Abstract
The Mediator (Med) complex relays regulatory information from DNA-bound transcription factors to the RNA polymerase II in eukaryotes. This macromolecular unit is composed of three core subcomplexes in addition to a separable kinase module. In this study, conservation of Meds has been investigated in 16 plant species representing seven diverse groups across the plant kingdom. Using Hidden Markov Model-based conserved motif searches, we have identified all the known yeast/metazoan Med components in one or more plant groups, including the Med26 subunits, which have not been reported so far for any plant species. We also detected orthologs for the Arabidopsis (Arabidopsis thaliana) Med32, -33, -34, -35, -36, and -37 in all the plant groups, and in silico analysis identified the Med32 and Med33 subunits as apparent orthologs of yeast/metazoan Med2/29 and Med5/24, respectively. Consequently, the plant Med complex appears to be composed of one or more members of 34 subunits, as opposed to 25 and 30 members in yeast and metazoans, respectively. Despite low similarity in primary Med sequences between the plants and their fungal/metazoan partners, secondary structure modeling of these proteins revealed a remarkable similarity between them, supporting the conservation of Med organization across kingdoms. Phylogenetic analysis between plant, human, and yeast revealed single clade relatedness for 29 Med genes families in plants, plant Meds being closer to human than to yeast counterparts. Expression profiling of rice (Oryza sativa) and Arabidopsis Med genes reveals that Meds not only act as a basal regulator of gene expression but may also have specific roles in plant development and under abiotic stress conditions.
The proper functioning and development of an organism is orchestrated by a large number of transcription factors (TFs), which in turn regulate the expression of several downstream genes affecting various regulatory and metabolic pathways. Mediator (Med) is a multiprotein complex that acts as an interface to pass on the message from the TFs to the basal transcriptional unit assembled at the core promoter, bringing about either transcriptional activation or repression (Björklund and Gustafsson, 2005; Conaway et al., 2005b; Kornberg, 2005). Discovered in the budding yeast Saccharomyces cerevisiae, for its ability to respond to a transcriptional activator (Kelleher et al., 1990; Flanagan et al., 1991), Med was later shown to participate in the transcription of a majority of yeast genes (Holstege et al., 1998).
Electron microscopy studies have shown Med structure to form an unfolded arc around the RNA polymerase II (Pol II; Asturias et al., 1999), with the dense regions corresponding to three distinct units, namely, the head, the middle, and the tail modules. Yeast Med is a complex of 25 subunits, where eight subunits form the head module, seven constitute the middle module, and six constitute the tail. In addition, a fourth regulatory module comprising two Med subunits along with a cyclin-dependent kinase, Cdk8, and its associated cyclin, CycC, collectively called the kinase or Cdk8-cyclin C module, is also part of the Med complex. In contrast to the above model, Baidoobonso et al. (2007) reported an intact and stable complex comprising only the tail and head modules without the middle module. Until recently, the metazoan Med complexes were known to share 22 Med subunits with S. cerevisiae (Bourbon et al., 2004), having eight subunits (Med23 to -30) unique to them. Recent in silico studies, however, have established that orthologs of all yeast Med components are indeed represented in metazoans (Bourbon, 2008).
Mediators facilitate transcription by increasing the efficiency/rate of Pol II preinitiation complex formation at the promoters (Cantin et al., 2003) and activating transcription from promoters with stalled Pol II (Lee et al., 2010). The recruitment of Pol II is proposed to be achieved by direct contacts between the head and the middle Med modules and the C-terminal domain of the Rpb1 subunit (Asturias et al., 1999; Davis et al., 2002). Pol II C-terminal domain phosphorylation has been shown to be established in a mediator-dependent fashion (Boeing et al., 2010). The kinase module phosphorylates subunits of the general transcription factor (GTF) TFIID and Med2 (Hallberg et al., 2004; Liu et al., 2004) and facilitates reinitiation of the preinitiation complex (Yudkovsky et al., 2000). The Med complex has the flexibility to acquire different structures upon binding of different activators to different/same subunits (Taatjes et al., 2002, 2004). These distinct activator-Med structures differentially affect Pol II activity (Meyer et al., 2010) and regulate Med function in gene-specific ways (Ebmeier and Taatjes, 2010). Although the function of Med as a GTF has been widely accepted (Takagi and Kornberg, 2006), its role as a global regulator of transcription has been questioned in some recent reports (Deato et al., 2008; Thiaville et al., 2008). However, Ansari et al. (2009) have shown Med to be a direct requirement for Pol II association at constitutively transcribed genes in yeast, justifying Med as a GTF. Convincing evidence has also been provided for the role of the Med complex in recruiting the cohesin protein complex, which in turn promotes and/or stabilizes the physical proximity between enhancers and promoters (Kagey et al., 2010). Kim and coworkers (2011) have expanded the role of the Med complex in the Pol II-mediated intergenic transcription of small and long noncoding RNAs.
Meds have been biochemically identified in several fungi like S. cerevisiae (Kim et al., 1994; Li et al., 1995; Myers et al., 1998) and Schizosaccharomyces pombe (Spåhr et al., 2000), metazoans including mammals (Fondell et al., 1996; Jiang et al., 1998; Malik and Roeder, 2000; Sato et al., 2003), Drosophila melanogaster, an insect (Park et al., 2001), and worms like Caenorhabditis elegans (Kwon et al., 1999). Med homologs have also been identified in several eukaryotes by homology-based methods (Bourbon, 2008). The identification of Med subunits in various organisms has also resulted in different nomenclatures for homologs of the same subunit. To bring uniformity, Bourbon and coworkers (2004) proposed a common unified nomenclature for different Med subunits across species. Biochemical purification of the first plant Med complex in Arabidopsis (Arabidopsis thaliana) identified 21 conserved and six Arabidopsis-specific Med subunits (Bäckström et al., 2007). The study identified STRUWWELPETER (SWP) as Med14 and PHYTOCHROME AND FLOWERING TIME1 (PFT1) as Med25 subunits. Later, Med21 was shown to interact with HISTONE MONOUBIQUITINATION1 (Dhawan et al., 2009) and the Med12-Med13 pair to regulate pattern formation timing during embryogenesis in Arabidopsis (Gillmor et al., 2010).
Although a wealth of information on fungal and metazoan Med structure and function is available, a similar understanding of this complex in plants is still in its infancy. The past decade has witnessed a rapid increase in the number of genome sequences for several plant species. This study expands the search for Med genes across different groups of the plant kingdom, from algae to higher angiosperms, using in silico approaches. We find that all the reported Med subunits are present in one or the other plant group. Our study also establishes that despite the low sequence similarity between plant, fungal, and metazoan homologs of the same Med subunits, these proteins exhibit considerable similarity in their secondary structures. Expression profiling supports the fundamental role for some Med genes in transcriptional regulation and also highlights the role of other Meds in regulating development- and stress-specific expression in rice (Oryza sativa) and Arabidopsis.
RESULTS AND DISCUSSION
Identification of Med Proteins in Plants
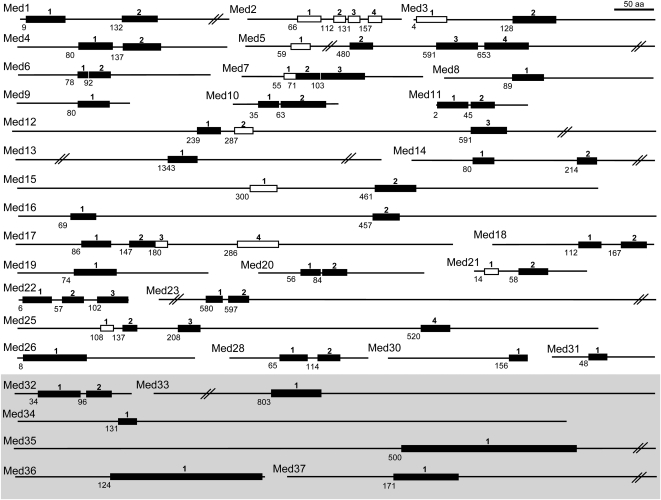
Med subunits have been biochemically identified in several fungi and metazoans but only for one plant, Arabidopsis (Bäckström et al., 2007). In this study, putative Meds have been identified in seven groups of the plant kingdom by Hidden Markov Model (HMM) methods. Using a dual approach, both full-length (FL) and high-homolgy (HH) HMM profiles were generated using complete sequences and highly conserved regions across metazoan, fungi, and plant kingdoms. This exercise identified 67 HH regions across all Meds, with more than one HH region in 22 Med subunits (Fig. 1). This intensive methodology enabled us to identify novel Meds possessing one or more HH regions. Furthermore, we singled out HH regions that define highly conserved regions in plants (marked as black boxes in Fig. 1) for each Med subunit owing to their presence in all/most of the predicted plant sequences (Supplemental Table S1). Cdk8 and CycC counterparts were inferred using only the FL-HMMs with very high expect values, to segregate them from various other Cdk and cyclin family members. Bioinformatics analysis has previously established yeast Med2, -3, and -5 as orthologs of the metazoan Med29, -27, and -24 subunits, respectively (Bourbon, 2008). Therefore, the yeast nomenclature has been retained in this study. Bäckström et al. (2007) had identified six new plant-specific Meds through biochemical analysis, which were numbered Med32 to -37. To find orthologs of these Meds, Position-Specific Iterative (PSI)-BLAST, HMM search, and the presence of common Pfam protein domains with the bona fide Arabidopsis Meds helped us to identify putative Med32 to -37 in all the plant groups. Our in silico search revealed that all the predicted plant Med33s were the same as those we predicted as Med5 (Supplemental Table S1). In addition, several putative Med32 proteins were found to have a conserved Med29 Pfam domain (Supplemental Table S1), and all of the 18 Med32 predictions were supported by PSI-BLAST analyses that were jump-started using Med2/29 alignment as query sequences (Supplemental Table S2). These results corroborated earlier predictions of plant Med32 and Med33 to be apparent homologs of Med2/29 and Med5/24, respectively (Bourbon, 2008). Consequently, all Med32 and Med33 hits in this study were annotated as Med2 and Med5, respectively. After removing all the redundancy in various Med subunits, the Med complex was finally found to have 32 unique Med subunits (Med1 to -23, -25, -26, -28, -30, -31, and Med34 to -37) along with Cdk8 and CycC components of the kinase module. All the genes of the Med complex will henceforth be collectively called MCC (for Med, Cdk8, and CycC) genes.
