Abstract
Legumes host their Rhizobium spp. symbiont in novel root organs called nodules. Nodules originate from differentiated root cortical cells that dedifferentiate and subsequently form nodule primordia, a process controlled by cytokinin. A whole-genome duplication has occurred at the root of the legume Papilionoideae subfamily. We hypothesize that gene pairs originating from this duplication event and are conserved in distinct Papilionoideae lineages have evolved symbiotic functions. A phylogenetic strategy was applied to search for such gene pairs to identify novel regulators of nodulation, using the cytokinin phosphorelay pathway as a test case. In this way, two paralogous type-A cytokinin response regulators were identified that are involved in root nodule symbiosis. Response Regulator9 (MtRR9) and MtRR11 in medicago (Medicago truncatula) and an ortholog in lotus (Lotus japonicus) are rapidly induced upon Rhizobium spp. Nod factor signaling. Constitutive expression of MtRR9 results in arrested primordia that have emerged from cortical, endodermal, and pericycle cells. In legumes, lateral root primordia are not exclusively formed from pericycle cells but also require the involvement of the root cortical cell layer. Therefore, the MtRR9-induced foci of cell divisions show a strong resemblance to lateral root primordia, suggesting an ancestral function of MtRR9 in this process. Together, these findings provide a proof of principle for the applied phylogenetic strategy to identify genes with a symbiotic function in legumes.
Most legumes (Fabaceae) can establish a unique endosymbiosis with nitrogen-fixing soil bacteria, collectively named Rhizobium spp.. Rhizobium spp. bacteria grant their hosts access to combined nitrogen. To achieve this, root nodules are formed, which are unique plant organs that provide optimal conditions for Rhizobium spp. to fix nitrogen. The Rhizobium spp.-legume symbiosis is set in motion by bacterial signal molecules, named Nod factors. Nod factors are perceived by plant-specific LysM domain transmembrane receptors, which in turn activate downstream signaling networks essential for nodule organogenesis (Kouchi et al., 2010). Among the downstream signaling networks is the cytokinin phosphorelay pathway (Frugier et al., 2008). How legumes have recruited such genes to function in symbiosis remains largely unknown. Recently, it was shown that legumes of the large Papilionoideae subfamily (papilionoids) underwent a whole-genome duplication (WGD; Cannon et al., 2006). This duplication event occurred early in papilionoid evolution; it is estimated to have occurred 56 to 65 million years ago (Fawcett et al., 2009; Cannon et al., 2010). Papilionoids represent all major legume crops, and Rhizobium spp. symbiosis is common to most of the approximately 13,000 species (Gepts et al., 2005). We hypothesize that the papilionoid-specific WGD has contributed substantially to the makeup of root nodules in this subfamily, even though Rhizobium spp. symbiosis itself possibly evolved at an earlier time point (Fawcett et al., 2009; Cannon et al., 2010). To test this hypothesis, we focused on the cytokinin phosphorelay signaling pathway.
The role of cytokinin signaling in root nodule symbiosis is demonstrated by physiological and molecular genetic studies. Early studies showed that, in some legume species, initiation of nodule organogenesis could be mimicked by external cytokinin application. For example, in alfalfa (Medicago sativa), lotus (Lotus japonicus), and white clover (Trifolium repens), the formation of nodule-like structures can be triggered with an architecture similar to Nod factor-induced nodules (Cooper and Long, 1994; Mathesius at al., 2000a; Heckmann et al., 2011). In addition, in many legume species, it is shown that externally applied cytokinin leads to induction of symbiotic genes, which can also be activated by Nod factors (Frugier et al., 2008). Genetic integration of the cytokinin phosphorelay pathway in Nod factor signaling is best demonstrated by gain-of-function and loss-of-function mutants of the His kinase cytokinin receptor (HK) LjLHK1/MtCRE1 in lotus and medicago (Medicago truncatula). A functional LjLHK1/MtCRE1 gene is indispensable for nodule formation, and a dominant positive mutation in the receiver domain even leads to spontaneous nodule formation (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007; Ovchinnikova et al., 2011; Plet et al., 2011). Spontaneous nodulation driven by the gain-of-function HK mutant requires other components of the Nod factor-induced signaling pathway (e.g. NSP2 and NIN), which highlights the intertwining of both networks. LjLHK1/MtCRE1 also functions in lateral root formation, indicating that the symbiotic activity of these HKs is derived from this nonsymbiotic process (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007; Plet et al., 2011). Examples of cytokinin signaling related to root development are the control meristem size, cell differentiation, vasculature development, and lateral root primordium initiation (Bishopp et al., 2009). The latter process generally is considered to occur in the root pericycle, whereas in legumes root nodules, primordia are largely formed from root cortical cells (Laplaze et al., 2007; Crespi and Frugier 2008; Péret et al., 2009).
The cytokinin phosphorelay pathway consists of four signaling components: HKs, phosphotransfer proteins (HPs), and two types of response regulators (RRs). Upon activation, HK phosphorylates an HP. Subsequently, the HP migrates to the nucleus and transfers the phosphate to a type-B RR, which in turn acts as a transcriptional activator. Among the primary targets of type-B RRs are so-called type-A RRs. Both RR types are homologous in sequence, although type-A RRs lack a putative DNA-binding domain. It is generally assumed that type-A RRs act as negative regulators of cytokinin signaling (Müller and Sheen, 2007). In line with the symbiotic role of LjHK1/MtCRE1, it can be anticipated that other components of the cytokinin phosphorelay pathway have also evolved to function in symbiotic signaling. One such gene is the A-type RR MtRR4, which functions downstream of MtCRE1 (Plet et al., 2011).
