Abstract
The SET domain-containing protein, pTAC14, was previously identified as a component of the transcriptionally active chromosome (TAC) complexes. Here, we investigated the function of pTAC14 in the regulation of plastid-encoded bacterial-type RNA polymerase (PEP) activity and chloroplast development. The knockout of pTAC14 led to the blockage of thylakoid formation in Arabidopsis (Arabidopsis thaliana), and ptac14 was seedling lethal. Sequence and transcriptional analysis showed that pTAC14 encodes a specific protein in plants that is located in the chloroplast associated with the thylakoid and that its expression depends on light. In addition, the transcript levels of all investigated PEP-dependent genes were clearly reduced in the ptac14-1 mutants, while the accumulation of nucleus-encoded phage-type RNA polymerase-dependent transcripts was increased, indicating an important role of pTAC14 in maintaining PEP activity. pTAC14 was found to interact with pTAC12/HEMERA, another component of TACs that is involved in phytochrome signaling. The data suggest that pTAC14 is essential for proper chloroplast development, most likely by affecting PEP activity and regulating PEP-dependent plastid gene transcription in Arabidopsis together with pTAC12.
The chloroplast is a semiautonomous organelle that depends on the coordinated expression of nuclear and chloroplast genes (Mullet, 1988). It contains more than 120 genes important for its development and function (Sugita and Sugiura, 1996; Sato et al., 1999). The transcriptional machinery plays an important role in the regulation of chloroplast development at different developmental stages (Mullet, 1993; Emanuel et al., 2004; Zoschke et al., 2007). Higher plant cells possess at least two types of RNA polymerases: a plastid-encoded bacterial-type RNA polymerase (PEP), which is a multisubunit eubacterium-type enzyme, and a nucleus-encoded phage-type RNA polymerase (NEP) that resembles the single-subunit type shared with phage T3/T7 and mitochondrial enzymes (Hu and Bogorad, 1990; Igloi and Kössel, 1992; Lerbs-Mache, 1993; Pfannschmidt and Link, 1997; Liere and Maliga, 2001). The chloroplast-encoded photosynthetic genes are exclusively transcribed by PEP (class I genes). A few genes, mostly encoding components of the transcription/translation apparatus, are exclusively transcribed by NEP (class III genes), and nonphotosynthetic housekeeping genes are mostly transcribed by both PEP and NEP (class II genes). NEP promoters are more active in the youngest, nongreen tissues early in leaf development, while PEP increases its activity during the maturation of chloroplasts (Allison et al., 1996; Hajdukiewicz et al., 1997). The complexity of promoter specificity and chloroplast transcription initiated by PEP is further increased by multiple nucleus-encoded σ factors, whose expression is spatially and temporally regulated by environmental cues (Hess and Börner, 1999; Allison, 2000; Shiina et al., 2005; Loschelder et al., 2006). There are six chloroplast σ factors (SIG1–SIG6) in Arabidopsis (Arabidopsis thaliana). In the SIG6 knockout line in Arabidopsis, a high molecular transcript spans the atpB-atpE operon and extends far upstream of atpB (Schweer et al., 2006). This region represents the NEP promoter. In PEP-deficient mutants, transcripts initiated by NEP would cover the entire plastome in situations where no functional σ factor is available for PEP.
The complex plastid gene expression machinery requires the involvement of proteins encoded by the nuclear genes. Subunits of the PEP core are present in two plastid protein preparations, the transcriptionally active chromosome (TAC) and the soluble RNA polymerase. TAC is a type of chloroplast multisubunit complex enzyme isolated in the form of large DNA-protein complexes. TACs contain polypeptides involved in replication, transcription, translation, detoxification, protein modification, and plastid metabolism (Pfalz et al., 2006). Pfalz et al. (2006) isolated the TAC from Arabidopsis and mustard (Sinapis alba) chloroplasts and identified 35 of the components. They named 18 components, which were not studied in depth at that time, as pTACs (for plastid TAC proteins). Among the recent advances made in the study of TACs, several components responsible for the albino or pale-green phenotype have been described, such as heteromeric FSD2 and FSD3 (Myouga et al., 2008), Whirly proteins (Maréchal et al., 2009), pTAC4/Vipp1 (Kroll et al., 2001; Aseeva et al., 2007), pTAC2/6/12 (Pfalz et al., 2006), AtMurE (Garcia et al., 2008), TRXz (Arsova et al., 2010), and FLN1 and FLN2 (Arsova et al., 2010). Recently, Chen et al. (2010) found that HEMERA, previously called pTAC12, had a function in the nucleus, where it acted specifically in phytochrome signaling. It is a specific and early PHY signal transduction component in the nucleus that is required for the light-dependent proteolysis of PHYA, PIF1, and PIF3, implicating a direct role for HMR in proteolysis. However, the actual role of HMR in the chloroplast was still unknown. Although TACs have been isolated from several organisms and a few components have been studied previously, the interactions between each component and the effects of the deletion of a single component on TAC stability have not yet been investigated in depth.
Histone methylation is one type of epigenetic marker that serves an essential regulatory function in the organization of chromatin structure and genome function (Yu et al., 2009b; Liu et al., 2010). Enzymes that catalyze histone Lys methylation contain an evolutionarily conserved SET domain (Tschiersch et al., 1994). There are at least 47 SET proteins present in Arabidopsis, which can be classified into seven distinct phylogenetic groups: class I, E(z) class; class II, ASH1 class; class III, Trithorax class; class IV, proteins with a SET domain and a PHD domain; class V, Su(var) class; class VI, proteins with an interrupted SET domain; and class VII, nonhistone methyltransferases and related proteins (Ng et al., 2007). So far, studies indicate that SET domain proteins are involved in chromatin structure, gene silencing, transcriptional activation, plant metabolism, and other processes (Trievel et al., 2002; Yu et al., 2009b; Liu et al., 2010). However, studies on these SET domain proteins are still limited, and the biological functions of a large number of SET domain-containing proteins in chloroplast development remain a mystery.
In this study, we characterize the pTAC14 gene encoding a SET domain protein associated with the thylakoid. The knockout of pTAC14 leads to the arrest of thylakoid membrane formation in the Arabidopsis chloroplast. In the ptac14 mutant, the transcription level of PEP-dependent chloroplast genes is decreased. Moreover, pTAC14 interacts with pTAC12, another component of the TACs. These data suggest that pTAC14, a functional component of the TACs, regulates chloroplast gene expression and chloroplast development by interacting with pTAC12 in Arabidopsis.