Figure 1.
The distribution of the conserved HH regions of the Med complex proteins among various eukaryotes. The HH regions (boxes) are numbered from the N terminus onward for each Med protein, represented as horizontal lines. The HH regions defining the most conserved regions in plants are shown as black boxes. The number below each HH region shows its start position relative to human or Arabidopsis (marked in the shaded area) Med proteins.
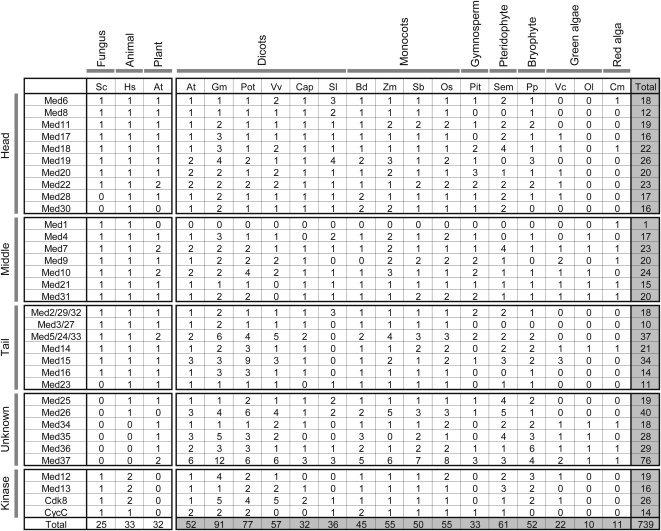
Of the 739 MCC genes identified in the study (Fig. 2), 699 were Meds, 26 were Cdk8 orthologs, and 14 were CycC components. Our study establishes, to our knowledge for the first time, that at least one homolog for all the animal/fungal Med subunits is represented in the plant kingdom. We have predicted 52 Arabidopsis MCC proteins (Fig. 2). These include more paralogs for six Meds (Med15, -20, -19, and -35 to -37) as well as orthologs for Med26, Med30, and the kinase module subunits not identified earlier in the biochemical screen (Bäckström et al., 2007). We have identified SENSITIVE TO FREEZING6 (SFR6) as Med16, a nucleus-localized protein with roles in cold acclimation (Knight et al., 2009) as well as photoperiodic and clock gene expression (Knight et al., 2008). In addition, two of the Arabidopsis Med33 subunits (AT2G48110 and AT3G23590; Bäckström et al., 2007) are annotated as REDUCED EPIDERMAL FLUORESCENCE4 (REF4) and REF4-RELATED1, respectively (The Arabidopsis Information Resource [TAIR]). The dominant mutations in REF4 lead to reduced accumulation of phenylpropanoid end products and affect plant growth (Stout et al., 2008). Although the exact function of REF4 has not been ascertained, the authors argue against the function of REF4 as a TF, owing to putative membrane-spanning domains in the protein. However, REF4 is not a part of any membrane proteome (Stout et al., 2008; TAIR) and has been isolated as a component of the Med complex in a biochemical screen (Bäckström et al., 2007), making it a legitimate transcriptional regulator. On the other hand, the rice counterparts of Med5/33 have been annotated as “structural constituent of ribosome” in the Rice Genome Annotation Project (RGAP) database. This seems to be an anomaly in the annotation. Domain identification for these proteins in the Pfam, SMART, as well as InterProScan databases did not provide information on any specific protein signatures for the three rice Med5 paralogs. In fact, in Pfam, there was no information on any conserved protein domains for any of the 37 putative Med5 proteins. Our results further contribute toward enriching the current annotation of several plant proteomes by annotating several genes of undefined functions as Meds. These include Arabidopsis (8 of 52 = 15%), Cyanidioschyzon merolae (8 of 11 = 73%), rice (20 of 55 = 36%), and Vitis vinifera (100%; Supplemental Table S3).
Figure 2.
The distribution of putative Med proteins across the plant kingdom. The Med subunits are grouped as head, middle, tail, and kinase module according to Bourbon (2008). The unknown group has members whose positions in the complex are unassigned. The left panel lists the number of biochemically purified Med proteins in S. cerevisiae (Sc), human (Hs), and Arabidopsis (At). The right panel reports the number of homologs predicted in the study for diverse plants belonging to various groups (marked at the top of each respective group) of the plant kingdom. The total numbers of Med proteins identified for an organism and for an individual Med subunit are represented in shaded boxes at the ends of the columns and rows, respectively. Organisms are as follows: G. max (Gm), P. trichocarpa (Pot), V. vinifera (Vv), C. papaya (Cap), S. lycopersicum (Sl), B. distachyon (Bd), Z. mays (Zm), S. bicolor (Sb), O. sativa (Os), P. taeda (Pit), S. moellendorffii (Sem), P. patens (Pp), V. carteri (Vc), O. lucimarinus (Ol), and C. merolae (Cm).
Evidence of Animal Med26 in Plants
Orthologs of metazoan Med26 have not been reported for plants in any study so far. Med26, a subunit of the Cofactor Required for SP1 Activation complex (Ryu et al., 1999), is a target of a zinc finger TF and is involved in epigenetic silencing of neuronal gene expression along with Med19 (Ding et al., 2009). Our intensive HH study helped us to identify novel Med26 in all land plants; however, they were not detected in the algal group. Of the six Med26 rice and Arabidopsis proteins, four (two from each species) are annotated as transcriptional elongation factors and one each as a transcriptional regulator (in Arabidopsis) and a hypothetical protein (in rice) in TAIR and RGAP, respectively. Moreover, these six Med26 proteins showed more than 90% probability of localizing to the nucleus (data not shown). In addition, all the 40 predicted Med26 subunits showed the Pfam Med26 domain having the conserved TFIIS helical bundle domain, a conserved N-terminal region found in the transcription elongation factors TFIIS and Elongin A (Booth et al., 2000). Although the role of Med26 in plants remains to be established, it appears that Med26 may play a role in transcription elongation, as has been reported for the Cdk8 submodule in the serum response network (Donner et al., 2010) and in p53-directed Pol II elongation via the Med complex in humans (Meyer et al., 2010).
Conservation, Loss, and Duplication of Fungal/Animal Med Subunits in Plants
For each core module of the Med (head, middle, and tail), a high conservation except for one or two subunits was observed across kingdoms. All the 34 MCCs could be identified in one or the other species studied here (Fig. 2), with the fungal/metazoan Med7, -18, -21, and -31 being represented in all seven groups. In yeast, Med7 and -21 are important for cell viability and constitute the core along with eight other subunits upon which other Med subunits assemble (Björklund et al., 2001; Kang et al., 2001; Guglielmi et al., 2004). High diversity in Med subunits was observed for the plant-specific Meds and those with unknown functions (Fig. 2). Algal Meds (including plant-specific subunits) were predominantly divergent from those in the other classes of plants; however, as in yeast, algae also lacked Med23, -25, -26, and -30 subunits. Except for Volvox carteri, all the algal members also lacked the kinase module counterparts. Functional Med complexes lacking the kinase module have been well documented in various cell types (Sato et al., 2004). Despite a rigorous search, only the red alga C. merolae had a detectable Med1 subunit in the plant kingdom. Med1 is the target for the aryl-hydrocarbon receptor (Wang et al., 2004) and several liganded nuclear receptors via its LxxLL (Leu-X-X-Leu-Leu) motif (Yuan et al., 1998; Malik et al., 2002). CmMed1_1 has two LxxLL motifs at positions 219 and 384. In mammals, Med1 exists only in a subpopulation of the total Med complexes (Zhang et al., 2005), and in yeast, it is not critical for cell survival (Balciunas et al., 1999). It appears that Med1 functions have either been lost in higher plants or the roles have been acquired by other Med subunits/transcription regulators during the course of evolution.
The yeast Med complex has single homologs of all the MCC components, while the more complex metazoans possess additional Med subunits as well as more than one member in the kinase module. However, wherever detected, only Med21 and -23 showed a single homolog across all plant species studied. And within the submodules, only Carica papaya exhibited a single homolog in the head and the middle modules while Brachypodium distachyon, Physcomitrella patens, Ostreococcus lucimarinus, and C. merolae had a single homolog only in the middle module. In this collection of plant Meds, Populus trichocarpa had the highest number of paralogs (nine for Med15) among all the yeast/metazoan Meds. In addition, several other plant species showed high numbers of Med15 paralogs. Med15 physically interacts with several disparate TFs both in metazoans and S. cerevisiae (Kato et al., 2002; Yang et al., 2006; Thakur et al., 2009). Consistent with this, an increased number of plant Med15 paralogs could bestow additional regulatory potential to the Med complex to interact with diverse signals.
