Abstract
The ubiquitin (Ub)-26S proteasome pathway is implicated in various cellular processes in higher plants. AtAIRP1, a C3H2C3-type RING (for Really Interesting New Gene) E3 Ub ligase, is a positive regulator in the Arabidopsis (Arabidopsis thaliana) abscisic acid (ABA)-dependent drought response. Here, the AtAIRP2 (for Arabidopsis ABA-insensitive RING protein 2) gene was identified and characterized. AtAIRP2 encodes a cytosolic C3HC4-type RING E3 Ub ligase whose expression was markedly induced by ABA and dehydration stress. Thus, AtAIRP2 belongs to a different RING subclass than AtAIRP1 with a limited sequence identity. AtAIRP2-overexpressing transgenic (35S:AtAIRP2-sGFP) and atairp2 loss-of-function mutant plants exhibited hypersensitive and hyposensitive phenotypes, respectively, to ABA in terms of seed germination, root growth, and stomatal movement. 35S:AtAIRP2-sGFP plants were highly tolerant to severe drought stress, and atairp2 alleles were more susceptible to water stress than were wild-type plants. Higher levels of drought-induced hydrogen peroxide production were detected in 35S:AtAIRP2-sGFP as compared with atairp2 plants. ABA-inducible drought-related genes were up-regulated in 35S:AtAIRP2-sGFP and down-regulated in atairp2 progeny. The positive effects of AtAIRP2 on ABA-induced stress genes were dependent on SNF1-related protein kinases, key components of the ABA signaling pathway. Therefore, AtAIRP2 is involved in positive regulation of ABA-dependent drought stress responses. To address the functional relationship between AtAIRP1 and AtAIRP2, FLAG-AtAIRP1 and AtAIRP2-sGFP genes were ectopically expressed in atairp2-2 and atairp1 plants, respectively. Constitutive expression of FLAG-AtAIRP1 and AtAIRP2-sGFP in atairp2-2 and atairp1 plants, respectively, reciprocally rescued the loss-of-function ABA-insensitive phenotypes during germination. Additionally, atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1 complementation lines were more tolerant to dehydration stress relative to atairp1 and atairp2-2 single knockout plants. Overall, these results suggest that AtAIRP2 plays combinatory roles with AtAIRP1 in Arabidopsis ABA-mediated drought stress responses.
Dehydration and continuous water deficit drastically hinder plant growth and development. To survive under such severe environmental conditions, sessile plants have developed adaptive strategies that involve integrated molecular, cellular, and metabolic programs (Fujita et al., 2006; Yoo et al., 2009; Ahuja et al., 2010; Hirayama and Shinozaki, 2010; Hummel et al., 2010). Inhibition of plant growth and development by water stress conditions is directly correlated with worldwide reductions in crop yields. Thus, elucidation of stress defense mechanisms and the development of resistant transgenic crops against drought have attracted much interest for many years.
The plant stress hormone abscisic acid (ABA) regulates drought stress responses as a main modulator. In particular, ABA induces stomata closing to mitigate undesirable transpirational water loss and activates various gene groups to initiate rapid and efficient defense programs (Xiong et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2006; Tuteja, 2007; Cho et al., 2009; Kim et al., 2010b; Raghavendra et al., 2010). Among the diverse gene sets induced by ABA are the E3 ubiquitin (Ub) ligases. This suggests the existence of a functional network between ABA-mediated stress responses and Ub-dependent protein degradation.
Ub is a conserved 76-amino acid polypeptide that functions as a posttranslational protein tag. Ub-mediated protein modification is ubiquitously found in eukaryotic cells (Dye and Schulman, 2007; Hunter, 2007; Vierstra, 2009). In higher plants, the ubiquitination system is associated with many cellular processes as diverse as environmental stress responses, circadian rhythms, cell cycles, and hormone signaling (Moon et al., 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007; Vierstra, 2009; Lee and Kim, 2011). Ub tagging of target proteins is performed by three consecutive actions of E1 Ub-activating enzymes, E2 Ub-conjugating enzymes, and E3 Ub ligases (Glickman and Adir, 2004; Smalle and Vierstra, 2004). Polyubiquitinated substrate proteins are degraded by the 26S proteasome complex, while monoubiquitination or multiubiquitination confers nonproteolytic functions, such as DNA repair, protein trafficking, protein activity, and protein-protein interactions (Mukhopadhyay and Riezman, 2007; Jacobson et al., 2009).
Approximately 6% of the Arabidopsis (Arabidopsis thaliana) proteome is involved in the Ub 26S proteasome pathway; in particular, there are more than 1,400 different E3 Ub ligase genes in the Arabidopsis genome (Smalle and Vierstra, 2004; Vierstra, 2009). Proteins encoded by Ub ligase genes contain distinct functional motifs, such as RING (for Really Interesting New Gene), U Box, HECT (for Homology to E6-AP Carboxyl Terminus), SCF (for Skp1-Cullin-F Box), or APC (for Anaphase-Promoting Complex). Arabidopsis contains at least 477 RING domain-containing E3 Ub ligase genes (Kraft et al., 2005; Stone et al., 2005; Vierstra, 2009). These RING E3 Ub ligase members play various physiological roles, including hormonal perception and signaling, seed germination and seedling development, and nitrogen and sugar responses (Xie et al., 2002; Zhang et al., 2005; Stone et al., 2006; Peng et al., 2007; Bu et al., 2009; Huang et al., 2010; Liu and Stone, 2010). Particularly, RING E3 proteins are shown to function as key mediators in defense mechanisms against salt and osmotic stresses by increasing ABA biosynthesis (Ko et al., 2006) and ABA-dependent drought signaling (Zhang et al., 2007, 2008). Furthermore, DRIP-RING E3 Ub ligase functions as a negative regulator in the drought stress response by ubiquitinating the drought-induced DREB2A transcription factor (Qin et al., 2008), whereas the ER-localized RING E3 Rma1H1 is a positive regulator that induces Ub/26S proteasome-dependent degradation of a water channel protein, aquaporin PIP2;1 (Lee et al., 2009). Recently, it was reported that RHA2a and RHA2b RING E3s play positive roles in ABA signaling and drought responses (Li et al., 2011). These studies indicate that different isoforms of RING E3 Ub ligases are crucially involved either positively or negatively in Arabidopsis drought stress responses. In addition, RING E3 Ub ligases function in drought stress responses in rice (Oryza sativa), a monocot model crop (Liu et al., 2008; Park et al., 2010; Bae et al., 2011; Ning et al., 2011).
Because ABA is a well-characterized plant stress hormone (Xiong et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2006; Tuteja, 2007; Cutler et al., 2010; Hubbard et al., 2010; Kim et al., 2010b), we wanted to elucidate the functional relationship between ABA and RING E3 Ub ligases in response to dehydration stress in Arabidopsis. Previously, we identified 100 RING E3 Ub ligase genes, which appeared to be up-regulated in response to abiotic stresses based on in silico data (http://www.genevestigator.com). Mutant seeds with T-DNA insertion knockouts of these selected genes were obtained from the Arabidopsis Biological Resource Center and were screened for ABA sensitivity during the germination stage (Ryu et al., 2010). Several mutants that displayed ABA-insensitive phenotypes in comparison with wild-type plants were isolated. One of these mutants was named atairp1 (for Arabidopsis ABA-insensitive RING protein 1). Phenotypic analysis suggested that AtAIRP1, a C3H2C3-type RING E3 Ub ligase, is a positive regulator in the Arabidopsis ABA-dependent drought response.
In this study, another loss-of-function mutant that was insensitive to ABA during the germination stage was characterized. This mutant was referred to as atairp2. The AtAIRP2 gene encodes a C3HC4-type RING E3 Ub ligase, and its expression was markedly induced in response to ABA and a broad spectrum of abiotic stresses, including drought, cold, and high salt levels. AtAIRP2 overexpressors and atairp2 loss-of-function mutant plants exhibited inverse phenotypes in terms of ABA-responsive seed germination, root growth, and stomatal movement. Furthermore, 35S:AtAIRP2-sGFP transgenic plants were highly tolerant of severe drought stress; in contrast, atairp2 alleles were more susceptible to mild water stress than were wild-type plants. These results suggest that AtAIRP2, an Arabidopsis C3HC4-type RING E3 Ub ligase, is involved in positively regulating ABA-dependent drought stress responses. To address the functional relationship between AtAIRP1 and AtAIRP2, the FLAG-AtAIRP1 and AtAIRP2-sGFP genes were ectopically expressed in atairp2 and atairp1 mutant plants, respectively. These complementation transgenic (atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1) plants were subsequently used for the analysis of ABA-related phenotypes. The results showed that constitutive expression of FLAG-AtAIRP1 and AtAIRP2-sGFP in atairp2 and atairp1, respectively, reciprocally rescued the loss-of-function ABA-insensitive phenotypes. Collectively, the results presented in this report suggest that the RING E3 Ub ligase AtAIRP2 plays combinatory roles with AtAIRP1 in ABA-mediated drought stress responses in Arabidopsis.
RESULTS
Identification of AtAIRP2 Encoding a C3HC4-Type RING E3 Ub Ligase in Arabidopsis
Germination tests of 100 different T-DNA insertion loss-of-function Arabidopsis mutants, in which RING E3 Ub ligase genes were silenced, revealed that the #72 mutant seedlings were significantly less sensitive to ABA as compared with the wild-type seedlings. In terms of cotyledon greening, germination percentages of wild-type seedlings were clearly reduced in response to ABA. In our experimental conditions, less than 20% of the wild-type cotyledons were able to green and expand in the 7-d incubation period in the presence of 0.5 μm ABA (Supplemental Fig. S1A). However, approximately 44% of the #72 mutant seedlings exhibited normal greening and expanded cotyledons 7 d after germination. Mutant #72 was subsequently referred to as atairp2-1 (SAIL_686_G08). Search of the Arabidopsis Biological Resource Center database (http://abrc.osu.edu) identified the second allele of the mutant (atairp2-2; Salk_005082). The atairp2-2 mutant seedlings also displayed an ABA-insensitive phenotype highly similar to the atairp2-1 mutant at the germination stage (Supplemental Fig. S1B).
