.
Quantitative Real-Time Reverse Transcription-PCR
Total RNA was isolated from wheat, tobacco, and Arabidopsis leaves using TRI reagent (Sigma) according to the supplier’s instructions. First-strand cDNA synthesis of 2 μg of total RNA in a final volume of 20 μL was performed with RevertAid H minus Moloney murine leukemia virus reverse transcriptase (Fermentas) according to the supplier’s protocol using random hexamer primers. Control reactions were performed by omitting reverse transcriptase. For quantitative real-time reverse transcription (QRT)-PCR, 9 μL of 1:45 diluted cDNA was mixed with Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies), 5 pmol of forward primer, and 5 pmol of reverse primer (as listed below) in a final volume of 20 μL in three replicates. No-template controls were included. QRT-PCR was done using an ABI Prism 7000 Sequence Detection System (Applied Biosystems) with the following protocol for wheat and Arabidopsis: 95°C for 10 min, 45 cycles at 95°C for 15 s, followed by 60°C for 1 min; the protocol for tobacco was as follows: 95°C for 10 min, 45 cycles at 95°C for 15 s, followed by 64°C for 1 min. The specificity of the QRT-PCR amplification was confirmed by the following criteria: (1) a single peak in the melting temperature curve analysis of real-time PCR-amplified products (ABI Prism Dissociation Curve Analysis Software); (2) a single band on an agarose gel. Primer pairs were designed to detect relative expression levels of the pds genes of wheat and tobacco (see below). The expression level of each group was normalized to (1) ubiquitin as a housekeeping gene and (2) the initial amounts of pds, cab, and psbA transcripts in the control samples of wheat, tobacco, and Arabidopsis. The relative transcript levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
For QRT-PCR, the following pairs of primers were used: wheat ubiquitin forward, 5′-CTGGCGAGGATATGTTCCAT-3′, and wheat ubiquitin reverse, 5′-TCGGATGGAAGACCTTTGTC-3′; wheat pds forward, 5′-GAAATACCTGGCTTCCATGG-3′, and wheat pds reverse, 5′-CGGGACAGCATCTTAGAATCC-3′; wheat cab forward, 5′-GATCGTCGACCCACTCTAC-3′, and wheat cab reverse, 5′-ACAATGGCCTGCACAAAG-3′; tobacco ubiquitin forward, 5′-TCCAGGACAAGGAGGGTATCC-3′, and tobacco ubiquitin reverse, 5′-TAGTCAGCCAAGGTCCTTCCAT-3′; tobacco pds forward, 5′-CAGATTTCTTCAGGAGAAACATGGTTCA-3′, and tobacco pds reverse, 5′-CCACAATCGGCATGCAAAGTCTC-3′; Arabidopsis ubiquitin forward, 5′-AACCCTAACGGGAAAGACGATTA, and Arabidopsis ubiquitin reverse, 5′-TGAGAACAAGATGAAGGGTGGAC-3′; Arabidopsis psbA forward, 5′-TGATTGTATTCCAGGCTGAGCA-3′, and Arabidopsis psbA reverse, 5′-TGCCCGAATCTGTAACCTTCAT-3′.
Carotenoid and Chl Content Determination
Analysis of total carotenoid in plant tissue was performed in N,N-dimethylformamide (Inskeep and Bloom, 1985). The optical density values were measured at 663, 646, and 470 nm using a spectrophotometer (Hitachi U-2900), and the Chl content were determined according to Porra (2002). The total carotenoid content was determined according to Lichtenthaler (1987).
