Abstract
DNA-binding protein phosphatases (DBPs) have been identified as a novel class of plant-specific regulatory factors playing a role in plant-virus interactions. NtDBP1 from tobacco (Nicotiana tabacum) was shown to participate in transcriptional regulation of gene expression in response to virus infection in compatible interactions, and AtDBP1, its closest relative in the model plant Arabidopsis (Arabidopsis thaliana), has recently been found to mediate susceptibility to potyvirus, one of the most speciose taxa of plant viruses. Here, we report on the identification of a novel family of highly conserved small polypeptides that interact with DBP1 proteins both in tobacco and Arabidopsis, which we have designated DBP-interacting protein 2 (DIP2). The interaction of AtDIP2 with AtDBP1 was demonstrated in vivo by bimolecular fluorescence complementation, and AtDIP2 was shown to functionally interfere with AtDBP1 in yeast. Furthermore, reducing AtDIP2 gene expression leads to increased susceptibility to the potyvirus Plum pox virus and to a lesser extent also to Turnip mosaic virus, whereas overexpression results in enhanced resistance. Therefore, we describe a novel family of conserved small polypeptides in plants and identify AtDIP2 as a novel host factor contributing to resistance to potyvirus in Arabidopsis.
DNA-binding protein phosphatases (DBPs) are a unique family of plant-specific protein phosphatases of the 2C class that are capable of binding DNA (Carrasco et al., 2003). These two activities lie in separate structural domains. Thus, the N-terminal region is responsible for DNA binding, whereas protein phosphatase activity resides in the C-terminal domain (Carrasco et al., 2003, 2005). DBP factors have only been found in plants and are present throughout the plant kingdom, where they seem to have undergone functional diversification (Carrasco et al., 2005).
Tobacco (Nicotiana tabacum) DBP1 (NtDBP1), the first member of this family to be identified, was shown to participate in the transcriptional regulation of a defense-related gene in the context of compatible plant-virus interactions (Carrasco et al., 2003). More recently, the closest DBP1 relative in the model species Arabidopsis (Arabidopsis thaliana; AtDBP1) has been found to contribute to susceptibility to Plum pox virus (PPV; Castelló et al., 2010).
Potyviruses are single-stranded positive sense RNA viruses that represent one of the most speciose taxa of plant viruses and infect a broad range of plant species, including most crop plants, causing severe losses in agriculture worldwide (Gibbs and Ohshima, 2010). The potyviral genome possesses a 3′ poly(A) tail but lacks a cap structure at the 5′ end, which is instead covalently bound to a virus-encoded protein termed VPg. Resistance against potyviruses is frequently recessive, pointing to host factors that are necessary for completion of the virus infective cycle. Among these proteins, translation initiation factors eIF4E and eIF(iso)4E are key to successful viral infection and have been repeatedly found to be associated with both natural and induced resistance to potyviruses (Díaz-Pendón et al., 2004; Robaglia and Caranta, 2006; Maule et al., 2007; Trüniger and Aranda, 2009; Hébrard et al., 2010; Le Gall et al., 2011; Nieto et al., 2011). However, additional host components might be participating in the interaction with potyviruses, either promoting infection as susceptibility factors or mediating resistance. Only recently, some of these factors have been discovered, like a Cys-rich VPg-interacting protein of unknown function involved in potyvirus movement (Dunoyer et al., 2004), a DEAD box RNA helicase also interacting with VPg (Huang et al., 2010), heat shock protein HSP70 (Hafrén et al., 2010; Jungkunz et al., 2011), and proteasome components (Jin et al., 2007; Dielen et al., 2011), although the roles they play and the underlying mechanisms remain obscure in most cases. Another example is AtDBP1, whose function aids potyviruses like PPV and Turnip mosaic virus (TuMV) to successfully infect the plant, likely through its interaction with eIF(iso)4E (Castelló et al., 2010).
AtDBP1 involvement in potyvirus infection suggests that factors regulating AtDBP1 might also have an effect in plant-potyvirus interactions. Here, we report on the identification of NtDIP2 (for DBP1-interacting protein 2), an NtDBP1 interactor that belongs to a novel family of conserved plant small polypeptides. AtDIP2, the Arabidopsis ortholog, interacts in vivo and functionally interferes with AtDBP1. Changes in AtDIP2 gene expression result in significant differences in the accumulation of the potyvirus PPV and to a lesser extent of TuMV, another potyvirus that is more virulent to Arabidopsis and causes more severe symptoms in the plant. Thus, an AtDIP2-deficient mutant shows increased susceptibility to PPV and TuMV, whereas AtDIP2 overexpression leads to lower viral accumulation. Therefore, AtDIP2 seems to represent a novel class of host factors enhancing tolerance to potyvirus infection.
RESULTS AND DISCUSSION
Identification of NtDIP2
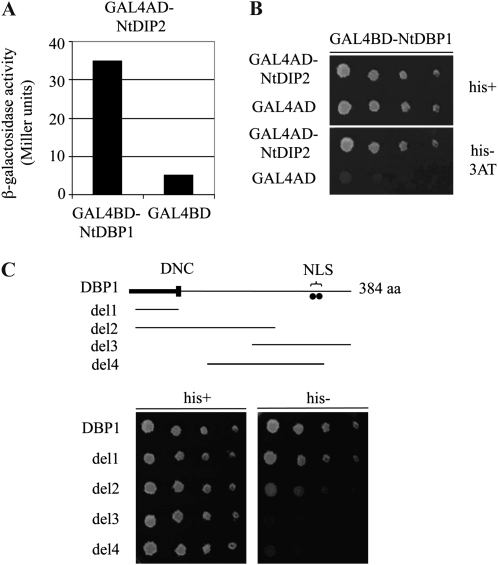
NtDBP1 was the first member of the DBP family to be isolated. With the aim of identifying proteins interacting with tobacco DBP1 that could provide an insight into the function and regulation of this novel class of proteins, a two-hybrid screen was accomplished using full-length NtDBP1 as a bait (Carrasco et al., 2006). In this screen, besides 14-3-3G, which was found to mediate nucleocytoplasmic shuttling of NtDBP1 (Carrasco et al., 2006), a second protein was identified that specifically activated expression of the LacZ (Fig. 1A) and HIS3 (Fig. 1B) reporter genes. We named this protein DIP2. Deletion analysis showed that the region of NtDBP1 engaged in the interaction with NtDIP2 was the N-terminal domain (Fig. 1C), as only deletion constructs bearing this domain (Carrasco et al., 2006) were able to mediate the interaction. The fact that del2 protein renders weaker reporter gene activation than del1, while both constitute the N-terminal domain, may be due to lower stability and/or structural constraints derived from improper folding or reduced accessibility of the referred domain in the case of del2 protein. In support of this explanation, the interaction with 14-3-3G, also mediated by the DBP1 N-terminal domain, was previously shown to be again partially compromised in del2 (Carrasco et al., 2006), suggesting that this effect is intrinsic to the del2 construct regardless of the interacting partner involved. These two interactions point to the N-terminal domain of DBP factors as an important regulatory domain that provides docking sites for proteins modulating DBP function. Moreover, this region was previously shown to be responsible for binding DNA (Carrasco et al., 2005). Thus, the possibility arises that some interference may exist between the described interactions and that NtDIP2 may not be able to interact with NtDBP1 when bound to DNA or to 14-3-3G. Alternatively, NtDIP2 might displace DNA or 14-3-3G from their binding sites in NtDBP1. To ascertain whether this is the case will require further investigation.
