Abstract
Catharanthus roseus produces a large array of terpenoid indole alkaloids (TIAs) that are an important source of natural or semisynthetic anticancer drugs. The biosynthesis of TIAs is tissue specific and induced by certain phytohormones and fungal elicitors, indicating the involvement of a complex transcriptional control network. However, the transcriptional regulation of the TIA pathway is poorly understood. Here, we describe a C. roseus WRKY transcription factor, CrWRKY1, that is preferentially expressed in roots and induced by the phytohormones jasmonate, gibberellic acid, and ethylene. The overexpression of CrWRKY1 in C. roseus hairy roots up-regulated several key TIA pathway genes, especially Tryptophan Decarboxylase (TDC), as well as the transcriptional repressors ZCT1 (for zinc-finger C. roseus transcription factor 1), ZCT2, and ZCT3. However, CrWRKY1 overexpression repressed the transcriptional activators ORCA2, ORCA3, and CrMYC2. Overexpression of a dominant-repressive form of CrWRKY1, created by fusing the SRDX repressor domain to CrWRKY1, resulted in the down-regulation of TDC and ZCTs but the up-regulation of ORCA3 and CrMYC2. CrWRKY1 bound to the W box elements of the TDC promoter in electrophoretic mobility shift, yeast one-hybrid, and C. roseus protoplast assays. Up-regulation of TDC increased TDC activity, tryptamine concentration, and resistance to 4-methyl tryptophan inhibition of CrWRKY1 hairy roots. Compared with control roots, CrWRKY1 hairy roots accumulated up to 3-fold higher levels of serpentine. The preferential expression of CrWRKY1 in roots and its interaction with transcription factors including ORCA3, CrMYC2, and ZCTs may play a key role in determining the root-specific accumulation of serpentine in C. roseus plants.
Catharanthus roseus (Madagascar periwinkle) produces a large array of terpenoid indole alkaloids (TIAs). Many TIAs are defense molecules in response to biotic and abiotic factors (Aerts et al., 1994; Roepke et al., 2010). Some of these TIAs, in their natural or semisynthetic forms, are of pharmaceutical importance, as exemplified by the anticancer agents vinblastine, vincristine, vindesine, and vinorelbine, as well as the antihypertensive compounds ajmalicine and serpentine. Accumulation of TIAs is tissue specific: vindoline and catharanthine, precursors for the assembly of vinblastine, accumulate in leaf cells and leaf exudates, respectively, whereas ajmalicine and serpentine are mainly found in roots (Roepke et al., 2010). TIAs purified from C. roseus are expensive due to low yields and variations associated with environmental effects. Intensive efforts to engineer C. roseus for increased TIA production have shown limited success (for review, see O’Connor and Maresh, 2006; El-Sayed and Verpoorte, 2007; Zhou et al., 2009). TIA engineering is impeded by the reality that the multistep, branched biosynthetic pathway (Supplemental Fig. S1) is only partially characterized. Furthermore, regulation of the pathway is highly temporal and spatial, often in response to developmental and environmental signals in a tissue-specific manner. Nevertheless, from a biological chemistry perspective, the complexity of the TIA pathway, which involves more than 20 enzymes and perhaps a larger number of gene regulators, makes C. roseus an excellent system for studying alkaloid biosynthesis (Facchini and De Luca, 2008).
A number of genes encoding TIA pathway enzymes and transcriptional regulators from C. roseus have been isolated and characterized (Liu et al., 2007; Costa et al., 2008). Attempts to increase TIA production by ectopic expression of genes encoding several rate-limiting enzymes have met with less than satisfactory results. For instance, despite the observation that overexpression of the Tryptophan Decarboxylase (TDC) gene has led to a moderate increase of alkaloid accumulation (Hong et al., 2006), it is generally believed that TDC activity does not correlate positively with TIA production (Goddijn et al., 1995; El-Sayed and Verpoorte, 2007). A more promising approach is the ectopic expression of transcription factors (TFs) that regulate the TIA pathway in order to enhance alkaloid production (Memelink and Gantet, 2007). Specific TFs are often capable of coordinating the transcription of multiple biosynthetic pathway genes, making them particularly effective in metabolic pathway engineering. A significant increase of ajmalicine production in C. roseus cells has been achieved by the overexpression of an Arabidopsis (Arabidopsis thaliana) TF, Agamous-like 12 (Montiel et al., 2007). Increased catharanthine accumulation is detected in C. roseus hairy roots upon coexpression of the TF ORCA3 and the pathway enzyme Geraniol 10-Hydroxylase (G10H) but not in roots expressing these genes individually (Wang et al., 2010a). TIA biosynthesis responds strongly to a group of chemical or fungal elicitors that are known to regulate gene transcription, providing an explanation for why the manipulation of TFs often results in substantial effects to the TIA pathway (El-Sayed and Verpoorte, 2007). Promoter analyses of several structural and TF genes related to the C. roseus TIA pathway have revealed the presence of chemical- and elicitor-responsive cis-elements, leading to the cloning of jasmonate-responsive TFs including the AP2-like ORCA2 and ORCA3 (Menke et al., 1999; Ouwerkerk and Memelink, 1999; van der Fits and Memelink, 2000), the G box-binding factors CrGBF1 and CrGBF2 (Sibéril et al., 2001), the P box-binding factor CrBPF1 (van der Fits et al., 2000), the zinc-finger repressors ZCT1 (for zinc-finger C. roseus transcription factor 1), ZCT2, and ZCT3 (Pauw et al., 2004), and the basic helix-loop-helix TF CrMYC2 (Zhang et al., 2011).
Recently, we analyzed all available promoters of the TIA pathway genes and found the W box (TTGACC/T) element in almost all of these promoters. The number of W boxes in each promoter varies, ranging from one in G10H to two in Cytochrome P450 Reductase (CPR) and four in TDC and Deacetylvindoline-4-O-Acetyltransferase (Ouwerkerk and Memelink, 1999; Suttipanta et al., 2007; Wang et al., 2010b). The W box is a cognate binding site for WRKY TFs. The characteristic of WRKY TFs is a conserved WRKY domain that consists of the peptide motif WRKYGQK and a zinc finger (Yamasaki et al., 2005). WRKY TFs form a large, plant-specific TF family and play dynamic roles in, among other biological processes, biotic and abiotic stress responses (for review, see Rushton et al., 2010). WRKY TFs function alone or in combination with other regulators to activate, repress, or derepress transcription. WRKY TFs are known to be involved in alkaloid biosynthesis (Kato et al., 2007). In addition, when Medicago truncatula is exposed to fungal elicitors or methyl jasmonate (MJ), a large number of WRKY genes are up-regulated, some of which are involved in the production of defense compounds such as flavonoids and terpenoids (Naoumkina et al., 2008). Response to fungal elicitors and phytohormones such as MJ is a hallmark of TIA pathway regulation. Therefore, we hypothesized that WRKY TFs are involved in the transcriptional regulation of the C. roseus TIA pathway. However, despite three reported sequences of putative C. roseus WRKY genes, obtained from EST sequencing (Murata et al., 2006), no WRKY TF from C. roseus has been functionally characterized to date.