RESULTS
Sequence and Phylogenetic Analysis of the Thylakoid-Associated pTAC14 Protein
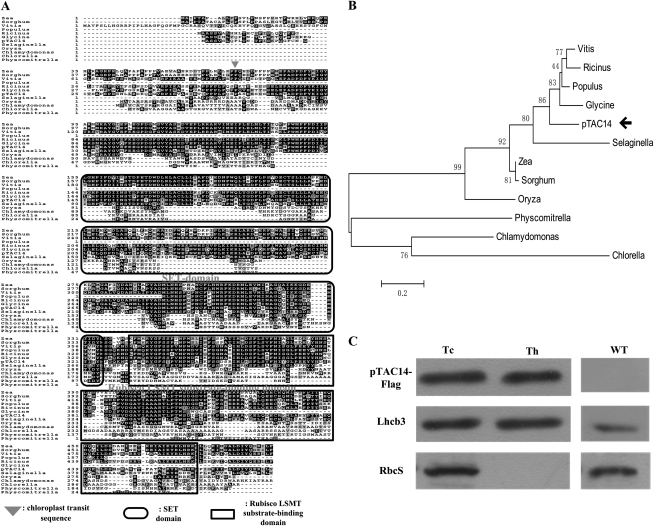
The locus At4G20130 encodes an Arabidopsis SET domain protein, which was previously designated as pTAC14 and was originally identified from TACs (Pfalz et al., 2006). Its open reading frame encodes a polypeptide of 483 amino acids with a calculated molecular mass of 53.24 kD. Sequence analysis indicated that pTAC14 contains two domains (Fig. 1A). Amino acids 101 to 325 define the SET domain, which is suggested to be responsible for the possible methylation function of the pTAC14 protein. Amino acids 332 to 466 comprise a Rubisco LSMT substrate-binding domain. BLAST searching the complete Arabidopsis genome revealed that only one copy of the pTAC14 gene is present in the nuclear genome. Sequence alignment revealed that pTAC14 homologs exist in various plants, including Ricinus communis (XP_002528137.1; 99% similarity), Orzya sativa (EEC79774.1; 88% similarity), Zea mays (NP_001146209.1; 89% similarity), Vitis vinifera (XP_002275523.1; 89% similarity), Sorghum bicolor (XP_002440355.1; 92% similarity), Glycine max (ACU20841.1; 67% similarity), Selaginella moellendorffii (EFJ04899.1; 89% similarity), Populus trichocarpa (XP_002304603.1; 85% similarity), Chlorella variabilis (EFN53459.1; 40% similarity), Chlamydomonas reinhardtii (XP_001698255.1; 66% similarity), and Physcomitrella patens (XP_001759585.1; 85% similarity; Fig. 1A). However, no homologs were found in photosynthetic bacteria. These data indicated that the pTAC14 protein is highly conserved and specific in plants. To investigate the relationship between pTAC14 homologs in evolutionary history, a phylogenetic analysis was performed. As shown in Figure 1B, the proteins from lower plants and higher plants are clearly differentiated from one another. Proteins from algae form one subclade. Moss, as the lowest group of the higher plants, is close to algae. In the higher plant clade, pteridophyte and angiosperm proteins form one subclade. The proteins from monocots and dicots are divided clearly among the angiosperm plants. Our data show that the pTAC14 protein is a conserved protein in plants.
Figure 1.
Sequence alignment, phylogenetic analysis, and subcellular location of the pTAC14 protein. A, Alignment analysis between pTAC14 and its homologs from R. communis, O. sativa, Z. mays, V. vinifera, S. bicolor, G. max, S. moellendorffii, P. trichocarpa, C. variabilis, C. reinhardtii, and P. patens. The arrowhead denotes the putative cleavage site of the transit peptide in the pTAC14 protein. The rounded rectangular box denotes the SET domain, and the square rectangular box denotes Rubisco LMST substrate-binding domains of the pTAC14 protein. B, Phylogenetic analysis of the pTAC14 protein and its homologs supports a close relationship among R. communis, O. sativa, Z. mays, V. vinifera, S. bicolor, G. max, S. moellendorffii, P. trichocarpa, C. variabilis, C. reinhardtii, and P. patens. C, pTAC14 is located in the chloroplast and is associated with the thylakoid. The fractionation of transgenic Arabidopsis seedling chloroplasts was performed with anti-FLAG, anti-Lhcb3, and anti-RbcS antibodies. Wild-type chloroplasts (WT) were used as controls. Tc, Total chloroplast protein; Th, thylakoid fraction.
The pTAC14 protein contains a chloroplast transit peptide at its N-terminal sequence, as predicted by TargetP (Emanuelsson et al., 2000) and Predotar (Small et al., 2004). These data are consistent with those from proteomic studies (Pfalz et al., 2006). To further demonstrate its subcellular location in plant cells, we generated transgenic Arabidopsis in a wild-type background (Columbia) with the pTAC14 full-length coding sequence (CDS) fused with a FLAG tag. Immunoblot analysis was performed using an anti-FLAG antibody on chloroplast fractions isolated from the transgenic plants. The purity of chloroplast and thylakoid fractions was assessed using antibodies against corresponding marker proteins of a thylakoid lumenal protein, light-harvesting complex of PSII (Lhcb3), and the stromal Rubisco complex (RbcS). As shown in Figure 1C, RbcS was only present in the chloroplast, and pTAC14-FLAG displayed the same fraction pattern as Lhcb3 (i.e. present in both the chloroplast and the thylakoid). These results confirmed that pTAC14 is a chloroplast-localized protein associated with the thylakoid.
The pTAC14 Gene Expression Pattern
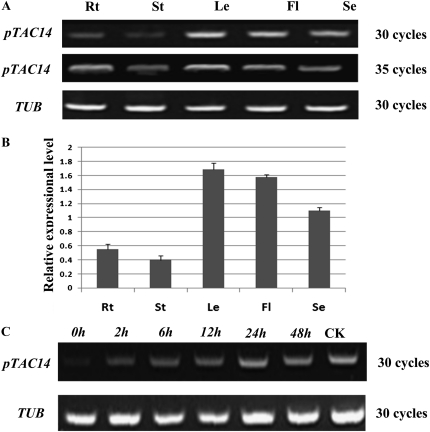
Expression data from Genevestigator showed that the pTAC14 gene is widely expressed in various Arabidopsis tissues (Zimmermann et al., 2004; http://www.genevestigator.com). We extracted RNA from root, stem, leaf, flower, and seedling tissues in order to perform reverse transcription (RT)-PCR and analyze the expression of pTAC14 in these tissues. β-Tubulin was used as a control for normalization of the data set. Our results confirmed that pTAC14 was present in all investigated tissues; it was highly expressed in leaves but minimally expressed in stems and roots (Fig. 2A). Quantitative real-time RT-PCR analysis further confirmed its expression pattern in these tissues (Fig. 2B). These results revealed that the pTAC14 gene is widely expressed in different tissues, with the highest expression in leaves.
Figure 2.
pTAC14 expression pattern in different Arabidopsis tissues. A and B, RT-PCR analysis (A) and quantitative real-time RT-PCR analysis (B) of the pTAC14 gene in the root (Rt), stem (St), leaf (Le), flower (Fl), and seedling (Se). C, Expression of pTAC14 was regulated by light. TUB, TUBULIN expression as a control.