Figure 1.
Interaction of NtDBP1 and NtDIP2 in the yeast two-hybrid system. A, Activation of lacZ reporter gene expression. β-Galactosidase activity was measured from yeast liquid cultures coexpressing NtDBP1 (bait) and NtDIP2 (prey) fused to the GAL4 DNA-binding (BD) and activation (AD) domains, respectively. The empty bait vector was used as a control in conjunction with the NtDIP2-GAL4AD fusion. B, Activation of HIS3 reporter gene expression. Serial dilutions of yeast liquid cultures expressing the referred prey constructs along with the bait GAL4BD-NtDBP1 fusion were plated on medium containing His (his+) and medium lacking His (his−) supplemented with 5 mm 3-amino-triazole (3AT), a competitive inhibitor of the HIS3 gene product, used to eliminate leaky expression of the reporter gene. C, Identification of the domain in NtDBP1 responsible for the interaction with AtDIP2. Different bait constructs comprising different deletions of the NtDBP1 protein were analyzed in the yeast two-hybrid system for the activation of HIS3 reporter gene expression when coexpressed with the GAL4AD-NtDIP2 fusion protein. Yeast growth in the absence of His was observed only in the presence of the N-terminal region of NtDBP1 in the bait construct. aa, Amino acids; DNC, DBP N-terminal core, a characteristic motif of DBP factors involved in DNA binding; NLS, putative bipartite nuclear localization signal.
Sequence analysis revealed that the cDNA borne by the rescued library plasmid encodes a 52-amino acid polypeptide with a predicted molecular mass of 5.9 kD. Recent works point out the relevance of small peptides in plant biology (Fukuda and Higashiyama, 2011). Peptides have been shown to play important roles in cell-cell communication and signaling processes (Farrokhi et al., 2008). Although many of these peptides undergo proteolytic processing and are secreted to the extracellular space, there also exist intracellular peptides that do not show apparent secretion or processing, like peptides of the ROT4 family (Ikeuchi et al., 2011). NtDIP2 represents a novel class of intracellular peptides, since, according to the SignalP program (Bendtsen et al., 2004), it lacks any signal sequence. The deduced amino acid sequence shows no significant homology to proteins of known function or identified functional domains. As in the case of DBP1, counterparts were only found in plant species and appear to be encoded by single-copy genes. Deduced DIP2 polypeptide sequences exhibit a remarkable degree of similarity that likely denotes functional conservation (Fig. 2), suggesting that they might have developed to meet specific regulatory needs derived from the evolution of DBP function.
Figure 2.
Sequence homology of DIP2 proteins. Alignment of deduced amino acid sequences of plant DIP2 proteins. Black background denotes identical residues, and gray background denotes conservative changes. Numbers on the right indicate size in amino acids. Nt, Nicotiana tabacum; Nb, Nicotiana benthamiana; St, Solanum tuberosum; Ca, Capsicum annuum; Gm, Glycine max; Mt, Medicago truncatula; At, Arabidopsis thaliana; Bn, Brassica napus; Cr, Catharanthus roseus; Pa, Petunia axillaris; Vv, Vitis vinifera; Md, Malus domestica; Cm, Cucumis melo; Cc, Citrus clementina; Gh, Gossypium hirsutum; Pt, Populus tremuloides. The bar at the bottom indicates the region of highest similarity among DIP2 proteins.
AtDBP1 Also Interacts with the DIP2 Ortholog in Arabidopsis
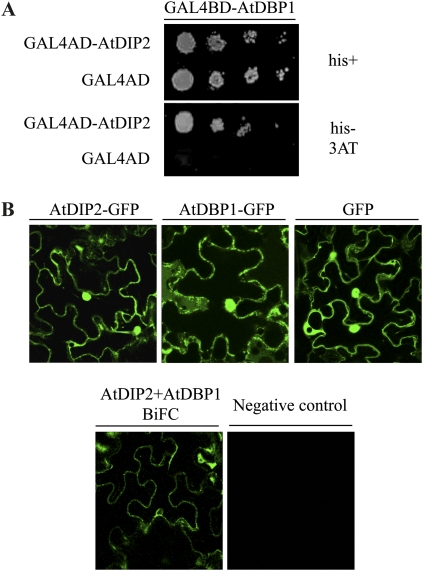
AtDBP1 is the closest relative to NtDBP1 in the plant model system Arabidopsis (Carrasco et al., 2005). As shown in Figure 2, we also found a putative DIP2 ortholog in Arabidopsis. Therefore, we sought to test whether Arabidopsis DIP2 protein was capable of interacting with AtDBP1. Using the two-hybrid system, we could indeed show that this is the case (Fig. 3A), further supporting an evolutionarily conserved relationship between DBP factors and DIP2 polypeptides.
Figure 3.
In vivo interaction of AtDBP1 and AtDIP2. A, AtDBP1 interacts with AtDIP2 in the yeast two-hybrid system. Serial dilutions of yeast liquid cultures expressing the indicated constructs were plated on medium containing His (his+) and medium lacking His (his−) supplemented with 5 mm 3-amino-triazole (3AT). B, Top panels, confocal microscopy images showing the subcellular localization of AtDIP2 and AtDBP1 when fused to GFP and transiently expressed in leaves of N. benthamiana. GFP alone was included as a control. Bottom panels, BiFC assay. AtDBP1 and AtDIP2 were fused to N-terminal and C-terminal fragments of YFP and transiently expressed in N. benthamiana leaves by agroinfiltration. Left panel, confocal microscopy of leaves expressing YFPNt-AtDIP2 and YFPCt-AtDBP1; right panel, negative control consisting of AtDBP1 separately fused to both fragments. [See online article for color version of this figure.]