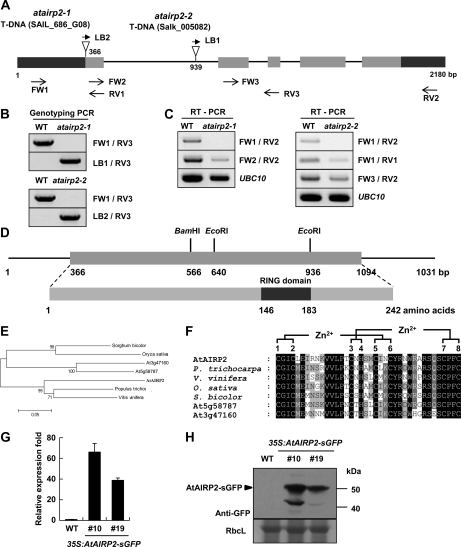
The AtAIRP2 gene (At5g01520; GenBank accession no. NM_120230) is located on chromosome 5 and is composed of 2,180 bp with five exons and four introns. The T-DNA insertions were mapped to the first exon (atairp2-1) and the first intron (atairp2-2) in AtAIRP2 (Fig. 1A). Homozygous mutant plants were selected based on genotyping PCR using primer sets FW1/RV3 and LB_6313R/RV3 (Fig. 1B). Reverse transcription (RT)-PCR revealed that, while partial AtAIRP2 transcripts were detected in the atairp2-1 and atairp2-2 mutant seedlings, full-length mRNAs were undetectable in both alleles, indicating that expression of functional AtAIRP2 mRNAs was repressed in the atairp2 mutant plants (Fig. 1C).
Figure 1.
Identification of atairp2 loss-of-function mutants, sequence analysis of the AtAIRP2 gene, and construction of AtAIRP2 overexpressors. A, Schematic representation of the atairp2-1 (SAIL_686_G08) and atairp2-2 (Salk_005082) alleles with T-DNA insertions. Gray bars indicate coding regions, black bars indicate the 5′ and 3′ untranslated regions, and solid lines represent introns of the AtAIRP2 gene (GenBank accession no. NM_120230). T-DNA insertions are indicated by triangles. T-DNA-specific (LB1 and LB2) and gene-specific (FW1, FW2, FW3, RV1, RV2, and RV3) primers used in genotyping PCR and RT-PCR are indicated with arrows. B, Genotyping PCR of the two atairp2 T-DNA insertion mutant alleles (atairp2-1 and atairp2). Gene-specific and T-DNA-specific primer sets used for genomic PCRs are indicated on the right. WT, Wild type. C, Expression levels of AtAIRP2 transcripts in wild-type and atairp2 mutant plants. Gene-specific primer sets for RT-PCR are indicated on the right. Constitutively expressed UBC10 (for E2 ubiquitin-conjugating enzyme) mRNA was used as a loading control. Primer sequences are listed in Supplemental Table S1. D, Schematic structure of the full-length AtAIRP2 cDNA clone and its deduced protein. The gray bar indicates the coding region, and solid lines represent the 5′ and 3′ untranslated regions. The C-terminal C3HC4-type RING domain is indicated by the black bar. E, Phylogenetic analysis of the seven AtAIRP2 homologs from Arabidopsis (At5g58787 and At3g47160), rice (GenBank accession no. NP_001060539), poplar (XP_002309135), grape (XP_002280008), and sorghum (XP_002447334). F, Amino acid sequence alignment of the RING motifs of AtAIRP2 and other C3HC4-type RING proteins. Potential Zn2+-interacting amino acid residues (C-X2-C-X11-C-X1-H-X2-C-X2-C-X10-C-X2-C) are indicated. Amino acid residues identical in all seven RING domains are shown in black, and those conserved in at least four of the seven sequences are shaded. G, Real-time qRT-PCR analysis of the wild type and AtAIRP2 overexpressors. Expression levels of AtAIRP2 transcripts in wild-type and T3 35S:AtAIRP2-sGFP transgenic (independent lines 10 and 19) plants were determined by real-time qRT-PCR using gene-specific primer sets. UBC10 mRNA levels were used as a loading control. H, Immunoblot analysis of wild-type and AtAIRP2-sGFP (lines 10 and 19) plants. Expression levels of the AtAIRP2-sGFP fusion protein were determined using an anti-GFP antibody. Rubisco large subunit (RbcL) was used as a loading control.
The predicted AtAIRP2 protein consists of 242 amino acids (molecular mass of 28 kD) with a single RING domain in its C-terminal region (Fig. 1D). Consistent with the notion that RING E3 Ub ligases are encoded by a multigene family (Kraft et al., 2005; Stone et al., 2005; Vierstra, 2009), the amino acid sequence identity of AtAIRP2 to other Arabidopsis RING proteins was relatively low (63% identical to At5g58787 and 60% identical to At3g47160; Fig. 1E). Additionally, AtAIRP2 is 64% to 77% identical to rice, poplar (Populus trichocarpa), grape (Vitis vinifera), and sorghum (Sorghum bicolor) RING proteins whose cellular functions are unknown (Supplemental Fig. S2). The Cys-X2-Cys-X11-Cys-X1-His-X2-Cys-X2-Cys-X10-Cys-X2-Cys motif is conserved in the C-terminal RING domain of AtAIRP2, indicating that AtAIRP2 is a C3HC4-type RING E3 Ub ligase (Fig. 1F). Recently, AtAIRP1 (NM_118474) was identified as a stress- and ABA-inducible E3 Ub ligase (Ryu et al., 2010). AtAIRP1 contains a C3H2C3-RING motif with a predicted molecular mass of 16.9 kD (153 amino acids), which is significantly smaller than that of AtAIRP2. AtAIRP2 is only 13% identical to AtAIRP1; therefore, they belong to different subclasses of the RING multigene family.
AtAIRP2 Expression Is Induced in Response to ABA and Other Abiotic Stresses
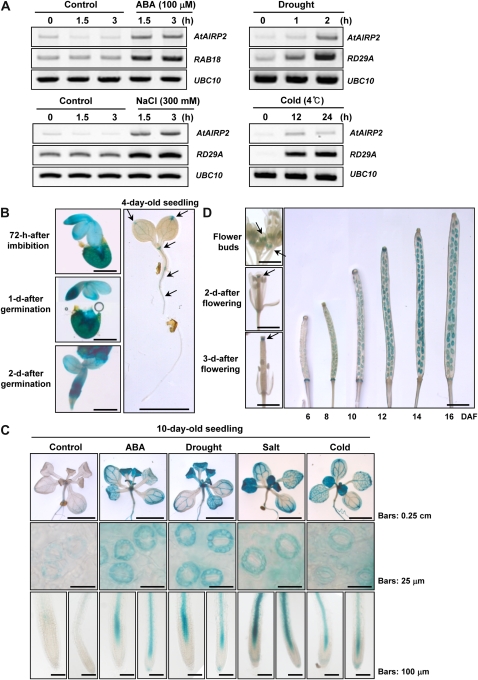
AtAIRP2 was initially considered an ABA- and abiotic stress-induced gene based on the microarray data (http://www.genevestigator.com). To address the in planta induction patterns of AtAIRP2, light-grown 10-d-old seedlings were subjected to ABA treatment or various environmental stresses. Total RNA was isolated from the treated tissues and used for RT-PCR. The results in Figure 2A demonstrate that steady-state levels of AtAIRP2 mRNAs were heightened in response to ABA (100 μm for 1.5–3 h), drought (1–2 h), high salinity (300 mm NaCl for 1.5–3 h), and cold temperature (4°C for 12–24 h; Fig. 2A). The induction kinetics of AtAIRP2 was comparable to those of the marker genes (RAB18 for ABA and RD29A for abiotic stress).
Figure 2.
Expression profiles of AtAIRP2 in response to ABA and different abiotic stress conditions. A, Light-grown, 10-d-old Arabidopsis seedlings were treated with 100 μm ABA (1.5–3 h), drought (1–2 h), high salinity (300 mm NaCl for 1.5–3 h), or cold (4°C for 12–24 h). Total RNA was isolated from the treated tissues and used for RT-PCR. The RAB18 and RD29A genes were positive controls for ABA and abiotic stress responses, respectively. UBC10 was used as a loading control. B to D, AtAIRP2 promoter activity. AtAIRP2-promoter:GUS transgenic T3 plants were incubated with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid for 12 h. AtAIRP2 promoter activity was visualized by GUS-specific staining. B, Histochemical localization of GUS activity in young seedlings (72 h after imbibition, 1 d after germination, 2 d after germination, and 4-d-old seedlings). Arrows indicate GUS signals. Bars = 0.25 cm. C, GUS-specific staining patterns in 10-d-old seedlings in response to ABA, drought, salt, and cold treatments. GUS signals were markedly induced in guard cells in rosette leaves and roots. Bar lengths are indicated to the right. D, GUS activity in mature plants. GUS signals were detected in anthers (flower buds), upper region of stigma (2–3 d after flowering [DAF]), and siliques (6–16 d after flowering). Bars = 0.25 cm. [See online article for color version of this figure.]
To further analyze the AtAIRP2 expression profile, a transcriptional fusion of the 1.3-kb AtAIRP2 upstream region with the GUS reporter gene was constructed and introduced into Arabidopsis. GUS activity was monitored in T3 transgenic plants. AtAIRP2 promoter activity was detected in embryos and testa in very young seedlings 72 h after imbibition and 1 to 2 d after germination, respectively (Fig. 2B). In 4-d-old light-grown seedlings, AtAIRP2 expression was low and restricted to limited areas, including leaf hydathodes, shoot apical meristems, and vascular tissues of shoots and roots (Fig. 2B). In 10-d-old plants, a low basal level of the promoter activity was markedly induced by both ABA and abiotic stresses throughout the plant tissues. ABA- and stress-induced gene expression was clearly identified in guard cells (Fig. 2C). This raises the possibility that AtAIRP2 may play a role in ABA-mediated stomatal movement. A significant amount of GUS staining was also observed in floral organs from fully matured plants, such as anthers, upper stigma regions, and siliques (Fig. 2D). Taken together, gene expression studies suggest that AtAIRP2 is indeed an ABA- and abiotic stress-inducible gene in Arabidopsis.