In this study, the cytokinin phosphorelay components from three papilionoid legume species for which substantial genome information is available, namely medicago, lotus, and soybean (Glycine max), were analyzed to find gene pairs that were maintained from the papilionoid-specific WGD. We used as criterion that both gene copies should be maintained in all three legume species and that the timing of the duplication should match the WGD event. One such gene pair, encoding type-A RRs, was found. Functional studies revealed that these genes are transcriptionally induced upon Nod factor signaling in both medicago and lotus. For the medicago genes MtRR9 and MtRR11, we show that their induction depends on the nuclear localized calcium calmodulin kinase (CCaMK), which is a key element in Nod factor signaling (Lévy et al., 2004; Mitra et al., 2004a; Smit et al., 2005). Ectopic expression of MtRR9 results in arrested lateral primordia that are associated with multiple cortical and pericycle cell divisions. These data provide a proof of principle for the phylogenetic strategy based on a legume-specific WGD to identify genes involved in Rhizobium spp. symbiosis.
RESULTS
One Gene Pair of Type-A RRs Is Maintained upon Papilionoid-Specific WGD
The genes encoding components of the cytokinin phosphorelay pathway are well characterized in Arabidopsis (Arabidopsis thaliana), which facilitated the identification of legume genes of this pathway (Supplemental File S1). To test whether some of these genes are specifically duplicated in papilionoid legumes, we performed a phylogenetic analysis to identify gene pairs that originate from the papilionoid-specific WGD (Supplemental Fig. S1). The genomes of three legumes, medicago, lotus, and soybean) and three nonlegumes, Arabidopsis, black cottonwood poplar (Populus trichocarpa), and grapevine (Vitis vinifera), were analyzed.
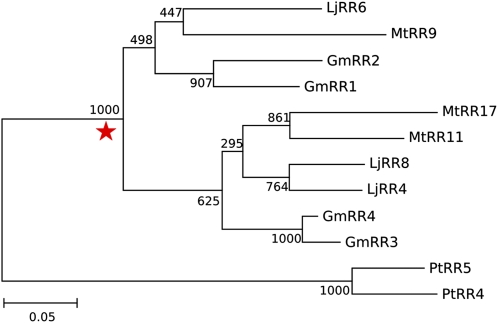
Only one clade displayed a legume-specific duplication maintained in all three legume species (Fig. 1). This clade belongs to the type-A RR gene family and is referred to as orthology group 2.4 in Supplemental Figure S1. To date this duplication, we used a maximum likelihood estimation based on the molecular clock hypothesis (Kimura, 1969). The duplication was estimated to have occurred 61 million years ago, which falls within the confidence interval for the papilionoid-specific WGD (Fawcett et al., 2009). Besides this duplication event, lineage-specific duplications occurred in all species investigated. In the cases of soybean and black cottonwood poplar, this is likely the result of more recent WGDs (Shoemaker et al., 2006; Fawcett et al., 2009; Schmutz et al., 2010). The medicago type-A RR genes in orthology group 2.4 were named MtRR9, MtRR11, and MtRR17, the latter of which represents a pseudogene caused by a frame-shift mutation. The soybean genes were named GmRR1 to GmRR4, whereas nomenclature for lotus and black cottonwood poplar was adopted from the literature: LjRR4, LjRR6, LjRR8, PtRR4, and PtRR5 (Ramírez-Carvajal et al., 2008; Ishida et al., 2009; Fig. 1).
Figure 1.
Maintained duplication of type-A RR genes in legumes. Red star, Legume-specific duplication in this selection of orthology group 2.4 (complete tree of type-A RRs is shown in Supplemental Figure S1). Tree is constructed using maximum likelihood phylogeny (PhyML version 3.0; Anisimova and Gascuel, 2006) and branch support test from 1,000 bootstrap repetitions. Gm, Soybean; Lj, lotus; Mt, medicago; Pt, black cottonwood poplar. [See online article for color version of this figure.]
Nod Factor-Induced Expression of Duplicated Type-A RR Genes
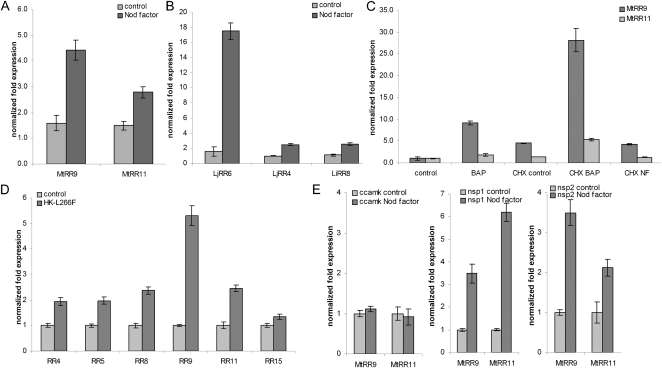
Transcriptional regulation upon Nod factor application was tested to investigate whether the identified paralogous pair of RR genes could play a role in Rhizobium spp. symbiosis. To investigate the extent of Nod factor induction of type-A RR genes, we included all 12 medicago type-A RRs in this analysis. For six genes, expression in roots could be detected, of which three were transcriptionally activated 3 h after application of Sinorhizobium meliloti Nod factors, including MtRR9 and MtRR11 (Fig. 2A; Supplemental Fig. S2). This suggests that both genes could have a function early in symbiotic signaling. Besides MtRR9 and MtRR11, MtRR8 was also strongly induced, whereas MtRR5 was down-regulated (Supplemental Fig. S2). MtRR8 is the putative ortholog of AtARR5 (Supplemental Fig. S1), a gene widely used as a cytokinin-responsive marker in a diverse range of species, including legumes (D’Agostino et al., 2000; Lohar et al., 2004). It is noteworthy that we did not find Nod factor-induced transcriptional activation of MtRR4, a type-A RR that is transcriptionally activated upon Rhizobium spp. inoculation within 24 h (Gonzalez-Rizzo et al., 2006; Plet et al., 2011).
Figure 2.