To test the effects of light on the expression of pTAC14, we treated 7-d-old wild-type Arabidopsis etiolated seedlings exposed to light for 0, 2, 6, 12, 24, and 48 h. Total RNA extracted from these seedlings was analyzed by RT-PCR to detect the expression levels of pTAC14. RNA from the seedlings grown in standard light intensities for 7 d was used as the control. Our results indicated that pTAC14 was weakly expressed in etiolated seedlings, and the gene expression level was gradually increased with increasing light exposure, reaching a stable level when the plants were exposed to light for 24 h (Fig. 2C). Therefore, these results confirmed that light plays a role in regulating the expression of pTAC14 during plastid development.
The Null Mutation of pTAC14 Leads to the Albino Phenotype
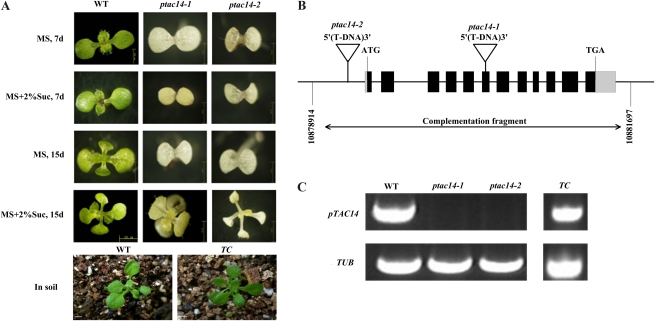
To evaluate the putative role of the pTAC14 gene in chloroplast development, we obtained two independent T-DNA insertion lines, SAIL_566_F06 and SALK_005814, from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu; Alonso et al., 2003). In the SAIL_566_F06 line, the T-DNA is inserted 1,438 bp downstream of the translation start site (the sixth exon of pTAC14). In the SALK_005814 line, the T-DNA is inserted 135 bp upstream of the ATG codon of pTAC14. Both mutants displayed albino cotyledons and were unable to grow photoautotrophically. If supplemented with Suc as a carbon source, the two mutants could produce albino primary leaves, but they were still seedling lethal (Fig. 3A). Genetic analysis indicated that the progeny from one heterozygote line (SAIL_566_F06) showed segregation between green and albino (green:albino = 190:58; χ20.05 = 0.18, P > 0.05). Investigation of about 100 homozygote mutants indicated that all analyzed mutants were linked to the T-DNA insertion, and the albino phenotype was caused by a single, recessive Mendelian locus. Furthermore, RT-PCR results indicated that the pTAC14 transcript was absent in both homozygous mutants (Fig. 3C). Because the T-DNA insertion position of SAIL_566_F06 was in the exon, we chose this mutant line for further analysis and designated it as ptac14-1 and designated SALK_005814 as ptac14-2.
Figure 3.
Visible phenotypes of wild-type (WT) plants and ptac14 mutants grown on MS medium with or without 2% Suc. A, Photographs of seedling growth on MS medium with or without 2% Suc for 7 or 15 d and wild-type and ptac14-1 complementary (TC) plants grown in soil. All plants were grown under standard light intensities. Bars = 2 mm. B, Positions of the two T-DNA insertions in the pTAC14 gene. SAIL_566_F_06 was named ptac14-1 and SALK_005814 was named ptac14-2. Exons, the untranslated region, and introns are indicated by black boxes, gray boxes, and lines, respectively. C, pTAC14 gene expression levels in wild-type, ptac14-1, and ptac14-2 plants as well as the complementary lines.
To further confirm that the phenotype of the ptac14 mutant was caused by the T-DNA insertion in pTAC14, we performed genomic complementation analysis of the mutant phenotype with the wild-type pTAC14 genomic sequence. We cloned a 4,656-bp wild-type genomic fragment containing the pTAC14 gene, as well as the 1,613-bp upstream and 259-bp downstream sequences, into the binary pCAMBIA1300 vector (CAMBIA; http://www.cambia.org.au). After introducing the cloned plasmids into heterozygous plants (pTAC14-1/ptac14-1) via Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998), we obtained a total of 81 independent T1 transgenic plants with resistance to hygromycin. We found that seven of them were of the ptac14-1/ptac14-1 background. All of these seven lines were of a normal phenotype that was similar to the wild-type plants. RT-PCR analysis confirmed that the transcripts of pTAC14 were present in these plants (Fig. 3C). Furthermore, transmission electron microscopy (TEM) observations showed that the chloroplasts in these plants contained well-organized thylakoid membranes similar to those found in wild-type chloroplasts (Fig. 4B). These results indicate that the 4,656-bp genomic fragment can successfully complement the mutated phenotype. In addition, we introduced the construct of pTAC14 CDS fused to the FLAG tag under the control of the cauliflower virus 35S promoter into the pTAC14 heterozygous background and obtained 15 independent T1 transgenic plants; one of them was found to have a homozygous knockout background (ptac14-1/ptac14-1). This transgenic line grew normally with a wild-type phenotype. Thus, the fusion protein could complement the mutated phenotype and function normally. Taken together, all of these data demonstrate that the mutated phenotype of ptac14 is caused by the inactivation of pTAC14 and that pTAC14 is essential for Arabidopsis chloroplast development.
Figure 4.
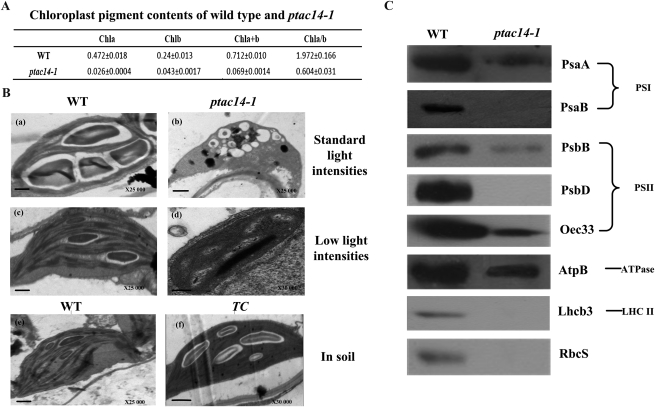
Transmission electron micrographs of plastid structures, and immunoblot analysis of photosynthetic proteins in the wild type (WT) and ptac14-1 mutants. A, Chloroplast pigment content of the wild type and ptac14-1. B, Ultrastructure of chloroplasts in leaves from wild-type and ptac14-1 plants and the ptac14-1 complementary plants (TC). Chloroplasts are from 14-d-old leaves of wild-type and ptac14-1 plants grown on MS medium supplemented with 2% Suc under standard and low light intensities and from leaves of the wild type and complementary lines grown in the soil. Bars = 0.5 μm. C, Total protein extracts were prepared from 14-d-old seedlings and then separated by SDS-PAGE. Western-blot analysis was carried out with the specific antibodies mentioned previously. A total of 40 μg of total protein was loaded per lane.
Chloroplast Development Is Defective in ptac14 Mutants
Since both ptac14-1 and ptac14-2 displayed albino cotyledons without the primary leaves and were unable to grow photoautotrophically, we quantified the content of chlorophyll a and b in the mutant and wild-type plants. Total chlorophyll was decreased in ptac14-1 (0.069 ± 0.001 mg mL−1, fresh weight) compared with that in the wild type (0.712 ± 0.01 mg mL−1, fresh weight).