The largest numbers of MCC homologs were identified in Glycine max (91 proteins). Interestingly, G. max had one or more additional paralogs for each MCC family than Arabidopsis, except Med6 to -8, -10, -13, -15, -20 to -23, -25, -34, and CycC, where the homolog numbers for both the organisms were the same. G. max has a highly duplicated genome (Walling et al., 2006), with nearly 75% of the genes present in multiple copies (Schmutz et al., 2010). The expansion in the genome size in G. max and also P. trichocarpa (Tuskan et al., 2006), which has the second highest number of predicted MCC paralogs (77) in our repertoire of Meds, may account for the increased number of detectable MCC paralogs in these dicots as compared with Arabidopsis. Likewise, C. papaya, reported to have fewer genes than any other sequenced angiosperm (Ming et al., 2008), had the least number of MCC paralogs among dicots. Similarly, in monocots, the reported similar gene numbers across a broad diversity of grasses (International Brachypodium Initiative, 2010) was reflected in the number of MCC paralogs identified. Dicots showed a wider variation in the total numbers of MCC proteins (32–91) for various subunits than monocots (45–55).
Detection of all known metazoan/fungal Meds in plants (except Med1 in higher plants) suggests a conservation of Med function across kingdoms. In addition, the presence of novel plant-specific Meds along with an enhanced number of paralogs for several Meds suggests that these subunits may have been acquired to perform a specialized role in plants. Lack of several Meds and the presence of Med1 in algae suggest a variable composition of the Med complex in certain groups. However, it is prudent to say that with the availability of more sequence (genome/transcriptome) data on the species studied and complete genome sequences of more organisms, further insights can be obtained in the future.
Evolutionary Relatedness between Plant, Fungal, and Metazoan Mediators
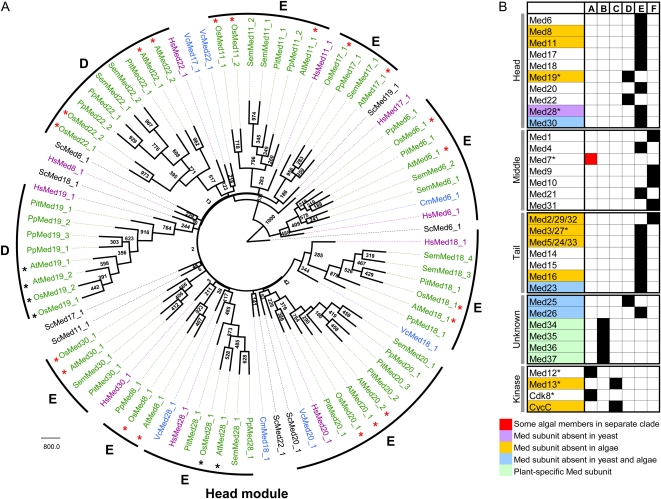
To assess the evolutionary relationship and conservation between homologs of various Med families, phylogenetic analysis was performed on sequences from the yeast, human, and one representative member from the seven plant groups selected in the study (Arabidopsis, rice, Pinus taeda, Selaginella moellendorffii, P. patens, V. carteri, and C. merolae). These sequences were divided according to the different Med modules followed by generating respective phylogenetic trees (Fig. 3A; Supplemental Figs. S1 and S2). Considering a bootstrap cutoff of at least 50% as significant, the cumulative depiction of different Med homologs is provided in Figure 3B. A single clade representing all the putative plant homologs as well as their fungal and metazoan counterparts (category A) was seen for three subunits: one belonging to the middle module and two to the kinase module. “All plants” refers to a group consisting of all the terrestrial plants (angiosperms, gymnosperms, pteridophytes, and bryophytes) in a single clade that also included all or at least one member of the algal taxa with high confidence within the same clade. In addition, four other subunits (category B) formed a single clade comprising all the plant homologs but lacking the yeast and human members. There were 22 subunits (categories C, D, and E) where all the terrestrial plants exhibited a high degree of conservation but their algal partners did not. Of note, these also included 12 Med subunits for which we did not find any detectable Meds in the algal group (see above). While both human and yeast Meds shared the clade with terrestrial plants in category C, both were absent in category E. Category D had the human Meds grouping with the terrestrial plant clade, while yeast did not. If the terrestrial plants themselves either formed more than one clade for a Med class (Med9) or had less than 50% bootstrap values (Med10, -31, and -2), they were put in category F. Med1, with no members in the land plants, was also assigned to category F.
Figure 3.
Phylogenetic relationship of Med subunits among various members of the yeast, metazoan, and plant kingdoms. Med proteins from one representative member of the seven plant groups, human (Hs), and S. cerevisiae (Sc) were assigned into the different modules of the Med complex. A, An unrooted tree of the head module constructed using the PHYLIP program by the neighbor-joining method. Numbers at the nodes represent bootstrap values from 1,000 replicates. A bootstrap value of at least 500 was used to define the Med subunits into groups A to G (see B). All the terrestrial plants (angiosperms, gymnosperms, pteridophytes, and bryophytes) are shown in green, algal members in blue, and Hs and Sc sequences in purple and black, respectively. Angiosperm-specific clades are marked by black asterisks and mixed clades by red asterisks. The scale bar represents amino acid substitutions per site. For the phylogenetic trees of the middle, tail, unknown, and kinase modules, see Supplemental Figures S1 and S2. B, Summary of the phylogenetic grouping of plants with Sc and Hs Med subunits. Groups A and B represent a single clade of all the putative Med proteins in terrestrial plants and algal members (black boxes) or at least one member of the algal group (red boxes) at 500 or greater bootstrap value. Groups C to E represent only the terrestrial plant members (either the algal members have not been predicted or do not group together with the land plants). Groups A and C include both the Sc and Hs members, while group D has only Hs members in the same clade as the plants. In groups B and E, the Sc and Hs members do not group together with plants. When the land plants did not group together to form a single clade, they were assigned to group F. Med subunits having angiosperm-specific clades are marked by asterisks.
The phylogenetic analysis highlighted that higher plants grouped together in a single clade for 29 MCC families (groups A–E). Apparently, the plant Meds were found to be closer to human Meds than their yeast counterparts. The former shared the plant clade for eight MCC families (groups A, C, and D) as opposed to just five (groups A and C) for yeast (Fig. 3B). Investigations on the relationship between rice and Arabidopsis revealed that they grouped together to form an angiosperm-specific clade with high confidence of 50% or greater in seven MCC families (black asterisks in Fig. 3 and Supplemental Figs. S1 and S2). Furthermore, lineage-specific clades were formed for both dicots (Med4, -18, -22, and -25) and monocots (Med11, -16, -20, and -30), pointing to independent expansion of Med gene families in the angiosperms.
Structural Conservation of Mediator Submodules in Plants
This study, along with a previous work (Bourbon, 2008), shows that Med sequences are only weakly conserved between different organisms. We reason that if the function of a Med has been conserved across kingdoms, then during evolution, the selection pressure would favor sustaining structural determinants that support intersubunit contacts. Therefore, we compared the protein secondary structures between yeast, human, and a plant (Arabidopsis) Meds for the defined submodules as well as those identified through yeast two-hybrid screens.
Med7 is the key architectural subunit of the middle module and forms subcomplexes with Med21 and -31; the Med7/21 dimer in turn binds to the Med10 C terminus (C) in yeast (Koschubs et al., 2009, 2010). Secondary structure modeling of these four Meds showed high conservation, with the exception of an extended Arabidopsis Med21 N terminus (N; Fig. 4). The two yeast Med7N poly-Pro stretches, which aid in wrapping around Med31 helices (Koschubs et al., 2009), were conserved in plants too (Supplemental Fig. S3A). Notably, like human Med7, there were fewer amino acids intervening in these Pro regions in plants than in yeast. The only plant lacking this region was P. patens, which appeared to be a partial sequence. We noted that the plant Med31C sequences exhibited poly-Pro regions followed by a nuclear localization signal domain (Supplemental Fig. S3B). Interestingly, these sequences were absent in yeast, human, as well as algae, highlighting that these sequences were very similar early in evolution, when lower plants diversified from the fungal/animal group, and the higher plants probably acquired them later.
Figure 4.
Secondary structure comparison among human, yeast, and Arabidopsis mediator sequences. The protein secondary structures were predicted using PSIPRED and superimposed on the alignments generated using MAFFT. The purple rectangles and green arrowheads denote the predicted protein helices and sheets, respectively. A solid black line indicates no secondary structure, and a dotted line denotes a gap in the alignment. Each red bar represents a length equivalent to 20 amino acids. Blue lines indicate the extent of interaction of Med4 with Med7 and Med21. Hs, Human; Sc, S. cerevisiae; At, Arabidopsis.
The middle module Med4/9 also forms a dimer (Koschubs et al., 2010). Secondary structure comparison of Med4 highlighted the high level of homology these proteins shared, including in the regions that have been shown to interact with Med7 and -21 (Guglielmi et al., 2004; Fig. 4). The yeast Med4C region required for cell viability (amino acid residues 194–250; Fig. 4) showed considerable structural similarity with plants; however, its human counterpart formed more sheets in this region. The Med9C region showed much more structural conservation between the three organisms as compared with highly divergent N termini (Fig. 4). This is not surprising, as in yeast, Med9N is not stably bound to the core complex and the predicted Med9 loop comprising amino acid residues 19 to 63 is not required for Med4/9 formation (Koschubs et al., 2010).
The yeast Med14N (amino acid residues 1–259) connects the tail to the middle module through Med10 (Guglielmi et al., 2004); moreover, in Arabidopsis, Med14N (amino acids 1–959) interacts with a transcription regulator (Gonzalez et al., 2007). The yeast and Arabidopsis Med14 sequences aligned only for 171 amino acids in the N-terminal region, showing only 21% identity and 41% similarity (data not shown). However, within this region, all three proteins showed the same number of helices (five) and sheets (three; Fig. 4). We note that Arabidopsis and human Med14C shared higher structural correlation than their yeast counterpart.