AtAIRP2 Has in Vitro E3 Ub Ligase Activity and Is Predominantly Localized to Cytosolic Fractions
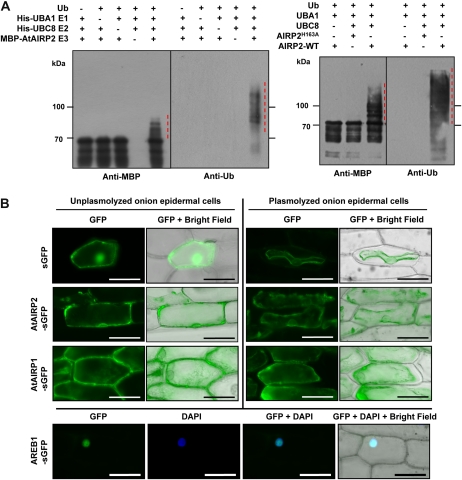
The deduced AtAIRP2 protein possesses a single C3HC4-type RING motif in its C-terminal region, suggesting that AtAIRP2 functions as an E3 Ub ligase. AtAIRP2 was expressed in Escherichia coli as a fusion protein with maltose-binding protein (MBP). The purified recombinant protein was subjected to an in vitro E3 Ub ligase assay. Incubation of MBP-AtAIRP2 with Ub, ATP, UBA1 (Arabidopsis E1), and UBC8 (Arabidopsis E2) at 30°C for 1 h gave rise to high-molecular-mass smearing ladders detected by either anti-MBP or anti-Ub antibody (Fig. 3A). In contrast, MBP-AtAIRP2 failed to display E3 activity in the absence of Ub, E1, or E2. Furthermore, a single-amino acid substitution derivative (MBP-AtAIRP2H163A), in which the conserved His-163 residue was modified to Ala-163, did not exhibit ligase activity (Fig. 3B). Thus, bacterially expressed AtAIRP2 possessed in vitro E3 Ub ligase activity.
Figure 3.
AtAIRP2 is a cytosolic RING E3 Ub ligase. A, In vitro E3 Ub ligase assay. Left panel, bacterially expressed MBP-AtAIRP2 was incubated with ATP in the presence or absence of Ub, Arabidopsis E1 (His-UBA1), and Arabidopsis E2 (His-UBC8) at 30°C for 2 h. Reaction mixtures were separated by SDS-PAGE and subjected to immunoblot analysis using either anti-MBP antibody or anti-Ub antibody. Right panel, MBP-AtAIRP2 and single-amino acid substitution mutant MBP-AtAIRP1H163A were incubated at 30°C for 2 h in the presence of ATP, Ub, E1, and E2. Ubiquitinated proteins were detected by either anti-MBP or anti-Ub antibody. WT, Wild type. B, Cytosolic localization of AtAIRP2. 35S:sGFP, 35S:AtAIRP2-sGFP, 35S:AtAIRP1-sGFP, and 35S:AREB1-sGFP gene constructs were transformed into onion epidermal cells using particle bombardment. Localization of the expressed proteins was visualized by fluorescence microscopy (dark and bright fields) in both unplasmolyzed and plasmolyzed onion cells. Arabidopsis AREB1 and RING E3 Ub ligase AtAIRP1 were used as specificity controls for nuclear and cytosolic proteins, respectively. DAPI, 4′,6-Diamino-phenylindole. Bars = 100 μm. [See online article for color version of this figure.]
To explore the subcellular localization of AtAIRP2, 35S:AtAIRP2-sGFP and control 35S:sGFP constructs were transformed into onion (Allium cepa) epidermal cells using particle bombardment. Localization of expressed fusion proteins was visualized by fluorescence microscopy under dark and bright fields. As shown in Figure 3B, the fluorescence signal of sGFP was uniformly distributed throughout the onion cells. The localization signal of the AtAIRP2-sGFP fusion protein was similar to that of the sGFP control, suggesting that AtAIRP2 is present in the cytosolic fractions. The cytosolic localization of AtAIRP2-sGFP was more evident in plasmolyzed onion epidermal cells. AtAIRP1 and Arabidopsis AREB1 (for ABA response element-binding protein 1) were used as specificity controls. AtAIRP1 is a cytosolic E3 Ub ligase (Ryu et al., 2010), while AREB1 is a nuclear transcription factor (Yoshida et al., 2010). Consistent with previous findings, AtAIRP1 was found in the cytosol of both unplasmolyzed and plasmolyzed cells, whereas AREB1 was exclusively detected in the nuclei (Fig. 3B). Collectively, it is concluded that AtAIRP2 is a cytosolic RING E3 Ub ligase.
Germination Percentages of Arabidopsis Seedlings Are Intimately Linked with the Expression Levels of AtAIRP2 in the Presence of ABA and NaCl Stress
Before exploring detailed phenotypes of the atairp2-1 and atairp2-2 mutant alleles, transgenic Arabidopsis plants that overexpressed the AtAIRP2 gene were constructed using the cauliflower mosaic virus 35S promoter. Several T3 transgenic plants were obtained based on resistance to the herbicide BASTA (glufosinate ammonium), and two independent transgenic lines (lines 10 and 19) were selected for further experiments (Supplemental Fig. S3). Real-time quantitative (q)RT-PCR and immunoblotting with an anti-GFP antibody indicated that both lines 10 and 19 effectively expressed AtAIRP1 mRNA and AtAIRP1-sGFP protein without stress treatments (Fig. 1, G and H).
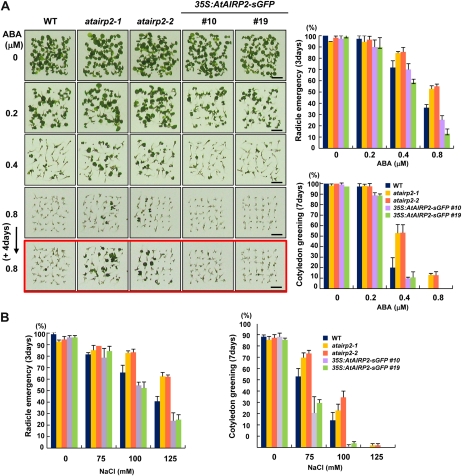
The atairp2 mutants were initially selected due to their ABA-insensitive phenotypes (Supplemental Fig. S1). For phenotypic analysis, seed germination percentages were examined in the presence or absence of ABA. Sterilized seeds from wild-type, atairp2 mutant, and 35S:AtAIRP2-sGFP plants were plated on Murashige and Skoog (MS) growth medium containing different concentrations (0, 0.2, 0.4, or 0.8 μm) of ABA. Germination rates were monitored in terms of radicle emergence and cotyledon greening 3 and 7 d after stratification, respectively. As ABA concentrations increased, radicle emergence rates of wild-type seeds concomitantly decreased from 97.2% (0.2 μm ABA) to 71.8% (0.4 μm ABA) and 37.6% (0.8 μm ABA) 3 d after germination (Fig. 4A). However, both atairp2-1 and atairp2-2 knockout mutant seeds showed hyposensitivity to ABA as compared with wild-type seeds. Approximately 85% of the mutant seeds were able to germinate on medium containing 0.4 μm ABA, and more than 55% of the mutants still germinated normally with 0.8 μm ABA. In contrast, AtAIRP2-overexpressing plants displayed a hypersensitive phenotype toward ABA. Only 26.2% (line 10) and 12.9% (line 19) of the 35S:AtAIRP2-sGFP seeds could germinate in the presence of 0.8 μm ABA (Fig. 4A).
Figure 4.
Germination rates of wild-type, atairp2, and 35S:AtAIRP2-sGFP plants in response to ABA and NaCl. A, ABA sensitivity of the wild type (WT), two atairp2 mutant alleles (atairp2-1 and atairp2-2), and AtAIRP2 overexpressors (transgenic lines 10 and 19) during the germination stage. Sterilized seeds were imbibed in water for 2 d at 4°C and incubated on MS medium in the presence of different concentrations of ABA (0, 0.2, 0.4, and 0.8 μm) at 22°C under a 16-h-light/8-h-dark photoperiod. Germination percentages were determined in terms of radical emergence 3 d after germination and cotyledon greening 7 d after germination. sd values were determined from four biological replicates (n > 36). Bars = 0.5 cm. B, NaCl sensitivity of the wild type, two atairp2 mutant alleles (atairp2-1 and atairp2-2), and AtAIRP2 overexpressors (transgenic lines 10 and 19) during the germination stage. Germination rates were determined in the presence of different concentrations of NaCl (0, 75, 100, and 125 mm) as described above. Data represent means ± sd (n > 36) from three independent experiments. [See online article for color version of this figure.]
Subsequently, cotyledon greening percentages were monitored 7 d after germination. The results indicate that mutant and overexpressing seedlings were hyposensitive and hypersensitive to ABA, respectively, relative to wild-type plants. Approximately 53% of the mutant and 10% of the 35S:AtAIRP2-sGFP seedlings developed true green cotyledons on medium supplemented with 0.4 μm ABA, while 20% of wild-type plants developed normal cotyledons (Fig. 4A). In the presence of 0.8 μm ABA, no wild-type or 35S:AtAIRP2-sGFP plants could display normal cotyledons. In contrast, 13% of both mutant alleles were still able to develop green cotyledons.
Germination tests were repeated in the presence of NaCl (0, 75, 100, and 125 mm). Again, mutant and overexpressing seedlings exhibited hyposensitivity and hypersensitivity to NaCl, respectively, relative to wild-type seedlings in both radicle emergence and cotyledon greening (Supplemental Fig. S4). Phenotypic differences became more evident as salinity increased. A significant number of atairp2 mutants (32%–44%) displayed normal cotyledons with 100 mm NaCl 7 d after germination, whereas the growth of most 35S:AtAIRP2-sGFP seedlings was arrested under the same conditions (Fig. 4B). Wild-type seedlings exhibited intermediate phenotypes between mutant and overexpressing plants under high-salinity conditions (Fig. 4B). Taken together, these results provide evidence that the atairp2 mutants and AtAIRP2 overexpressors have inverse phenotypes in response to ABA and NaCl during the germination stage, suggesting that AtAIRP2 is positively involved in Arabidopsis ABA-modulated germination processes.