To investigate whether pTAC14 is involved in the initial process of chloroplast development, we used TEM to examine morphological changes in the chloroplasts between wild-type plants and ptac14 mutants. As shown in Figure 4B, when grown under standard light intensities, the size of the chloroplasts in the ptac14-1 mutant was irregular and much smaller than that in the wild-type plant. Additionally, chloroplasts of the ptac14 mutant lacked internal membrane structures, such as stromal thylakoids or stacked grana thylakoids, but they contained oval-shaped vesicles. The results showed that the thylakoid membrane is severely impaired in ptac14-1 mutants. Because photooxidative stress can affect chloroplast development (Gutiérrez-Nava et al., 2004), we further studied whether the chloroplast development of the ptac14-1 mutant was affected by photooxidative damage. Under low light intensities, stacked thylakoid structures cannot be formed, although dispersive lamellar structures can be observed in the knockout mutant. These results showed that the pTAC14 gene is important for thylakoid biogenesis and that it has an important role in an early stage of chloroplast development. In addition, it appears that photooxidative stress may aggravate the damage of chloroplast development in the mutant under normal light intensities.
To investigate how the defects in thylakoid membrane development caused by the mutation of pTAC14 affect the accumulation of photosynthesis-related proteins, western-blot analyses were performed. For protein gel blotting, we used antibodies for two members of PSI complexes (PsaA and PsaB), three members of PSII complexes (PsbB, PsbD, and OEC33; Jain et al., 1998; Jeong et al., 2003), the photosynthetic and respiratory ATP synthase (AtpB; Inatomi, 1986), RbcS (Krebbers et al., 1988), and Lhcb3 (Jackowski et al., 2001). Our data showed that PsaB, PsbD, RbcS, and Lhcb3 proteins were undetectable in the ptac14-1 mutant, while the levels of PsaA, PsbB, and OEC33 proteins were significantly decreased in the ptac14-1 mutant compared with the wild-type plant, and the accumulation of AtpB was less affected in ptac14-1. These results indicate that the accumulation of most photosynthetic complexes and the Rubisco complex was affected in ptac14-1 mutants, which is possibly an indirect result of the failed formation of the thylakoid membrane in the mutant.
PEP Activity Was Decreased in ptac14-1
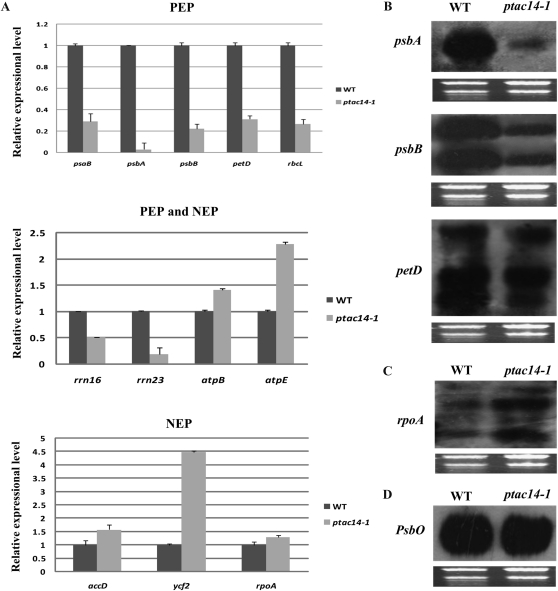
PEP is responsible for the transcription of photosynthetic genes and is important for chloroplast development. It is essential to maintain PEP activity; any factor affecting PEP activity may block chloroplast development. We detected the relative expression levels of chloroplast genes using quantitative real-time RT-PCR compared with those of the wild type. The psaB, psbA, psbB, petD, and rbcL genes were chosen as PEP-dependent genes (class I); rpoA, ycf2, and accD were chosen as NEP-dependent genes (class III); and rrn16, rrn23, atpB, and atpE were used as both PEP- and NEP-dependent genes (class II; Chateigner-Boutin et al., 2008; Myouga et al., 2008; Yu et al., 2009a). Our results showed that the transcription level of class I genes in ptac14-1 was clearly decreased by 70% to 95%. In the case of the class III genes, they were increased by varying degrees. As for the class II genes, the expression levels of rrn16 and rrn23 were down-regulated by over 50%, but the transcripts of atpB and atpE were clearly up-regulated. All of the transcripts that were more highly expressed in ptac14-1 are at least in part transcribed by the NEP, whereas those with low levels have been reported to be transcribed by PEP. This result indicated that the chloroplast expression system that utilizes PEP is deficient in the ptac14 mutants, and pTAC14 is important for the maintenance of PEP activity in Arabidopsis chloroplasts.
To further demonstrate the decreased PEP activity, we detected the abundance of the representative chloroplast transcripts in both wild-type and ptac14-1 plants. Our results showed that the PEP-dependent transcripts, psbA, psbB, and petD, were dramatically reduced in the ptac14-1 mutant compared with the wild type, but no major changes were found in their maturation patterns, whereas increased rpoA transcripts were observed (Fig. 5C). In contrast to the chloroplast genes, we examined the steady-state level produced by a photosynthesis-related nuclear gene, PsbO, which encodes a 34-kD protein of the subunit of the reaction center of PSII (OEC) localized in the thylakoid. We found that the nuclear transcript was unaffected by the ptac14-1 mutation (Fig. 5D). These results were in accordance with the real-time RT-PCR results and confirmed the decreased PEP activity in the ptac14-1 mutant.
Figure 5.
Chloroplast gene expression and northern-blot analysis for plastid- and nucleus-encoded genes in the wild type (WT) and ptac14-1 mutants. A, The transcript abundance of three types of chloroplast genes (class I, class II, and class III) was measured by quantitative real-time RT-PCR. Data are given as log2 of mutant-wild type ratios from at least three independent experiments. Class I genes refer to psaB, psbA, psbB, petD, and rbcL. Class II genes refer to rrn16, rrn23, atpB, and atpE. Class III genes refer to accD, ycf2, and rpoA. B to D, Northern-blot analysis of transcript levels for PEP-dependent (B) and NEP-dependent (C) chloroplast genes and a nucleus-encoded gene, PsbO (D), in the wild type and ptac14-1. Total RNA stained with ethidium bromide is shown as a loading control.