Med8N in S. cerevisiae binds to a TATA box-binding protein, and Med8C attaches the Med18/20 submodule to the mediator (Larivière et al., 2006, 2008). The domains of the Med8/18/20 triad are interdependent on each other for folding and proper complex formation (Shaikhibrahim et al., 2009). Surprisingly, the conserved Med8 domain could not be detected in any of the plant species (Supplemental Table S1), including the one identified via biochemical screening in Arabidopsis (Bäckström et al., 2007). Despite this, comparative modeling indicated that Arabidopsis Med8 as well as its interacting Med18/20 partners adopt spatial conformations significantly similar to those determined for the yeast proteins (Fig. 4). We observed that the putative plant Med8 sequences were nearly double in size than their yeast and human counterparts.
Med17 is the most important scaffolding subunit of the head module; its conditional knockout mutants in yeast are incapable of expressing most of the protein-coding genes (Thompson and Young, 1995). Comparative modeling confirmed the importance of Med17 in showing high structural similarity between the three organisms (Fig. 4). Yeast two-hybrid screens support the interaction of Med17 with Med8C and -22 as well as Med6 (Guglielmi et al., 2004). That study also identified interactions between Med22 and -11 as well as Med16 and -5. Structural comparisons revealed similar topology for Med5, -6, -11, -16, and -22 in the three organisms modeled (Fig. 4). Of note, the Arabidopsis Med5C formed more helices, owing to longer protein length.
The Med2/3 pair interacts with Med15 to form a triad in yeast (Zhang et al., 2004). Notably, the human and plant Med2 and -3 sequences are closer than their fungal counterpart, whereas, apart from Med15N and parts of its middle region, there was less apparent structural similarity between the three organisms for Med15 (Fig. 4). The yeast Med15N KIX domain (Novatchkova and Eisenhaber, 2004) is a bundle of three helices that forms the interface for transcriptional regulator binding (Thakur et al., 2009). The metazoan Med15N has an ARC105 domain, a three-helix structure with marked similarity to the KIX domain (Yang et al., 2006). The canonical Pfam KIX domain was not apparent in the bona fide Arabidopsis Med15 (Supplemental Table S1) but was represented in some dicots (C. papaya, G. max, and P. trichocarpa) and monocots (rice and B. distachyon). The structural comparison showed that several plant Med15s shared the three-helix structure; moreover, amino acid residues implicated in the interaction of the KIX domain (Thakur et al., 2009) were also conserved/similar between plants and yeast (Supplemental Fig. S4).
The ARM domain shared by elongation factor TFIIS and Med26 enables yeast TFIIS to interact with SAGA and Med13 (Diebold et al., 2010). Secondary structure analysis showed a similar pattern of conservation between the Arabidopsis and human Med26 ARM domains (Fig. 4). Moreover, the conserved “LFG” (Leu-Phe-Gly) motif between yeast Med13 and TFIIS had similar “P(Pro)FG” and “A(Ala)FG” motifs in human and Arabidopsis sequences, respectively (data not shown).
This extensive secondary structure analysis supports a conserved structural organization for the Med submodules across kingdoms. However, the presence of divergent regions in Meds may have evolved to facilitate interactions with different host-specific TFs in an organism-specific manner.
Differential Regulation of Meds during Plant Development
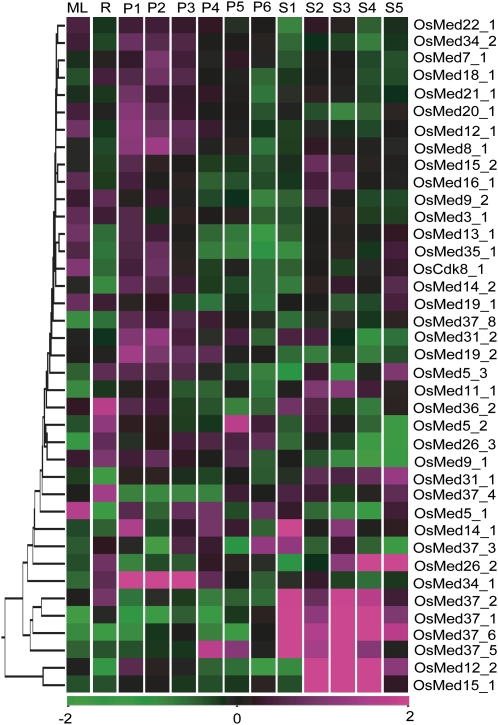
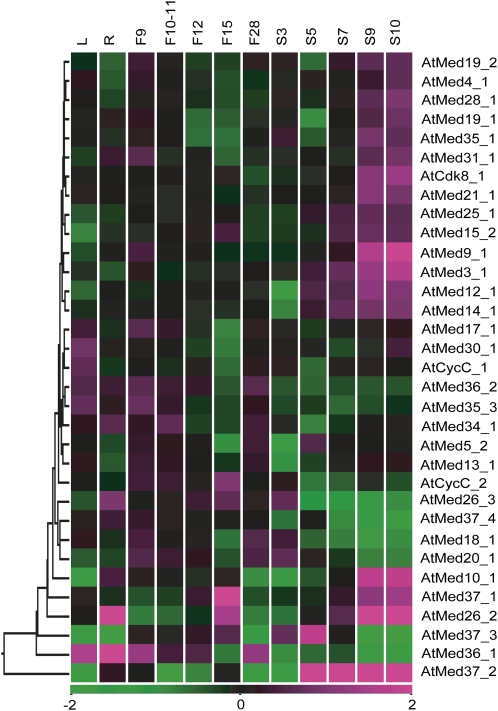
Out of the 52 rice and 43 Arabidopsis MCC genes represented on the microarray chips, 50 and 42 genes were differentially expressing at a statistically significant value of P ≤ 0.05 in at least one of the developmental stages analyzed in the monocot and the dicot, respectively (Supplemental Tables S4 and S5). Furthermore, we kept the criteria for significant differential expression to be 2-fold change with respect to the control. A total of 39 rice (Fig. 5) and 33 Arabidopsis (Fig. 6) genes showed more than 2-fold change in at least one reproductive substage in comparison with either of the vegetative stages. These gene numbers stood at 22 and 13 in rice and Arabidopsis (Fig. 7, A and B), respectively, when both the vegetative controls were assessed vis-a-vis the reproductive stages. Furthermore, OsMed5_1 showed opposite regulation in P1, S1, and S5 stages with respect to leaf and root controls. Detailed assessment between the various reproductive stages in comparison with both the vegetative controls revealed that the P1 stage in panicle and the S3 and S5 stages in seed for rice as well as the F15 stage in flower and the S9 and S10 stages in seed development for Arabidopsis had the most differentially regulated genes. The expression of none of the genes changed significantly in all flower/panicle substages with respect to the controls. However, OsMed37_1 and OsMed37_6 showed more than 2-fold induction, while the expression of AtMed36_1 was depressed in all the seed stages. The seed stages exhibited much more pronounced expression, highlighting that more MCC genes are directly involved during embryo development and seed maturation stages. A search for cis-elements in the promoter regions of these Med genes revealed several motifs found in the promoters of seed storage protein genes as well as those involved in embryo and/or endosperm development (Supplemental Table S6). In addition, elements involved in pollen development, the binding site of LEAFY (important for the transition from the vegetative to the reproductive phase) in OsMed31_2, as well as the CArG consensus sequence found in the promoter of the flowering pathway integrator gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 were also identified in regulatory regions of some of the genes that had high expression in panicle/flower development.
Figure 5.
Microarray-based expression analysis of selected Med genes in rice development stages. Expression profiles of at least 2-fold differentially regulated Med genes at P ≤ 0.05, with respect to the vegetative controls (mature leaf [ML] and root [R]), are shown. Developmental stages are listed at the top of each column in the temporal order of development. Reproductive stages comprise panicle (P1–P6) and five stages of seed (S1–S5) development. Hierarchical clustering of the expression profile was done with log-transformed average values, taking mature leaf as the baseline. The color scale at the bottom of the heat map is given in log2 intensity value, whereby green represents low-level expression, black shows medium-level expression, and magenta signifies high-level expression.
Figure 6.
Microarray-based expression analysis of selected Med genes in Arabidopsis development stages. Expression profiles of at least 2-fold differentially regulated Med genes at P ≤ 0.05, with respect to the vegetative controls (leaf [L] and root [R]), are shown. Developmental stages are listed at the top of each column in the temporal order of development. Reproductive stages comprise flower (F9–F28) and seed (S3–S10) stages. Hierarchical clustering of the expression profile was done with log-transformed average values, taking leaf as the baseline. The color scale at the bottom of the heat map is given in log2 intensity value, whereby green represents low-level expression, black shows medium-level expression, and magenta signifies high-level expression.
Figure 7.
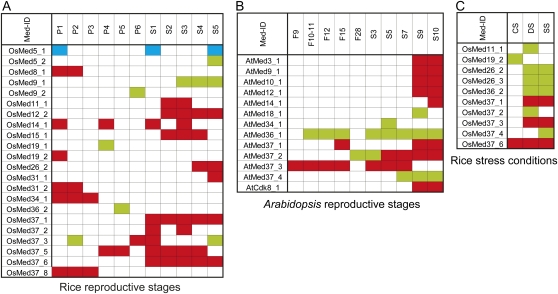
Expression analysis of rice and Arabidopsis Med genes using a microarray. A, Expression profiles for genes that are more than 2-fold differentially regulated at P ≤ 0.05 in rice panicle (P1–P6) and seed (S1–S5) stages with respect to both the vegetative controls, leaf and root. Red and green boxes represent up- and down-regulated genes, respectively. A box marked in blue defines an opposite regulation in that substage with respect to leaf and root controls. B, Expression profiles for Arabidopsis flower (F9–F28) and seed (S3–S10) stages. C, Differentially expressing Med genes in rice in abiotic stress stages. CS, Cold stress; DS, desiccation stress; SS, salt stress.