In this report, we describe the isolation and functional characterization of a WRKY TF, CrWRKY1, from C. roseus. CrWRKY1 is preferentially expressed in roots and induced by phytohormones such as MJ, GA, and ethylene. The overexpression of CrWRKY1 in C. roseus hairy roots up-regulates several key pathway genes, especially TDC, and the ZCT TFs, while it represses the transcriptional activators ORCA2, ORCA3, and CrMYC2. We show that CrWRKY1 activates the TDC gene by directly targeting its promoter. Moreover, transgenic CrWRKY1 hairy roots accumulate significantly higher levels of serpentine compared with controls. The preferential expression of CrWRKY1 in the root suggests that it is a key factor in determining the root-specific accumulation of serpentine in C. roseus plants. This work represents, to our knowledge, the first functional characterization of a C. roseus WRKY TF.
RESULTS
Isolation and Expression Profiling of CrWRKY1
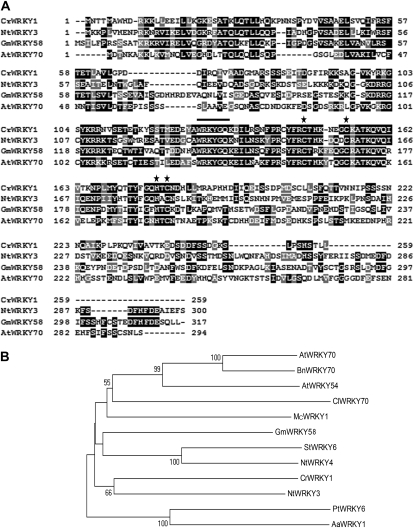
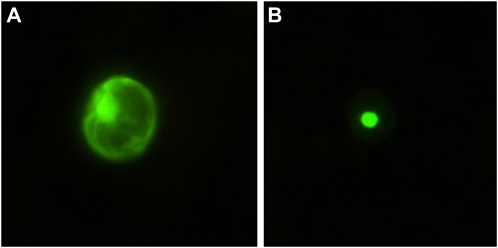
We aimed to isolate WRKY TFs that are induced by MJ and have corresponding expression profiles as key TIA pathway genes. Degenerate PCR primers (Supplemental Table S1), based on conserved sequences of WRKY domains, amplified bands of approximately 180 bp from mRNA isolated from MJ-treated C. roseus tissues. DNA sequencing yielded a total of six unique open reading frames containing the WRKYGQK sequence. Functional characterization of one of the WRKY genes, designated as CrWRKY1 (GenBank accession no. HQ646368), is described here. CrWRKY1 encodes a 259-amino acid protein, having a calculated molecular mass of 29.3 kD and a deduced pI of pH 9.4. Sequence alignment (Fig. 1A) revealed that CrWRKY1 shares high amino acid sequence identity with Nicotiana tabacum NtWRKY3 (42%), Glycine max GmWRKY58 (45%), and Arabidopsis AtWRKY70 (40%), all of which are classified as group III WRKY TFs (Zhang and Wang, 2005). Phylogenetic analysis of selected WRKY proteins placed CrWRKY1 close to NtWRKY3 (Fig. 1B). CrWRKY1 possesses one WRKY domain together with a zinc finger that is unique to group III WRKY proteins (Eulgem et al., 2000). A putative bipartite nuclear localization signal (RKKSAGVKYRKGSYKRR at amino acids 92–108) was also identified in CrWRKY1. To demonstrate that CrWRKY1 is indeed nucleus localized, we fused CrWRKY1 in frame to the enhanced GFP gene, eGFP, and expressed the fusion gene in C. roseus cell suspension-derived protoplasts. While control eGFP accumulated throughout the cell (Fig. 2A), the CrWRKY1-eGFP fusion protein was localized to the nucleus (Fig. 2B).
Figure 1.
Multiple sequence alignment and phylogenetic analysis of selected WRKY proteins. A, The deduced amino acid sequence of CrWRKY1 is aligned with homologs from N. tabacum (NtWRKY3; accession no. AF193770), G. max (GmWRKY54; accession no. EU375354), and Arabidopsis (AtWRKY70; accession no. AF421157). The WRKY signature motif (WRKYGQK) is indicated by the line, and the characteristic residues for the zinc-finger motif are marked by asterisks. B, A neighbor-joining phylogenetic tree of CrWRKY1 and selected WRKY domain proteins from other plant species constructed using the MEGA4 software. The statistical reliability of individual nodes of the newly constructed tree is assessed by bootstrap analyses with 1,000 replications. The WRKY proteins, their respective plant species, and GenBank accession numbers are as follows: AtWRKY70, Arabidopsis (AF421157; At3G56400); BnWRKY70, Brassica napus (FJ384113); AtWRKY54, Arabidopsis (NM_129637; At2G40750); ClWRKY70, Citrullus lanatus (GQ453670); McWRKY1, Matricaria chamomilla (AB035271); GmWRKY58, G. max (EU375354); StWRKY6, Solanum tuberosum (EU056918); NtWRKY3, N. tabacum (AF193770); CrWRKY1, C. roseus (HQ646368); NtWRKY4, N. tabacum (AF193771); PtWRKY6, Populus tomentosa × Populus bolleana (GQ377426); AaWRKY1, Artemisia annua (FJ390842).
Figure 2.
Subcellular localization of CrWRKY1 in C. roseus protoplasts. eGFP is accumulated throughout the cell (A), whereas CrWRKY1-eGFP is localized to the nucleus in C. roseus cells (B). This experiment was repeated two times, and a representative result is shown here.