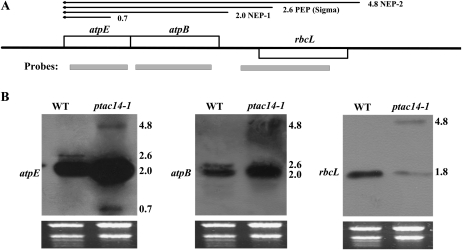
atpB-atpE Transcriptions Were Affected in the ptac14-1 Mutant
Many chloroplast genes are organized as clusters and are cotranscribed as polycistronic units. The chloroplast atp genes are organized into two transcriptional units in Arabidopsis. One cluster of genes has the order atpB-atpE. In the atpB-atpE operon, 2.6- and 2.0-kb transcripts are transcribed by PEP (σ dependent) and NEP, respectively (Schweer et al., 2006). We found that the expression patterns of atpB-atpE transcripts are specifically affected in the ptac14-1 mutant compared with the wild type. Using an atpB-specific probe, the 2.6-kb transcripts seen in the wild-type plant are not transcribed in the ptac14-1 mutant, whereas the 2.0-kb transcripts are increased compared with the wild type. In addition, a novel 4.8-kb transcript was visible in ptac14-1. A similar transcriptional pattern was obtained when using the atpE probe, except for an additional 0.7-kb small transcript that corresponds to the ε-subunit of monocistronic mRNA (Schweer et al., 2006; Fig. 6B). Thus, PEP deficiency in ptac14-1 was further confirmed by the transcription patterns of the atpB-atpE operon. Besides, we took note of the rbcL gene, as its transcriptional pattern was also strongly affected in ptac14-1 mutants (Fig. 6B). The 1.8-kb transcript was seen in both wild-type plants and ptac14-1 mutants, but the transcript was reduced in abundance in ptac14-1. However, a larger molecular transcript of approximately of 4.8 kb was detected only in ptac14-1. Our studies showed that this transcript had the same origin as the 4.8-kb transcript promoter of its adjacent atpB-atpE operon. The 4.8-kb transcript is transcribed by another NEP promoter element in the far upstream region of atpB. These data indicated that pTAC14 is also involved in regulating the transcription of the atpB-atpE operon.
Figure 6.
Transcript pattern analysis by probes spanning distinct portions of the atpE-rbcL region in the wild type (WT) and ptac14-1 mutants. A, Coding regions (boxes), positions of probes (gray bars), and transcripts (arrows in the 5′ to 3′ direction; sizes in kb). B, Transcripts after northern-blot hybridization analysis. A total of 10 μg of RNA was loaded per lane.
pTAC14 Interacts with pTAC12 in Yeast
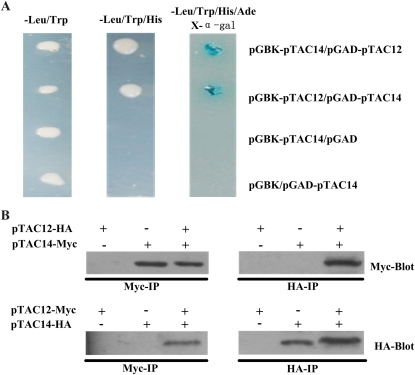
To obtain further insight into the function of pTAC14, a yeast two-hybrid screen was performed to identify its interacting proteins. The cDNA corresponding to the pTAC14 protein lacking 1 to 62 amino acids of the N-terminal region was fused with the pGBK-T7 vector and used as a bait protein. The prospective interaction partners were then screened from a prey pool composed of 35 TAC components fused with the pGAD-T7 vectors. A total of 30 clones were randomly selected for DNA sequencing, and the results indicated that the sequences of five clones correspond to pTAC12. This result suggested that pTAC14 may interact with pTAC12 in the chloroplast. pTAC12 is a component that is required for the proper transcription of PEP-dependent genes in TACs (Pfalz et al., 2006), and it acts specifically in phytochrome signaling in the nucleus (Chen et al., 2010). To further confirm the interaction between pTAC12 and pTAC14, we fused pTAC12 to pGBK-T7 to generate the bait and fused pTAC14 to pGAD-T7 to serve as the prey. The transformants can grow on selective medium plates. All positive clones we selected were further assayed by X-α-Gal for blue-white screening (Fig. 7A). All these results demonstrated the specific interaction between pTAC14 and pTAC12 in yeast.
Figure 7.
Detection of the interaction between pTAC14 and pTAC12. A, Interaction between pTAC14 and pTAC12 proteins in yeast two-hybrid assays. Growth of yeast cells expressing pTAC14 and pTAC12 truncated proteins without the chloroplast-targeting signal peptides grown at 30°C for 72 h on selected medium with dropout of several amino acids is shown. Media used to assess the interaction between bait (pGBK-T7) and prey (pGAD-T7) were as follows: SD-Leu-Trp, SD-Leu-Trp-His, and SD-Leu-Trp-His-Ade+X-α-Gal. Empty pGBK-T7 and pGAD-T7 vectors served as negative controls. B, Coimmunoprecipitation assay revealing in vitro interactions between pTAC14 and pTAC12.
To provide further evidence for the interaction between pTAC14 and pTAC12, coimmunoprecipitation experiments were performed. pTAC14 with a C-terminal Myc epitope tag (pTAC14-Myc) and pTAC12 with a C-terminal hemagglutinin (HA) epitope tag (pTAC12-HA) were cotransferred into yeast strain AH109. Yeast protein extracts were subsequently immunoprecipitated with the anti-Myc antibody. We found that pTAC14-Myc could be coimmunoprecipitated with pTAC12-HA by blotting with the anti-Myc antibody. Similar results were obtained by immunoprecipitating pTAC14-HA with the anti-Myc antibody and blotting with the anti-HA antibody when yeast expressing both pTAC14 with a C-terminal HA epitope tag (pTAC14-HA) and pTAC12 with a C-terminal Myc epitope tag (pTAC12-Myc) were used (Fig. 7B). These results further confirmed the physical interaction between pTAC14 and pTAC12.
DISCUSSION
In this study, we characterized a SET domain-containing TAC component protein, pTAC14, in plastid gene expression and chloroplast development. The lethal phenotype and the blockage of thylakoid formation caused by the absence of pTAC14 demonstrated its essential role during the early stage of chloroplast development. The transcriptional levels of all investigated PEP-dependent chloroplast genes were clearly decreased in the knockout lines, suggesting that PEP activity was deficient in ptac14. An in vitro assay demonstrated that pTAC14 interacted with pTAC12, another TAC component that acts specifically in phytochrome signaling.
pTAC14, a Thylakoid-Associated Protein, Is Essential for Chloroplast Development
pTAC14 was originally identified from chloroplasts as one component of TACs by gel filtration and affinity chromatography purification (Pfalz et al., 2006), but its molecular function remains unclear to date. Here, we characterized the two allelic knockout lines for pTAC14. Genetics analysis, genomics, and full-length pTAC14 CDS-fused FLAG tag complementation experiments confirmed that the albino and lethal phenotypes were caused by the absence of pTAC14, which is essential for early stages of seedling development. TEM observations showed that the chloroplast of ptac14 had an irregular shape and oval-shaped vesicles but did not have single stromal or stacked grana thylakoids (Fig. 4B). The lack of thylakoid membrane formation instantly affects the accumulation of photosynthetic proteins (Fig. 4C). These results show that pTAC14 plays important roles in thylakoid biogenesis in Arabidopsis. The ptac14 mutant cells did not contain intact plastids like those that are detected in other TAC mutants, such as ptac6 and ptac12 (Pfalz et al., 2006), atmurE (Garcia et al., 2008), trxz (Arsova et al., 2010), fln1 (Arsova et al., 2010), and fsd2 and fsd3 (Myouga et al., 2008). Similarly, differently sized vesicles can be observed in the chloroplasts of these mutants. The thylakoid membrane system of Δvipp1 mutants is also severely disturbed, and the chloroplasts in vipp1 cannot form vesicles (Kroll et al., 2001; Aseeva et al., 2007). These results suggest that chloroplast development in these mutants is arrested at the thylakoid formation stage, as in the ptac14 mutant. However, in ptac2 plants (Pfalz et al., 2006), grana structures are unequally expanded and grana-interlacing stromal thylakoids are completely missing. This finding indicates that chloroplast development in ptac2 is not as severely affected as in ptac14. The different stages of arrest during chloroplast development in these TAC mutants cause us to speculate that the TAC components may have different roles in chloroplast function.