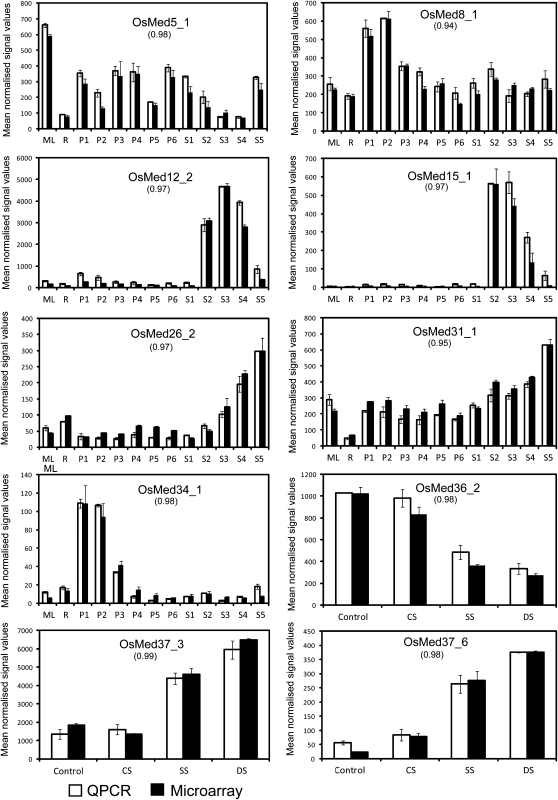
The normalized expression values of developmental stages for rice and Arabidopsis are provided in Supplemental Tables S7 and S8, respectively. A comparison of the expression patterns obtained for selected genes using quantitative real-time PCR (QPCR) exhibited a similar trend of expression to that observed for the microarray data, having correlation coefficients of more than 0.9 (Fig. 8).
Figure 8.
QPCR results for the expression of selected genes during development and stress and their correlation with microarray data. Three biological replicates were taken for both QPCR and microarrays. Three technical replicates were employed for each QPCR biological replicate. Error bars show se values for data obtained using both techniques. QPCR data were normalized to ease profile matching with each microarray’s data. Pearson correlation coefficients between QPCR and microarray data are indicated in parentheses. The y axis represents raw expression values obtained from microarray analysis; the x axis depicts various developmental stages or abiotic stress conditions. ML, Mature leaf; R, root; S1 to S5, seed development stages; CS, cold stress; SS, salt stress; DS, desiccation stress.
Limited Regulation of Plant Meds in Response to Abiotic Stress
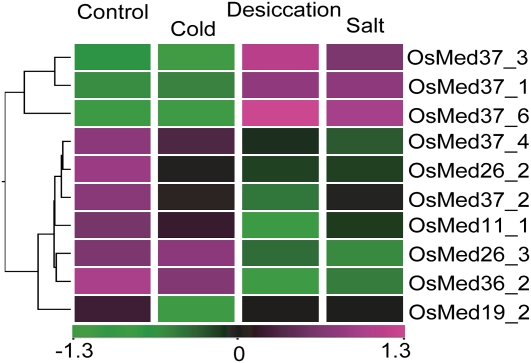
Microarray analysis in response to three stress conditions (desiccation, cold, and salt) was also performed. When the expression profiles were compared, 29 rice genes (Supplemental Table S4) and only four Arabidopsis genes (Supplemental Table S5) were differentially expressed in at least one of the stress phases as compared with the unstressed control. Furthermore, expression levels for 10 rice genes (Fig. 9) and none of the Arabidopsis MCC genes were significantly affected in response to any of the stress treatments. We noted that the expression profiles for all these 10 rice genes during desiccation and salt stress were comparable. This is in accordance with the similar responses these stresses are known to trigger in affected plants (Bartels and Sunkar, 2005). Only OsMed37_6 showed more than 2-fold change in transcript abundance in all three stress conditions tested. An in silico analysis showed the presence of abiotic stress recognition motifs in the promoters of some of these genes (Supplemental Table S6). We note that the expression of more rice genes (seven) decreased in response to at least one of the stresses than those that were augmented (three; Fig. 7C). It appears that thousands of years of domestication of rice have rendered it less tolerant to stresses and probably explains why it down-regulates its transcription machinery during stress. The expression profiles of three genes under stress conditions were also verified by QPCR (Fig. 8).
Figure 9.
Microarray-based expression analysis of selected Med genes in rice during abiotic stress conditions. Expression profiles of at least 2-fold differentially regulated Med genes at P ≤ 0.05, with respect to the control (untreated 7-d-old-seedling), are shown. Hierarchical clustering of the expression profile was done with log-transformed average values taking an untreated 7-d-old-seedling as the baseline. The color scale at the bottom of the heat map is given in log2 intensity value, whereby green represents low-level expression, black shows medium-level expression, and magenta signifies high-level expression.
Med genes have been widely accepted as GTFs. The ubiquitous expression of many MCC genes at similar levels during stress and development (Supplemental Tables S9 and S10) supports the basic role these genes play as components of the transcription machinery. However, differential expression of some Med genes in specific states provides ample possibilities for the differential regulation of plant processes as well.
Expression Profile of the Core Module Genes during Reproductive Development
In order to determine a role of the Med complex in reproductive development, we compared the expression patterns of various Med genes in a module. In the head module, Med17 (the most important scaffolding gene of this module), -6, -20, -22, -28, and -30 did not show significant differential expression in the reproductive stages vis-a-vis both the vegetative controls in both the angiosperms. The same was true for rice Med18 and Arabidopsis Med8 and -19; however, AtMed18_1 showed 4-fold down-regulation in the S9 stage with respect to the vegetative controls, and OsMed8_1 was up to 3-fold induced in the early panicle stages P1 and P2. Also, while OsMed19_1 showed down-regulation in the P4 stage, the expression rose in the P1 stage of OsMed19_2. In addition, OsMed11_1 showed more than 2-fold enhanced expression in the S2 and S3 stages of seed development, while the Arabidopsis counterpart could not be assessed, as it was not represented on the chip.
Expression profiles for Med7, the key architectural unit of the middle module, and Med4 showed that their expression was not significantly altered in reproductive stages in the two angiosperms. The expression data for AtMed21_1 showed its specific differential regulation in seed stages (i.e. approximately 2-fold from S7 to S9 and then staying high in S10; Supplemental Table S10). These seed stages in Arabidopsis correspond to embryo development and cotyledon expansion stages (Schmid et al., 2005), providing credence to the portrayed role of Med21 in embryo development (Dhawan et al., 2009). In rice, on the other hand, the expression of Med21 was 2-fold up-regulated in only the P1 stage of panicle development with respect to the vegetative controls, while in seeds, its expression levels remained similar to those in vegetative tissues (Supplemental Tables S4 and S9). Thus, it appears that Med21 plays a distinctive role in Arabidopsis seed development; however, in rice, its function might be more pronounced during early panicle development. The same study also suggested Med21 to be activated by microbial infection and also by factors involved in stress signaling; however, we did not find any positive correlation when assaying for the three abiotic stress responses in both the angiosperms (Supplemental Tables S4 and S5). It is possible, however, that Med21 is involved in stress signaling during reproductive stages and not in younger vegetative tissues. In rice, another Med (OsMed31_2) was up-regulated in the P1 and P2 stages of panicle development, while its paralog OsMed31_1 showed 3-fold enhanced regulation in leaf tissue as compared with root. However, in Arabidopsis, Med31 did not show any significant differential expression across the stages analyzed. Likewise, while the expression remained unchanged for Med9 in the flower stages of Arabidopsis with respect to the vegetative controls, OsMed9_2 was 2-fold down-regulated in the P6 stage. Of note, the expression of AtMed9_1 was augmented in the S9 and S10 stages, and the expression in OsMed9_1 declined in the equivalent substages. Of the remaining middle module gene, Med10, the lone rice homolog did not show differential expression in reproductive stages when compared with the vegetative stages; however, the Arabidopsis homolog AtMed10_1 exhibited up-regulation in the S9 and S10 stages. AtMed10_2 could not be assessed, as it was not represented on the chip.