Effect of DIP2 on DBP1 Protein Phosphatase Activity
In the absence of any functional indication on the role of DIP2 polypeptides, we considered the possibility that they could act as modulators of DBP protein phosphatase activity. To verify this hypothesis, we sought to test whether DIP2 interferes with DBP1 protein phosphatase activity. For that purpose, NtDBP1 was expressed in Escherichia coli translationally fused to the maltose-binding protein and subsequently purified by amylose-affinity chromatography. Similarly, hexa-His-tagged NtDIP2 (NtDBP1-His6) was also expressed in E. coli and purified. As shown in Supplemental Figure S1A, no effect was observed in the activity of NtDBP1 at stoichiometric NtDBP1:NtDIP2 ratios between 1:1 and 1:5 (see Supplemental Materials and Methods S1). However, we cannot rule out that tagging NtDIP2 could be interfering with a productive interaction. We also tested the protein phosphatase activity of similarly expressed AtDBP1 in the presence of different concentrations of a synthetic polypeptide encompassing the highly conserved central core region of DIP2 proteins (DIP2c; Fig. 2). Again, no significant effect was observed when DIP2c was added at the same stoichiometric ratios used above (Supplemental Fig. S1B). Therefore, these results seem to indicate that DIP2 does not modify DBP1 catalytic activity. However, as already suggested, tagging DIP2 might prevent an appropriate interaction, and on the other hand, the synthetic polypeptide used, DIP2c, consisting of an incomplete DIP2 protein, may not be active or stable. DIP2 could also be interfering with the binding of DBP1 to a particular substrate or positive modulator, thereby negatively regulating activity. Since no clear evidence supports that under the conditions tested both proteins are interacting properly, no definitive conclusion can be drawn in this respect. As Arabidopsis is a more amenable system, we continued to work on the AtDIP2-AtDBP1 interaction.
AtDIP2 and AtDBP1 Interact in Vivo
To validate in planta the interaction between AtDBP1 and AtDIP2, we used bimolecular fluorescence complementation (BiFC). This approach relies on the reconstitution of a fluorescent complex by two nonfluorescent fragments of the yellow fluorescent protein (YFP) brought together by the interaction between two proteins that are expressed as fusions to those fragments (Schütze et al., 2009). A prerequisite for two proteins to interact is that they must colocalize within the cell. We had previously shown that tobacco DBP1 accumulates both in the nucleus and the cytosol (Carrasco et al., 2006). A similar analysis of AtDBP1 and AtDIP2 subcellular localization as fused to the GFP after transient expression in Nicotiana benthamiana leaves revealed that both proteins are also distributed between the nucleus and the cytosol (Fig. 3B, top panels). GFP alone was used as a control and showed a similar localization pattern. The nuclear pore complex allows passive diffusion of small proteins such as GFP (Hicks, 2005). Due to the small size of AtDIP2, estimated to be around 7 kD, the AtDIP2-GFP fusion could also be capable of freely shuttling between the nucleus and the cytosol. However, that does not preclude a possible active mechanism of AtDIP2 nuclear import, as long known for other small proteins such as histones (Breeuwer and Goldfarb, 1990). An active import enables transport against a concentration gradient and is invoked in those cases as more efficient and more amenable to regulation than diffusion (Sekimoto et al., 2005). On the other hand, AtDBP1 also exhibits a nucleocytoplasmic distribution resembling that previously described for tobacco DBP1 (Carrasco et al., 2006).
To test their interaction in planta, AtDBP1 and AtDIP2 were fused to the N-terminal and C-terminal fragments of YFP in both combinations and again transiently expressed in leaves of N. benthamiana. Results are shown in Figure 3B (bottom panels) for the YFPNt-AtDIP2 and YFPCt-AtDBP1 fusions, but they were similar for the reciprocal combination. Fluorescence was clearly detected both in the nucleus and the cytosol, mirroring protein localization and indicating that AtDBP1 and AtDIP2 interact in planta. In contrast, a negative control consisting of fusions of both YFP fragments to AtDBP1 gave no significant signal. A closer examination of the fluorescence detected in the BiFC assay reveals that when compared with the single proteins fused to GFP (Fig. 3B, top panels), the signal seems to decrease in the nucleus relative to the cytosol, suggesting that the interaction may take place preferentially in the cytosol even though both proteins are also found in the nucleus. When present in the nucleus, at least a fraction of AtDBP1 will be presumably bound to DNA, and that, as discussed above, might affect the interaction with AtDIP2. This might explain the relatively lower incidence of the interaction in the nucleus compared with the cytosol.