The N-terminal sequence of pTAC14 possessed the attributes of a chloroplast transit peptide, indicating that the protein is located in this organelle. We attempted to generate a construct containing the full-length pTAC14 CDS fused to GFP and introduced it into Arabidopsis protoplasts by the polyethylene glycol transformation method; however, the GFP signal could not be detected in the protoplasts. Thus, chloroplast fractionation and immunoblot analysis were used to confirm its subcellular position and showed that pTAC14 was localized to the chloroplast associated with the thylakoid compartment (Fig. 1C). This distinct location of pTAC14 is reminiscent of reports of thylakoid-associated proteins, including MPF1 (Jeong et al., 2003), PEND (Terasawa and Sato, 2009), sulfite reductase (Sekine et al., 2007), CND41 (Murakami et al., 2000), and other characterized TAC components, such as FSD3 (Myouga et al., 2008), FLN1, and FLN2 (Arsova et al., 2010). These proteins were suggested to be tightly associated with plastid nucleoids. There have been indications that the plastid transcriptional apparatus and the plastid nucleoids are supposed to be membrane associated (Sato, 2001; Sato et al., 2003; Karcher et al., 2009; Schweer et al., 2010a), while the plastid nucleoids are believed to be associated with the inner envelope in developing plastids and with the thylakoid membranes in mature chloroplasts (Jeong et al., 2003). Considering pTAC14 as one component of TACs, we thought that pTAC14 possibly functions at the nucleoids and was closely DNA binding in mature chloroplast, similar to FSD3, another TAC component.
pTAC14 Is Important for PEP Activity
Pfalz et al. (2006) hypothesized that nucleus-encoded components required for transcription and translation in plastids might be associated with the TACs. Recent reports have revealed that mutations in several of the genes encoding TAC components cause deficiencies in PEP-dependent transcription (Pfalz et al., 2006; Garcia et al., 2008; Myouga et al., 2008). As shown in Figure 5, real-time RT-PCR and northern-blot hybridization showed that the expression pattern of plastid genes in ptac14-1 mutants resembles that observed in Δrpo mutants (Serino and Maliga, 1998; Krause et al., 2000) and in mutants with lesions in PEP function (Hess et al., 1993; Allison et al., 1996; Hajdukiewicz et al., 1997; Chateigner-Boutin et al., 2008; Chi et al., 2008). These results suggest that pTAC14 is indispensable for the proper function of the PEP transcription machinery.
As for the atpB-atpE operon, the 2.0-kb transcript was observed in both the wild type and the mutant, while the 2.6-kb transcript disappeared and a novel 4.8-kb transcript accumulated in ptac14-1 (Fig. 6B). A similar transcriptional pattern was observed in the SIG6 knockout mutant (Loschelder et al., 2006; Schweer et al., 2006) and other PEP-deficient mutants, such as ptac2/6/12 (Pfalz et al., 2006), clb19 (Chateigner-Boutin et al., 2008), dg1 (Chi et al., 2008), and ys1 (Zhou et al., 2008). In addition, a 4.8-kb transcript was also detected for rbcL in ptac14-1 (Fig. 6B). The probe used for rbcL was included in the upstream sequence of the 4.8-kb transcript of atpB (Fig. 6A). Thus, we considered that the 4.8-kb transcript detected by the rbcL probe is the same as that detected by the atpB-atpE probes (Fig. 6A). SIG6 was reported to be essential for PEP to recognize the promoter region of the 2.6-kb transcript (Loschelder et al., 2006; Schweer et al., 2006). In ptac14-1, the 2.6-kb transcript cannot be detected (Fig. 6B). This finding suggested that pTAC14 might be involved in SIG6 function. However, there exists another possibility for pTAC14 to regulate PEP activity. Even in wild-type plants, the 2.6-kb transcript is weakly transcribed by PEP compared with the 2.0-kb transcript transcribed by NEP (Fig. 6B). In ptac14 mutants, PEP activity was clearly decreased (Fig. 5). It is quite possible that the low PEP activity in ptac14 is not enough to initiate the transcription of the 2.6-kb transcript.
pTAC14 Interacts with pTAC12 to Regulate PEP Activity
Although TACs have been isolated from several organisms, the interaction between each component is still not well documented. In this study, we found that pTAC14 interacts with pTAC12 in yeast (Fig. 7A). This result was further confirmed in vitro by coimmunoprecipitation (Fig. 7B). Previous reports indicated that the null pTAC12 mutation in Arabidopsis displayed a striking albino phenotype and that the chloroplasts of ptac12 contained oval-shaped vesicles, which were similar to those in the ptac14-1 mutant (Figs. 3 and 4). Moreover, both ptac14 and ptac12 mutants displayed very similar and characteristic changes in transcription patterns of chloroplast genes. This finding was also corroborated by northern analysis for the atpB, atpE, and rbcL genes (Pfalz et al., 2006; Fig. 6). Therefore, the similarities in chloroplast development status and expression patterns support the direct interaction between pTAC14 and pTAC12 in TACs.
pTAC12 was previously identified as HEMERA and localized in both the nucleus and chloroplast, and it was considered as a proteolysis-related protein involved in phytochrome signaling in the nucleus (Chen et al., 2010). However, its function in the chloroplast still remains unknown. Both pTAC12 and pTAC14 are shown to be essential components of the PEP complex (Steiner et al., 2011). In this work, our results demonstrated that pTAC14 is directly associated with pTAC12 in the chloroplast. Both pTAC12 (Pfalz et al., 2006) and pTAC14 (Fig. 5) are essential for PEP activities. The PEP complex contains more than 35 components (Pfalz et al., 2006), and many of them are important for PEP activities. Different environmental factors such as light will affect PEP activity and plastid gene expression. It is possible that each component in the PEP complex may play a role in which environmental factors affect PEP activity. pTAC12 might be a key component of the phytochrome signaling pathway to regulate PEP activity and plastid gene expression. Protein methylation is involved in multiple biological processes, such as transcriptional regulation and RNA processing (Deng et al., 2010). pTAC14 is a member of the class VII subgroup of the SET family, with a truncated SET domain and a Rubisco LSMT substrate-binding domain (Ng et al., 2007; Fig. 1). The class VII SET protein is now recognized as having methyltransferase activity targeted to nonhistone proteins (Ng et al., 2007), and the protein with a Rubisco substrate-binding domain is related to a non-histone-specific methylation enzyme in the large or small subunit of the Rubisco holoenzyme (Ying et al., 1999; Couture et al., 2006). Several Lys and Arg methylation sites were identified in pTAC12 based on the predictions of the program MeMo (http://www.bioinfo.tsinghua.edu.cn/~tigerchen/memo/form.html). Thus, pTAC14 may regulate the activities of pTAC12 and/or other TAC components through methylation to affect PEP activity in the chloroplast. Nevertheless, this needs to be confirmed in future studies.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana Columbia) and mutant Arabidopsis seeds were grown in soil with a photon flux density of 120 mmol m−2 s−1 at 22°C. For growth on agar plates, seeds were sown on Murashige and Skoog (MS) medium containing 2% Suc. After they were vernalized under 4°C conditions for 3 d, the seedlings were transferred to grow in a chamber at 22°C with a 16-h-light/8-h-dark cycle.