Four of the seven Med genes in the tail module showed appreciable differential transcript abundance in at least one stage of reproductive development in either Arabidopsis or rice with respect to the vegetative controls. These were Med3, -5, -14, and -15 (Fig. 7, A and B). For Med3, only the Arabidopsis homolog showed up-regulation in the S9 and S10 stages of seed development. In Arabidopsis, SWP (Med14) has been shown to interact with the corepressor LEUING (Gonzalez et al., 2007) and its expression levels are important in defining the duration of cell proliferation during leaf development; thus, it plays a role in pattern formation at the meristem (Autran et al., 2002) as well as in the regulation of root elongation by repressing the root-specific gene Lateral Root Primordium1 via histone deacetylation (Krichevsky et al., 2009). We identified two Med14 paralogs in rice, of which only OsMed14_1 had significant differential expression in some reproductive substages (P1, P4, S1, and S3) with respect to both the vegetative controls. Notably, there was an 11-fold up-surge in its transcript abundance soon after fertilization in the S1 stage, and in the subsequent developmental stage, the expression levels came down to levels similar to vegetative stages. OsMed14_2, on the other hand, did not show any differential transcript accumulation in any of the reproductive stages vis-a-vis the vegetative controls. AtMed14_1 showed specific enhanced expression only in the S9 and S10 stages. Among the three rice Med5 homologs and two members in Arabidopsis, only OsMed5_2 showed significant change in its mRNA levels in a reproductive stage (S5; down-regulation) in comparison with both the vegetative controls. Of the two Med15 homologs in rice, only OsMed15_1 showed high-level expression in some seed stages, the expression being up-regulated by 75-, 60-, and 5-fold in the S2, S3, and S4 stages, respectively, signifying a possible role in rice embryo development. Of the three Arabidopsis Med15 paralogs, AtMed15_1 is not represented on the chip and the other two did not show appreciable differential expression in reproductive stages with respect to leaf and root. Med16 is a coactivator of lipopolysaccharide- and heat shock-induced transcriptional activator in Drosophila (Kim et al., 2004) and is known as SFR6 in Arabidopsis. SFR6 is part of a complex network regulating the photoperiodic pathway and circadian clock gene expression in response to external and internal changes such as light, metabolic status, and temperature (Knight et al., 2008). The rice as well as Arabidopsis Med16 genes did not display much variation in expression patterns across all the developmental stages. The sfr6 mutant was isolated for its inability to cold acclimate to freezing temperatures (Warren et al., 1996). Surprisingly, we did not see any differential response in both plant species even under cold stress (Supplemental Tables S4 and S5). However, this is in accordance with the reported limited transcriptional regulation of the gene by cold; SFR6, in fact, has been proposed to act posttranslationally on the C box-binding factor TFs to modulate their activity to provide cold tolerance (Knight et al., 2009).
Expression analysis between comparable reproductive stages of the dicots and the monocots highlights that not only do some members of the same gene family exhibit variable expression patterns between the two angiosperms but also among its paralogs. This not only provides evidence for tissue- or cell-specific dynamics in the constitution of Med complexes, as has been well documented in mammals (Conaway et al., 2005a; Taatjes, 2010), but also differential regulation of Med in a species-specific manner.
Expression Profile of the Kinase Module and Other Associated Med Genes during Reproductive Development
The kinase module of the Med complex can repress as well as activate transcription (Taatjes, 2010), where Med12 and Med13 subunits play a critical role for the subcomplex-dependent repression (Knuesel et al., 2009). Med12 promotes the epigenetic silencing of target genes by recruiting a histone methyltransferase and methylating chromatin (Ding et al., 2008). In Arabidopsis, Med12 (CENTER CITY [CCT]) and Med13 (GRAND CENTRAL [GCT]) exert a transient repression on the transcriptional machinery that interferes with embryo development (Gillmor et al., 2010). Mutations in these genes delay the specification of cell identity and the globular-to-heart transition but have minimal effects on the initial growth rate of the embryo. In our microarray analysis, this expected enhancement in transcript levels was clearly visible for the Arabidopsis homolog and one rice paralog (OsMed12_2). The expression levels were several-fold higher in the equivalent Arabidopsis S5 and rice S2 stages as compared with the preceding seed development stages (Supplemental Tables S9 and S10). It is noteworthy that these stages represent the late globular embryo stage of seed development (Schmid et al., 2005; Agarwal et al., 2007). While the expression continued to be high in the S3 and S4 stages in rice, dipping only in the S5 stage, the Arabidopsis homolog exhibited highest expression in the S9 and S10 stages, which are maturation stages of embryo development. Apparently, the other rice paralog does not seem to participate in this activity. In contrast to Med12, the Med13 genes of both the angiosperms did not show any significant differential expression during stages of reproductive development; however, OsMed13_1 showed approximately 3-fold up-regulation in leaf with respect to root. Between the CyclinC-Cdk8 genes, all the CycC homologs in Arabidopsis and rice as well as OsCdk8_1 were not significantly differentially regulated in reproductive stages when compared with leaf as well as root. However, the expression increased in S9 and S10 for AtCdk8_1, while OsCdk8_1 was up-regulated by more than 3-fold in only leaf in comparison with root.
There are several Meds whose positions in the complex are uncertain. One of them is Med25, which is absent in yeast but plays a role in the regulation of xenobiotic metabolism and lipid homeostasis in human liver (Rana et al., 2011). A biochemical screen identified Arabidopsis PFT1 as Med25 (Bäckström et al., 2007). The PFT1 negatively regulates the phytochrome signaling pathway (Wollenberg et al., 2008), regulating flowering in response to light quality (Cerdán and Chory, 2003) and by acting as a positive regulator of jasmonic acid signaling, which regulates plant defense responses during fungal pathogen infection (Kidd et al., 2009). The expression profile of AtMed25_1 revealed very similar transcript abundance in flower as compared with vegetative tissues, augmenting by approximately 2-fold in the S9 and S10 seed stages. The rice homolog had comparable transcript levels in the reproductive and vegetative stages. This is not surprising, since PFT1 is known to exert its effect on flowering only in response to a suboptimal light environment (Cerdán and Chory, 2003), specific conditions not analyzed in this study. Moreover, the role of PFT1 in chromatin remodeling via histone acetylases like human Med25 (Lee et al., 2007) cannot be underestimated.
The other animal-specific Med in this category is Med26. Of the three rice Med26 paralogs, OsMed26_1 did not show any significant differential expression (Supplemental Table S9), OsMed26_2 transcripts increased in the S3, S4, and S5 stages, and OsMed26_3 transcripts dipped by more than 4-fold in leaf tissue in comparison with roots (Supplemental Table S4). Arabidopsis also has three Med26 members, of which AtMed26_1 was not represented on the chip, while AtMed26_2 and AtMed26_3 were not significantly differentially regulated in reproductive stages in comparison with both the vegetative stages. However, like OsMed26_3, the expression of these genes was down-regulated in leaf in comparison with roots. Several of the plant-specific Med34, -35, -36, and -37 homologs showed high expression in both the reproductive stages as compared with the vegetative stages, signifying the important roles these genes may play in regulating plant-specific functions.
CONCLUSION
Among eukaryotes, plants are unique in being sessile, enduring the vagaries of their surrounding environment, both biotic and abiotic. In addition, several processes like flowering and response to the quality as well as the quantity of light in several metabolic as well as developmental processes are specific to plants. Thus, it is not surprising that several plant-specific TF families have emerged during the course of evolution (Riechmann et al., 2000). Consequently, one would expect a similar expansion in the Med components that act as a link between these TFs and the Pol II core complex. Our in-depth in silico prediction for these subunits revealed more than one homolog for various Med subunits in different plants. This expands the possibility of the number of potential Med complexes that might form and, in turn, regulate myriad functions during different developmental stages as well as in response to external environmental cues. In this regard, the identification of several Med subunits as components of the plant machinery that participate in cell proliferation (SWP), embryo development (CCT and GCT), chromatin modification (Med21), and response to external stimuli like light quality, which in turn regulates genes involved in flowering (PFT1) and circadian rhythms (SFR6), is beginning to unravel. This view is strengthened by the genome-wide expression analysis using microarrays of Med genes in two angiosperms, a dicot and a monocot, in various plant developmental stages and during abiotic stress conditions in this study. The differential transcript abundance of several Med genes (or their paralogs) in both/either angiosperms highlights their importance in regulating plant development in a redundant as well as organism-specific manner. These observations also present the possibility that alternative forms of Med complexes may be functional in various cell types during different developmental stages. We identify 34 unique Med components, including their kinase and cyclin counterparts in plants. The data show that mediators are conserved across the plant kingdom and suggest that all the yeast/metazoan Med subunits may have been incorporated within the Med complex before the plants diverged from fungus/animal groups; however, it appears that as the plants evolved and attained specialized functions and structures, some of these Meds were lost (Med1) while others were gained (Med34 to -37). Secondary structure modeling of Arabidopsis proteins and its comparison with yeast and human counterparts revealed that despite high sequence variation, the functions of Meds may have been maintained across eukaryotes by adopting similar structures. However, some subunits may have gained additional regulatory power either by acquiring new regions or by forming new conformations to interact with TFs or chromatin-modifying proteins. Future studies will be required not only to confirm the existence of these predicted Med homologs as structural components of the Med complex but also to ascertain their biological functions.
MATERIALS AND METHODS
Sequence Retrieval
In this study, putative Med subunits have been identified from 16 plant species belonging to various groups. Protein sequences were downloaded for the angiosperms Arabidopsis (Arabidopsis thaliana), Brachypodium distachyon, Carica papaya, Glycine max, rice (Oryza sativa), Populus trichocarpa, Sorghum bicolor, Vitis vinifera, and Zea mays, the pteridophyte Selaginella moellendorffii, the bryophyte Physcomitrella patens, the green algae Volvox carteri f. sp. nagariensis and Ostreococcus lucimarinus, and the red alga Cyanidioschyzon merolae strain 10D. EST clusters were downloaded for Solanum lycopersicum (an angiosperm) and Pinus taeda (a gymnosperm) and translated with ESTScan-3.0.2 using plant-specific matrices (Iseli et al., 1999). The details of the source databases are provided in Supplemental Table S11. For promoter analysis, 2-kb sequences upstream of the translational initiation sites of some rice and Arabidopsis Med genes were downloaded from the RGAP and TAIR databases, respectively.