Fitness-Based Interferential Genetics Reveals a Functional Interaction between AtDIP2 and AtDBP1
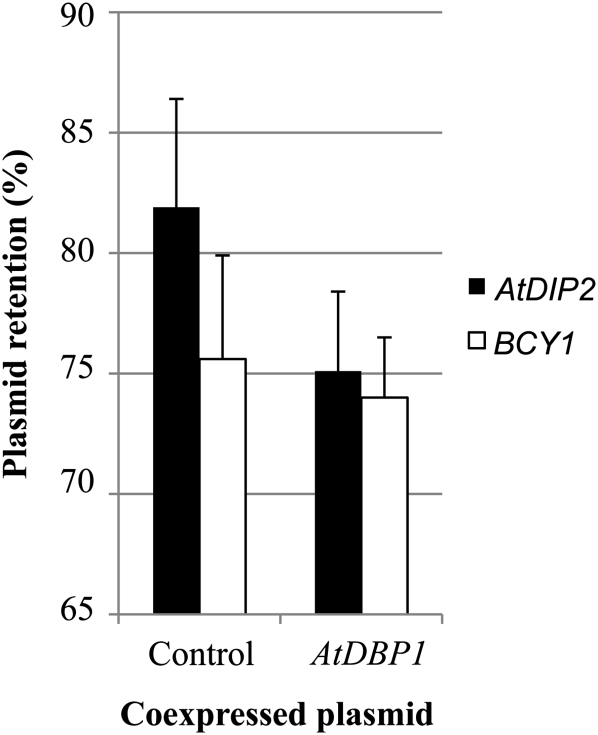
The described physical interaction between DIP2 and DBP1 proteins suggests a role of DIP2 as a modifier of DBP1 function. To test this hypothesis and reveal a possible impact of this interaction on AtDBP1 function, we used fitness-based interferential genetics (FIG), a verified approach to show a functional link between proteins (Daniel, 1996b, 2008). FIG is based on the finding that overexpression of virtually any gene in yeast compromises cell fitness (Daniel, 1996a). By using high gene dosages obtained by multicopy plasmids, FIG allows the demonstration of an in vivo functional interaction between two proteins by monitoring the rate of plasmid loss in the absence of selective pressure for one of the proteins, typically the interfering protein, while retaining high-level expression of the second protein, usually that exhibiting the target activity. Following this approach, we expressed AtDIP2 and AtDBP1 in yeast strain C90-A, as described in “Materials and Methods,” using the empty AtDBP1-expressing plasmid as a control. If not imposing the relevant selection, a plasmid will be lost over generations. The rate at which this will happen will be proportional to the magnitude of the deleterious effects the plasmid may have for the cell. When, in the absence of selection, retention of the plasmid driving the expression of AtDIP2 was estimated according to Daniel (2008), a significant deviation was observed between cells simultaneously expressing AtDBP1 and those bearing the relevant empty vector (Fig. 4, black bars). The plasmid was lost at a higher rate (i.e. the percentage of yeast cells retaining the plasmid was lower) when coexpressing AtDBP1 (Fig. 4, AtDBP1 column) than when using the corresponding empty plasmid (Fig. 4, control column). That this effect was specific to the AtDIP2/AtDBP1 match was shown by replacing AtDIP2 by an unrelated yeast gene, BCY1 (encoding the regulatory subunit of protein kinase A). Under these conditions, a much smaller change was found between the BCY1/AtDBP1 match and the BCY1/vector control (Fig. 4, white bars). Also, no change was observed in a vector/AtDBP1 match as compared with the vector/vector control (data not shown). Thus, these results indicate a functional cross talk between AtDBP1 and AtDIP2 proteins that increases the negative impact of AtDIP2 expression on yeast cell physiology and accelerates plasmid loss. To our knowledge, this is the first time that the FIG approach has been applied to plants, but the difference with respect to the control empty plasmid (i.e. 6.8 ± 3.3) is within the range found in other FIG assays reported for yeast proteins (Daniel, 2007). Thus, beyond the physical interaction shown in the two-hybrid assay, this result unveils a functional interplay between AtDBP1 and AtDIP2 and suggests a role for AtDIP2 in modulating AtDBP1 biological activity. However, from this FIG assay using a doubly heterologous gene system, no definitive conclusion can be drawn regarding the direction of this effect in plants, whether it is stimulating or inhibitory, since the nature of the particular toxicity in yeast of each protein is indeterminate.
Figure 4.
Functional interference between AtDIP2 or an unrelated yeast gene, BCY1, and AtDBP1. Data shown are percentages of colonies retaining the plasmid driving the expression of AtDIP2 (black bars) or BCY1 (white bars) in the two-streak test as described in “Materials and Methods.” The plasmid bears the ADE2 marker, so cells that lose the plasmid (and thus their resulting colonies) appear red due to the accumulation of derivatives of the substrate of the ADE2 enzyme, whereas cells harboring the plasmid and therefore endowed with ADE2 activity appear white. The proportion of white colonies in the population when coexpressed with AtDBP1 or with the corresponding empty vector is shown as the mean ± sd of 12 experiments in the case of AtDIP2 and at least four experiments for BCY1.
Modifying AtDIP2 Gene Expression Alters Susceptibility to Potyvirus
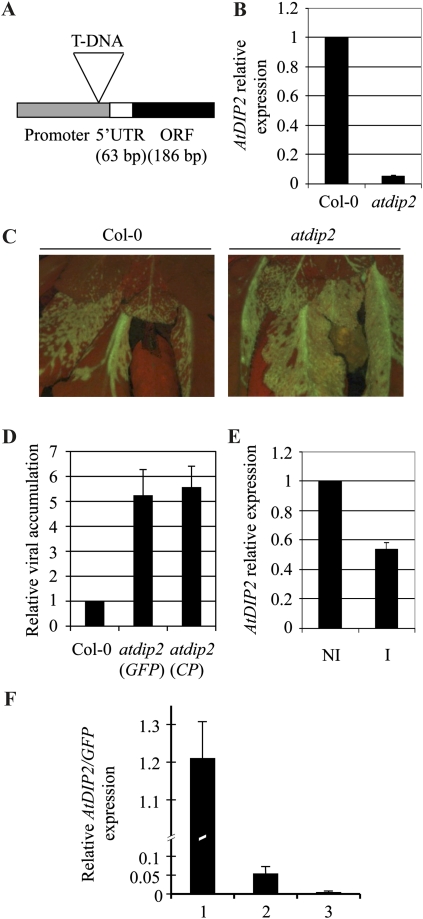
As a newly described family, DIP2 polypeptides are orphan of function. We have recently reported the involvement of AtDBP1 in plant-potyvirus interactions (Castelló et al., 2010). Down-regulating AtDBP1 gene expression leads to enhanced resistance to potyviruses like PPV and TuMV. Since AtDIP2 seems to modify AtDBP1 function, it might also play a role in potyvirus infection. To test this hypothesis, we selected homozygous plants for a T-DNA insertion in the AtDIP2 gene from the SALK collection (SALK_131929; Alonso et al., 2003). The dip2 mutant turned out to be a knockdown mutant, since, although the insertion significantly reduces transcript accumulation, as determined by reverse transcription-quantitative PCR (RT-qPCR), it does not abolish expression of the gene (Fig. 5B). This is likely due to a position effect, since, as depicted in Figure 5A, the T-DNA was inserted in the proximal promoter of the gene. Down-regulation of AtDIP2 does not lead to any apparent alteration in morphology and development, and mutant plants are phenotypically indistinguishable from wild-type plants.
Figure 5.
Molecular and phenotypic characterization of the atdip2 mutant. A, Diagram showing the position of the T-DNA insertion in the AtDIP2 gene relative to the promoter of the gene and the transcribed region in the SALK_131929 line. ORF, Open reading frame; UTR, untranslated region. B, RT-qPCR with primers specific for the AtDIP2 gene. RNA was extracted from rosette leaves of 4-week-old plants. Expression data were normalized using ACT2/8 and referred to Col-0. The average ± sd of three experiments is shown. C, Fluorescence microscopy image of Col-0 (left panel) and atdip2 (right panel) plants infected with a GFP-tagged PPV infectious clone at 21 d post inoculation (dpi). D, Accumulation of viral RNA in the atdip2 mutant relative to Col-0 plants as measured by RT-qPCR with primers specific both for the GFP gene and the viral CP gene. RNA was extracted from the rosettes of infected plants at 21 dpi. Data were normalized using ACT2/8 and referred to Col-0. The average ± sd of three experiments is shown. E, AtDIP2 gene expression in PPV-infected Col-0 plants at 21 dpi analyzed by RT-qPCR. Data were normalized using ACT2/8 and referred to the expression level in noninoculated plants. NI, Noninoculated plants; I, inoculated plants at 21 dpi. The average ± sd of three experiments is shown. F, AtDIP2 gene expression referred to the accumulation of viral RNA during the PPV infection process. Expression was measured by RT-qPCR, normalized using ACT2/8 expression, and referred to the similarly normalized data of viral GFP RNA. RNA was extracted from infected leaf material showing low (bar 1), medium (bar 2), and high (bar 3) PPV accumulation, as detected by fluorescence microscopy. [See online article for color version of this figure.]