Chloroplast and Thylakoid Fractionation
Chloroplast isolation was performed according to Ketcham et al. (1984). Chloroplast proteins of seedling material were homogenized in STN solution (50 mmol L−1 Tris-HCl, pH 7.6, 5 mmol L−1 MgCl2·6H2O, 10 mmol L−1 NaCl, 0.4 mol L−1 Suc, and 0.1% bovine serum albumin [BSA]), crude filtered, and centrifuged at 1,000g to eliminate tissue deposition. Samples were then centrifuged again at 2,000g to pellet the chloroplasts. STN solution was used to resuspend the chloroplasts. Thylakoid isolation was performed according to Dunahay et al. (1984). The Arabidopsis seedling was precooled and homogenized in B1 buffer (0.4 mol L−1 Suc, 2 mmol L−1 MgCl2, 0.2% BSA, and 20 mmol L−1 Tricine, pH 8.0) in ice-bath conditions. It was then crude filtered and centrifuged at 300g to eliminate tissue deposition. The liquid supernatant was centrifuged at 4,000g, and the pellet was resuspended in B2 buffer (0.15 mol L−1 Suc, 5 mmol L−1 MgCl2, 0.2% BSA, and 20 mol L−1 Tricine, pH 8.0) and centrifuged at 4,000g. Again, the pellet was suspended in B3 buffer (15 mmol L−1 NaCl, 5 mmol L−1 MgCl2, and 20 mmol L−1 MES, pH 6.5). The experimental procedure was carried out under 4°C or ice-bath conditions.
Identification of T-DNA Insertional Lines and Complementation of the pTAC14 Mutant Phenotype
Identification of Salk T-DNA insertion lines was performed according to Alonso et al. (2003). The primers used for the SAIL T-DNA and SALK T-DNA insertion lines are listed in Supplemental Table S1.
The full-length CDS of pTAC14 fused with the 35S promoter and the FLAG tag was introduced into wild-type plants (Columbia) and heterozygous pTAC14/ptac14 plants, respectively. The pTAC14 genomic fragment with the 1,613-bp upstream and 259-bp downstream sequences was amplified using KOD plus polymerase (TOYOBO; http://www.toyobo.co.jp), subcloned into pCAMBIA1300, and introduced into heterozygous pTAC14/ptac14 plants by Agrobacterium tumefaciens-mediated transformation. The genomic backgrounds of these independent hygromycin-resistant transgenic plants were further analyzed with the gene-specific primers listed in Supplemental Table S1.
Chlorophyll Determination and Chloroplast Ultrastructure Observation
Total chlorophyll content was determined with 14-d-old wild-type and mutant Arabidopsis seedlings grown on MS medium with 2% Suc under normal conditions, according to our previously described protocol (Yu et al., 2009a). Chloroplasts from ptac14-1 mutants were grown under normal light intensities for 14 d, and the ultrastructure micrographs were obtained as described in the same protocol (Yu et al., 2009a).
Protein Extraction and Immunological Analysis of Chloroplast Proteins
Total proteins of 14-d-old wild-type and mutant plants were homogenized in media A buffer (25 mmol L−1 Tris-HCl, pH 8.6, 1.46 g of NaCl, 0.08 g of NH4Cl, and 3.43 g of MgCl2·6H2O) and 25% (v/v) Triton X-100 at 4°C, and the total protein content was assayed using a modified Bradford assay (Bio-Rad DC Protein Assay; http://www.bio-rad.com). A total of 40 μg of total protein was used per lane. After separation by 12% SDS-PAGE, proteins were transferred electrophoretically to nitrocellulose filter membranes and then blotted with various specific antibodies. Antibodies were detected using a modified chemiluminescence system (General Electric; http://www.ge.com). Photosynthesis-related antibodies were obtained from Agrisera (http://www.agrisera.com), and the anti-FLAG antibody was obtained from Sigma (http://www.sigmaaldrich.com).
RNA Isolation, cDNA Synthesis, RT-PCR, and Quantitative Real-Time RT-PCR
Procedures for the purification of total RNAs for cDNA synthesis, RT-PCR, and quantitative real-time RT-PCR have been described previously (Yu et al., 2009a).
RNA Gel-Blot Analysis
A total of 15 μg of each RNA sample was electrophoresed on 1.2% agarose-formaldehyde gels and transferred onto nylon membranes (Pall). They were then hybridized with the specific gene fragment labeled with digoxigenin. All hybridization and chemiluminescence detections were carried out according to the Roche protocol (http://www.roche.com). Primer pairs are listed in Supplemental Table S1.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed using the Clontech (www.clontech.com) two-hybrid system according to a modified version of the manufacturer’s instructions. A fragment of the pTAC14 protein without the 62-amino acid transit peptide was amplified and cloned into the pGBKT7 vector as the bait. The fragments corresponding to the 35 TAC members were cloned into pGADT7 individually, and the obtained plasmids were mixed into a prey pool. The pTAC14 bait was cotransformed into the yeast strain AH109 with the prey pool. The transformants were screened on supplemented synthetic dextrose (SD) medium lacking Leu and Trp or on supplemented SD medium lacking Leu, Trp, His, and adenine hemisulfate salt with X-α-Gal. The positive clones were then amplified with AD LD-insert screening amplimers (Clontech, www.clontech.com; Supplemental Table S1) and sequenced.
Coimmunoprecipitation Assay
For coimmunoprecipitation, pTAC14 with a C-terminal Myc epitope tag (pTAC14-Myc) and pTAC12 with a C-terminal Myc tag (pTAC12-Myc) were cotransferred into yeast strain AH109. Following treatment with DNase and phenylmethylsulfonyl fluoride, the total yeast protein was incubated with antibodies coupled covalently to Protein A- and G-Sepharose beads for 1.5 h at 4°C. After the Sepharose beads were washed three times with coimmunoprecipitation buffer (50 mmol L−1 Tris-Cl, pH 7.5, 15 mmol L−1 EDTA, 100 mmol L−1 NaCl, 0.1% [v/v] Triton X-100, protease inhibitor mixture, 5 mg mL−1 chymostatin, 5 mg mL−1 pepstatin A, and 5 mg mL−1 leupeptin), the precipitated proteins were dissociated in SDS sample buffer by heating at 95°C for 5 min. Following SDS-PAGE, the precipitated proteins were immunoblotted using specific antibodies against the tagged proteins.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NP_193746.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Oligonucleotides used in this work.