Western-Blot Analysis
Four-centimeter leaf segments cut from wheat leaves and Arabidopsis leaf discs were frozen in liquid nitrogen, ground to a fine powder, and then homogenized in 500 μL of Laemmli buffer. The homogenates were incubated at 90°C for 5 min followed by a 20-min incubation at 37°C, and then proteins were separated by 15% denaturing SDS-PAGE. The proteins were blotted on nitrocellulose membranes using a semidry blotting system with methanol-containing buffer. The nitrocellulose membranes were blocked using 5% skim milk powder in Tris-buffered saline plus Tween 20 (TBST) buffer (10 mm Tris, pH 8.0, 0.15 m NaCl, and 0.1% Tween 20) for 2 h and incubated with primary antibodies raised against Lhcb1, -2, -3, and -6, Lhca1, -2, and -3, and psbA for 2 h in TBST buffer with 5% milk powder. The membranes were washed three times for 5 min in TBST buffer and incubated with goat anti-rabbit IgG horseradish peroxidase conjugate (Millipore) at a 1:5,000 dilution in TBST buffer with 5% milk powder for 2 h. Immunoblotted membranes were incubated for 5 min in ECL Plus horseradish peroxidase substrate (GE Healthcare Bio-Sciences), and chemiluminescence was detected on Hyperfilm ECL photographic film (GE Healthcare Bio-Sciences). The developed film was digitalized and analyzed by 1D Scan software.
Acknowledgments
We thank Mónika Pummer and Katalin László (Biological Research Centre) for their technical help during the synthesis of ODNs. We thank Drs. Éva Hideg and László Kozma-Bognár (Biological Research Centre) for stimulating discussions.
References
- Agrawal S, Temsamani J, Tang JY. (1991) Pharmacokinetics, biodistribution, and stability of oligodeoxynucleotide phosphorothioates in mice. Proc Natl Acad Sci USA 88: 7595–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley GE, Scolnik PA. (1995) Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke MA. (2008) Chemical modification of siRNAs for in vivo use. Oligonucleotides 18: 305–319 [DOI] [PubMed] [Google Scholar]
- Bennett CF, Cowsert LM. (1999) Antisense oligonucleotides as a tool for gene functionalization and target validation. Biochim Biophys Acta 1489: 19–30 [DOI] [PubMed] [Google Scholar]
- Campbell JM, Bacon TA, Wickstrom E. (1990) Oligodeoxynucleoside phosphorothioate stability in subcellular extracts, culture media, sera and cerebrospinal fluid. J Biochem Biophys Methods 20: 259–267 [DOI] [PubMed] [Google Scholar]
- Cazenave C, Frank P, Büsen W. (1993) Characterization of ribonuclease H activities present in two cell-free protein synthesizing systems, the wheat germ extract and the rabbit reticulocyte lysate. Biochimie 75: 113–122 [DOI] [PubMed] [Google Scholar]
- Chamovitz D, Sandmann G, Hirschberg J. (1993) Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J Biol Chem 268: 17348–17353 [PubMed] [Google Scholar]
- Chan JH, Lim S, Wong WS. (2006) Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol 33: 533–540 [DOI] [PubMed] [Google Scholar]
- Crooke ST. (1999) Molecular mechanisms of action of antisense drugs. Biochim Biophys Acta 1489: 31–44 [DOI] [PubMed] [Google Scholar]
- Crooke ST. (2004) Progress in antisense technology. Annu Rev Med 55: 61–95 [DOI] [PubMed] [Google Scholar]
- Dagle JM, Weeks DL. (2001) Oligonucleotide-based strategies to reduce gene expression. Differentiation 69: 75–82 [DOI] [PubMed] [Google Scholar]
- Damha MJ, Noronha AM, Wilds CJ, Trempe JF, Denisov A, Pon RT, Gehring K. (2001) Properties of arabinonucleic acids (ANA & 20’F-ANA): implications for the design of antisense therapeutics that invoke RNase H cleavage of RNA. Nucleosides Nucleotides Nucleic Acids 20: 429–440 [DOI] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman FA. (2008) The dynamics of photosynthesis. Annu Rev Genet 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Eckstein F. (1985) Nucleoside phosphorothioates. Annu Rev Biochem 54: 367–402 [DOI] [PubMed] [Google Scholar]