A and B, Relative expression of RR genes was determined using quantitative RT-PCR after 3-h application of Nod factors (10−9 m) for medicago (A) and lotus (B). C, Relative expression levels in medicago roots of MtRR9 and MtRR11 in the absence or presence of CHX during exposure to Nod factors (3 h) or BAP (10−8 m; 1 h). D, Relative expression levels in medicago control roots (empty vector) versus roots harboring the Mt35S:CRE1*[L267F] construct. E, Relative expression levels of MtRR9 and MtRR11 in medicago mutant ccamk, nsp1, and nsp2 roots. Quantification was normalized using stable expressed reference genes MtGAPDH, MtPTB, LjATPS, and LjUBQ. Bars, sd of three technical repeats.
To determine whether the Nod factor-induced expression of the duplicated gene pair in orthology group 2.4 is conserved in legumes, we studied the expression of the orthologous lotus genes LjRR4, LjRR6, and LjRR8 (Fig. 1). To this end, Nod factors of Sinorhizobium sp. NGR234, a symbiont of lotus, were applied to lotus roots for 3 h. This revealed that, in lotus, mainly LjRR6 is activated, which is in line with the findings in medicago, where the orthologous gene, MtRR9, also is most strongly induced (Fig. 2, A and B).
Because type-A RRs are primary targets of cytokinin signaling in Arabidopsis (To and Kieber, 2008), we also studied the regulation of MtRR9 and MtRR11 upon cytokinin and Nod factor application in the presence of the protein synthesis blocker cycloheximide (CHX). Both genes were induced by cytokinin (6-benzylaminopurine [BAP] 10−8 m), also in the presence of CHX (Fig. 2C). This is in contrast with Nod factor-induced expression, in which protein synthesis was essential for transcriptional activation of both RRs (Fig. 2C). To further support that medicago type-A RR genes are targets of the cytokinin phosphorelay pathway, we isolated RNA from medicago roots transformed with the gain-of-function MtCRE1 construct (35S:MtCRE1*[L267F]), which causes spontaneous nodule formation (Ovchinnikova et al., 2011). Quantitative reverse transcriptase (RT)-PCR on root RNA showed that, of all type-A RR genes, MtRR9 was most strongly induced and also that MtRR4, MtRR5, MtRR8, and MtRR11 were activated (Fig. 2D). These results show that these five genes are indeed primary targets of cytokinin signaling downstream of MtCRE1 and suggest that their expression is under direct control of a type-B RR.
In legumes, Nod factor signaling is achieved by a conserved signaling pathway that contains several key proteins, including a nuclear localized CCaMK and two GRAS-type transcription factors, NSP1 and NSP2 (Mitra et al., 2004a; Kaló et al., 2005; Smit et al., 2005). CCaMK, NSP1, and NSP2 are reported to be essential for the induction of nearly all symbiotic genes by Nod factors (Mitra et al., 2004b). We studied the transcriptional regulation of MtRR9 and MtRR11 upon Nod factor application in the medicago Nod factor signaling knockout mutants Mtdmi3 (ccamk), Mtnsp1, and Mtnsp2 to determine whether induction depends on these key symbiotic genes. This revealed that the induction of MtRR9 and MtRR11 was dependent on CCaMK but could be triggered in both nsp mutants (Fig. 2E). This suggests that Nod factor activation of the cytokinin phosphorelay pathway can occur independently from both GRAS-type regulators, resulting in bifurcation of Nod factor-induced signaling downstream of CCaMK. A similar bifurcation of Nod factor signaling downstream of CCaMK has also been shown in lotus (Madsen et al., 2010). All together, these studies suggest that legume type-A RR genes have gained a function in Nod factor-induced root nodule formation.
MtRR9 Is Induced in the Nod Factor-Susceptible Zone
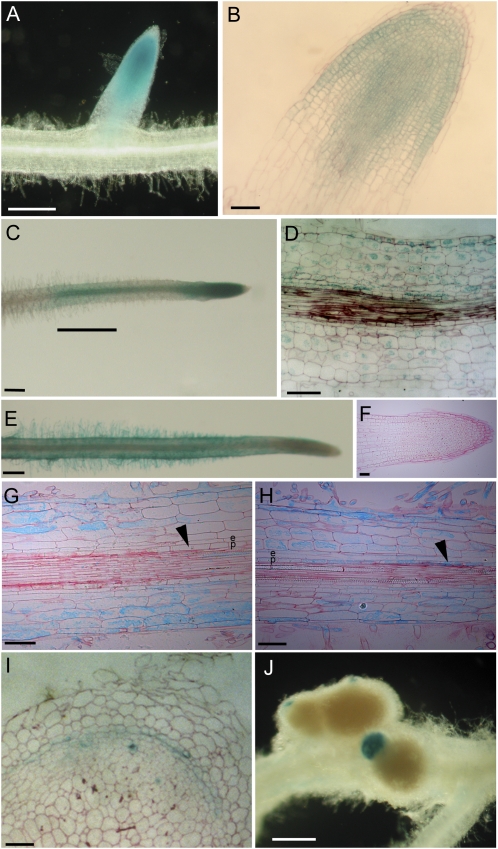
To study the symbiotic regulation of MtRR9 and MtRR11 in more detail, the spatial expression pattern of both genes was determined using GUS reporter constructs. For both genes, ∼2,500 bp upstream of the transcriptional start site was used as a putative promoter. In medicago roots, the nonsymbiotic expression pattern of pMtRR9::GUS was found exclusively in the root meristematic zone (Fig. 3, A and B). pMtRR11::GUS was found not to be expressed in the meristem but in the epidermis, cortex, and endodermis of the differentiation zone, including the zone susceptible to Nod factors (Fig. 3, E–G). Upon local application of Nod factors to the susceptible zone, theMtRR9 promoter activity was induced in all cell layers within 3 h (Fig. 3, C and D). Such elevated expression in the epidermis and cortex was less obvious for pMtRR11::GUS because nonsymbiotic expression was already present, and GUS is not very suitable for quantitative interpretations. However, upon application of Nod factors, the MtRR11 promoter was found to be elevated in the pericycle (Fig. 3, G and H). In root nodules, both genes were found to be expressed in the apical region of differentiated nodules, a region similar to that observed for MtCRE1 expression (Fig. 3, I and J; Plet et al., 2011).