AtAIRP2 Is Positively Involved in ABA-Mediated Root Growth Inhibition and Stomatal Closure
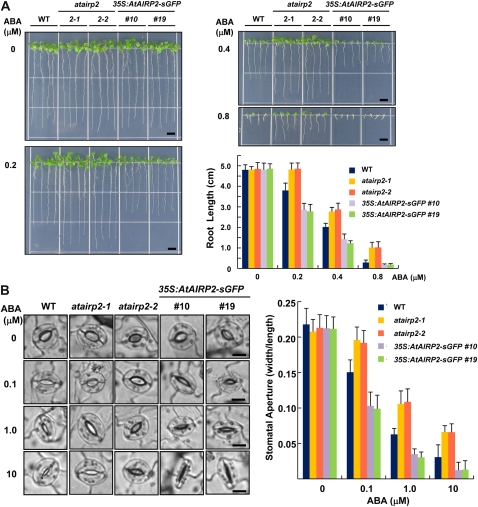
Since reverse effects of ABA on seed germination were clearly identifiable in atairp2 mutants and AtAIRP2 overexpressors (Fig. 4), we next investigated the effects of ABA on postgermination growth. Inhibition of seedling root growth is a typical action of ABA (Quiroz-Figueroa et al., 2010; Ryu et al., 2010). As shown in Figure 5A, atairp2 and 35S:AtAIRP2-sGFP plants displayed hyposensitivity and hypersensitivity, respectively, to ABA in terms of young root growth. When wild-type, atairp2 allele, and 35S:AtAIRP2-sGFP (lines 10 and 19) seedlings were grown for 10 d with 0.2 μm ABA, the mutant root growth appeared to be unaffected. In contrast, elongation of 35S:AtAIRP2-sGFP roots was significantly reduced by approximately 43% under the same ABA concentration (Fig. 5A). With 0.4 μm ABA, growth of 35S:AtAIRP2-sGFP roots was inhibited by 74.7%, while that of loss-of-function mutant roots was reduced by only 42.4%. 35S:AtAIRP2-sGFP root growth was severely impaired and elongation ceased in the presence of 0.8 μm ABA. However, mutant roots were still alive and growing under the same conditions. Wild-type roots showed intermediate phenotypes in response to all of the different ABA concentrations examined (Fig. 5A).
Figure 5.
Root growth and stomatal aperture of wild-type, atairp2, and 35S:AtAIRP2-sGFP plants in response to ABA treatment. A, Root-growth phenotypes of the wild type (WT), two atairp2 mutant alleles (atairp2-1 and atairp2-2), and AtAIRP2 overexpressors (transgenic lines 10 and 19) in response to different concentrations (0, 0.2, 0.4, and 0.8 μm) of ABA. Sterilized seeds were imbibed in water for 2 d and grown vertically on MS medium supplemented with the indicated concentrations of ABA for 10 d. Root growth patterns were monitored and analyzed using Scion Image software. Data represent means ± sd (n = 20). Bars = 0.5 cm. B, Stomatal aperture of the wild type, two atairp2 mutant alleles (atairp2-1 and atairp2-2), and AtAIRP2 overexpressors (transgenic lines 10 and 19) in response to different concentrations (0, 0.1, 1.0, and 10 μm) of ABA. Mature leaves from wild-type, atairp2 allele, and AtAIRP2-overexpressing plants were treated with a stomatal opening solution for 2 h and incubated with the indicated concentrations of ABA for 2 h. Stomata on abaxial surfaces were photographed by light microscopy. Bars = 10 μm. Stomatal aperture (the ratio of width to length) was quantified using at least 30 guard cells from each sample. Data represent means ± sd (n = 30). [See online article for color version of this figure.]
ABA-dependent stomatal closure was next examined in wild-type, atairp2, and 35S:AtAIRP2-sGFP plants. Light-grown 4-week-old rosette leaves were pretreated with stomatal opening solution to induce full opening of the guard cells (Kwak et al., 2003). The leaves were subsequently incubated with different concentrations of ABA (0, 0.1, 1.0, and 10 μm) for 2 h and stomatal behavior was monitored. While stomatal apertures in all leaves examined were indistinguishable without ABA, there were clear differences in response to ABA. In the presence of 0.1 μm ABA, average stomatal apertures (the ratio of width to length) of wild-type, atairp2-1, and 35S:AtAIRP2-sGFP (line 10) plants were 0.15 ± 0.02, 0.20 ± 0.02, and 0.10 ± 0.02, respectively (Fig. 5B). Differences in average stomatal apertures of these plants became progressively more evident as ABA concentrations increased. With 1 μm ABA, the stomatal apertures of the atairp2-1 mutant and AtAIRP2 overexpressor line 10 were 0.11 ± 0.02 and 0.03 ± 0.01, respectively. With 10 μm ABA, the stomatal aperture of mutant leaves was 0.07 ± 0.01, which was approximately 7-fold greater than that of the overexpressors (0.01 ± 0.01; Fig. 5B). In addition, stomatal movement of the atairp2-2 allele and 35S:AtAIRP2-sGFP line 19 displayed similar opposite profiles in response to ABA. Stomatal behavior patterns in wild-type leaves were intermediate between the mutants and overexpressors under all ABA concentrations examined (Fig. 5B). These results indicate that ABA-mediated stomatal closure in atairp2 mutant leaves was markedly hindered as compared with wild-type and AtAIRP2 overexpressors. Because morphological differences between wild-type, atairp2, and 35S:AtAIRP2-sGFP plants were undetectable in the absence of exogenously applied ABA, the data presented in Figures 4 and 5 strongly suggest that AtAIRP2 is positively involved in ABA responses during germination and postgermination growth.
Expression Levels of AtAIRP2 Are Closely Associated with Drought Tolerance in Arabidopsis
Several recent studies reported that ectopic expression of drought-induced RING E3 Ub ligases results in a tolerant phenotype to water stress in an ABA-dependent or ABA-independent manner. For example, transgenic Arabidopsis plants that constitutively express the hot pepper (Capsicum annuum) RING E3 Ub ligase Rma1H1 are highly tolerant to severe dehydration stress via an ABA-independent pathway (Lee et al., 2009). On the other hand, overexpression of SDIR1, AtAIRP1, or RHA2a, all of which encode Arabidopsis RING E3 ligases, conferred resistance to water deficit in an ABA-dependent fashion (Zhang et al., 2007; Ryu et al., 2010; Li et al., 2011). AtAIRP2 was induced by ABA as well as drought stress (Fig. 2). Furthermore, AtAIRP2 was positively involved in ABA-mediated responses, including seed germination (Fig. 4), seedling root growth (Fig. 5A), and stomatal movement (Fig. 5B). Thus, it is postulated that AtAIRP2 participates in the ABA-dependent drought response.
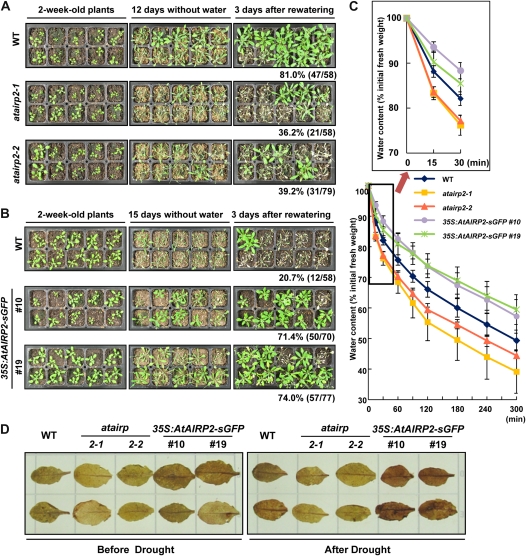
Light-grown, 2-week-old, healthy wild-type and atairp2 mutant (atairp2-1 and atairp2-2) plants were further grown for 12 d under normal conditions without irrigation. These water-stressed plants were then irrigated, and their survival ratios were determined after 3 d of irrigation. As shown in Figure 6A, 81.0% (47 of 58) of the wild-type plants grew normally after reirrigation. On the other hand, significantly lower percentages of mutant plants (36.2% of atairp2-1 and 39.2% of atairp2-2 mutants) resumed their growth. Therefore, atairp2 knockout mutant alleles were more susceptible than were the wild-type plants to mild drought conditions. Subsequently, 2-week-old wild-type and 35S:AtAIRP2-sGFP (lines 10 and 19) plants were grown for 15 d without irrigation. This drought condition resulted in complete drying of the potted soil and induced severe dehydration stress. Survival rates were then estimated 3 d after reirrigation. AtAIRP2-overexpressing plants displayed a markedly resistant phenotype, and their survival percentages reached 71.5% (50 of 70 for line 10) and 74.0% (55 of 77 for line 19; Fig. 6B). The survival rate of wild-type plants was only 20.7% (12 of 58). Thus, AtAIRP2 overexpressors were more tolerant of severe water deficits; in contrast, atairp2 mutants were more sensitive to the stress than were the wild-type plants.
Figure 6.
AtAIRP2 expression levels were closely associated with drought tolerance. A, atairp2 loss-of-function mutants were more sensitive to drought than were wild-type (WT) plants. Light-grown, 2-week-old wild-type and atairp2 mutant allele (atairp2-1 and atairp2-2) plants were further grown for 12 d under normal conditions but without irrigation. The water-stressed plants were irrigated, and their survival ratios were determined after 3 d of irrigation. B, Overexpression of AtAIRP2 conferred tolerance to drought stress. Light-grown, 2-week-old wild-type and 35S:AtAIRP2-sGFP (lines 10 and 19) plants were grown for 15 d without irrigation. Survival percentages were determined 3 d after irrigation. C, Water loss rates of detached rosette leaves. Mature rosette leaves from 2-week-old wild-type, atairp2 allele, and 35S:AtAIRP2-sGFP lines were detached, and their fresh weights were measured at the indicated time points. Water loss rates were calculated as the percentage of fresh weight of the excised leaves. Data represent means ± sd (n = 7) from eight independent experiments. D, H2O2 production in response to drought stress. Control and water-stressed rosette leaves from wild-type, atairp2-1, atairp2-2, and 35S:AtAIRP2-sGFP plants were stained with 100 μg mL−1 DAB overnight. Levels of drought-induced H2O2 production were visualized as a dark brown color. [See online article for color version of this figure.]