- Eckstein F. (2000) Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev 10: 117–121 [DOI] [PubMed] [Google Scholar]
- Galarneau A, Min KL, Mangos MM, Damha MJ. (2005) Assay for evaluating ribonuclease H-mediated degradation of RNA-antisense oligonucleotide duplexes. Methods Mol Biol 288: 65–80 [DOI] [PubMed] [Google Scholar]
- Gewirtz AM, Sokol DL, Ratajczak MZ. (1998) Nucleic acid therapeutics: state of the art and future prospects. Blood 92: 712–736 [PubMed] [Google Scholar]
- Ghosh MK, Ghosh K, Cohen JS. (1993) Phosphorothioate-phosphodiester oligonucleotide co-polymers: assessment for antisense application. Anticancer Drug Des 8: 15–32 [PubMed] [Google Scholar]
- Giles RV, Spiller DG, Green JA, Clark RE, Tidd DM. (1995) Optimization of antisense oligodeoxynucleotide structure for targeting bcr-abl mRNA. Blood 86: 744–754 [PubMed] [Google Scholar]
- Gillardon F, Moll I, Uhlmann E. (1995) Inhibition of c-Fos expression in the UV-irradiated epidermis by topical application of antisense oligodeoxynucleotides suppresses activation of proliferating cell nuclear antigen. Carcinogenesis 16: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Gleave ME, Monia BP. (2005) Antisense therapy for cancer. Nat Rev Cancer 5: 468–479 [DOI] [PubMed] [Google Scholar]
- Govindjee (2004) Chlorophyll a fluorescence: a bit of basics and history. Papageorgiou GC, Govindjee, , Chlorophyll a Fluorescence: A Signature of Photosynthesis. Advances in Photosynthesis and Respiration, Vol 19. Springer, Dordrecht, The Netherlands, pp 1–42 [Google Scholar]
- Hideg E, Barta C, Kálai T, Vass I, Hideg K, Asada K. (2002) Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photoinhibition or UV radiation. Plant Cell Physiol 43: 1154–1164 [DOI] [PubMed] [Google Scholar]
- Hoke GD, Draper K, Freier SM, Gonzalez C, Driver VB, Zounes MC, Ecker DJ. (1991) Effects of phosphorothioate capping on antisense oligonucleotide stability, hybridization and antiviral efficacy versus herpes simplex virus infection. Nucleic Acids Res 19: 5743–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Bally MB, Madden TD. (2002) Subcellular trafficking of antisense oligonucleotides and down-regulation of bcl-2 gene expression in human melanoma cells using a fusogenic liposome delivery system. Nucleic Acids Res 30: 3632–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inskeep WP, Bloom PR. (1985) Extinction coefficients of chlorophyll a and b in N,N-dimethylformamide and 80% acetone. Plant Physiol 77: 483–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S. (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Junge W. (1977) Membrane potentials in photosynthesis. Annu Rev Plant Physiol 28: 503–536 [Google Scholar]
- Kramer DM, Avenson TJ, Edwards GE. (2004) Dynamic flexibility in the light reactions of photosynthesis governed by both electron and proton transfer reactions. Trends Plant Sci 9: 349–357 [DOI] [PubMed] [Google Scholar]
- Krumova SB, Laptenok SP, Kovács L, Tóth T, van Hoek A, Garab G, van Amerongen H. (2010) Digalactosyl-diacylglycerol-deficiency lowers the thermal stability of thylakoid membranes. Photosynth Res 105: 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai MH, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill LK. (1995) Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci USA 92: 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurreck J. (2003) Antisense technologies: improvement through novel chemical modifications. Eur J Biochem 270: 1628–1644 [DOI] [PubMed] [Google Scholar]
- Lazár D, Schansker G. (2009) Models of chlorophyll a fluorescence transients. Laisk A, Govindjee L Nedbal, , Photosynthesis in Silico: Understanding Complexity from Molecules to Ecosystems. Advances in Photosynthesis and Respiration, Vol 29. Springer, Dordrecht, The Netherlands, pp 85–123 [Google Scholar]