Figure 3.
Spatial expression pattern of MtRR9 (A–D and I) and MtRR11 (E–H and J). pMtRR9::GUS/pMtRR11::GUS transformed and histochemically stained roots. A, Untreated root meristem. B, Microsection of untreated root meristem. C, Root locally treated with Nod factors (10−9 m). Large bar, Location of Nod factor containing agarose slice. D, Microsection of root at location of Nod factor (10−9 m) exposure (3 h). E, Untreated root. F, Microsection of untreated root meristem. G, Microsection of zone susceptible to Nod factors of untreated root. Arrowhead points to the pericycle. H, Microsection of the root after Nod factor (10−9 m) exposure (3 h). Arrowhead points to the pericycle. I, Expression of pMtRR9::GUS in nodule, microsection. J, Expression of pMtRR11::GUS in nodule. e, Endodermis; p, pericycle. Bars in A, C, E, and J, 400 μm; bars in B, D, and F to I, 100 μm.
Based on this study, we conclude that upon symbiotic signaling, the spatial expression patterns of both genes largely overlap. The induction of MtRR9 upon Nod factor signaling in the epidermis, cortex, endodermis, and pericycle, and of MtRR11 in the pericycle of the susceptible zone before the occurrence of symbiotic cell divisions, suggests that both genes function in root nodule primordium formation. Because MtRR9 and its lotus ortholog LjRR6 are most strongly induced by Nod factors, MtRR9 is most strongly induced in 35S:MtCRE1*[L267F] roots, and MtRR9 is activated in the zone susceptible to Nod factors, we focused on MtRR9 for further functional studies.
Ectopic Expression of MtRR9 Results in Arrested Primordia
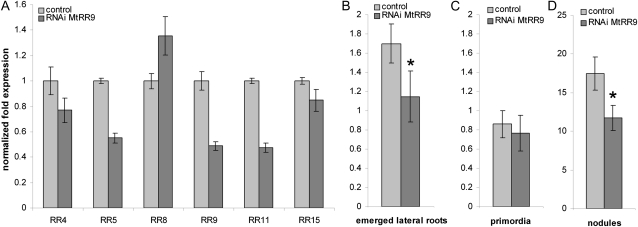
To investigate the role of MtRR9 in root nodule primordium formation, we conducted ectopic expression as well as RNA interference (RNAi) experiments. First, we made an RNAi construct to target MtRR9 and introduced it into medicago roots by Agrobacterium rhizogenes-mediated transformation. Because MtRR9 is highly homologous to MtRR11 as well as several other type-A RRs, we determined the specificity of this targeting construct. Therefore, the expression of all six root-expressed type-A RRs was quantified by RT-PCR. Analysis showed that this RNAi construct affects MtRR9 and MtRR11 but also MtRR5. The latter is the closest homolog of MtRR9 and MtRR11, although it showed opposite regulation by Nod factors when compared with MtRR9/MtRR11 (Supplemental Figs. S1 and S2). mRNA levels of all three genes show a knockdown level of approximately 50% in medicago RNAi roots (Fig. 4A). We searched for a primordium formation phenotype in the RNAi plants and noted that the RNAi roots had approximately 33% fewer emerged lateral roots when compared with the wild-type plants (n = 46, Mann-Whitney U test, P < 0.05) (Fig. 4, B and C). Inoculation of these RNAi roots also resulted in a decreased nodulation efficiency of approximately 33% of the average number of nodules per transgenic root (n = 37, Mann-Whitney U test, P < 0.05) (Fig. 4D). These findings indicate that the type-A RR genes MtRR9 and MtRR11, and possibly MtRR5, are required both for nodule organogenesis and for lateral root formation.
Figure 4.
A, Quantification of root-expressed type-A RRs. Relative expression levels in medicago pooled control roots (empty vector) versus roots harboring the MtRR9-RNAi construct. Quantification was normalized using stably expressed reference genes MtGAPDH and MtPTB. Bars, sd of three technical repeats. B and C, Number of emerged lateral roots (B) and lateral root primordia (C) per transgenic root of medicago plants harboring either a control (empty vector) or the MtRR9-RNAi construct. An asterisk indicates that the difference in number of emerged lateral roots between control and MtRR9-RNAi is statistically significant (Mann-Whitney U test, P < 0.05). Error bars, se (n = 46). D, Number of nodules per transgenic root of medicago plants harboring either a control (empty vector) or the MtRR9-RNAi construct. An asterisk indicates that the difference in nodule number between control and MtRR9-RNAi is statistically significant (Mann-Whitney U test, P < 0.05). Error bars, se (n = 37). Two independent biological replicates were performed for all experiments (A–d).
Next, we ectopically expressed MtRR9 in medicago roots using the constitutive cauliflower mosaic virus (CaMV) 35S promoter. For Arabidopsis, it is reported that ectopic expression of different type-A RRs results in the formation of more lateral roots (Ren et al., 2009). Similarly, pCaMV35S::MtRR9-expressing medicago roots showed an increased number of emerged lateral roots (Supplemental Fig. S3). Furthermore, such transgenic roots also contained primordia-like structures that were positioned in between emerged lateral roots (Fig. 5A; Supplemental Figs. S4 and S5). These could either represent arrested lateral root primordia or mimic de novo-induced root nodule primordia. Microscopic analysis of sections of these MtRR9-induced primordia showed that cell divisions had occurred in the pericycle, endodermis, and root cortex (Fig. 5B). In medicago, such cell divisions can also be triggered by Nod factors (Timmers et al., 1999), which might suggest that the MtRR9-induced primordia have a symbiotic nature.