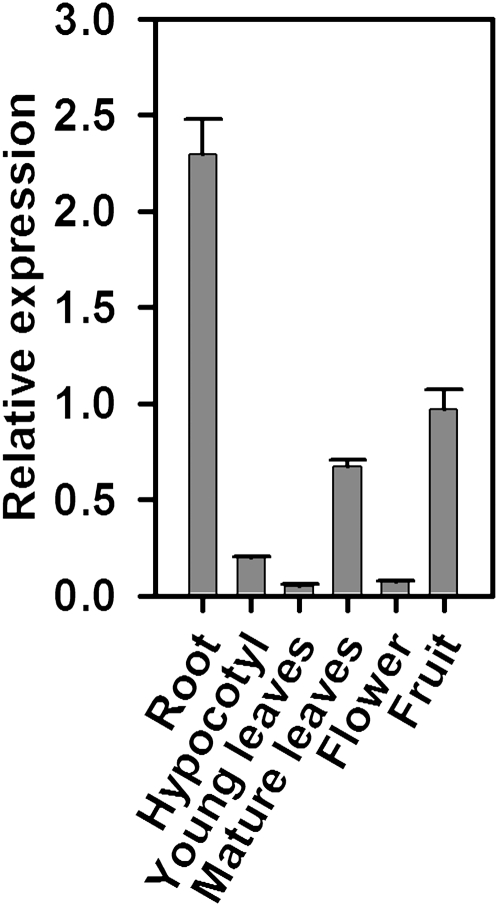
The expression of CrWRKY1 in various C. roseus tissues was examined using quantitative (q)PCR (Fig. 3). Significant expression of CrWRKY1 could be detected in fruit tissue and mature leaf, whereas the expression was barely detectable in hypocotyl, young leaf, and flower. However, the highest expression of CrWRKY1 (approximately 2.5-fold of that in fruit) was found in the root.
Figure 3.
qRT-PCR analysis of CrWRKY1 expression levels in different tissues of C. roseus. The values expressed are relative to the expression level in fruit. Data are means and sd from three biological replicates.
Induction of CrWRKY1 Expression by MJ, Ethylene, and GA
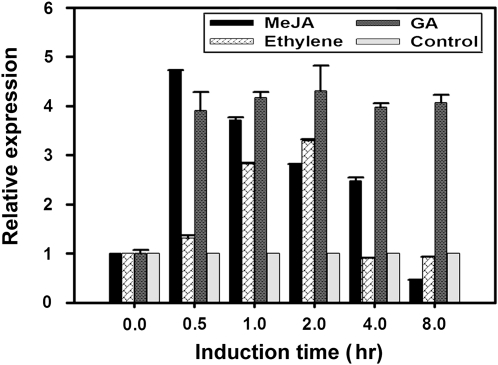
To investigate the effect of phytohormones on CrWRKY1 expression, 10-d-old C. roseus seedlings were treated with MJ, ethylene, and GA in a time-course study. The expression of CrWRKY1 was measured by qPCR using RNA isolated from treated plants (Fig. 4). Compared with the uninduced control, CrWRKY1 expression increased approximately 5-fold within 30 min after exposure to 100 μm MJ and gradually decreased during the next 8 h. Effects of ethylene (1 μL L−1) on CrWRKY1 expression reached its peak in 2 h and returned to the basal level in the next 2 h. By contrast, the effect of GA (10 μm) on CrWRKY1 expression was longer lasting, reaching a more than four times increase within 30 min and maintaining the high expression level for at least 8 h (Fig. 4). The JA induction pattern of CrWRKY1 closely resembled that of TDC (Supplemental Fig. S2).
Figure 4.
qRT-PCR analysis of CrWRKY1 expression levels in C. roseus seedlings upon induction by MJ (MeJA), GA, and ethylene. Seedlings without phytohormone induction served as controls. All experiments were performed in triplicate, and error bars represent sd.
Activation of the TDC Gene by CrWRKY1 in Transgenic Hairy Roots
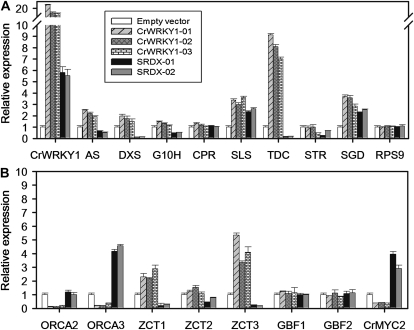
We generated a number of independent hairy root lines expressing CrWRKY1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The frequency of cotransformation of Ri T-DNA and transgene T-DNA was approximately 25%. The presence of both pCAMBIA and Ri plasmid T-DNAs in independent CrWRKY1 overexpression and empty vector control hairy root lines was verified by PCR using gene-specific primers (Supplemental Fig. S3C). Two empty vector controls and three overexpression lines were chosen for further analysis (Fig. 5; Supplemental Fig. S4, A and B). In the CrWRKY1-overexpressing hairy roots, the CrWRKY1 gene was expressed 17- to 22-fold higher than the empty vector control (Fig. 5A). The expression of several other pathway genes was also up-regulated; among them, the most striking was the TDC gene, which increased from 7- to 9-fold in the hairy roots compared with the control. Although to a lesser extent than TDC, the expression of Anthranilate Synthase (AS), 1-Deoxy-d-Xylulose-5-Phosphate Synthase (DXS), Secologanin Synthase (SLS), and Strictosidine β-d-Glucosidase (SGD) increased in CrWRKY1-overexpressing hairy roots by an average of 1.2-, 0.7-, 2.3-, and 2.4-fold, respectively, compared with the control (Fig. 5A). Overexpression of CrWRKY1 did not significantly affect the expression of other pathway genes, including G10H, CPR, and Strictosidine Synthase (STR). We also measured the expression of several TF genes known to be involved in the TIA pathway in CrWRKY1 hairy roots. The expression of ORCA2, ORCA3, and CrMYC2 was suppressed by an average of 90%, 80%, and 60%, respectively, compared with that of the vector-only control (Fig. 5B). Overexpression of CrWRKY1 also led to increases in the expression of ZCT1, ZCT2, and ZCT3 by an average of 1.4-, 0.3-, and 3.2-fold and had little effect on the expression of the two G box-binding factors, GBF1 and GBF2 (Fig. 5B).
Figure 5.
Relative expression levels of selected TIA pathway genes in CrWRKY1 and CrWRKY1-SRDX hairy roots. Expression levels of these genes are normalized to the empty vector control line. The Rps9 gene was used as an internal control. Relative expression levels of structural genes (A) and TF genes (B) in CrWRKY1 and CrWRKY1-SRDX hairy roots were measured by qPCR. Each relative gene expression represents the average of three measurements, with error bars representing sd.
Because TDC is the gene most affected in the CrWRKY1 hairy roots, we applied gene-silencing technology to further clarify the role of CrWRKY1 in the regulation of TDC. We employed chimeric repressor-silencing technology (Hiratsu et al., 2003), in which the SRDX repression domain was fused to the C terminus of CrWRKY1, and obtained hairy roots overexpressing CrWRKY1-SRDX. The transgenic status of the CrWRKY1-SRDX lines was verified by PCR amplifying the transgenes from genomic DNA using gene-specific primers (Supplemental Fig. S3C). Two empty vector control and two CrWRKY1-SRDX hairy root lines were chosen for further analysis. qPCR was performed to measure the expression of pathway and TF genes using RNA isolated from the CrWRKY1-SRDX hairy roots (Fig. 5; Supplemental Fig. S4, A and B). In general, an opposite expression pattern was observed for each gene compared with the CrWRKY1 overexpression lines (Fig. 5). Compared with the empty vector control, TDC expression in the CrWRKY1-SRDX lines decreased by an average of 83%, while the expression of AS and DXS decreased by an average of 47% and 88%, respectively (Fig. 5A). In the same repression hairy root lines, expression of the three transactivator genes, ORCA2, ORCA3, and CrMYC2, was 1-, 4-, and 3-fold of that of the vector-only control, while expression of the three transrepressors, ZCT1, ZCT2, and ZCT3, decreased by an average of 71%, 33%, and 72%, respectively (Fig. 5B).