The observed 20-fold reduction in the AtDIP2 transcript level prompted us to analyze the performance of the dip2 mutant upon potyvirus infection. Mutant and ecotype Columbia (Col-0) wild-type plants were inoculated with a GFP-tagged PPV infectious clone as described in “Materials and Methods.” The progress of infection was monitored by detection of GFP by fluorescence microscopy at different time points. As shown in Figure 5C, 3 weeks after PPV inoculation, dip2 plants consistently exhibited more extensive infection and higher viral accumulation in infected tissue. For a more precise comparison between the two genotypes, viral RNA accumulation was measured by RT-qPCR using primers specific for the GFP gene and found to be about 5-fold higher in dip2 plants as compared with wild-type plants (Fig. 5D). Similar results were obtained when the qPCR amplification was carried out using primers targeting the viral coat protein (CP) gene to exclude the possibility that the virus might get rid of the foreign GFP sequence and still replicate and spread in the plant. Therefore, reducing AtDIP2 expression leads to enhanced susceptibility. This inverse relationship between AtDIP2 gene expression and susceptibility to PPV correlates with the decrease in AtDIP2 transcript accumulation that occurred in infected wild-type Col-0 plants in comparison with uninfected plants (Fig. 5E), using the housekeeping ACT2/8 gene as a reference gene. Since the whole rosette was used for RNA extraction, this repression might actually be higher in the directly infected tissue. To establish a more precise relationship between AtDIP2 gene expression and the progression of viral infection, we analyzed the accumulation of AtDIP2 mRNA relative to the viral GFP transcript, here used as a measure of the degree of infection. For that purpose, infected leaves were divided into three categories according to the amount of fluorescence detected with the microscope. Thus, leaf material showing low, medium, and high viral accumulation was harvested and analyzed for AtDIP2 and GFP gene expression by RT-qPCR relative to the ACT2/8 reference gene. Then, the ratio between normalized AtDIP2 and viral GFP gene expression was determined. As depicted in Figure 5F, AtDIP2 gene expression sharply declines as viral RNA accumulates, indicating a repression of AtDIP2 expression during the course of PPV infection.
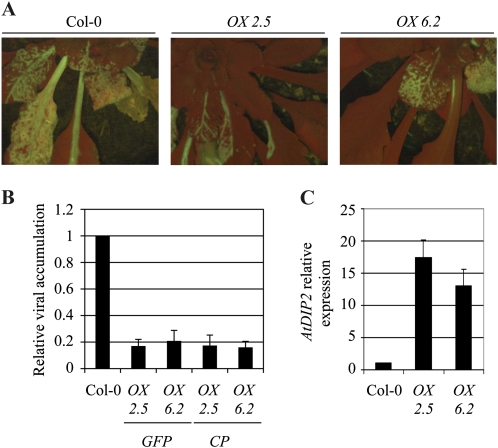
To complement this analysis, transgenic Arabidopsis plants were generated where AtDIP2 expression was driven by the cauliflower mosaic virus 35S promoter, aimed at generating plants with a constitutively high expression of the AtDIP2 gene. Two independent, homozygous, single-insertion lines, namely 2.5 and 6.2, were analyzed in detail. AtDIP2 expression in these lines was found to exceed that in Col-0 plants by about 17- and 11-fold, respectively, as determined by RT-qPCR (Fig. 6C). As previously observed for the knockdown mutant plants, overexpression does not cause any visible change in plant architecture and growth. However, when inoculated with PPV, plants of these lines restricted viral progression with higher efficiency than wild-type plants (Fig. 6A). Determining PPV accumulation by RT-qPCR corroborated this observation. Plants of both overexpressing lines exhibited a significant reduction in both GFP and CP RNA accumulating in leaves of infected plants 3 weeks postinoculation (Fig. 6B), with an average of 5- to 6-fold less viral RNA in plants of lines 2.5 and 6.2 as compared with Col-0 wild-type plants. Therefore, while down-regulating AtDIP2 expression leads to higher PPV accumulation, increased expression confers enhanced resistance to infection, thereby demonstrating a role of AtDIP2 in the interaction.
Figure 6.
Analysis of PPV infection in AtDIP2-overexpressing plants. A, Fluorescence microscopy image of representative plants of Col-0 (left panel) and of two independent AtDIP2-overexpressing lines (middle and right panels), infected with a GFP-tagged PPV infectious clone, at 21 d post inoculation (dpi). B, RT-qPCR with primers specific for the GFP and CP genes. RNA was extracted from the rosettes of infected plants 21 dpi. Data were normalized using ACT2/8 and referred to Col-0. The average ± sd of three experiments is shown. C, AtDIP2 gene expression in the overexpressing plants analyzed by RT-qPCR. Data were normalized using ACT2/8 and referred to Col-0. The average ± sd of three experiments is shown. [See online article for color version of this figure.]
A similar trend was observed when plants with altered AtDIP2 expression were inoculated with another potyvirus, TuMV (Supplemental Fig. S3). atdip2 mutant plants showed higher viral accumulation than Col-0 wild-type plants, whereas AtDIP2 overexpression resulted in increased resistance. Only quantitative differences were found as compared with PPV, which might be due to the different virulence exhibited by these two potyviruses.