Figure 5.
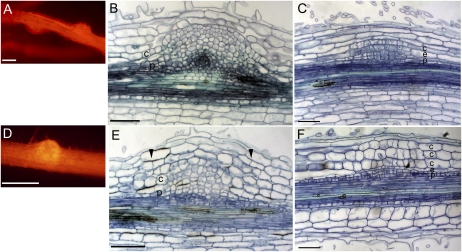
. MtRR9 constitutive expression results in primordia. A and D, Primordia on roots of medicago (A) and lotus (D) as a result of constitutive expression of MtRR9 (pCaMV35S::MtRR9). Transgenic roots were selected based on DsRed fluorescence. B, Microsection of a medicago primordium shows cell divisions in the inner cortical cell layers and pericycle. E, Microsection of a lotus primordium with cell divisions in inner and outer cortical cell layers and pericycle. Microsection of young lateral root primordia of medicago (E) and lotus (F). Divisions in the outer cortex are marked with arrowheads. c, Cortex; e, endodermis; p, pericycle. Bars in A and C, 400 μm; bars in B and D to F, 100 μm.
To determine whether the capacity to induce such primordia is specific for legume type-A RR genes of orthology group 2.4, we conducted the same experiment with PtRR5, the putative ortholog of black cottonwood poplar. Transgenic medicago roots ectopically expressing PtRR5 also formed such primordia, although to a lesser extent (Supplemental Fig. S4). This indicates that the MtRR9-encoded protein has not specifically evolved to fulfill such function.
In contrast with medicago, lotus root nodule primordia originate from the middle and outer cortical layers (Szczyglowski et al., 1998; van Spronsen et al., 2001). To determine whether ectopic expression of MtRR9 can mitotically activate outer cortical cells, we introduced CaMV35Sp::MtRR9 into lotus roots. Also, lotus primordia are induced, as in medicago (Fig. 5D). Sectioning revealed that in lotus, not only pericycle and inner cortical cells divided but also cells in the middle and outer cortical layers (Fig. 5E). This shows that the location of MtRR9-induced cell divisions coincides with the spatial position of symbiotic divisions in the cortex. Furthermore, it indicates that there is not a specific function of MtRR9 dedicated to indeterminate-type nodulation.
The general view is that lateral root primordia develop from the pericycle and endodermal cell layers (Péret et al., 2009). However, for many species, including some legumes, it has been reported that cortical cell divisions can also accompany lateral root development (Tschermak-Woess and Dolezal, 1953; Mallory et al., 1970; McCully, 1975; Bryne et al., 1977; Casero et al., 1996; Mathesius et al., 2000b). To better compare the MtRR9-induced primordia, we studied the involvement of cortical cells during lateral root primordium formation in lotus and medicago. In both species, we observed that the formation of a lateral root primordium in the pericycle cell layer is associated with cell divisions in the endodermal and cortical cell layers (Fig. 5, C and F).Interestingly, in lotus, the cell divisions in the cortex were extended more to the outer cortical cell layers when compared with medicago, similar to what was observed upon Nod factor-induced nodule primordium formation (Szczyglowski et al., 1998; van Spronsen et al., 2001). Because both species display root cortical cell divisions during lateral root formation, it suggests that ectopic expression of MtRR9 results in arrested primordia that are the result of activation of shared developmental programs essential for nodule as well as lateral root formation.
DISCUSSION
In this study, we present a phylogenetic strategy to identify genes originating from the papilionoid-specific WGD that have gained a function in Rhizobium spp. symbiosis. To test this strategy, we focused on the cytokinin phosphorelay pathway because it is presumed to be an integrative part of Rhizobium spp.-induced signaling (Frugier et al., 2008). A total of 22 orthology groups were investigated, which resulted in the identification of a single conserved gene pair originating from this WGD. We demonstrate that the encoded type-A RRs are part of the Nod factor-induced symbiotic signaling cascade. This shows that despite massive gene loss upon the WGD event that occurred early in the evolution of the papilionoid subfamily, duplicated gene pairs can be identified that have contributed to the evolution of the Rhizobium spp. symbiosis in this subfamily. Therefore, the presented phylogenetic approach can be a useful tool to identify novel genes that function in Rhizobium spp. symbiosis.
Type-A RRs are generally considered to be negative regulators of cytokinin phosphorelay signaling (Hwang and Sheen, 2001; Osakabe et al., 2002; Kiba et al., 2003, 2004; To et al., 2004; Hirose et al., 2007). In accordance with this view, we hypothesize that MtRR9 and MtRR11 are also part of a negative feedback mechanism on the cytokinin phosphorelay signaling. Cytokinin plays a negative role in lateral root initiation, and it has been shown that Arabidopsis type-A RR mutants may fulfill a key function to control this inhibition (To et al., 2004; Ren et al., 2009). The function of MtRR9 as a negative regulator is based on the finding that ectopic MtRR9 expression results in more lateral roots, which is in line with similar findings for ectopic expression of type-A RRs in Arabidopsis and with the fact that lowered endogenous levels of cytokinin lead to a higher lateral root density (Laplaze et al., 2007; Nibau et al., 2008; Ren et al., 2009). Furthermore, we observed an increased number of arrested lateral primordia. Besides its negative role in the initiation of lateral roots, cytokinin is known to regulate, antagonistically to auxin, the proper patterning of the embryonic root meristem (Müller and Sheen, 2008). Whether similar genetic mechanisms regulate lateral root meristem development is unknown. We anticipate that ectopic expression of a negative regulator of cytokinin signaling may disturb proper lateral root meristem patterning, resulting in arrested lateral primordia.