Increase of TDC Activity in CrWRKY1-Overexpressing Hairy Roots
TDC can convert the phytotoxic Trp analog 4-methyl tryptophan (4-MT) into the nontoxic 4-methyl tryptamine and thus is a useful selectable marker for plant cells (Goddijn et al., 1993). To test whether CrWRKY1 hairy roots, in which the TDC gene is highly expressed, confer 4-MT resistance, we screened the hairy roots for growth in the presence or absence of 4-MT (Fig. 6). At 250 μm 4-MT, CrWRKY1 hairy roots sustained growth and formed new roots (Fig. 6B), whereas empty vector control roots were severely stunted (Fig. 6E). Even at 500 μm 4-MT, CrWRKY1 hairy roots, although somewhat stunted, continued their growth (Fig. 6C), while the control roots did not survive (Fig. 6F). These results indicate that elevated TDC activity is associated with the CrWRKY1 hairy roots.
Figure 6.
Inhibition of CrWRKY1-overexpressing and empty vector control hairy roots by 4-MT. Hairy roots were grown for 90 d on one-third-strength Schenk and Hilderbrandt medium containing 0 μm (A, CrWRKY1; D, control), 250 μm (B, CrWRKY1; E, control), and 500 μm (C, CrWRKY1; F, control) 4-MT.
Soluble proteins were extracted from hairy roots grown on medium without 4-MT and assayed for TDC enzyme activity using Trp as a substrate. TDC activities in empty vector control, CrWRKY1, and CrWRKY1-SRDX hairy roots were 21.6 ± 3.8, 37.7 ± 2.2, and 17.3 ± 5.3 pm tryptamine s−1 mg−1 total soluble protein, respectively, representing a 73% increase of TDC activity in CrWRKY1 hairy roots and a 20% decrease of the enzyme activity in CrWRKY1-SRDX hairy roots. Correspondingly, tryptamine content in control, CrWRKY1, and CrWRKY1-SRDX hairy roots was 30.1 ± 2.3, 76.0 ± 9.9, and 23.3 ± 4.6 μg g−1 dry weight, respectively, representing a 150% increase in CrWRKY1 hairy roots and a 23% decrease in CrWRKY1-SRDX hairy roots compared with the control.
Binding of CrWRKY1 to the W Boxes in the TDC Promoter
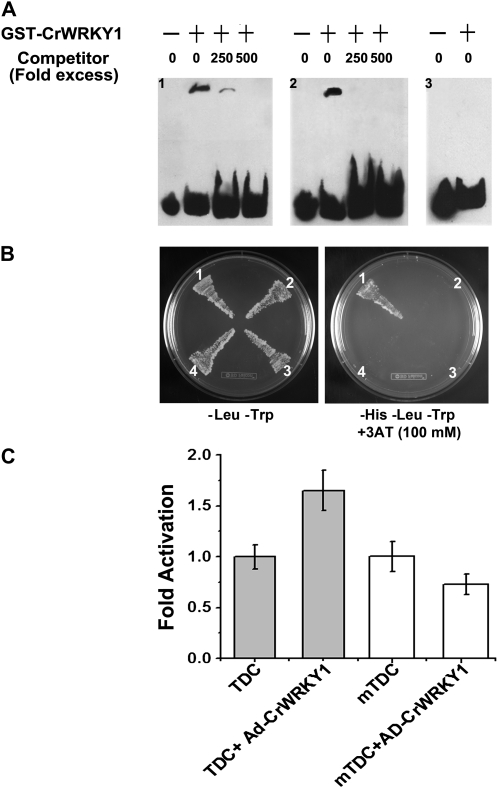
Electrophoretic mobility shift assay (EMSA) was used to determine the binding affinity of CrWRKY1 to the TDC promoter. We first purified the recombinant, glutathione S-transferase (GST)-tagged CrWRKY1 protein (GST-CrWRKY1) from Escherichia coli using GST affinity chromatography, which yielded approximately 160 μg of purified protein per liter of cell culture. For EMSA experiments, three DNA probes, with or without biotin labeling, were synthesized based on the W box elements of the TDC promoter. Probe 1 (5′-CAAAAATTTGACTATACTTGACTATTAGTG-3′) is identical to the TDC promoter region (–1,179 to –1,150 relative to the transcription start site [TSS]), where two W boxes (underlined) are adjacent to one another. Probe 2 (5′-GACTGCATGTTGACCTAAAATTATG-3′) is a native TDC promoter sequence (–583 to –559 relative to the TSS) containing a single W box. The third probe, mProbe 2 (5′-GACTGCATGTTAATCTAAAATTATG-3′), is identical to probe 2 except that the W box sequence was destroyed by mutation in the W box core sequence. As shown in Figure 7A, the recombinant CrWRKY1 bound to probe 1 and probe 2 and resulted in mobility shifts (Fig. 7A, gels 1 and 2, lane 2); however, the protein did not bind mProbe 2 (Fig. 7A, gel 3, lane 2). The W box-binding specificity of GST-CrWRKY1 was further confirmed by a competition experiment showing that binding of the labeled probes by CrWRKY1 could be eliminated by increasing concentrations (250× and 500× in excess) of unlabeled probe 1 and probe 2 (Fig. 7A, gels 1 and 2, lanes 2 and 3).
Figure 7.