DIP2-Mediated Enhanced Potyvirus Resistance Is Independent of eIF(iso)4E
Loss of AtDBP1 function is accompanied by a decrease in eIF(iso)4E protein accumulation likely due to a higher degradation rate by the proteasome in the absence of AtDBP1 (Castelló et al., 2010). Lack of a specific antiserum precluded a similar analysis of AtDIP2 protein level in the atdbp1 mutant background. Thus, we cannot rule out the possibility that AtDBP1, in contrast to the stabilizing effect observed on eIF(iso)4E protein, could instead promote AtDIP2 degradation. As mentioned above, the translation initiation factors eIF4E and its plant-specific isoform eIF(iso)4E are key factors in potyvirus resistance. Null eIF(iso)4E mutants are resistant to PPV, TuMV, and Lettuce mosaic virus, whereas mutations in eIF4E lead to resistance to Clover yellow vein virus (Duprat et al., 2002; Lellis et al., 2002; Sato et al., 2005; Decroocq et al., 2006). Both eIF(iso)4E and eIF4E interact with the viral protein VPg, and the ability of these proteins to interact correlates with virus infectivity (Léonard et al., 2000; Kang et al., 2005). Furthermore, Charron et al. (2008) showed evidence for coevolution between eIF4E and VPg. Therefore, the reduced eIF(iso)4E protein level has been suggested to account for the enhanced potyvirus resistance of the atdbp1 mutant (Castelló et al., 2010). For that reason, we examined by western blot the accumulation of eIF(iso)4E in DIP2 knockdown and overexpressing plants. As shown in Supplemental Figure S2, no significant change in the amount of eIF(iso)4E protein was observed in any of the mutants as compared with wild-type Col-0 plants. Without disregarding the crucial and determinant role of eIF4E and eIF(iso)4E factors in potyvirus susceptibility, this result suggests that besides the recently reported modulation of eIF(iso)4E stability, AtDBP1 would be involved in additional eIF(iso)4E-independent pathways contributing to potyvirus susceptibility, which would be negatively modulated by DIP2.
The results obtained with knockdown and overexpressing plants suggest a role for AtDIP2 in the Arabidopsis response to PPV infection, establishing an inverse relationship between AtDIP2 gene expression and PPV susceptibility, which would also be consistent with the reduction in expression that we observed in susceptible wild-type plants upon infection. This does not necessarily mean that AtDIP2 is per se directly involved in halting virus progression as part of the defense mechanisms of the plant against PPV. Alternatively, AtDIP2 might be directly or indirectly hindering virus access to host functions and/or components important for completion of the viral cycle. Because of their limited genetic complement, viruses need to recruit numerous host factors to be able to replicate and move inside the infected plant. This is exemplified by genome-wide screens performed in yeast, in which over 100 genes with diverse functions and involved in a variety of cellular processes turned out to have an effect on the replication and recombination of Tomato bushy stunt virus (TBSV; Panavas et al., 2005; Serviene et al., 2005, 2006). TBSV belongs to the Tombusviridae family, whose members, like potyviruses, contain linear single-stranded (+)RNA genomes. These results suggest that the host-virus interaction is very complex, with viral replication becoming affected by many factors and pathways inside the host cell. Their identification could pave the way to new strategies for improving plant resistance with a minor impact on plant development and physiology.
Searching for modulators of AtDBP1 function, which was recently described as a novel factor playing a role in potyvirus infections, has allowed us to identify AtDIP2 as a novel component of plant resistance against PPV. This is a notable result, since it assigns a biological context to a novel family of small polypeptides of unknown function and also identifies a new player in plant-potyvirus interactions. Although we could not show evidence for a direct effect of DIP2 on DBP1 protein phosphatase activity, taken together, our results suggest that AtDIP2 functionally interferes with AtDBP1 during PPV infection and quantitatively contributes to resistance. Besides the unquestionable key role of eIF4E and eIF(iso)4E, which are essential factors in potyvirus infection, this supports the notion that resistance to potyvirus is a multifaceted response resulting from a complex balance between the attempt of the virus to use plant components to its own benefit and the defense mechanisms of the plant, with multiple host factors being involved in such an interaction. In accordance with this idea, symptoms induced by PPV infection in Arabidopsis show high variability among different accessions and also within specific susceptible ecotypes such as Col-0 (Decroocq et al., 2006; Sicard et al., 2008). The differences in expression levels and/or in the regulation of protein functions involved may account, at least partly, for this variability in a quantitative manner. Our results open the way to the identification of additional host factors engaged in the plant-potyvirus interaction by searching for modulators and/or targets of AtDBP1 function as well as the development of antiviral drugs targeting AtDBP1 to modify its activity.
MATERIALS AND METHODS
Yeast Two-Hybrid System
The coding sequence of tobacco (Nicotiana tabacum) NtDBP1 was cloned into the yeast shuttle vector pAS2-1 (Clontech) in frame with the GAL4 DNA-binding domain. The resulting construct was used to transform the yeast strain PJ69-4A (James et al., 1996). A tobacco cDNA library constructed in the vector pAD-GAL4-2.1 (Stratagene) was screened following the polyethylene glycol/LiAc/single-stranded DNA protocol developed by Gietz et al. (1997). Positive interacting clones were selected on His-lacking medium supplemented with 5 mm 3-amino-triazole (a competitive inhibitor of the HIS3 reporter gene product) and analyzed for activation of the second reporter gene, the lacZ gene, by β-galactosidase filter lift assays. The veracity and specificity of the interaction were confirmed by retransformation of the library plasmids into the original bait strain used in the screening and additional control strains. β-Galactosidase activity was measured by a quantitative liquid culture assay using a standard protocol.
RNA Extraction, RT, and qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s recommendations and further purified by lithium chloride precipitation. For RT, the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas Life Sciences) was used. qPCR amplifications and measurements were performed using an ABI PRISM 7000 sequence detection system and SYBR-Green (Perkin-Elmer Applied Biosystems). The oligonucleotides utilized in the analyses were as follows: GFP-FW, 5′-ACGTAAACGGCCACAAGTTC-3′; GFP-RV, 5′-AAGTCGTGCTGCTTCATGTG-3′; PPV-CP-FW, 5′-GACTACGGCGTCAATGCTCAAC-3′; PPV-CP-RV, 5′-GTTTGCAGTTGAGGTCCTGACAC-3′; ACTIN2/8-FW, 5′-GGTAACATTGTGCTCAGTGGTGG-3′; ACTIN2/8-RV, 5′-AACGACCTTATCTTCATGCTGC-3′; DIP2-FW, 5′-TGGAGGCTGTTCCTCCACAAATC-3′; DIP2-RV, 5′-TCAAGAATCGGAGAAACCGATGGC-3′; TuMV-FW, 5′-GGCAAGGATGTTGCACAAGA-3′; TuMV-RV, 5′-TTCCAGAGGTTCCAGCGTTT-3′.