In contrast with ectopic expression of type-A RR genes, type-A RR Arabidopsis mutants have fewer lateral roots, which reflect the negative role of cytokinin in lateral root initiation. Type-A RRs function redundantly because inhibitory effects were only observed when multiple members were knocked out (To et al., 2004). Our RNAi construct was designed with the intention only to target MtRR9, but because of high homology, other type-A RRs were also knocked down. Therefore, the observed lowered amount of emerged lateral roots on the RNAi roots are probably due to the combined knockdown of multiple type-A RRs, suggesting redundant functioning of these genes. Notably, a decrease in nodule number is also observed in knockdown roots. This seems like a paradox; because cytokinin is promoting root nodule formation, one would anticipate that downregulation of cytokinin inhibitor genes would promote root nodule formation. So far, we do not have a mechanistic explanation for this finding, although we anticipate that the positive effect of cytokinin on root nodule formation acts only transiently, subsequently resulting in a new auxin maximum in the developing root nodule primordium (Plet et al., 2011). The three genes targeted by RNAi show opposite regulation by Nod factors; MtRR9 and MtRR11 were transcriptionally activated, whereas MtRR5 was down-regulated. This may provide an explanation for the observed phenotype because precise cytokinin signaling may be crucial for nodule development. Constitutive knockdown of MtRR9, MtRR11, and MtRR5 therefore may act negatively on root nodule formation as well.
Strikingly, the arrested primordia that are obtained in MtRR9 overexpression roots are composed of cells that originate from the cortex, endodermis, and pericycle. This observation made us to investigate the ontogeny of lateral root primordia in lotus and medicago. It was found that in both legumes, the cortical ground tissue also contributes substantially to this developmental process. Furthermore, we noticed that, to some extent, the spatial position of mitotically active cortical cells in lateral root primordia coincides with the spatial position of nodule primordia. This suggests that the potential to mitotically reactivate cortical cells is not an exclusive characteristic of Rhizobium spp. Nod factor-induced signaling but an intrinsic feature of these cells. However, the fact that root cortical cells are mitotically activated during lateral root development is not a legume-specific characteristic because it is reported for several nonlegume species as well (Tschermak-Woess and Dolezal, 1953; Mallory et al., 1970; McCully, 1975; Casero et al., 1996). The precise function of the dividing cortical cells during lateral root primordium formation remains unknown. Two possible functions can be hypothesized. Either the redifferentiated cortical cells become an integrative part of the primordium; alternatively, these divisions facilitate lateral root emergence through the cortex. The latter hypothesis is proposed for plants that have multiple cortex layers (Péret et al., 2009), including medicago and lotus, which have at least five cortical cell layers.
Besides MtRR9, other type-A RRs were shown to function in Rhizobium spp. symbiosis (Gonzalez-Rizzo et al., 2006; Vernié et al., 2008). Although their exact molecular functioning remains elusive, we can now position MtRR9 and MtRR11 in the Nod factor signaling network. We demonstrate that these genes are transcriptionally activated upon Rhizobium spp. Nod factor signaling. This Nod factor-induced expression is not dependent on the GRAS-type transcription factor complex MtNSP1-MtNSP2, whereas it requires MtCCaMK/MtDMI3, a nuclear localized and calcium-regulated kinase that functions upstream of the MtNSP1-MtNSP2 transcription factor complex (Kouchi et al., 2010). Furthermore, we found that Nod factor-induced MtRR9 and MtRR11 expression requires de novo protein synthesis, indicating that Nod factor-regulated gene products have a positive effect on the cytokinin signaling pathway. These could be newly synthesized enzymes involved in reallocation or metabolism of bioactive cytokinin. Such a presumed cytokinin signal is then likely perceived by the HK receptor MtCRE1, which has several type-A RRs among its downstream targets, including MtRR4, MtRR45, MtRR48, MtRR49, and MtRR411 (Plet et al., 2011).
The identification of a novel gene pair involved in the Rhizobium spp.-legume symbiosis by using a phylogenetic approach based on the papilionoid-specific WGD provides a proof of principle for the feasibility of this approach. Therefore, we propose that this phylogenetic strategy can be used on a genome-wide scale to identify new (candidate) genes involved in Rhizobium spp. symbiosis, even when such gene pairs share redundant functions, which hampers their identification by forward genetic screens.
MATERIALS AND METHODS
Vectors and Constructs
MtRR9 and PtRR5 full-length genomic sequence and MtRR9 RNAi target sequence were derived by PCR amplification using the primers listed in Supplemental Table S1. The genes were cloned into a pENTR-d-Topo vector (Invitrogen), creating pENTR1-2_MtRR9 and pENTR1-2_PtRR5. The CaMV35S promoter and terminator were cloned into a pENTR4-1 and pENTR2-3 (Invitrogen), thereby creating two modified pENTR clones: pENTR4-1_p35S and pENTR2-3_T35S. All three pENTR vectors were combined into the binary destination vector pKGW-RR-MGW by a multisite gateway reaction (Invitrogen). pKGW-RR-MGW contains pAtUBQ10::DsRED1 of pRedRoot as selection marker (Limpens et al., 2004). The MtRR9 RNAi target sequence was cloned into the DsRed modified gateway vector pK7GWIWG2(II) driven by the CaMV35S promoter as described by Limpens et al. (2005). 35S:MtCRE1*[L267F] was used as described by Ovchinnikova et al. (2011).
The putative promoter region of MtRR9 and MtRR11, approximately 2,500 bp upstream of the translational start site, was PCR amplified using primers listed in Supplemental Table S1. The putative promoters were cloned into a pENTR-d-Topo, thereby creating pENTR1-2_pMtRR9 and pENTR1-2_pMtRR11. Subsequently, each promoter was recombined into pKGWFS7-RR containing a GUS-GFP fusion reporter as well as pAtUBQ10::DsRed1 as a selectable marker (Karimi et al., 2002). All cloning vectors and constructs are available upon request from our laboratory or via the Functional Genomics unit of the Department of Plant Systems Biology (Vlaams Instituut voor Biotechnologie-Ghent University).