Binding of CrWRKY1 to W box elements in the TDC promoter. A, Autoradiographs show up-shifted bands of the EMSA using purified GST-CrWRKY1 protein and probe 1 (containing two W boxes), probe 2 (containing one W box), or mProbe 2 (containing a mutated W box). Gel 1 shows the products of DNA-binding reactions in the absence and presence of GST-CrWRKY1 or labeled and unlabeled probe 1. Gel 2 is the same as gel 1 except that labeled and unlabeled probe 2 were used. Gel 3 shows the reaction products of mProbe 2 in the absence and presence of GST-CrWRKY1 proteins. B, Yeast one-hybrid assays demonstrate CrWRKY1 activation of the C. roseus TDC promoter. The plasmid harboring a GAL4 activation domain/CrWRKY1 fusion (pAD-CrWRKY1) was cotransformed with the pTDC-HIS2 or pmTDC-HIS2 reporter plasmid. The transformants were grown in either double selection medium (SD-Leu-Trp) or triple selection medium (SD-His-Leu-Trp) with 100 mm 3-AT. Sector 1, pHIS2-TDC + pAD-CrWRKY1; sector 2, pHIS2-mTDC + pAD-CrWRKY1; sector 3, pHIS2-TDC + pAD; sector 4, pHIS2-mTDC + pAD. C, CrWRKY1 binds to W box elements of the TDC promoter in C. roseus protoplasts. The reporter plasmid (TDC), consisting of the Luciferase gene under the control of tandem repeats of W box elements upstream of a minimal 35S promoter, was either transformed alone or cotransformed with AD-CrWRKY1 into C. roseus protoplasts. Another reporter plasmid (mTDC), which is identical to the TDC reporter except that the W box sequence is mutated, was also transformed alone or together with AD-WRKY1 into the protoplasts. The relative levels of activation of the Luciferase gene are presented. Experiments were performed in triplicate.
Binding of CrWRKY1 to the TDC promoter was also verified in vivo using a yeast one-hybrid assay. The reporter plasmid used in the yeast one-hybrid assay contains three tandem repeats of the W box-containing region of the TDC promoter (–559 to –583 relative to the TSS), driving expression of the HIS3 selection gene. The W box core sequence was mutated in one reporter plasmid to serve as a control. The chimeric CrWRKY1 fused with the yeast GAL4 activation domain (AD-CrWRKY1) was cotransformed into yeast cells with the individual reporter plasmid. Yeast colonies expressing the TDC W box reporter grew on triple (-His-Leu-Trp) selection medium with 3-amino-1,2,4-triazole (3-AT; Fig. 7B), indicating transactivation of the TDC promoter by AD-CrWRKY1. The yeast cells transformed by AD-CrWRKY1 and the reporter with the mutated W box failed to grow on the triple selection medium, indicating that the W-box elements are essential for binding and activation by AD-CrWRKY1.
Binding of CrWRKY1 to the TDC promoter was further confirmed by a transient protoplast assay (Fig. 7C). Luciferase reporter plasmids containing tandem repeats of the TDC W box or the mutated W box sequence (mTDC) were electroporated into C. roseus protoplasts alone or with AD-CrWRKY1. The luciferase activity in protoplasts electroporated with the TDC reporter + AD-CrWRKY1 was approximately 1.7-fold of that in the protoplasts electroporated with the reporter alone. Furthermore, AD-CrWRKY1 failed to activate the mTDC luciferase reporter containing the mutated W-box sequence (Fig. 7C).
Alkaloid Accumulation in Hairy Roots Overexpressing or Repressing CrWRKY1
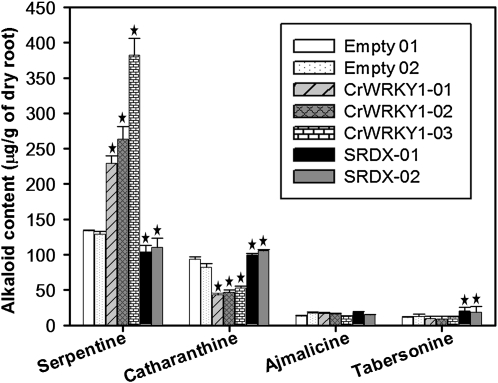
Quantification of alkaloids in vector-only control, CrWRKY1, and CrWRKY1-SRDX hairy roots was measured on an equal dry weight basis. Based on the color reaction with ceric ammonium sulfate and UV absorbance characteristics, the major alkaloids were identified by thin-layer chromatography (TLC) as serpentine, ajmalicine, catharanthine, and tabersonine. These alkaloids were subsequently analyzed by reverse-phase HPLC with a photodiode array detector in combination with liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS; Supplemental Figs. S5–S7). On average, the two independent lines of control hairy roots produced 131.5 ± 23.7, 87.5 ± 6.5, 12.8 ± 1.4, and 11.2 ± 3.0 μg g−1 dry roots of serpentine, catharanthine, ajmalicine, and tabersonine, respectively (Fig. 8). In contrast, the three independent CrWRKY1 lines showed variable but significant increases of serpentine (291.5 ± 73.2 μg g−1 dry roots) and ajmalicine (15.4 ± 1.6 μg g−1) as well as significant decreases of catharanthine (45.5 ± 5.1 μg g−1) and tabersonine (9.0 ± 3.0 μg g−1). The three independent lines of CrWRKY1-SRDX produced comparable levels of serpentine and ajmalicine to those of the control lines; however, they produced significantly higher levels of catharanthine (100.2 ± 15.1 μg g−1) and tabersonine (19.3 ± 1.6 μg g−1).
Figure 8.
Accumulation of serpentine, catharanthine, ajmalicine, and tabersonine in two empty vector control, three independent CrWRKY1, and three independent CrWRKY1-SRDX hairy root lines. On an equal dry weight basis, alkaloid extracts from control, CrWRKY1, and CrWRKY1-SRDX lines were analyzed by HPLC-diode array detection and LC-MS/MS, and the levels of serpentine, ajmalicine, and catharanthine were estimated based on peak areas. All experiments were performed in triplicate. Error bars represent sd. Stars above the bars indicate statistically significant increases or decreases in measurements compared with the control (P < 0.05, t test).
DISCUSSION
The Phytohormone-Inducible CrWRKY1 Belongs to the Group III WRKY Superfamily and Preferentially Expresses in Roots
JA induction of a number of genes encoding C. roseus TIA pathway enzymes and TFs is well established. Phytohormones, including JA and GA, have been shown to induce the expression of certain WRKY genes in other species (Naoumkina et al., 2008; Ramamoorthy et al., 2008). The presence of W boxes in the majority of characterized promoters of TIA-associated genes suggests that WRKY genes are involved in regulating these genes. In this study, we functionally and biochemically characterize the C. roseus WRKY gene, CrWRKY1. CrWRKY1 possesses the characteristics of group III WRKY, featuring a single WRKY domain in the center of the polypeptide and the unique zinc-finger motif C-X6-C-X23-H-XC (Fig. 1). The lack of both activation or repression activities, in protoplast and yeast one-hybrid assays, suggests that it mainly functions by recruiting a transcriptional activator or repressor to the targeted promoters. Although WRKY TFs can bind to W boxes as a monomer, they are also known to form homodimers as well as heterodimers with other TFs. In Arabidopsis, different WRKY18, WRKY40, and WRKY60 complexes play distinct roles in response to pathogens (Xu et al., 2006). Similar to these Arabidopsis WRKY factors, CrWRKY1-GFP is also localized in the nucleus (Fig. 2).