FIG Assay
The following plasmids were constructed for the FIG assay. The AtDBP1-encoding sequence was introduced into vector YEp21bA-PGK1p-ubi-URA3 by marked homologous recombination in yeast (Daniel, 1995) using PCR fragments (containing, in addition to the AtDBP1-encoding sequence, appropriate small linker sequences) to give plasmid YEp21bA-PGK1p-ubi-AtDBP1 (markers ADE2 and LEU2). From the resulting plasmid and vector YEp352, plasmid YEp352-PGK1p-ubi-AtDBP1 (marker URA3) was constructed by homologous recombination in yeast using their common flanking β-galactosidase small DNA sequences. To obtain plasmid YEp21bA-GPD1p-AtDIP2 (markers ADE2 and LEU2), a BamHI-SalI PCR fragment containing the AtDIP2-encoding sequence was introduced into vector YEp21bA-GPD1p. Gene toxicity (Daniel, 1996a) was measured for each Arabidopsis (Arabidopsis thaliana) gene thus expressed in yeast and found to be 10% for AtDBP1 and 18% for AtDIP2.
The FIG assay was carried out basically as described previously (Daniel, 1996b, 2008, 2009; see “Results and Discussion” and Fig. 4 legend). Briefly, yeast cells containing both plasmids YEp21bA-GPD1p-AtDIP2 and YEp352-PGK1p-ubi-AtDBP1 (by having selected for both ADE2/LEU2 and URA3 markers) were first grown in the absence of uracil (thus selecting for YEp352-PGK1p-ubi-AtDBP1) but in the presence of adenine and Leu (i.e. nonselective condition for YEp21bA-GPD1p-AtDIP2) until the stationary phase and then streaked in a solid medium containing Leu and limiting concentrations of adenine but no uracil, so as to yield many isolated colonies. The proportion of white colonies, which result from the presence of plasmid YEp21bA-GPD1p-AtDIP2 in the cell at the origin of their formation, was measured relative to the total (i.e. white colonies plus small red colonies), and this value was compared with that obtained under the same conditions with the control vector YEp352 instead of YEp352-PGK1p-ubi-AtDBP1.
Constructs for AtDIP2 Overexpression and BiFC Analysis
Constructs were generated using Gateway technology (Invitrogen) following the manufacturer’s recommendations. The AtDIP2 open reading frame was amplified by PCR using specific oligonucleotides bearing attB sites, then cloned into pDONR207 (Invitrogen), and finally transferred to the pMDC32 plasmid (Curtis and Grossniklaus, 2003) under the transcriptional control of two copies of the 35S promoter of Cauliflower mosaic virus. Similarly, for BiFC analysis, coding sequences of the relevant proteins were cloned into pYFN43 and pYFC43 plasmids created by A. Ferrando (Instituto de Biología Molecular y Celular de Plantas; http://www.ibmcp.upv.es) to generate C-terminal translational fusions to the N-terminal and C-terminal fragments of YFP, respectively.
Transient Expression in Nicotiana benthamiana Leaves
N. benthamiana plants were grown in a phytochamber under short-day conditions at 23°C/19°C. Mid, almost fully expanded, leaves were infiltrated with a suspension of Agrobacterium tumefaciens C58 bearing the relevant construct in 10 mm MES, pH 5.6, 10 mm MgCl2, and 150 μm acetosyringone at an optical density at 600 nm of 0.5. After 3 d, fluorescence was analyzed in infiltrated leaves by confocal microscopy. For coinfiltration, Agrobacterium cultures grown separately and adjusted to an optical density of 0.5 were mixed. Agrobacterium expressing the viral silencing suppressor P19 was included in all infiltrations.
Viral Inoculation
PPV and TuMV (Castelló et al., 2010) were inoculated by gently making a single puncture on the distal part of a leaf with a sterile toothpick soaked in a suspension of Agrobacterium bearing an infectious cDNA clone at an optical density at 600 nm of 1. The Agrobacterium suspension was prepared as described above.
Fluorescence Microscopy
GFP/YFP fluorescence in inoculated plants was monitored using Nikon SMZ800 and Leica MZ16F microscopes.
The NtDIP2 sequence has been deposited in GenBank with accession number JF951856.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein phosphatase activity assay.
Supplemental Figure S2. Detection of eIF(iso)4E in protein extracts by western blot.
Supplemental Figure S3. Viral accumulation in the atdip2 mutant (atdip2) and AtDIP2-overexpressing plants (OX 2.5 and OX 6.2) inoculated with the potyvirus TuMV.
Acknowledgments
Polyclonal antiserum against eIF(iso)4E from Arabidopsis was kindly provided by Karen Browning (University of Texas, Austin).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Breeuwer M, Goldfarb DS. (1990) Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell 60: 999–1008 [DOI] [PubMed] [Google Scholar]
- Carrasco JL, Ancillo G, Castelló MJ, Vera P. (2005) A novel DNA-binding motif, hallmark of a new family of plant transcription factors. Plant Physiol 137: 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco JL, Ancillo G, Mayda E, Vera P. (2003) A novel transcription factor involved in plant defense endowed with protein phosphatase activity. EMBO J 22: 3376–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco JL, Castelló MJ, Vera P. (2006) 14-3-3 mediates transcriptional regulation by modulating nucleocytoplasmic shuttling of tobacco DNA-binding protein phosphatase-1. J Biol Chem 281: 22875–22881 [DOI] [PubMed] [Google Scholar]
- Castelló MJ, Carrasco JL, Vera P. (2010) DNA-binding protein phosphatase AtDBP1 mediates susceptibility to two potyviruses in Arabidopsis. Plant Physiol 153: 1521–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron C, Nicolaï M, Gallois JL, Robaglia C, Moury B, Palloix A, Caranta C. (2008) Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J 54: 56–68 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. (1995) DNA insertion system for complex yeast shuttle vectors. Curr Genet 27: 309–311 [DOI] [PubMed] [Google Scholar]
- Daniel J. (1996a) Measuring the toxic effects of high gene dosage on yeast cells. Mol Gen Genet 253: 393–396 [DOI] [PubMed] [Google Scholar]
- Daniel J. (1996b) Detection of antagonistic cellular regulatory functions by the gene-gene interference method in yeast. Curr Genet 29: 114–121 [DOI] [PubMed] [Google Scholar]
- Daniel J. (2007) Direct in vivo access to potential gene targets of the RPD3 histone deactylase using fitness-based interferential genetics. Yeast 24: 575–587 [DOI] [PubMed] [Google Scholar]
- Daniel JH. (2008) A potentially general method for the in vivo selection of inhibitory peptides targeted at a specific protein using yeast. Curr Genet 53: 373–379 [DOI] [PubMed] [Google Scholar]