Plant Materials and Treatments
For the quantitative RT-PCR on type-A RR genes, medicago (Medicago truncatula) and lotus (Lotus japonicus) germinated seedlings were grown vertically on modified Fåhraeus medium agar plates with low nitrate [0.2 mm Ca(NO3)2] on top of filter paper for 48 h (Fahraeus, 1957). Then, water-dissolved Nod factors (approximately 10−9 m; Sinorhizobium NGR234 Nod factors for lotus and Sinorhizobium meliloti Nod factors for medicago) or water as a control were pipetted on top of every root (Hussain et al., 1998). Roots were exposed for 3 h; subsequently, 1-cm root pieces were cut just above the root tip and were snap-frozen (n = 15). For CHX experiments, plants were grown in modified Fåhraeus slides using modified liquid Fåhraeus medium (Heidstra et al., 1994) with low nitrate [0.2 mm Ca(NO3)2]. A single germinated seedling was placed in each slide, and medium was exchanged every 24 h. Experiments were done with plants grown for 48 h in Fåhraeus slides. Plants in the slides were treated either with BAP (10−8 m)-purified Nod factors (approximately 10−9 m), 50 μm CHX, 50 μm CHX + Nod factors (approximately 10−9 m), 50 μm CHX + 10−8 m BAP for 3 h, or Fåhraeus medium as a control. Subsequently, root pieces were snap-frozen as described above. For all experiments, plants were grown in an environmentally controlled growth chamber at 20°C with a 16-h-light/8-h-dark cycle and 70% relative humidity.
Quantitative RT-PCR
RNA was isolated from snap-frozen root samples using the plant RNA kit (E.Z.N.A, Omega Bio-Tek) as described in the manufacturer’s protocol. Complementary DNA was synthesized from 1 μg of total RNA using the iScript cDNA synthesis kit (Bio-Rad) as described in the manufacturer’s protocol. Quantitative RT-PCR was performed using SYBR Green-based detection (Eurogentec). Experimental setup and execution were conducted using a MyIQ optical cycler, according to protocol provided by the manufacturer (Bio-Rad). All primers, including the genes used for normalization (MtGAPDH, MtPTB, LjATPS, and LjUBQ) are given in Supplemental Table S1. As a control for the experimental set up of each Nod factor-induced sample, the induction of NIN in both medicago and lotus were checked and confirmed (data not shown). Data analysis was performed using Bio-Rad iQ5 software. Baselines were set at 100 relative fluorescence units to calculate the threshold cyclevalues. Threshold cycle values of 31 and higher were excluded from the analysis, although they were still checked for transcriptional induction (see Supplemental Table S1). A representative sample of three independent biological replicates is shown in all figures.
Plant Transformation and Nodulation Assay
Agrobacterium rhizogenes-mediated hairy roots transformation was used to transform medicago (Jemalong A17) as described in (Limpens et al., 2004), with the adaptation that 0.2 mm Ca2(NO3)2 was used in Fåhraeus medium instead. Transgenic roots were selected based on DsRED1 expression. Three weeks after transformation, transgenic roots from promoter studies and ectopic expression studies were transferred to low-nitrate Fåhraeus plates [0.2 mm Ca(NO3)2]. MtRR9 RNAi and empty vector control plants were investigated for lateral roots and primordia 10 d after transfer to Fåhraeus plates. After transformation, MtRR9 RNAi and empty vector control plants were grown and inoculated for 3 weeks in perlite as described by Limpens et al. (2004). pMtRR9::GUS- and pMtRR11::GUS-transformed plants were inoculated and grown in perlite in the same way. The differentiation zone (at approximately 0.7 cm above the tip) of pMtRR9::GUS and pMtRR11::GUS transgenic roots were exposed on Fåhraeus plates for 3 h to 2- to 3-mm-thin slices of Nod factor (10−9 m) or deionized water dissolved in low-melting point water-agarose, respectively. Afterward, these roots were fixed and sectioned as described by Limpens et al. (2005). Histochemical GUS staining was performed as described in Supplemental Protocols S1. pCaMV35S::MtRR9 and pCaMV35S::PtRR5 roots were investigated for lateral roots and primordia 10 d after transfer to Fåhraeus plates. Primordia were fixed and sectioned as described in Supplemental Protocols S1. All statistical tests were executed using SigmaStat software version 3.5 (SYSTAT Software).
Phylogeny
The phylogenetic trees were reconstructed using the maximum likelihood method implemented in the software PhyML version 3.0 (Guindon and Gascuel, 2003). More details are described in Supplemental Protocols S1.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JQ013379.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis of genes encoding components in the cytokinin phosphorelay pathway.
Supplemental Figure S2. Quantitative RT-PCR of medicago type-A RR genes upon Nod factor application.
Supplemental Figure S3. Number of emerged lateral roots per pCaMV35S::MtRR9 transgenic medicago root.
Supplemental Figure S4. Number of primordia per pCaMV35S::MtRR9 transgenic medicago root.
Supplemental Figure S5. Primordia on roots of medicago in between emerged lateral roots as a result of constitutive expression of MtRR9.
Supplemental Table S1. List of primers.
Supplemental Protocols S1. Supplemental protocols.
Supplemental File S1. File with sequences in Fasta Format “sequences.fas”.
Acknowledgments
We thank Eva Deinum for support with the statistical analyses and undergraduate students Hans van Kessel, Frank Leavis, and Ruben Higler for contributions to the project.