Consistent with the drought-tolerant phenotype, 2-week-old detached rosette leaves from 35S:AtAIRP2-sGFP plants lost water more slowly than did wild-type leaves. After a 5-h incubation under dim light at room temperature, 35S:AtAIRP2-sGFP leaves retained approximately 60% of their fresh weights and wild-type leaves retained approximately 55% of their fresh weights (Fig. 6C). However, atairp2 alleles retained only approximately 40% to 45% of their fresh weights after the 5-h incubation. It is worth noting that the reduction in fresh weight of the mutant leaves was more rapid and evident earlier during the initial stages of the incubation period (within 15 to 30 min) than later, suggesting that the mutants were susceptible to the initial stage of dehydration (Fig. 6C, inset).
Reactive oxygen species, such as hydrogen peroxide (H2O2), are critical participants in the ABA-mediated drought stress responses in guard cells (Wang and Song, 2008; Cho et al., 2009; Jammes et al., 2009; Song and Matsuoka, 2009). To evaluate the degree of H2O2 production in response to drought stress, normal and water-stressed leaves from wild-type, atairp2 allele, and 35S:AtAIRP2-sGFP lines were incubated with 3,3′-diaminobenzidine (DAB). DAB interacts with H2O2 in the presence of endogenous peroxidases and produces a dark-brown color (Thordal-Christensen et al., 1997). Figure 6D demonstrates that higher levels of drought-induced H2O2 were produced in 35S:AtAIRP2-sGFP (lines 10 and 19) rosette leaves relative to that of atairp2 mutant leaves. H2O2 levels in wild-type leaves were intermediate between those in overexpressor and mutant plants before and after drought treatments (Fig. 6D), indicating that AtAIRP2 is positively involved in drought-induced H2O2 production. Overall, 35S:AtAIRP2-sGFP and atairp2 plants exhibited inverse phenotypes toward drought, indicating that expression levels of AtAIRP2 are closely associated with drought tolerance in Arabidopsis. These results are consistent with the hypothesis that AtAIRP2 is a positive component of an ABA-dependent response to drought.
The Positive Role of AtAIRP2 in ABA Induction of Drought Stress-Related Genes Is Dependent on SnRK Protein Kinase Activities
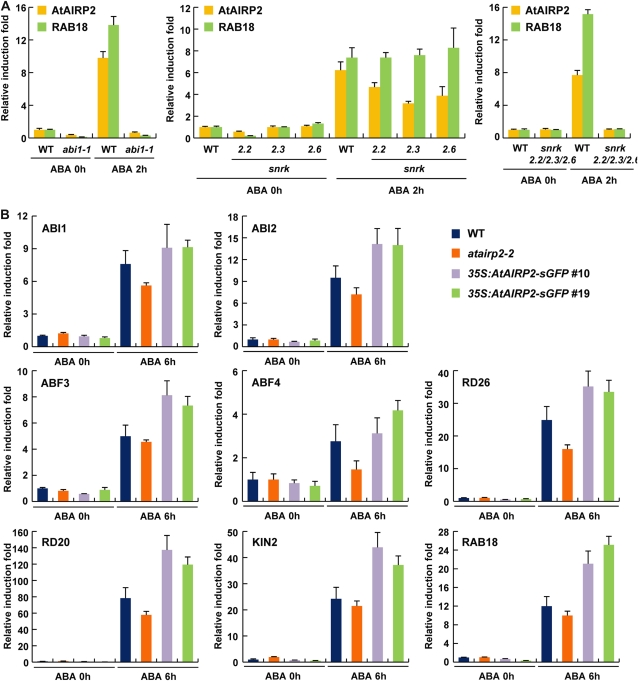
abi1-1 is an ABA-insensitive dominant mutant (Hubbard et al., 2010; Kim et al., 2010b; Raghavendra et al., 2010). As shown in Figure 7A, AtAIRP2 and RAB18, a marker gene for ABA induction, were not induced by exogenously applied ABA in abi1-1 mutant plants, confirming that AtAIRP2 is an ABA-induced gene.
Figure 7.
The positive role of AtAIRP2 in ABA induction of drought stress-related gene expression required SnRK protein kinase activity. A, ABA induction profiles of AtAIRP2 in wild-type (WT), abi1-1, snrk2.2, snrk2.3, and snrk2.6 single knockout mutant, and snrk2.2 snrk2.3 snrk2.6 triple mutant plants. Light-grown, 10-d-old wild-type and various snkr2 mutant seedlings were treated with 100 μm ABA. Total RNA was extracted from the treated tissues and analyzed by real-time qRT-PCR. RAB18 was a positive control for ABA induction, and UBC10 was used as a loading control. B, ABA induction profiles of drought-related genes in wild-type, atairp2-2, and AtAIRP2-overexpressing plants. Light-grown, 3-week-old plants were incubated with 100 μm ABA for 6 h. Induction patterns of various ABA- and drought-responsive genes (ABI1, ABI2, ABF3, ABF4, RD26, RD20, KIN2, and RAB18) were analyzed by real-time qRT-PCR. Data represent the fold induction of each gene by ABA (100 μm) relative to the control treatment (0 μm ABA). Mean values from three independent technical replicates were normalized to the levels of an internal control, glyceraldehyde-3-phosphate dehydrogenase C subunit mRNA. [See online article for color version of this figure.]
The SNF1-related protein kinase (SnRK) protein kinase family is a major component of the ABA signaling pathway and acts upstream of the AREB/ABF transcription factors (Hubbard et al., 2010; Kim et al., 2010b; Raghavendra et al., 2010). SnRKs were shown to be key positive regulators in ABA-dependent reactive oxygen species production and in water and osmotic stress responses (Mustilli et al., 2002; Fujita et al., 2009; Fujii et al., 2011). Consistent with their roles, triple knockout mutation of three SnRK genes (SnRK2.2, SnRK2.3, and SnRK2.6) greatly impaired ABA- and dehydration-induced gene expression (Fujii and Zhu, 2009; Fujita et al., 2009). Because AtAIRP2 was not only induced by ABA but was also positively involved in ABA-dependent responses, our next question was whether the mode of action of AtAIRP2 is downstream of SnRKs. To answer this question, ABA induction of AtAIRP2 was first examined in wild-type, snrk2.2, snrk2.3, and snrk2.6 single knockout mutant, and snrk2.2 snrk2.3 snrk2.6 triple mutant plants. Real-time qRT-PCR analysis indicated that ABA induction of AtAIRP2 in the single knockout mutants was very comparable to that in wild-type plants (Fig. 7A). In contrast, AtAIRP2 transcript levels remained unchanged before and after ABA treatment in the snrk2.2 snrk2.3 snrk2.6 triple loss-of-function mutant plants. Thus, the ABA induction of AtAIRP2 in the single snrk mutant lines may be due to the redundant functions of SnRK kinase family members. Similar induction profiles were also obtained for RAB18 (Fig. 7A). These results indicate that SnRK protein kinase activity is necessary for ABA-induced activation of AtAIRP2 as well as RAB18.
Expression patterns of various ABA-responsive genes were compared in wild-type, atairp2 mutant, and AtAIRP2-overexpressing plants using real-time qRT-PCR. As demonstrated in Figure 7B, following ABA induction, the gene expression of ABI1, ABI2, ABF3, and ABF4 was down-regulated and up-regulated in atairp2-2 mutant and AtAIRP2-overexpressing plants, respectively, relative to wild-type plants. ABI1 and ABI2 are ABA-responsive protein phosphatase 2C genes (Ma et al., 2009; Santiago et al., 2009), while ABF3 and ABF4 encode ABA-activated basic Leu zipper transcription factors (Finkelstein et al., 2002; Gómez-Porras et al., 2007; Lee et al., 2010). Furthermore, mRNA levels of various ABA- and stress-induced downstream marker genes (RD20, RD26, KIN2, and RAB18) were also lower in knockout mutant alleles and, in contrast, markedly higher in 35S:AtAIRP2-sGFP plants as compared with those in wild-type plants (Fig. 7B). Thus, AtAIRP2 positively regulates ABA induction of protein phosphatases, ABF/AREB transcription factors, and downstream marker gene expression. Collectively, the results presented in Figure 7 indicate that SnRK protein kinase activity is necessary for a positive role of AtAIRP2 in ABA induction of drought stress-related genes.
Functional Relationship of AtAIRP1 and AtAIRP2
In our previous study, AtAIRP1, a C3H2C3-type RING E3 Ub ligase, was shown to play a positive role in the ABA-dependent drought response in Arabidopsis. AtAIRP1-overexpressing and atairp1 loss-of-function mutant plants had opposite phenotypes, including germination rates, root elongation, stomatal closure, and tolerance to drought stress, following ABA-mediated responses (Ryu et al., 2010). AtAIRP1 belongs to a different RING subfamily than does AtAIRP2 (C3H2C3 type versus C3HC4 type), and its deduced molecular mass is quite smaller than that of AtAIRP2 (16.9 versus 28.0 kD; Fig. 1). However, both E3 ligases were predominantly localized to cytosolic fractions (Fig. 3). In addition, phenotypic properties of 35S:AtAIRP1-sGFP and atairp1 plants are reminiscent of those of 35S:AtAIRP2-sGFP and atairp2 plants, respectively, in terms of ABA-mediated responses (Figs. 4–6).