Expression analysis of CrWRKY1 in various tissues reveals that it preferentially expresses in roots of C. roseus (Fig. 3). The biosynthesis of TIAs is highly compartmentalized in C. roseus, presumably due to spatial and temporal transcriptional regulation. In situ RNA hybridization and immunocytochemical analyses show that TDC and STR mRNAs are heavily present in the epidermis of stems, leaves, and flower buds while also associating with cells around the apical meristem of root tips (St-Pierre et al., 1999). Hence, TFs associated with the TIA pathway are expected to display tissue- or cell-specific expression patterns.
In addition to the response to JA induction, TIA accumulation in C. roseus is also induced by other phytohormones such as ethylene and GA. The production of ajmalicine in cell suspensions, hairy roots, and shoot cultures of C. roseus is greatly enhanced by JA and ethylene but not by salicylic acid. The hormone-induced increases of the TIAs, in general, are accompanied by an increase of TDC activity (Vázquez-Flota et al., 2009). CrWRKY1 is highly up-regulated by MJ, ethylene, and GA; however, the different induction patterns of the three chemicals suggest that separate induction pathways are involved in governing CrWRKY1 expression.
CrWRKY1 Activates the TDC Gene by Targeting Its Promoter
TDC converts Trp to tryptamine, a starting substrate for the biosynthesis of TIAs. In a recent elegant endeavor to redirect the C. roseus TIA pathway to produce nonnatural TIAs, all natural TIAs were eliminated as a result of RNA interference gene silencing of TDC, creating a precursor-free system that can be explored for the production of novel TIAs by feeding unnatural starting substrates (Runguphan et al., 2009). TDC is clearly a key enzyme in the TIA pathway; thus, understanding the transcriptional regulation of TDC is important. Here, we present multiple lines of evidence to show that CrWRKY1 regulates TDC expression. First, TDC is highly up-regulated in CrWRKY1 hairy roots (Fig. 5). Second, consistent with increased TDC expression, TDC enzyme activity is elevated more than 73% in CrWRKY1 hairy roots compared with control roots. In addition, CrWRKY1 hairy roots are significantly more resistant to 4-MT inhibition than control roots (Fig. 6). Relying on the detoxification of 4-MT as selection has led to the isolation of ORCA3, an AP2-like TF that up-regulates TDC (van der Fits and Memelink, 2000). Overexpression of ORCA3 increases TDC activity and the accumulation of tryptamine in C. roseus suspension cells (van der Fits and Memelink, 2000) but not in hairy roots (Peebles et al., 2009). In contrast, CrWRKY1 hairy roots not only have increased TDC activity but also accumulate nearly 2-fold more tryptamine than control roots. In addition, CrWRKY1-SRDX expression results in an approximately 20% decrease of cellular TDC activity and a 23% reduction in tryptamine accumulation in hairy roots.
EMSA, yeast one-hybrid, and C. roseus protoplast transactivation assays were used to demonstrate the physical interaction of CrWRKY1 to W box elements in the TDC promoter (Fig. 7). Supporting evidence for the binding of CrWRKY1 to the TDC promoter in C. roseus cells also comes from CrWRKY1-SRDX hairy roots. TDC expression was reduced significantly in CrWRKY1-SRDX hairy roots (Fig. 5A), suggesting that the CrWRKY1-SRDX repressor binds to the TDC promoter, although the possibility that CrWRKY1 activates TDC by controlling the expression of an unknown TDC activator cannot be excluded. However, this seems unlikely, as it would require the existence of the same mechanism in yeast cells. Overexpression of CrWRKY1 does not affect the expression of STR, which lacks the W box consensus sequence in its promoter. Taken together, these results support that CrWRKY1 activates the TDC gene by targeting its promoter.
Overexpression or Repression of CrWRKY1 Affects the Accumulation of Serpentine in Hairy Roots
In roots of naturally grown C. roseus, serpentine and ajmalicine are the major alkaloids. The TIA pathway is highly branched and diversified from strictosidine onward (Supplemental Fig. S1). Enzymes and genes involved in the catharanthine pathway have yet to be isolated. The better-characterized vindoline pathway starts with the hydroxylation of tabersonine. Although no gene has been cloned for the serpentine pathway, it is believed that a cathenamine reductase converts cathenamine to ajmalicine, which is then metabolized to serpentine, putatively by a peroxidase. Overexpression of CrWRKY1 appears to preferentially activate the serpentine pathway at the expense of the catharanthine and tabersonine pathways, as indicated by the significant increase of serpentine and the reduced production of catharanthine and tabersonine in CrWRKY1 hairy roots. Consistent with this conclusion is the observation that the repression of CrWRKY1 by CrWRKY1-SRDX results in increases of catharanthine by approximately 15% and tabersonine by approximately 72% (Fig. 8). Feeding a synthetic tryptamine analog to the TDC-silenced C. roseus hairy roots results in a significant increase of unnatural ajmalicine, but not serpentine, indicating that the putative ajmalicine peroxidase is not sufficient to convert the increased ajmalicine to serpentine (Runguphan et al., 2009). This can either be a consequence of the inability of the peroxidase to catalyze the modified ajmalicine or its persistent low activity in the TDC-silenced lines. CrWRKY1 hairy roots accumulate considerably more serpentine than ajmalicine, indicating that the peroxidase is highly active in the conversion of ajmalicine to serpentine.