In Silico Identification of Mediator Subunits in Plants
The sequences obtained from published literature covering nearly 70 organisms (Bourbon, 2008) and Med sequences downloaded from the National Centre for Biotechnology Information (NCBI) by name search (Supplemental Table S12) and cross-verified to be “mediator specific” at a cutoff E value of less than e-20 in the SwissProt database (Boeckmann et al., 2003) were used to build HMM profiles. An abbreviation was specified for every organism (Supplemental Table S13). The sequences were categorized into different Med families, and HMM profiles were generated on alignments generated using ClustalW version 1.83 (Thompson et al., 1994) followed by identification of Meds using HMMER (http://hmmer.org/) in all the plant species. In the preliminary screening, protein hits having a cutoff E value of e−2 or less were selected as putative Meds; furthermore, Med sequences of metazoan, fungal, and plant origin were selected to construct FL-HMM. In order to pick distantly related Med homologs, HH regions for each Med subunit were defined. For this purpose, continuous stretches of at least seven amino acids having a quality score of 60 or more each were delineated over the entire length of an alignment, and the mean score for each block was calculated. The highest scoring amino acid stretch was singled out followed by the identification of other blocks having an average score within 3% (to account for a kingdom-specific bias [viz. metazoan, fungi, and plants]) of the highest HH score. These were collectively selected as HH regions and numbered from the N terminus onward; an example is shown in Supplemental Figure S5. Furthermore, the alignments corresponding to these HH regions were extracted using the multiple alignment editor Jalview (Waterhouse et al., 2009), HMM profiles (HH-HMMs) for Med1 to -31 were generated, and fresh HMMER searches were performed. Hits with E values of e−3 or less using FL-HMMs and E values of e−1 or less using HH-HMMs (expect values determined on bona fide Arabidopsis Med hits) were further screened on the basis of the most prevalent HH regions among hits and/or the presence of Med-specific Pfam domains (Finn et al., 2010; Supplemental Table S1). For identifying Cdk8 and CycC homologs, a stringent E value cutoff of e−90 or less was used after cross-referencing the hits using the Pfam database. For identifying Med32 to -37, reported only in Arabidopsis, initially, PSI-BLAST (Altschul et al., 1997) searches were performed on the protein sequences of the plant species selected for the study using the BLOSUM 62 substitution matrix and an iteration threshold of 0.001. The best hit from each plant species was selected and used to generate the FL- and HH-HMMs using the criteria described above. Med proteins with comparable expect values to bona fide Arabidopsis Med proteins and the presence of common Pfam domains were selected. The predicted plant Meds were named as follows: a rice Med11 gene having two paralogs were designated OsMed11_1 and OsMed11_2, respectively, numbered according to the increasing order of locus identifiers. To remove redundancy, all the splice variants of a locus were considered as a singular entry. All the putative plant Meds and an identity converter are listed in Supplemental Table S14.
Other Bioinformatics Tools Used
Protein domain identification was done using the Pfam, SMART (Letunic et al., 2009), and InterProScan (Quevillon et al., 2005) databases. Subcellular localization of proteins was performed using MultiLoc2 (Blum et al., 2009), and nucleus-localizing signals were identified using NLStradamus (Nguyen Ba et al., 2009). Protein secondary structures were predicted using PSIPRED (Bryson et al., 2005). The cis-elements in the promoter sequences were identified using the PLACE database (Higo et al., 1999). All these databases were queried at the default parameters.
Phylogenetic Analysis
Multiple alignments for Med proteins were generated on full-length amino acid sequences using the MAFFT iterative refinement method (1,000 iterations) incorporating local pairwise alignment (Katoh et al., 2005). The alignments were corrected manually, and a PHYLIP neighbor tree with 1,000 replicates defined as the bootstrap value was built using an online tool (http://bioweb2.pasteur.fr/phylogeny/). The tree was viewed in FigTree version 1.2.3 (available at http://tree.bio.ed.ac.uk/software/figtree/).
Microarray Data Analysis
Microarray analysis was performed on expression data obtained in our laboratory previously (Agarwal et al., 2007; Arora et al., 2007) on Affymetrix GeneChip Rice Genome Arrays representing 49,824 rice transcripts for three vegetative stages (mature leaf, 7-d-old seedling, and its root), six panicle development stages (all stages collected at a particular length [in cm] of panicles as follows: P1, 0–3; P2, 3–5; P3, 5–10; P4, 10–15; P5, 15–22; P6, 22–30), five seed development stages (S1, 0–2 d after pollination [DAP]; S2, 3–4 DAP; S3, 5–10 DAP; S4, 11–20 DAP; S5, 21–29 DAP), and three types of stress (cold, salt, and desiccation) in rice. The raw data (*.cel) files (platform accession no. GPL2025 under the series accession nos. GSE6893 and GSE6901, deposited at the Gene Expression Omnibus database of the NCBI) were imported to GeneSpring GX 10 software (Agilent Technologies) for detailed analysis. The redundancy in the probe sets was taken care of by preferring the 3′-most probe set for each gene (R. Sharma, P. Agarwal, S. Ray, P. Deveshwar, P. Sharma, N. Sharma, A. Nijhawan, M. Jain, A.K. Singh, V.P. Singh, J.P. Khurana, A.K. Tyagi, and S. Kapoor, unpublished data). Normalization of the raw data was performed using the Gene Chip Robust Multiarray Analysis algorithm (Wu et al., 2003), data were baseline transformed, following which signal intensity values were log transformed and averages of the three biological replicates for each sample were calculated. One-way ANOVA statistical analysis was used to perform differential expression analysis by taking vegetative controls as reference to identify genes expressing at greater than 2-fold in different stages of reproductive development (panicle and seed), with P ≤ 0.05. Similarly, for identifying differentially expressing stress-induced genes, analysis was performed by taking the untreated 7-d-old seedling as the reference.
The expression data for Arabidopsis was obtained from the Gene Expression Omnibus under the series accession numbers GSE5620, GSE5621, GSE5623, GSE5624, GSE5629, GSE5630, GSE5631, GSE5632, and GSE5634. These series represent Affymetrix GeneChip ATH1 Genome Arrays of stages comparable to those used for rice. A total of 55 *.cel files representing 21 stages of development as well as stress treatments were analyzed as described above for rice followed by generating heat maps for all/selected genes.
QPCR Analysis
QPCR was performed using three biological replicates. Each biological replicate in turn was assessed using three technical replicates. Primers were designed from the 3′ end of the selected genes using Primer Express (Applied Biosystems) at its default settings and checked for specificity by performing BLAST at the NCBI with other regions of the genome. DNase (Fermentas Life Sciences)-treated RNA was reverse transcribed to synthesize the first strand-cDNA (cDNA Archive kit; Applied Biosystems). SYBR Green PCR Master Mix (Applied Biosystems) was used to estimate the expression levels for the selected genes using the ABI Prism 7000 Sequence Detection System (Step One Plus; Applied Biosystems). The expression obtained using the ACTIN gene was used as the endogenous control to normalize the variation among different samples using cycle threshold (Ct value) as a discriminating criteria. The data were further normalized against the maximum average expression values obtained from the microarray.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic relationship of Med subunits for the middle and tail modules.
Supplemental Figure S2. Phylogenetic relationship of Med subunits for the kinase and unknown modules.
Supplemental Figure S3. Amino acid alignments of Med7N and Med31C for plant, yeast, and human Med sequences.
Supplemental Figure S4. The conserved Med15 KIX region.
Supplemental Figure S5. Defining the HH regions for a Med subunit.
Supplemental Table S1. Occurrence of the HH regions and Pfam domains in various Med gene families.
Supplemental Table S2. PSI-BLAST result of plant Med32, jump-started using Med2/Med29 aligned sequences.
Supplemental Table S3. List of Med genes with no definitive annotation in respective databases.
Supplemental Table S4. Differential expression analysis between vegetative stages, vegetative and reproductive stages, and stress conditions in rice.
Supplemental Table S5. Differential expression analysis between vegetative stages, vegetative and reproductive stages, and stress conditions in Arabidopsis.
Supplemental Table S6. Identification of cis-elements in promoter regions of selected genes differentially expressing in reproductive stages in rice and Arabidopsis as well as under stress conditions in rice.
Supplemental Table S7. Normalized expression values obtained for all rice Med genes as revealed by microarray analysis.
Supplemental Table S8. Normalized expression values obtained for all Arabidopsis Med genes as revealed by microarray analysis.
Supplemental Table S9. Differential expression analysis between all the reproductive stages in rice.
Supplemental Table S10. Differential expression analysis between all the reproductive stages in Arabidopsis.
Supplemental Table S11. Database sources of proteins for different organisms.
Supplemental Table S12. List of mediators from the NCBI used to build HMM profiles.
Supplemental Table S13. List of organisms and their abbreviations belonging to different groups.
Supplemental Table S14. List of putative plant mediators identified in this study.
Acknowledgments
Dr. Ramesh Hariharan of Strand Life Sciences, India, and Dr. Sanjeev Singh of Institute of the Informatics and Communication, University of Delhi South Campus, India, are thanked for useful discussions.