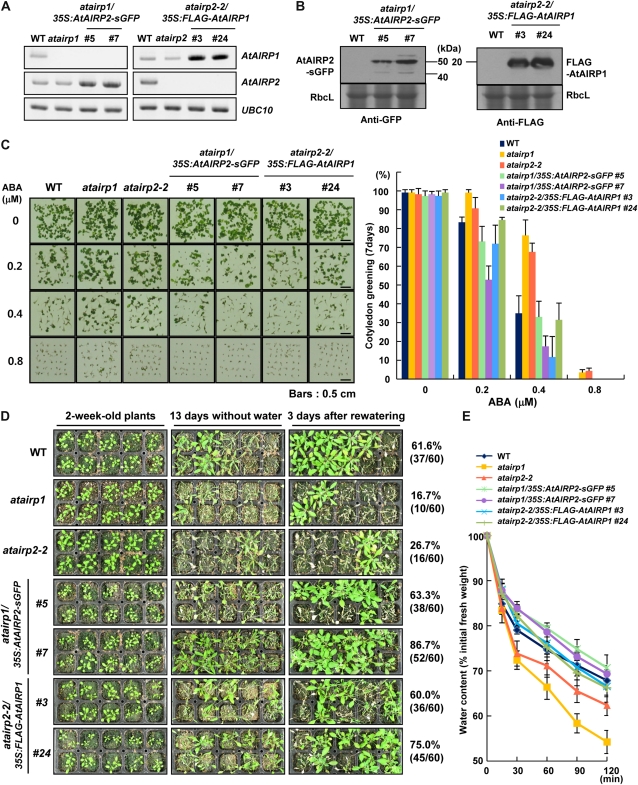
With these findings in mind, we hypothesized that AtAIRP1 and AtAIRP2 play a combinatory role in ABA-dependent drought stress responses. Alternatively, it is possible that AtAIRP1 and AtAIRP2 work independently. To test these possibilities, complementation tests were conducted. FLAG-AtAIRP1 and AtAIRP2-sGFP fusion genes were ectopically expressed in atairp2-2 and atairp1 mutant plants, respectively. Independent complementation lines were selected and confirmed by genomic Southern blotting (Supplemental Fig. S3). RT-PCR and immunoblot analyses revealed that AtAIRP2 and AtAIRP1 transgenes were clearly expressed in atairp1/35S:AtAIRP2-sGFP (lines 5 and 7) and atairp2-2/35S:FLAG-AtAIRP1 (lines 3 and 24) complementation T3 transgenic plants, respectively (Fig. 8, A and B). These T3 complementation lines were used to analyze ABA- and stress-related phenotypes to determine whether mutant phenotypes were reciprocally rescued. The results in Figure 8C show that both atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1 lines were more sensitive to ABA at all concentrations examined (0.2–0.8 μm) than were atairp1 and atairp2-2 single mutants, respectively, during the germination stage. For example, in the presence of 0.4 μm ABA, germination (cotyledon greening) percentages for wild-type, atairp1, atairp2-2, atairp1/35S:AtAIRP2-sGFP (lines 5 and 7), and atairp2-2/35S:FLAG-AtAIRP1 (lines 3 and 24) plants were 31.3%, 76.4%, 67.6%, 17.4% to 33.0%, and 11.7% to 31.5%, respectively. Thus, the degree of ABA sensitivity for both complementation progeny was approximately the same as the average for wild-type and overexpressing plants (compare Figs. 4 and 8). This indicates that the insensitive phenotypes of atairp1 and atairp2-2 young seedlings in response to ABA were efficiently rescued by ectopic expression of AtAIRP2 and AtAIRP1, respectively.
Figure 8.
Construction and characterization of atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1 complementation transgenic plants. A and B, RT-PCR and immunoblot analyses. AtAIRP2-sGFP and FLAG-AtAIRP1 fusion genes were ectopically expressed in atairp1 and atairp2-2 mutant plants, respectively. Transcript (A) and protein (B) levels of AtAIRP2-sGFP and FLAG-AtAIRP1 were examined in atairp1/35S:AtAIRP2-sGFP (lines 5 and 7) and atairp2-2/35S:FLAG-AtAIRP1 (lines 3 and 24) complementation T3 transgenic plants. Rubisco large subunit (RbcL) was used as a loading control. C, Phenotypic properties of atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1 complementation T3 transgenic plants during the germination stage. After imbibition in water for 2 d at 4°C, wild-type (WT), atairp1 and atairp2-2 mutant, and atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1 complementation T3 seeds were treated with different concentrations of ABA (0, 0.2, 0.4, and 0.8 μm) at 22°C under a 16-h-light/8-h-dark photoperiod. Germination percentages were determined in terms of cotyledon greening 7 d after germination. sd values were determined from four biological replicates (n = 40). Bars = 0.5 cm. D, Water stress tolerance of atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1 complementation T3 transgenic plants. Light-grown, 2-week-old wild-type, atairp1 and atairp2-2 mutant, and atairp1/35S:AtAIRP2-sGFP (lines 5 and 7) and atairp2-2/35S:FLAG-AtAIRP1 (lines 3 and 24) complementation T3 transgenic plants were further grown for 13 d without irrigation. Water-stressed plants were irrigated, and their survival ratios were determined after 3 d of irrigation. E, Water loss rates of detached rosette leaves. Mature rosette leaves from 2-week-old wild-type, atairp1 and atairp2-2 mutant, and atairp1/35S:AtAIRP2-sGFP (lines 5 and 7) and atairp2-2/35S:FLAG-AtAIRP1 (lines 3 and 24) complementation T3 transgenic plants were detached, and their fresh weights were measured at the indicated time points. Water loss rates were calculated as the percentage of fresh weight of the excised leaves. Data represent means ± sd (n = 7) from three independent experiments. [See online article for color version of this figure.]
Mature atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1 complementation lines were also markedly more tolerant to dehydration stress as compared with the atairp1 and atairp2-2 single knockout mutant plants. After 13 d of water stress, survival rates of wild-type, atairp1, and atairp2-2 progeny were determined to be 61.6%, 16.7%, and 26.7%, respectively, whereas those of atairp1/35S:AtAIRP2-sGFP (lines 5 and 7) and atairp2-2/35S:FLAG-AtAIRP1 (lines 3 and 24) plants were 63.3% to 86.7% and 60.0% to 75.0%, respectively (Fig. 8D). In addition, detached leaves from complementation lines lost water more slowly than those from single knockout mutant plants (Fig. 8E). Overall, these results strongly suggest that constitutive expression of AtAIRP1 and AtAIRP2 in atairp2-2 and atairp1 mutant plants, respectively, reciprocally rescued the loss-of-function ABA-insensitive phenotypes during both the germination and postgermination stages.
DISCUSSION
In this report, we identified atairp2 allele Arabidopsis mutants that were less sensitive to ABA treatment than were wild-type plants at the germination stage. The AtAIRP2 gene encodes a C3HC4-type RING E3 Ub ligase (Fig. 1). AtAIRP2 transcript levels were markedly heightened in response to ABA treatment and dehydration stress (Fig. 2). Consistent with other RING domain-containing proteins, bacterially expressed AtAIRP2 displayed in vitro E3 Ub ligase activity and was localized to cytosolic fractions of onion epidermal cells (Fig. 3). 35S:AtAIRP2-sGFP and atairp2 loss-of-function mutant plants were tested for a broad spectrum of ABA responsiveness, including seed germination, root growth, and stomatal movement. It was found that AtAIRP2 overexpressors and atairp2 alleles exhibited hypersensitive and hyposensitive phenotypes, respectively, toward ABA treatment during seed germination (Fig. 4A). Because high salinity (75–125 mm NaCl) exerted a similar opposite effect on seed germination in AtAIRP2 overexpressors and atairp2 mutants, AtAIRP2 likely controls early seedling development by inhibiting seed germination under unfavorable growth conditions, including high ABA concentrations and salt stress (Fig. 4B). Such an inhibitory function during seed germination was recently reported for the Arabidopsis PUB44/SAUL1 U-box E3 Ub ligase (Salt et al., 2011). PUB44/SAUL1 prevents seed germination under stress conditions, including those involving ABA, Glc, NaCl, and mannitol. In addition, 35S:AtAIRP2-sGFP and atairp2 progeny displayed opposite phenotypes in response to ABA treatment in all categories examined (Fig. 5).
The initial aim of this study was to illuminate the functional relationship between ABA and RING E3 Ub ligases in drought stress responses. 35S:AtAIRP2-sGFP transgenic plants were highly tolerant to severe drought stress; in contrast, atairp2 alleles were more susceptible to mild water stress than were wild-type plants (Fig. 6C). Higher levels of drought-induced H2O2 production were detected in AtAIRP2 overexpressors as compared with atairp2 alleles (Fig. 6D). Furthermore, ABA-induced drought-related gene expression was up-regulated in 35S:AtAIRP2-sGFP and down-regulated in atairp2 progeny (Fig. 7). The positive effects of AtAIRP2 on the ABA induction of stress genes were dependent on the protein kinase activity of SnRKs, a key component in the ABA signaling pathway (Fig. 7). Therefore, it is concluded that AtAIRP2 is involved in positive regulation of the ABA-dependent drought stress response in Arabidopsis.
RING E3 Ub ligase isoforms are implicated not only in normal growth and developmental processes but also in induced defense mechanisms against biotic and abiotic environmental stresses (Moon et al., 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007; Vierstra, 2009; Lee and Kim, 2011). However, the current understanding of the functional relationships between RING E3s and ABA-mediated drought stress responses is rudimentary. The Arabidopsis RING E3 Ub ligases XERICO and SDIR1 positively regulate drought responses by heightening ABA synthesis and acting upstream of ABA-responsive basic Leu zipper transcription factors, respectively (Ko et al., 2006; Zhang et al., 2007). In addition, the RING E3s AtAIRP1 and RHA2b are positive regulators of ABA signaling and drought responses (Ryu et al., 2010; Li et al., 2011). Through these studies and our data here, it is becoming increasingly apparent that there is a functional network(s) between RING E3 Ub ligases and the stress hormone ABA that helps plants fine-tune their cellular responses to dehydration stress, one of the most serious environmental stresses crop plants face. Overall, the results presented in this report implicate the RING E3 AtAIRP2 as a positive regulator of ABA-mediated drought stress responses in Arabidopsis.