The Roles of CrWRKY1 in TIA Pathway Regulation
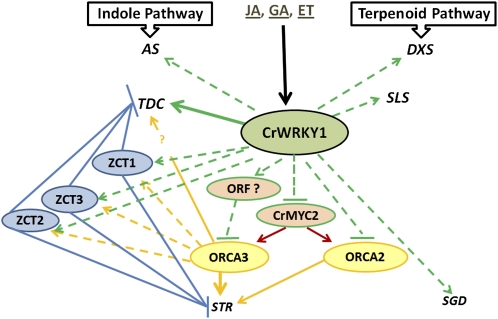
JA induces TIA-associated transcriptional activators such as CrMYC2 and ORCA3 that stimulate pathway genes, including TDC and STR (Zhang et al., 2011). However, JA also induces the repressors ZCT and GBF, proteins that are known to inhibit the expression of TDC and STR (Memelink and Gantet, 2007). The emerging picture of the transcriptional control of TIA biosynthesis suggests the simultaneous induction of activators and repressors by elicitors and developmental signals. These regulators form a dynamic network that fine-tunes the amplitude, timing, and tissue specificity of gene expression. The previously identified TIA-associated TFs include the ORCA proteins, ZCTs, GBFs, CrMYC2, and several AT-Hook DNA-binding proteins (Vom Endt et al., 2007). Here, we show that the newly characterized CrWRKY1 is a member of the TIA regulatory network. Our current understanding of CrWRKY1 illustrates the complex interaction and coregulation of the TIA pathway by multiple TFs (Fig. 9). Phytohormones (JA, GA, and ethylene) induce CrWRKY1 expression. Overexpression of CrWRKY1 up-regulates the DXS and SLS genes in the terpenoid pathway as well as the AS and TDC genes in the indole pathway. Based on the observation that CrWRKY1 by itself lacks transactivational activity, we anticipate that CrWRKY1 functionally interacts with other TFs. Overexpression of CrWRKY1 moderately up-regulates the ZCT genes but strongly represses ORCA3 and ORCA2. Expression of CrWRKY1-SRDX significantly up-regulates ORCA3 but not ORCA2 (Fig. 5B). We envision that CrWRKY1 represses ORCA3 by activating a yet to be identified ORCA3-repressing factor (ORF; Fig. 9). Memelink and colleagues propose that ORCA3 is dually controlled by sets of qualitative (on/off switch) and quantitative regulators (Vom Endt et al., 2007). In this scenario, quantitative regulators, such as the AT-Hook DNA-binding proteins, control the strength and timing of ORCA3 expression, while a potential repressor (ORF) acts as an on/off switch by occupying the ORCA3 promoter and preventing transactivation. CrWRKY1 may be an activator of ORF that is up-regulated when CrWRKY1 is overexpressed, resulting in the severe repression of ORCA3. The repression of ORF by CrWRKY1-SRDX derepresses the ORCA3 promoter, leading to the up-regulation of ORCA3. Alternatively, CrWRKY1 regulates ORCA3 by repressing CrMYC2, which controls the JA-responsive expression of ORCA3 (Zhang et al., 2011). Our results are in agreement with recent observations showing that JA induction increases TIA accumulation in C. roseus hairy roots, yet, overexpression of ORCA3 does not enhance TIA production (Peebles et al., 2009). Correspondingly, JA elicitation, but not ORCA3 overexpression, up-regulates TDC expression. In general, the transcript levels of TDC, DXS, G10H, SGD, and ORCA2 are lower in the ORCA3-overexpressing lines than in the JA-induced lines. The authors thus concluded that (1) a negative regulatory mechanism overrides the activation of these genes by ORCA3 and (2) another strong positive regulator of the TIA pathway exists in C. roseus (Peebles et al., 2009). Here, we demonstrate that CrWRKY1 is a negative regulator of ORCA3 and a positive regulator of the TIA pathway.
Figure 9.
Working model summarizing the interaction of CrWRKY1 with indole pathway genes, AS and TDC, terpenoid pathway genes, DXS and SLS, and downstream genes, STR and SGD, as well as the ORCA, CrMYC2, and ZCT regulators. Dashed lines represent interactions that may be direct or indirect. Solid lines indicate potentially direct interactions. Lines with arrows represent transactivation, and lines with bars represent transrepression. ET, Ethylene.
The genetic interaction of CrWRKY1 with ORCA3 is compelling. Overexpression of CrWRKY1 results in an up to 3-fold increase of serpentine in C. roseus hairy roots (Fig. 8); however, overexpression of TDC does not result in similar levels of serpentine production (Goddijn et al., 1995; Hughes et al., 2004). Therefore, up-regulation of TDC alone is evidently not sufficient to explain the large increase of serpentine induced by CrWRKY1. In addition, feeding the precursor, tryptamine, to C. roseus hairy roots does not lead to significant increases in serpentine or ajmalicine production (Goklany et al., 2009); simply increasing the concentration of tryptamine cannot explain how CrWRKY1 preferentially activates the serpentine branch of the pathway. We speculate that CrWRKY1 genetically interacts with other genes to regulate the TIA pathway (Fig. 9). The activator, ORCA3, apparently pushes the flux toward the vindoline and catharanthine pathways. Previous work has demonstrated that ORCA3 activates the vindoline pathway enzyme, tabersonine 16-hydroxylase (Memelink and Gantet, 2007), and when sufficient precursors from the terpenoid pathway become available, as the result of G10H overexpression, ORCA3 preferentially activates the catharanthine pathway (Wang et al., 2010a). Conversely, CrWRKY1 induces the serpentine pathway by repressing ORCA3 and activating the serpentine branch genes, including the putative peroxidase gene converting ajmalicine to serpentine. The reduction of catharanthine in CrWRKY1 hairy roots (Fig. 8) is also consistent with the repression of ORCA3 by CrWRKY1. In roots of naturally grown C. roseus, serpentine and its isomers account for approximately 65% of the alkaloids (Ferreres et al., 2010). This is consistent with our results showing high CrWRKY1 expression and relatively low ORCA3 expression in roots. Therefore, we believe that CrWRKY1 plays a key role in determining the preferential accumulation of ajmalicine and serpentine in C. roseus roots.
As a global regulator of the TIA pathway, CrWRKY1 possesses overlapping functions with the ORCA TFs. Similar to ORCA3, CrWRKY1 activates pathway enzymes including AS, DXS, SLS, SGD, and TDC as well as ZCT transrepressors (Fig. 5). These results point to the presence of cross talk within the TIA regulatory network. The picture of this network will no doubt become more complex as additional pathway enzymes and regulators are discovered and characterized. Nevertheless, the discovery and functional characterization of CrWRKY1 not only provides a biological explanation for the root-specific production of serpentine but also opens a new door to elucidating the molecular mechanisms underlying TIA biosynthesis. To the best of our knowledge, CrWRKY1 is the first C. roseus WRKY gene to be functionally characterized. Other C. roseus WRKY genes may also participate in the regulation of the TIA pathway. CrWRKY1 is capable of activating the serpentine branch to completion. This unique characteristic is biotechnologically relevant due to the pharmaceutical values of ajmalicine and serpentine. The genetic interaction between CrWRKY1 and ORCA3, as well as other regulators, can be explored for the engineering of selective TIAs. For example, silencing CrWRKY1 in C. roseus may help guide the pathway toward catharanthine and vindoline, the precursors of vinblastine and vincristine. Furthermore, CrWRKY1 hairy roots are an excellent system for gene discovery of the currently unknown enzymes that catalyze the serpentine pathway.