- Lebleu B, Moulton HM, Abes R, Ivanova GD, Abes S, Stein DA, Iversen PL, Arzumanov AA, Gait MJ. (2008) Cell penetrating peptide conjugates of steric block oligonucleotides. Adv Drug Deliv Rev 60: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin AA. (1999) A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta 1489: 69–84 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lindgren LO, Stålberg KG, Höglund AS. (2003) Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol 132: 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matsukura M, Shinozuka K, Zon G, Mitsuya H, Reitz M, Cohen JS, Broder S. (1987) Phosphorothioate analogs of oligodeoxynucleotides: inhibitors of replication and cytopathic effects of human immunodeficiency virus. Proc Natl Acad Sci USA 84: 7706–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A, Zhang L, Forestier M, Ghirardi ML, Seibert M. (2000) Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol 122: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Hunt T. (1986) The use of single-stranded DNA and RNase H to promote quantitative ‘hybrid arrest of translation’ of mRNA/DNA hybrids in reticulocyte lysate cell-free translations. Nucleic Acids Res 14: 6433–6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho A, Camacho L, Haley A, Pais MS, Trewavas A, Malhó R. (2001a) Antisense perturbation of protein function in living pollen tubes. Sex Plant Reprod 14: 101–104 [Google Scholar]
- Moutinho A, Hussey PJ, Trewavas AJ, Malhó R. (2001b) cAMP acts as a second messenger in pollen tube growth and reorientation. Proc Natl Acad Sci USA 98: 10481–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O. (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ. (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumley FG, Schmidt GW. (1987) Reconstitution of chlorophyll a/b light-harvesting complexes: xanthophyll-dependent assembly and energy transfer. Proc Natl Acad Sci USA 84: 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ. (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Ravichandran LV, Dean NM, Marcusson EG. (2004) Use of antisense oligonucleotides in functional genomics and target validation. Oligonucleotides 14: 49–64 [DOI] [PubMed] [Google Scholar]
- Rayburn ER, Zhang R. (2008) Antisense, RNAi, and gene silencing strategies for therapy: mission possible or impossible? Drug Discov Today 13: 513–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR. (2005) Fast-track applications: the potential for direct delivery of proteins and nucleic acids to plant cells for the discovery of gene function. Plant Methods 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani TD, Jolles B, Laigle A. (2001) Best minimally modified antisense oligonucleotides according to cell nuclease activity. Antisense Nucleic Acid Drug Dev 11: 129–136 [DOI] [PubMed] [Google Scholar]
- Sandy P, Ventura A, Jacks T. (2005) Mammalian RNAi: a practical guide. Biotechniques 39: 215–224 [DOI] [PubMed] [Google Scholar]
- Schansker G, Tóth SZ, Strasser RJ. (2005) Methylviologen and dibromothymoquinone treatments of pea leaves reveal the role of photosystem I in the Chl a fluorescence rise OJIP. Biochim Biophys Acta 1706: 250–261 [DOI] [PubMed] [Google Scholar]
- Scherer LJ, Rossi JJ. (2003) Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol 21: 1457–1465 [DOI] [PubMed] [Google Scholar]
- Shaw JP, Kent K, Bird J, Fishback J, Froehler B. (1991) Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res 19: 747–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Hoekstra D. (2004) Effective intracellular delivery of oligonucleotides in order to make sense of antisense. J Control Release 97: 189–209 [DOI] [PubMed] [Google Scholar]
- Sohail M, Southern EM. (2000) Selecting optimal antisense reagents. Adv Drug Deliv Rev 44: 23–34 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stein CA. (1995) Does antisense exist? Nat Med 1: 1119–1121 [DOI] [PubMed] [Google Scholar]