AtAIRP1 was previously reported to be a C3H2C3-type RING E3 Ub ligase that works as a positive mediator in the Arabidopsis ABA-dependent drought response (Ryu et al., 2010). The 35S:AtAIRP1-sGFP and atairp1 lines showed opposite germination and postgermination growth phenotypes in response to ABA treatment. Therefore, the phenotypic properties of the 35S:AtAIRP1-sGFP and atairp1 progeny were reminiscent of those of the 35S:AtAIRP2-sGFP and atairp2 lines, respectively. In this context, we theorized two possible modes of action for AtAIRP1 and AtAIRP2. The first possibility was that AtAIRP1 and AtAIRP2 play coordinate roles in ABA-dependent drought stress responses. The second possibility was that they work in nonoverlapping or parallel pathways. Our results demonstrated that overexpression of AtAIRP1 and AtAIRP2 reciprocally rescued the loss-of-function ABA-insensitive phenotypes of atairp2-2 and atairp1, respectively, in terms of seed germination and water stress tolerance (Fig. 8). Thus, it is highly likely that the Arabidopsis RING E3 Ub ligases AtAIRP1 and AtAIRP2 play combinatory roles in ABA-mediated drought stress responses. In addition, the Arabidopsis genome contains two AtAIRP2 homologs. At5g58787 and At3g47160 are 63% and 60% identical to AtAIRP2, respectively (Fig. 1E; Supplemental Fig. S2). Because the identities of these two homologs to AtAIRP2 are significantly higher than that of AtAIRP1, it is possible that At5g58787 and At3g47160 may also play combinatory roles with AtAIRP2 in ABA and drought stress responses. This possibility is currently under investigation.
The adaptive mechanisms that plants have developed in response to abiotic stresses are combinatorial and interconnected defensive webs that work coordinately for concomitant metabolic reprogramming (Ahuja et al., 2010; Hummel et al., 2010; Tardieu et al., 2011). In this sense, it is plausible that the E3 Ub ligase multigene family also functions in combination to cope with water deficit conditions. For example, two homologous Arabidopsis U-box E3 Ub ligases, AtPUB22 and AtPUB23, coordinately control a drought signaling pathway by sharing cytosolic RPN12a, a non-ATPase subunit of the 26S proteasome complex, as a substrate (Cho et al., 2008). Ubiquitination of RPN12a may result in a conformational change of the 26S proteasome complex, which in turn serves as a negative signal for the drought response. Similarly, the dehydration-responsive element-binding protein DREB2A is a common substrate for the homologous nuclear RING E3s DRIP1 and DRIP2. The DRIPs down-regulated dehydration stress-responsive gene expression by ubiquitinating and targeting DREB2A for 26S proteasome proteolysis (Qin et al., 2008). RHA2a and RHA2b may play redundant, yet distinguishable, roles in the control of ABA signaling and drought responses. RHA2a and RHA2b are plasma membrane and nuclear dual-localized RING E3s that act downstream of protein phosphatase 2C and ABI2 and in parallel with ABI3/4/5 (Li et al., 2011). Thus, it would not be uncommon to consider that E3 Ub ligase isoforms function in a coordinated manner to interact with their common target proteins more effectively. This notion is in agreement with the fact that plants contain over 1,400 E3 Ub ligases as compared with the approximately 600 human E3s (Vierstra, 2009; Liu and Walters, 2010). The combinatory work patterns of plant E3s may increase the efficiency of reprogramming metabolic responses to environmental stresses.
On the other hand, AtAIRP1 belongs to a different RING subfamily than does AtAIRP2 (C3H2C3 type versus C3HC4 type), and their deduced molecular masses are quite different (16.9 versus 28.0 kD), with limited deduced amino acid sequence identity (13%; Fig. 1). One notable common feature of AtAIRP1 and AtAIRP2 was that they were localized to cytosolic fractions (Fig. 3). Nevertheless, AtAIRP1 and AtAIRP2 were able to reciprocally complement loss-of-function ABA-insensitive mutant phenotypes (Fig. 8). One could thus postulate that AtAIRP1 and AtAIRP2 share a common substrate protein(s) that acts negatively during drought stress responses. Ubiquitinated negative regulators are targeted for 26S proteasome-dependent proteolysis, conferring tolerance to dehydration stress. This possibility, however, seems to be somewhat unlikely, because the structural properties of AtAIRP1 and AtAIRP2 may not be close enough for sharing common substrates. Alternatively, AtAIRP1 and AtAIRP2 could ubiquitinate different target proteins that are functionally interconnected. In this scenario, the output of two distinct ubiquitination pathways by AtAIRP1 and AtAIRP2 could influence each other by an as yet unknown metabolic mechanism, which in turn would increase ABA sensitivity and tolerance to water deficit. The results in Figure 8, C to E, indicate that the degrees of ABA sensitivity and drought tolerance for both complementation lines (atairp1/35S:AtAIRP2-sGFP and atairp2-2/35S:FLAG-AtAIRP1) were not as high as in overexpressors (35S:AtAIRP2-sGFP) but rather were approximately the same as the average for wild-type and overexpressing plants. These results may suggest that AtAIRP1 and AtAIRP2 ubiquitinate different target proteins rather than share a common substrate protein. Therefore, it is essential to identify the target proteins of AtAIRP1 and AtAIRP2 to decipher the dynamic mechanism and combinatory roles of these two cytosolic RING E3 Ub ligases.
Urbanization and global warming have had causal effects on the worldwide reduction of freshwater availability for crop plants. Continuously increasing human and industrial water consumption could pose a future threat to agricultural crop plants as well as humans (Hightower and Pierce, 2008; Yoo et al., 2009). Thus, it is of immense importance to develop drought-tolerant transgenic crops. In conclusion, the data presented in this report provide evidence that AtAIRP2 plays integrated roles with AtAIRP1 in ABA-mediated drought stress responses in Arabidopsis.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana ecotype Columbia-0) seeds were soaked in 30% bleach solution (1.5% sodium hypochlorite and 0.1% Triton X-100) for 10 min and washed 10 times with sterilized water. Young seedlings were grown in 1× MS medium (Duchefa Biochemie) supplemented with 1% to 3% Suc and 0.8% phytoagar (pH 5.7) or in soil (Sunshine Mix 5; Sun Gro) in a growth chamber at 22°C with a 16-h-light/8-h-dark cycle. The atairp2-1 (SAIL_686_G08) and atairp2-2 (Salk_005082) T-DNA insertion mutant alleles were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org).
Sequence Analysis
AtAIRP2 and its homologous proteins were identified with the WU-BLST program (http://arabidopsis.org/wublast/index2.jsp). Selected homolog protein sequences were analyzed with MEGA5 software (Tamura et al., 2007). Multiple sequence alignments were edited using the GeneDoc program (http://www.nrbsc.org/gfx/genedoc/). Phylogenetic trees were generated with MEGA5 software (Ryu et al., 2010).
RT-PCR and Real-Time qRT-PCR Analyses
Total RNA was isolated from abiotic stress- and ABA-treated 10-d-old seedlings using an RNA extraction kit (Intron Biotechnology) according to the manufacturer’s protocol. cDNA synthesis and RT-PCR were performed as described previously (Kim et al., 2010a). qRT-PCR was carried out using an IQ5 light cycler (Bio-Rad) with SYBR Premix Ex Taq II (Takara). qRT-PCR data were analyzed with Genex_Macro_IQ5_conversion_Template and Genex software (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase C subunit mRNA level was used as an internal control for qRT-PCR data normalization.
Histochemical GUS Assay
Arabidopsis genomic DNA was amplified using the AtAIPR2 pro-GUS FW and pro-GUS RV primer set (Supplemental Table S1). PCR products were inserted into a pCAMBIA1381 vector. The AtAIRP2 promoter-GUS construct was transformed into wild-type Arabidopsis using the Agrobacterium tumefaciens-mediated floral dip method as described by Joo et al. (2006). For the histochemical GUS assay, transgenic plant tissues were immersed in a GUS staining solution containing 2 mm X-GlcA (cyclohexylammonium salt; Duchefa Biochemie), 0.5 mm K3Fe(CN)6, and 0.5 mm K4Fe(CN)6 in 50 mm sodium phosphate buffer (pH 7.2) and incubated for 12 h at 37°C (Joo et al., 2004). To remove chlorophyll after GUS staining, GUS-stained tissues were incubated in 70% ethanol for several hours.
In Vitro Self-Ubiquitination Assay
The full-length coding region of AtAIRP2 cDNA was amplified with the XbaI-FW and PstI-RV primer set (Supplemental Table S1). PCR products were restricted by XbaI and PstI and inserted into pMAL C2 vectors (New England Biolabs). The single amino acid substitution derivative (AtAIRP2H163A) of wild-type AtAIRP2 was generated using the QuikChange site-directed mutagenesis kit (Stratagene) and the H163A-FW and H163A-RV primer set. MBP-AtAIRP2 and MBP-AtAIRP2H163A fusion proteins (500 ng) were expressed in Escherichia coli strain BL21 and purified using amylose resin (New England Biolabs). In vitro self-ubiquitination assays were conducted as described previously (Cho et al., 2006a). Immunoblot analyses were carried out with anti-MBP antibody (New England Biolabs) or anti-Ub antibody (Santa Cruz Biotechnology) according to Ryu et al. (2009). All primers used in this study are provided in Supplemental Table S1.
Subcellular Localization
The 35S:sGFP, 35S:AtAIRP2-sGFP, 35S:AtAREB1-sGFP, and 35S:AtAIRP1-sGFP plasmids were inserted into pBI221 transient expression vectors. The fusion constructs were introduced into onion (Allium cepa) epidermal cells by means of the particle bombardment method described by Lee and Kim (2003). Transiently expressed GFP signals were detected using a fluorescence microscope (BX51; Olympus). Images were acquired with a 1600 CCD camera (PCO) and analyzed using Image Pro Plus software (Media Cybernetics). AREB1-sGFP and AtAIRP1-sGFP were used as controls for nuclear and cytosolic localization, respectively. All primers used are listed in Supplemental Table S1.