- Daniel JH. (2009) A fitness-based interferential genetics approach using hypertoxic/inactive gene alleles as references. Mol Genet Genomics 281: 437–445 [DOI] [PubMed] [Google Scholar]
- Decroocq V, Sicard O, Alamillo JM, Lansac M, Eyquard JP, García JA, Candresse T, Le Gall O, Revers F. (2006) Multiple resistance traits control Plum pox virus infection in Arabidopsis thaliana. Mol Plant Microbe Interact 19: 541–549 [DOI] [PubMed] [Google Scholar]
- Díaz-Pendon JA, Truniger V, Nieto C, García-Mas J, Bendahmane A, Aranda MA. (2004) Advances in understanding recessive resistance to plant viruses. Mol Plant Pathol 5: 223–233 [DOI] [PubMed] [Google Scholar]
- Dielen AS, Sassaki FT, Walter J, Michon T, Ménard G, Pagny G, Krause-Sakate R, Maia IdeG, Badaoui S, Le Gall O, et al. (2011) The 20S proteasome α5 subunit of Arabidopsis thaliana carries an RNase activity and interacts in planta with the lettuce mosaic potyvirus HcPro protein. Mol Plant Pathol 12: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Thomas C, Harrison S, Revers F, Maule A. (2004) A cysteine-rich plant protein potentiates potyvirus movement through an interaction with the virus genome-linked protein VPg. J Virol 78: 2301–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C. (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32: 927–934 [DOI] [PubMed] [Google Scholar]
- Farrokhi N, Whitelegge JP, Brusslan JA. (2008) Plant peptides and peptidomics. Plant Biotechnol J 6: 105–134 [DOI] [PubMed] [Google Scholar]
- Fukuda H, Higashiyama T. (2011) Diverse functions of plant peptides: entering a new phase. Plant Cell Physiol 52: 1–4 [DOI] [PubMed] [Google Scholar]
- Gibbs A, Ohshima K. (2010) Potyviruses and the digital revolution. Annu Rev Phytopathol 48: 205–223 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Triggs-Raine B, Robbins A, Graham KC, Woods RA. (1997) Identification of proteins that interact with a protein of interest: applications of the yeast two-hybrid system. Mol Cell Biochem 172: 67–79 [PubMed] [Google Scholar]
- Hafrén A, Hofius D, Rönnholm G, Sonnewald U, Mäkinen K. (2010) HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébrard E, Poulicard N, Gérard C, Traoré O, Wu HC, Albar L, Fargette D, Bessin Y, Vignols F. (2010) Direct interaction between the Rice yellow mottle virus (RYMV) VPg and the central domain of the rice eIF(iso)4G1 factor correlates with rice susceptibility and RYMV virulence. Mol Plant Microbe Interact 23: 1506–1513 [DOI] [PubMed] [Google Scholar]
- Hicks GR. (2005) Nuclear import of plant proteins. Tzfira T, Citovsky V, , Nuclear Import and Export in Plants and Animals. Landes Bioscience/Kluwer Academic Publishers, Georgetown, TX, pp 61–82 [Google Scholar]
- Huang TS, Wei T, Laliberté JF, Wang A. (2010) A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol 152: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Yamaguchi T, Kazama T, Ito T, Horiguchi G, Tsukaya H. (2011) ROTUNDIFOLIA4 regulates cell proliferation along the body axis in Arabidopsis shoot. Plant Cell Physiol 52: 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YS, Ma DY, Dong JL, Jin JC, Li DF, Deng CW, Wang T. (2007) HC-Pro protein of Potato virus Y can interact with three Arabidopsis 20S proteasome subunits in planta. J Virol 81: 12881–12888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungkunz I, Link K, Vogel F, Voll LM, Sonnewald S, Sonnewald U. (2011) AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J 66: 983–995 [DOI] [PubMed] [Google Scholar]
- Kang B-C, Yeam I, Frantz JD, Murphy JF, Jahn MM. (2005) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J 42: 392–405 [DOI] [PubMed] [Google Scholar]
- Le Gall O, Aranda MA, Caranta C. (2011) Plant resistance to viruses mediated by translation initiation factors. Caranta C, Aranda MA, Tepfer M, López-Moya JJ, , Recent Advances in Plant Virology. Caister Academic Press, Norfolk, UK, pp 177–194 [Google Scholar]
- Lellis AD, Kasschau KD, Whitham SA, Carrington JC. (2002) Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 12: 1046–1051 [DOI] [PubMed] [Google Scholar]
- Léonard S, Plante D, Wittmann S, Daigneault N, Fortin MG, Laliberté JF. (2000) Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J Virol 74: 7730–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ, Caranta C, Boulton MI. (2007) Sources of natural resistance to plant viruses: status and prospects. Mol Plant Pathol 8: 223–231 [DOI] [PubMed] [Google Scholar]
- Nieto C, Rodríguez-Moreno L, Rodríguez-Hernández AM, Aranda MA, Truniger V. (2011) Nicotiana benthamiana resistance to non-adapted Melon necrotic spot virus results from an incompatible interaction between virus RNA and translation initiation factor 4E. Plant J 66: 492–501 [DOI] [PubMed] [Google Scholar]
- Panavas T, Serviene E, Brasher J, Nagy PD. (2005) Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci USA 102: 7326–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Caranta C. (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11: 40–45 [DOI] [PubMed] [Google Scholar]
- Sato M, Nakahara K, Yoshii M, Ishikawa M, Uyeda I. (2005) Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett 579: 1167–1171 [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C. (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Pfannschmidt T, , Plant Signal Transduction, Vol 479. Humana Press, Totowa, NJ, pp 189–202 [DOI] [PubMed] [Google Scholar]
- Sekimoto T, Katahira J, Yoneda Y. (2005) Nuclear import and export signals. Tzfira T, Citovsky V, , Nuclear Import and Export in Plants and Animals. Landes Bioscience/Kluwer Academic Publishers, Georgetown, TX, pp 50–60 [Google Scholar]
- Serviene E, Jiang Y, Cheng CP, Baker J, Nagy PD. (2006) Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J Virol 80: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. (2005) Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci USA 102: 10545–10550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard O, Loudet O, Keurentjes JJB, Candresse T, Le Gall O, Revers F, Decroocq V. (2008) Identification of quantitative trait loci controlling symptom development during viral infection in Arabidopsis thaliana. Mol Plant Microbe Interact 21: 198–207 [DOI] [PubMed] [Google Scholar]
- Trüniger V, Aranda MA. (2009) Recessive resistance to plant viruses. Adv Virus Res 75: 119–159 [DOI] [PubMed] [Google Scholar]