- Stein CA. (2001) The experimental use of antisense oligonucleotides: a guide for the perplexed. J Clin Invest 108: 641–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA, Tonkinson JL, Zhang LM, Yakubov L, Gervasoni J, Taub R, Rotenberg SA. (1993) Dynamics of the internalization of phosphodiester oligodeoxynucleotides in HL60 cells. Biochemistry 32: 4855–4861 [DOI] [PubMed] [Google Scholar]
- Sun C, Höglund AS, Olsson H, Mangelsen E, Jansson C. (2005) Antisense oligodeoxynucleotide inhibition as a potent strategy in plant biology: identification of SUSIBA2 as a transcriptional activator in plant sugar signalling. Plant J 44: 128–138 [DOI] [PubMed] [Google Scholar]
- Sun C, Ridderstråle K, Höglund AS, Larsson LG, Jansson C. (2007) Sweet delivery: sugar translocators as ports of entry for antisense oligodeoxynucleotides in plant cells. Plant J 52: 1192–1198 [DOI] [PubMed] [Google Scholar]
- Tao X, Zhou X. (2004) A modified viral satellite DNA that suppresses gene expression in plants. Plant J 38: 850–860 [DOI] [PubMed] [Google Scholar]
- Tsutsumi N, Kanayama K, Tano S. (1992) Suppression of alpha-amylase gene expression by antisense oligodeoxynucleotide in barley cultured aleurone layers. Jpn J Genet 67: 147–154 [DOI] [PubMed] [Google Scholar]
- Uhlmann E, Peyman A. (1990) Antisense oligonucleotides: a new therapeutic principle. Chem Rev 90: 543–584 [Google Scholar]
- Uhlmann E, Peyman A, Ryte A, Schmidt A, Buddecke E. (2000) Use of minimally modified antisense oligonucleotides for specific inhibition of gene expression. Methods Enzymol 313: 268–284 [DOI] [PubMed] [Google Scholar]
- Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. (2003) Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents: a comparative analysis. J Biol Chem 278: 7108–7118 [DOI] [PubMed] [Google Scholar]
- von Lintig J, Welsch R, Bonk M, Giuliano G, Batschauer A, Kleinig H. (1997) Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings. Plant J 12: 625–634 [DOI] [PubMed] [Google Scholar]
- Wang M, Wang G, Ji J. (2010) Suppression of the phytoene desaturase gene influence on the organization and function of photosystem II (PSII) and antioxidant enzyme activities in tobacco. Environ Exp Bot 67: 460–466 [Google Scholar]
- Wang M, Wang G, Ji J, Wang J. (2009) The effect of pds gene silencing on chloroplast pigment composition, thylakoid membrane structure and photosynthesis efficiency in tobacco plants. Plant Sci 177: 222–226 [Google Scholar]
- Wang T, Iyer LM, Pancholy R, Shi X, Hall TC. (2005) Assessment of penetrance and expressivity of RNAi-mediated silencing of the Arabidopsis phytoene desaturase gene. New Phytol 167: 751–760 [DOI] [PubMed] [Google Scholar]
- Wetzel CM, Rodermel SR. (1998) Regulation of phytoene desaturase expression is independent of leaf pigment content in Arabidopsis thaliana. Plant Mol Biol 37: 1045–1053 [DOI] [PubMed] [Google Scholar]
- Wilds CJ, Damha MJ. (2000) 2′-Deoxy-2′-fluoro-beta-D-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res 28: 3625–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Lima WF, Zhang H, Fan A, Sun H, Crooke ST. (2004) Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J Biol Chem 279: 17181–17189 [DOI] [PubMed] [Google Scholar]
- Yang L, Li J, Zhou W, Yuan X, Li S. (2004) Targeted delivery of antisense oligodeoxynucleotides to folate receptor-overexpressing tumor cells. J Control Release 95: 321–331 [DOI] [PubMed] [Google Scholar]
- Zamaratski E, Pradeepkumar PI, Chattopadhyaya J. (2001) A critical survey of the structure-function of the antisense oligo/RNA heteroduplex as substrate for RNase H. J Biochem Biophys Methods 48: 189–208 [DOI] [PubMed] [Google Scholar]
- Zamecnik PC, Stephenson ML. (1978) Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA 75: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]