Construction of 35S:AtAIRP2-sGFP, atairp2-2/35S:FLAG-AtAIRP1, and atairp1/35S:AtAIRP2-sGFP Transgenic Plants
Full-length AtAIRP2 and AtAIRP1 cDNAs were PCR amplified using AtAIRP2-specific primers (AtAIRP2 SacI-FW and AtAIRP2 SacI-RV) and AtAIRP1-specific primers (AtAIRP1 EcoRI-FW and AtAIRP1 XbaI-RV), respectively (Supplemental Table S1). PCR products were digested with each restriction enzyme and inserted into modified pENTR vectors (Invitrogen). AtAIRP2-sGFP and FLAG-AtAIRP1 clones were subsequently integrated into pEarlygate 100 destination vectors using LR Clonase II (Invitrogen) and transformed into wild-type, atairp1, or atairp2-2 plants using an Agrobacterium-mediated floral dip method (Joo et al., 2006). Transformed seeds were selected on MS plates containing 25 μg mL−1 BASTA. Expression of each transgene was examined by genomic Southern-blot, RT-PCR, and immunoblot analyses as described by Lee et al. (2009). Homozygous T3 lines were selected through self-crossing and were subsequently used in phenotypic analyses.
Seed Germination Assay
Seed germination assays were performed with greater than 36 seeds and repeated three times. Seeds, 3 d after imbibition, from wild-type, atairp1, atairp2-1, atairp2-2, 35S:AtAIRP2-sGFP, atairp2-2/35S:FLAG-AtAIRP1, and atairp1/35S:AtAIRP2-sGFP plants were grown on 1× MS medium supplemented with different concentrations (0, 0.2, 0.4, or 0.8 μm) of ABA (Sigma-Aldrich) at 22°C with a 16-h-light/8-h-dark photoperiod. The rates of radicle emergence and cotyledon greening were measured after 3 and 7 d, respectively.
Root Growth and Stomatal Aperture Measurements
To measure seedling root growth, seeds were vertically grown for 10 d on 1× MS medium containing 0.2 to 0.8 μm ABA, and root elongation was monitored and analyzed using Scion Image software (www.scioncorp.com). Mature rosette leaves from light-grown 4-week-old wild-type, atairp2-1, atairp2-2, and 35S:AtAIRP2-sGFP plants were detached and incubated in a stomatal opening solution (10 mm KCl, 100 μm CaCl2, and 10 mm MES, pH 6.1) for 2 h at 22°C (Kwak et al., 2003). Treated leaves were transferred to a stomatal opening solution containing ABA (0, 0.1, 1, or 10 μm) for 2 h. Epidermal strips were observed using a light microscope (Olympus BX51). Stomatal aperture was measured using Multigauge version 3.1 software (Fujifilm) as described by Ryu et al. (2010).
Drought Phenotype Analysis
Wild-type, atairp2-1, atairp2-2, 35S:AtAIRP2-sGFP, atairp2-2/35S:FLAG-AtAIRP1, and atairp1/35S:AtAIRP2-sGFP plants were grown for 2 weeks under normal growth conditions and then subjected to dehydration stress by ceasing irrigation for 12 to 15 d (Cho et al., 2006b). Three days after rewatering, surviving plants were counted as described previously (Kim et al., 2010a). Cut rosette water loss experiments were performed according to the method described by Ryu et al. (2010). For DAB staining, light-grown 2-week-old plants were treated with drought stress for 10 d, and rosette leaves were incubated with 100 μg mL−1 DAB solution as described previously (Ryu et al., 2010).
Sequence data used in this report are found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AtAIRP2 (At5g01520), AtAIRP1 (At4g23450), AtAREB1 (At1g45249), two Arabidopsis homologs (At5g58787 and At3g47160), Oryza sativa (NP_001060539), Populus trichocarpa (XP_002309135), Vitis vinifera (XP_002280008), and Sorghum bicolor (XP_002447334).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of atairp2 T-DNA insertion loss-of-function mutant alleles.
Supplemental Figure S2. Amino acid sequence comparison of seven AtAIRP2 homologs.
Supplemental Figure S3. Genomic Southern blot analysis of wild-type and T3 35S:AtAIRP2-sGFP, atairp2-2/35S:FLAG-AtAIRP1, and atairp1/35S:AtAIRP2-GFP transgenic Arabidopsis plants.
Supplemental Figure S4. Germination rates of wild-type, atairp2, and 35S:AtAIRP2-sGFP plants in response to NaCl.
Supplemental Table S1. PCR primer sequences used for this article.
References
- Ahuja I, de Vos RCH, Bones AM, Hall RD. (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15: 664–674 [DOI] [PubMed] [Google Scholar]
- Bae H, Kim SK, Cho SK, Kang BG, Kim WT. (2011) Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci 180: 775–782 [DOI] [PubMed] [Google Scholar]
- Bu Q, Li H, Zhao Q, Jiang H, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Wang D, et al. (2009) The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol 150: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DS, Shin DJ, Jeon BW, Kwak JM. (2009) ROS-mediated ABA signaling. J Plant Biol 52: 102–113 [Google Scholar]
- Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk Y-Y, Kim J, Pai HS, Kim WT. (2006a) Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol 142: 1664–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Kim JE, Park J-A, Eom TJ, Kim WT. (2006b) Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580: 3136–3144 [DOI] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. (2008) Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20: 1899–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 36: 131–150 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu J-K. (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA 108: 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu J-K. (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Adir N. (2004) The proteasome and the delicate balance between destruction and rescue. PLoS Biol 2: E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Porras JL, Riaño-Pachón DM, Dreyer I, Mayer JE, Mueller-Roeber B. (2007) Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics 8: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower M, Pierce SA. (2008) The energy challenge. Nature 452: 285–286 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li CY, Pattison DL, Gray WM, Park S, Gibson SI. (2010) SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response. Plant Physiol 152: 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, Christophe A, Pervent M, Bouteillé M, Stitt M, et al. (2010) Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol 154: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J, Liu CW. (2009) The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 S proteasome. J Biol Chem 284: 35485–35494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, et al. (2009) MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S, Park KY, Kim WT. (2004) Light differentially regulates the expression of two members of the auxin-induced 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.) seedlings. Planta 218: 976–988 [DOI] [PubMed] [Google Scholar]
- Joo S, Seo YS, Kim SM, Hong DK, Park KY, Kim WT. (2006) Brassinosteroid-induction of AtACS4 encoding an auxin-responsive 1-aminocyclopropane-1-carboxylate synthase 4 in Arabidopsis seedlings. Physiol Plant 126: 592–604 [Google Scholar]
- Kim EY, Seo YS, Lee H, Kim WT. (2010a) Constitutive expression of CaSRP1, a hot pepper small rubber particle protein homolog, resulted in fast growth and improved drought tolerance in transgenic Arabidopsis plants. Planta 232: 71–83 [DOI] [PubMed] [Google Scholar]
- Kim T-H, Böhmer M, Hu H, Nishimura N, Schroeder JI. (2010b) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH. (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47: 343–355 [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei Z-M, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JDG, Schroeder JI. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang IH, Kim WT. (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Kim WT. (2003) Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiol 132: 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim WT. (2011) Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cells 31: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kang JY, Park HJ, Kim MD, Bae MS, Choi HI, Kim SY. (2010) DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol 153: 716–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Jiang H, Bu Q, Zhao Q, Sun J, Xie Q, Li C. (2011) The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol 156: 550–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Walters KJ. (2010) Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci 35: 352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stone SL. (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Yang Y, Li G, Yang Y, Wang X, Basnayake BM, Li D, Song F. (2008) Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol Biol 68: 17–30 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Mustilli A-C, Merlot S, Vavasseur A, Fenzi F, Giraudat J. (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Jantasuriyarat C, Zhao Q, Zhang H, Chen S, Liu J, Liu L, Tang S, Park CH, Wang X, et al. (2011) The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol 157: 242–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GG, Park JJ, Yoon J, Yu SN, An G. (2010) A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol Biol 74: 467–478 [DOI] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ. (2007) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz-Figueroa F, Rodríguez-Acosta A, Salazar-Blas A, Hernández-Domínguez E, Campos ME, Kitahata N, Asami T, Galaz-Avalos RM, Cassab GI. (2010) Accumulation of high levels of ABA regulates the pleiotropic response of the nhr1 Arabidopsis mutant. J Plant Biol 53: 32–44 [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Ryu MY, Cho SK, Kim WT. (2009) RNAi suppression of RPN12a decreases the expression of type-A ARRs, negative regulators of cytokinin signaling pathway, in Arabidopsis. Mol Cells 28: 375–382 [DOI] [PubMed] [Google Scholar]
- Ryu MY, Cho SK, Kim WT. (2010) The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol 154: 1983–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt JN, Yoshioka K, Moeder W, Goring DR. (2011) Altered germination and subcellular localization patterns for PUB44/SAUL1 in response to stress and phytohormone treatments. PLoS ONE 6: e21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park S-Y, Márquez JA, Cutler SR, Rodriguez PL. (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Song X-J, Matsuoka M. (2009) Bar the windows: an optimized strategy to survive drought and salt adversities. Genes Dev 23: 1709–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tardieu F, Granier C, Muller B. (2011) Water deficit and growth: co-ordinating processes without an orchestrator? Curr Opin Plant Biol 14: 283–289 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tuteja N. (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2: 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang P, Song CP. (2008) Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol 178: 703–718 [DOI] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH. (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yoo CY, Pence HE, Hasegawa PM, Mickelbart MV. (2009) Regulation of transpiration to improve crop water use. Crit Rev Plant Sci 28: 410–431 [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH. (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Li Y, Gao T, Zhu H, Wang DJ, Zhang HW, Ning YS, Liu LJ, Wu YR, Chu CC, et al. (2008) Arabidopsis SDIR1 enhances drought tolerance in crop plants. Biosci Biotechnol Biochem 72: 2251–2254 [DOI] [PubMed] [Google Scholar]