References
- Anisimova M, Gascuel O. (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55: 539–552 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Help H, Helariutta Y. (2009) Cytokinin signaling during root development. Int Rev Cell Mol Biol 276: 1–48 [DOI] [PubMed] [Google Scholar]
- Bryne JM, Pesacreta TC, Fox JA. (1977) Development and structure of the vascular connection between the primary and secondary root of Glycine max (L.) Merr. Am J Bot 64: 946–959 [Google Scholar]
- Cannon SB, Ilut D, Farmer AD, Maki SL, May GD, Singer SR, Doyle JJ. (2010) Polyploidy did not predate the evolution of nodulation in all legumes. PLoS ONE 5: e11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Schiex T, et al. (2006) Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci USA 103: 14959–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero PJ, Casimiro I, Lloret PG. (1996) Pericycle proliferation pattern during the lateral root initiation in adventitious roots of Allium cepa. Protoplasma 191: 136–147 [Google Scholar]
- Cooper JB, Long SR. (1994) Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-Zeatin secretion. Plant Cell 6: 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M, Frugier F. (2008) De novo organ formation from differentiated cells: root nodule organogenesis. Sci Signal 1: re11. [DOI] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus G. (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA 106: 5737–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier F, Kosuta S, Murray JD, Crespi M, Szczyglowski K. (2008) Cytokinin: secret agent of symbiosis. Trends Plant Sci 13: 115–120 [DOI] [PubMed] [Google Scholar]
- Gepts P, Beavis WD, Brummer EC, Shoemaker RC, Stalker HT, Weeden NF, Young ND. (2005) Legumes as a model plant family. Genomics for food and feed report of the Cross-Legume Advances Through Genomics Conference. Plant Physiol 137: 1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Sandal N, Bek AS, Madsen LH, Jurkiewicz A, Nielsen MW, Tirichine L, Stougaard J. (2011) Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol Plant Microbe Interact. 24: 1385–1395 [DOI] [PubMed] [Google Scholar]
- Heidstra R, Geurts R, Franssen H, Spaink HP, Van Kammen A, Bisseling T. (1994) Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol 105: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539 [DOI] [PubMed] [Google Scholar]
- Hussain AKM, Jiang Q, Broughton WJ, Gresshoff PM. (1998) Lotus japonicus nodulates and fixes nitrogen with the broad host range Rhizobium sp. NGR234. Plant Cell Physiol 40: 894–899 [Google Scholar]
- Hwang I, Sheen J. (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Ishida K, Niwa Y, Yamashino T, Mizuno T. (2009) A genome-wide compilation of the two-component systems in Lotus japonicus. DNA Res 16: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kiba T, Aoki K, Sakakibara H, Mizuno T. (2004) Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol 45: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T. (2003) The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44: 868–874 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1969) The rate of molecular evolution considered from the standpoint of population genetics. Proc Natl Acad Sci USA 63: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al. (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R. (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM. (2004) Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J 38: 203–214 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory TE, Chiang SH, Cutter EG, Gifford EM., Jr (1970) Sequence and pattern of lateral root formation in 5 selected species. Am J Bot 57: 800–809 [Google Scholar]
- Mathesius U, Charon C, Rolfe BG, Kondorosi A, Crespi M. (2000a) Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol Plant Microbe Interact 13: 617–628 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Weinman JJ, Rolfe BG, Djordjevic MA. (2000b) Rhizobia can induce nodules in white clover by “hijacking” mature cortical cells activated during lateral root development. Mol Plant Microbe Interact 13: 170–182 [DOI] [PubMed] [Google Scholar]
- McCully ME. (1975) The development of lateral roots. Torrey JG Clarkson DT, , The Development and Function of Roots. Academic Press, London, pp 105–124 [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR. (2004a) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Shaw SL, Long SR. (2004b) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2007) Advances in cytokinin signaling. Science 318: 68–69 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC. (2008) Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 179: 595–614 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Miyata S, Urao T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2002) Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem Biophys Res Commun 293: 806–815 [DOI] [PubMed] [Google Scholar]
- Ovchinnikova E, Journet EP, Chabaud M, Cosson V, Ratet P, Duc G, Fedorova E, Liu W, den Camp RO, Zhukov V, et al. (2011) IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol Plant Microbe Interact 24: 1333-1344 [DOI] [PubMed] [Google Scholar]
- Péret B, Larrieu A, Bennett MJ. (2009) Lateral root emergence: a difficult birth. J Exp Bot 60: 3637–3643 [DOI] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F. (2011) MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J 65: 622–633 [DOI] [PubMed] [Google Scholar]
- Ramírez-Carvajal GA, Morse AM, Davis JM. (2008) Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol 177: 77–89 [DOI] [PubMed] [Google Scholar]
- Ren B, Liang Y, Deng Y, Chen Q, Zhang J, Yang X, Zuo J. (2009) Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res 19: 1178–1190 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shoemaker RC, Schlueter J, Doyle JJ. (2006) Paleopolyploidy and gene duplication in soybean and other legumes. Curr Opin Plant Biol 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo FB, de Bruijn FJ. (1998) Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact 11: 684–697 [Google Scholar]
- Timmers AC, Auriac MC, Truchet G. (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Kieber JJ. (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Tschermak-Woess E, Dolezal R. (1953) Durch Seitenwurzelbildung induzierte und spontane Mitosen in den Dauergeweben der Wurzel. Oesterr. Bot. Z. 100: 358–402 [Google Scholar]
- van Spronsen PC, Grønlund M, Pacios Bras C, Spaink HP, Kijne JW. (2001) Cell biological changes of outer cortical root cells in early determinate nodulation. Mol Plant Microbe Interact 14: 839–847 [DOI] [PubMed] [Google Scholar]
- Vernié T, Moreau S, de Billy F, Plet J, Combier JP, Rogers C, Oldroyd G, Frugier F, Niebel A, Gamas P. (2008) EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20: 2696–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]