References
- Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, Kapoor S, Tyagi AK. (2007) Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol 65: 467–485 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari SA, He Q, Morse RH. (2009) Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci USA 106: 16734–16739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. (1999) Conserved structures of Mediator and RNA polymerase II holoenzyme. Science 283: 985–987 [DOI] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J. (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. (2007) Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Baidoobonso SM, Guidi BW, Myers LC. (2007) Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae Mediator complex. J Biol Chem 282: 5551–5559 [DOI] [PubMed] [Google Scholar]
- Balciunas D, Gälman C, Ronne H, Björklund S. (1999) The Med1 subunit of the yeast Mediator complex is involved in both transcriptional activation and repression. Proc Natl Acad Sci USA 96: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24: 23–58 [Google Scholar]
- Björklund S, Buzaite O, Hallberg M. (2001) The yeast Mediator. Mol Cells 11: 129–136 [PubMed] [Google Scholar]
- Björklund S, Gustafsson CM. (2005) The yeast Mediator complex and its regulation. Trends Biochem Sci 30: 240–244 [DOI] [PubMed] [Google Scholar]
- Blum T, Briesemeister S, Kohlbacher O. (2009) MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics 10: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeing S, Rigault C, Heidemann M, Eick D, Meisterernst M. (2010) RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a Mediator-dependent fashion. J Biol Chem 285: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth V, Koth CM, Edwards AM, Arrowsmith CH. (2000) Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70. J Biol Chem 275: 31266–31268 [DOI] [PubMed] [Google Scholar]
- Bourbon HM. (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional Mediator complex. Nucleic Acids Res 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, et al. (2004) A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell 14: 553–557 [DOI] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. (2005) Protein structure prediction servers at University College London. Nucleic Acids Res 33: W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin GT, Stevens JL, Berk AJ. (2003) Activation domain-Mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc Natl Acad Sci USA 100: 12003–12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Conaway JW, Florens L, Sato S, Tomomori-Sato C, Parmely TJ, Yao T, Swanson SK, Banks CA, Washburn MP, Conaway RC. (2005a) The mammalian Mediator complex. FEBS Lett 579: 904–908 [DOI] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. (2005b) The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci 30: 250–255 [DOI] [PubMed] [Google Scholar]
- Davis JA, Takagi Y, Kornberg RD, Asturias FA. (2002) Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell 10: 409–415 [DOI] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. (2008) MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell 32: 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T. (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the Mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold ML, Koch M, Loeliger E, Cura V, Winston F, Cavarelli J, Romier C. (2010) The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26. EMBO J 29: 3979–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Boyer TG. (2009) MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J Biol Chem 284: 2648–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, Joseph SM, Friez MJ, Schwartz CE, Pradhan S, et al. (2008) Mediator links epigenetic silencing of neuronal gene expression with X-linked mental retardation. Mol Cell 31: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. (2010) CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol 17: 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeier CC, Taatjes DJ. (2010) Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci USA 107: 11283–11288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan PM, Kelleher RJ, III, Sayre MH, Tschochner H, Kornberg RD. (1991) A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350: 436–438 [DOI] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA 93: 8329–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor CS, Park MY, Smith MR, Pepitone R, Kerstetter RA, Poethig RS. (2010) The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. (2007) The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and Mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol 27: 5306–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. (2004) A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res 32: 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg M, Polozkov GV, Hu GZ, Beve J, Gustafsson CM, Ronne H, Björklund S. (2004) Site-specific Srb10-dependent phosphorylation of the yeast Mediator subunit Med2 regulates gene expression from the 2-μm plasmid. Proc Natl Acad Sci USA 101: 3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Iseli C, Jongeneel CV, Bucher P. (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 138–148 [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. (1998) Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci USA 95: 8538–8543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. (2001) The structural and functional organization of the yeast Mediator complex. J Biol Chem 276: 42003–42010 [DOI] [PubMed] [Google Scholar]
- Kato Y, Habas R, Katsuyama Y, Näär AM, He X. (2002) A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature 418: 641–646 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, III, Flanagan PM, Kornberg RD. (1990) A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61: 1209–1215 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. (2009) The Mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Kwon YJ, Kim JM, Song YH, Kim SN, Kim YJ. (2004) MED16 and MED23 of Mediator are coactivators of lipopolysaccharide- and heat-shock-induced transcriptional activators. Proc Natl Acad Sci USA 101: 12153–12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Mugford Nee Garton SG, Ulker B, Gao D, Thorlby G, Knight MR. (2009) Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J 58: 97–108 [DOI] [PubMed] [Google Scholar]
- Knight H, Thomson AJ, McWatters HG. (2008) SENSITIVE TO FREEZING6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol 148: 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. (2009) The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev 23: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. (2005) Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30: 235–239 [DOI] [PubMed] [Google Scholar]
- Koschubs T, Lorenzen K, Baumli S, Sandström S, Heck AJ, Cramer P. (2010) Preparation and topology of the Mediator middle module. Nucleic Acids Res 38: 3186–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschubs T, Seizl M, Larivière L, Kurth F, Baumli S, Martin DE, Cramer P. (2009) Identification, structure, and functional requirement of the Mediator submodule Med7N/31. EMBO J 28: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A, Zaltsman A, Kozlovsky SV, Tian GW, Citovsky V. (2009) Regulation of root elongation by histone acetylation in Arabidopsis. J Mol Biol 385: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JY, Park JM, Gim BS, Han SJ, Lee J, Kim YJ. (1999) Caenorhabditis elegans Mediator complexes are required for developmental-specific transcriptional activation. Proc Natl Acad Sci USA 96: 14990–14995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larivière L, Geiger S, Hoeppner S, Röther S, Strässer K, Cramer P. (2006) Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat Struct Mol Biol 13: 895–901 [DOI] [PubMed] [Google Scholar]
- Larivière L, Seizl M, van Wageningen S, Röther S, van de Pasch L, Feldmann H, Strässer K, Hahn S, Holstege FC, Cramer P. (2008) Structure-system correlation identifies a gene regulatory Mediator submodule. Genes Dev 22: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Park UH, Kim EJ, Um SJ. (2007) MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J 26: 3545–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Fletcher AG, Zhang L, Chen X, Fischbeck JA, Stargell LA. (2010) Activation of a poised RNAPII-dependent promoter requires both SAGA and Mediator. Genetics 184: 659–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37: D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bjorklund S, Jiang YW, Kim YJ, Lane WS, Stillman DJ, Kornberg RD. (1995) Yeast global transcriptional regulators Sin4 and Rgr1 are components of Mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA 92: 10864–10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. (2004) Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol 24: 1721–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci 25: 277–283 [DOI] [PubMed] [Google Scholar]
- Malik S, Wallberg AE, Kang YK, Roeder RG. (2002) TRAP/SMCC/Mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol 22: 5626–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Lin SC, Bernecky C, Gao Y, Taatjes DJ. (2010) p53 activates transcription by directing structural shifts in Mediator. Nat Struct Mol Biol 17: 753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD. (1998) The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev 12: 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. (2009) NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M, Eisenhaber F. (2004) Linking transcriptional mediators via the GACKIX domain super family. Curr Biol 14: R54–R55 [DOI] [PubMed] [Google Scholar]
- Park JM, Gim BS, Kim JM, Yoon JH, Kim HS, Kang JG, Kim YJ. (2001) Drosophila Mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol Cell Biol 21: 2312–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana R, Surapureddi S, Kam W, Ferguson S, Goldstein JA. (2011) Med25 is required for RNA polymerase II recruitment to specific promoters, thus regulating xenobiotic and lipid metabolism in human liver. Mol Cell Biol 31: 466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Ryu S, Zhou S, Ladurner AG, Tjian R. (1999) The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397: 446–450 [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Banks CA, Sorokina I, Parmely TJ, Kong SE, Jin J, Cai Y, Lane WS, Brower CS, et al. (2003) Identification of mammalian Mediator subunits with similarities to yeast Mediator subunits Srb5, Srb6, Med11, and Rox3. J Biol Chem 278: 15123–15127 [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, et al. (2004) A set of consensus mammalian Mediator subunits identified by multidimensional protein identification technology. Mol Cell 14: 685–691 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shaikhibrahim Z, Rahaman H, Wittung-Stafshede P, Björklund S. (2009) Med8, Med18, and Med20 subunits of the Mediator head domain are interdependent upon each other for folding and complex formation. Proc Natl Acad Sci USA 106: 20728–20733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spåhr H, Bève J, Larsson T, Bergström J, Karlsson KA, Gustafsson CM. (2000) Purification and characterization of RNA polymerase II holoenzyme from Schizosaccharomyces pombe. J Biol Chem 275: 1351–1356 [DOI] [PubMed] [Google Scholar]
- Stout J, Romero-Severson E, Ruegger MO, Chapple C. (2008) Semidominant mutations in reduced epidermal fluorescence 4 reduce phenylpropanoid content in Arabidopsis. Genetics 178: 2237–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ. (2010) The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci 35: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ, Näär AM, Andel F, III, Nogales E, Tjian R. (2002) Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295: 1058–1062 [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Schneider-Poetsch T, Tjian R. (2004) Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat Struct Mol Biol 11: 664–671 [DOI] [PubMed] [Google Scholar]
- Takagi Y, Kornberg RD. (2006) Mediator as a general transcription factor. J Biol Chem 281: 80–89 [DOI] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Chau KH, Wagner G, Näär AM. (2009) Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem 284: 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville MM, Dudenhausen EE, Awad KS, Gjymishka A, Zhong C, Kilberg MS. (2008) Activated transcription via mammalian amino acid response elements does not require enhanced recruitment of the Mediator complex. Nucleic Acids Res 36: 5571–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Young RA. (1995) General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA 92: 4587–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Walling JG, Shoemaker R, Young N, Mudge J, Jackson S. (2006) Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 172: 1893–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ge K, Roeder RG, Hankinson O. (2004) Role of Mediator in transcriptional activation by the aryl hydrocarbon receptor. J Biol Chem 279: 13593–13600 [DOI] [PubMed] [Google Scholar]
- Warren G, McKown R, Marin AL, Teutonico R. (1996) Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg AC, Strasser B, Cerdán PD, Amasino RM. (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol 148: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F. (2003) A Model Based Background Adjustment for Oligonucleotide Expression Arrays. Technical report. Department of Biostatistics Working Papers, Baltimore, MD [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. (2006) An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442: 700–704 [DOI] [PubMed] [Google Scholar]
- Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. (1998) The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA 95: 7939–7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S. (2000) A transcription reinitiation intermediate that is stabilized by activator. Nature 408: 225–229 [DOI] [PubMed] [Google Scholar]
- Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. (2004) A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol 24: 6871–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. (2005) MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell 19: 89–100 [DOI] [PubMed] [Google Scholar]