Acknowledgments
We thank ABRC Bioresources, which kindly offered the transgenic Arabidopsis lines (SAIL_566_F06 and SALK_005814). We are grateful to Mrs. Hui-Qi Zhang from Shanghai Normal University and Mr. Kai-He Du from Nanjing Normal University for their skillful technical assistance in TEM.
References
- Allison LA. (2000) The role of sigma factors in plastid transcription. Biochimie 82: 537–548 [DOI] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P. (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F. (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aseeva E, Ossenbühl F, Sippel C, Cho WK, Stein B, Eichacker LA, Meurer J, Wanner G, Westhoff P, Soll J, et al. (2007) Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol Biochem 45: 119–128 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, et al. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J. (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Ma JF, Zhang DY, Guo JK, Chen F, Lu CM, Zhang LX. (2008) The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol 147: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. (2006) Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J Biol Chem 281: 19280–19287 [DOI] [PubMed] [Google Scholar]
- Deng X, Gu LF, Liu CY, Lu TC, Lu FL, Lu ZK, Cui P, Pei YX, Wang BC, Hu SN, et al. (2010) Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunahay TG, Staehelin LA, Seibert M. (1984) Structural, biochemical and biophysical characterization of four oxygen evolving photosystem II preparations from spinach. Biochim Biophys Acta 764: 179–193 [Google Scholar]
- Emanuel C, Weihe A, Graner A, Hess WR, Börner T. (2004) Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J 38: 460–472 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H. (2008) An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 53: 924–934 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Nava MdeL, Gillmor CS, Jiménez LF, Guevara-García A, León P. (2004) CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol 135: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PTJ, Allison LA, Maliga P. (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess WR, Börner T. (1999) Organellar RNA polymerases of higher plants. Int Rev Cytol 190: 1–59 [DOI] [PubMed] [Google Scholar]
- Hess WR, Prombona A, Fieder B, Subramanian AR, Börner T. (1993) Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J 12: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Bogorad L. (1990) Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci USA 87: 1531–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL, Kössel H. (1992) The transcriptional apparatus of chloroplasts. CRC Crit Rev Plant Sci 10: 525–558 [Google Scholar]
- Inatomi K. (1986) Characterization and purification of the membrane-bound ATPase of the archaebacterium Methanosarcina barkeri. J Bacteriol 167: 837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski G, Kacprzak K, Jansson S. (2001) Identification of Lhcb1/Lhcb2/Lhcb3 heterotrimers of the main light-harvesting chlorophyll a/b-protein complex of photosystem II (LHC II). Biochim Biophys Acta 1504: 340–345 [DOI] [PubMed] [Google Scholar]
- Jain PK, Kochhar A, Khurana JP, Tyagi AK. (1998) The psbO gene for 33-kDa precursor polypeptide of the oxygen-evolving complex in Arabidopsis thaliana: nucleotide sequence and control of its expression. DNA Res 5: 221–228 [DOI] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Meier I. (2003) MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D, Köster D, Schadach A, Klevesath A, Bock R. (2009) The Chlamydomonas chloroplast HLP protein is required for nucleoid organization and genome maintenance. Mol Plant 2: 1223–1232 [DOI] [PubMed] [Google Scholar]
- Ketcham SR, Davenport JW, Warncke K, McCarty RE. (1984) Role of the γ subunit of chloroplast coupling factor 1 in the light-dependent activation of photophosphorylation and ATPase activity by dithiothreitol. J Biol Chem 259: 7286–7293 [PubMed] [Google Scholar]
- Krause K, Maier RM, Kofer W, Krupinska K, Herrmann RG. (2000) Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol Gen Genet 263: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Krebbers E, Herdies L, De Clercq A, Seurinck J, Leemans J, Van Damme J, Segura M, Gheysen G, Van Montagu M, Vandekerckhove J. (1988) Determination of the processing sites of an Arabidopsis 2S albumin and characterization of the complete gene family. Plant Physiol 87: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. (2001) VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA 98: 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S. (1993) The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc Natl Acad Sci USA 90: 5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K, Maliga P. (2001) Plastid RNA polymerases in higher plants. Aro E-M, Andersson B, , Regulation of Photosynthesis. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 29–49 [Google Scholar]
- Liu C, Lu F, Cui X, Cao X. (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Loschelder H, Schweer J, Link B, Link G. (2006) Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant Physiol 142: 642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Véronneau-Lafortune F, Joyeux A, Lang BF, Brisson N. (2009) Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci USA 106: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet JE. (1988) Chloroplast development and gene expression. Annu Rev Plant Physiol Plant Mol Biol 39: 475–502 [Google Scholar]
- Mullet JE. (1993) Dynamic regulation of chloroplast transcription. Plant Physiol 103: 309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Kondo Y, Nakano T, Sato F. (2000) Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Lett 468: 15–18 [DOI] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K. (2008) A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20: 3148–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC. (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769: 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G. (1997) The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol Gen Genet 257: 35–44 [DOI] [PubMed] [Google Scholar]
- Sato N. (2001) Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci 6: 151–155 [DOI] [PubMed] [Google Scholar]
- Sato N, Terasawa K, Miyajima K, Kabeya Y. (2003) Organization, developmental dynamics, and evolution of plastid nucleoids. Int Rev Cytol 232: 217–262 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 6: 283–290 [DOI] [PubMed] [Google Scholar]
- Schweer J, Loschelder H, Link G. (2006) A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett 580: 6617–6622 [DOI] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Kolpack A, Link G. (2010a) Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription: recent lessons from Arabidopsis thaliana. Eur J Cell Biol 89: 940–946 [DOI] [PubMed] [Google Scholar]
- Sekine K, Fujiwara M, Nakayama M, Takao T, Hase T, Sato N. (2007) DNA binding and partial nucleoid localization of the chloroplast stromal enzyme ferredoxin:sulfite reductase. FEBS J 274: 2054–2069 [DOI] [PubMed] [Google Scholar]
- Serino G, Maliga P. (1998) RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol 117: 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. (2005) Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol 244: 1–68 [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T. (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Sugiura M. (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Terasawa K, Sato N. (2009) Plastid localization of the PEND protein is mediated by a noncanonical transit peptide. FEBS J 276: 1709–1719 [DOI] [PubMed] [Google Scholar]
- Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. (2002) Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111: 91–103 [DOI] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. (1994) The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J 13: 3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying ZT, Mulligan RM, Janney N, Houtz RL. (1999) Rubisco small and large subunit N-methyltransferases: bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J Biol Chem 274: 36750–36756 [DOI] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN. (2009a) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Yu Y, Bu ZY, Shen WH, Dong AW. (2009b) An update on histone lysine methylation in plants. Prog Nat Sci 19: 407–413 [Google Scholar]
- Zhou WB, Cheng YX, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, Small I, Huang JR. (2008) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 58: 82–96 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T. (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50: 710–722 [DOI] [PubMed] [Google Scholar]