MATERIALS AND METHODS
Plant Materials, Isolation of WRKY cDNA, and Quantification of the CrWRKY1 Gene
Catharanthus roseus ‘Cooler Apricot’ (Swallowtail Garden) seeds were surface sterilized and germinated on half-strength Murashige and Skoog (MS) basal medium. Ten-day-old seedlings were treated with 100 μm MJ and used for total RNA isolation as described previously (Suttipanta et al., 2007). Partially degenerate PCR primers (Supplemental Table S1) were designed based on the highly conserved WRKY domain. A 180-bp product was PCR amplified by the degenerate primers using first-strand cDNA synthesized from RNA isolated from MJ-induced seedlings and cloned into the pGEM-T Easy vector (Promega) followed by sequencing. To isolate the full-length cDNA of the C. roseus WRKY genes, 5′ and 3′ RACE were applied as described previously (Suttipanta et al., 2007). The predicted polypeptide sequences of the C. roseus WRKY and other available WRKY proteins were compared using the BLAST service (http://www.ncbi.nlm.nih.gov/BLAST/). Alignment of nucleotide and deduced amino acid sequences was performed using the ClustalW program (Larkin et al., 2007) with the default parameters through the service of the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw). A phylogenetic tree was constructed and visualized using the neighbor-joining method through MEGA version 4.1 software (Tamura et al., 2007). The statistical reliability of individual nodes of the newly constructed tree was assessed by bootstrap analyses with 1,000 replications.
To study the effects of phytohormones on the expression of CrWRKY1, 10-d-old seedlings (100 mg fresh weight) were exposed to 100 μm MJ, 1 μL L−1 ethylene, or 10 μm GA following reported procedures (El-Sayed and Verpoorte, 2004). After the treatments, the seedlings were harvested at 0 and 30 min and 1.0, 2.0, 4.0, and 8.0 h and immediately frozen in liquid nitrogen. cDNA synthesis using total RNA from hypocotyl, young and mature leaf, root, and fruit was performed as described earlier (Suttipanta et al., 2007).
Reverse transcription (RT)-PCR and qPCR were performed as described by Pattanaik et al. (2010). The primers used in qPCR for several pathway genes are listed in Supplemental Table S1. The comparative threshold cycle method (Applied Biosystems bulletin) was used to measure the transcript levels. The C. roseus 40S Ribosomal Protein S9 (Rps9) gene was used as an internal control in qPCR to normalize the amount of total mRNA in all samples. The expression of Rps9 in different tissues was tested using another housekeeping gene, Elongation Factor1α (Wei, 2010), as a reference (Supplemental Fig. S8). All PCRs were performed in triplicate and repeated at least twice.
Plant Expression Vectors and Hairy Root Transformation
The CrWRKY1 gene was PCR amplified and cloned into a modified pCAMBIA2301 vector, containing the CaMV 35S promoter and the rbcS terminator, to generate pCrWRKY1 plasmid (Supplemental Fig. S3A). C. roseus hairy roots were developed as described previously (Choi et al., 2004; Suttipanta et al., 2007). The pCrWRKY1 plasmid was mobilized into Agrobacterium rhizogenes R1000 by the freeze-thaw method. After removal of the roots, 10-d-old seedlings were immersed in the Agrobacterium suspension for 30 min and then transferred onto solid MS basal medium for cocultivation. The pCAMBIA2301 vector was used as a vector-only control. After 48 h, the explants were transferred to half-strength MS basal medium containing 400 mg L−1 cefotaxime. Hairy roots usually formed at the cut surface of the explants in 3 weeks. After 6 to 7 weeks, root tips of approximately 5 mm in length were excised and transferred to one-third-strength Schenk and Hilderbrandt basal medium containing 100 mg L−1 kanamycin and 400 mg L−1 cefotaxime. Three independent hairy root lines were chosen for detailed analysis.
For the construction of the CrWRKY1-SRDX plasmid, sequence encoding the SRDX repressor domain (LDLELRLGFA) was fused to the C-terminal end of CrWRKY1. CrWRKY1-SRDX was subsequently cloned into the modified pCAMBIA2300 vector under the control of the CaMV 35S promoter and the rbcS terminator (Supplemental Fig. S3B).
Genomic DNA Isolation and PCR
Genomic DNA was isolated from independent CrWRKY1 overexpression, CrWRKY1-SRDX, and empty vector control hairy roots using the DNeasy Plant mini kit (Qiagen). To verify the presence of both pCAMBIA and Ri T-DNAs in the transgenic lines, CrWRKY1, Kanamycin, GUS, rolB, and rolC genes were amplified from the genomic DNA using gene-specific primers (Supplemental Table S1). All PCRs were performed using iProof High-Fidelity DNA polymerase (Bio-Rad) following the thermal cycling conditions recommended by the manufacturer.
Recombinant Protein Production and EMSA
The CrWRKY1 gene was cloned into the BamHI/EcoRI sites of pGEX 4T-1 vector (Amersham) to generate a GST fusion protein. The constructs were verified by DNA sequencing and transformed into Escherichia coli BL21 (DE3) cells. Cell cultures at an optical density at 600 nm of approximately 0.6 were induced by adding isopropyl b-d-thiogalactopyranoside to a final concentration of 1 mm. After induction for 3 h at 30°C, the cells were harvested by centrifugation and lysed using CelLytic B (Sigma) according to the manufacturer’s instructions. The GST fusion proteins were bound to Glutathione Sepharose 4B columns (Amersham) and eluted using 50 mm Tris-HCl, pH 7.2, 150 mm NaCl, and 30 mm glutathione.
For EMSA experiments, three DNA probes, with or without biotin labeling, were synthesized based on the W box elements of the TDC promoter (GenBank accession no. X67662): probe 1 (5′-CAAAAATTTGACTATACTTGACTATTAGTG-3′) is identical to the TDC promoter region (–1,179 to –1,150 relative to the TSS) with two W boxes (underlined); probe 2 (5′-GACTGCATGTTGACCTAAAATTATG-3′) is a native TDC promoter sequence (–583 to –559 relative to the TSS) containing a single W box; and mProbe 2 (5′-GACTGCATGTTAATCTAAAATTATG-3′) is identical to probe 2 except that the W box core sequence was destroyed by mutation. Complementary oligonucleotides, biotin labeled at the 5′ end of each strand, were synthesized by Integrated DNA Technologies and annealed to produce double-stranded probes for EMSA. The DNA-binding reactions were carried out in 10 mm Tris-HCl, pH 7.5, 50 mm KCl, 1 mm dithiothreitol, 5% glycerol, and 100 ng of poly(dI:dC) in a final volume of 20 μL. Purified proteins were incubated with 0.25 to 0.5 nm DNA probe at room temperature for 90 min. The DNA-protein complexes were resolved by electrophoresis on 6% nondenaturing polyacrylamide gels and then transferred to BiodyneB modified membrane (0.45 μm; Pierce). The band shifts were detected by a chemiluminescent nucleic acid detection module (Pierce) and exposed to x-ray films.
Yeast One-Hybrid Assay
The pHIS2 vector (Clontech), containing the HIS3 nutritional reporter gene downstream of a multiple cloning site and the minimal promoter of the HIS3 locus, was used for making the reporter plasmid. Three tandem copies of the W box-containing region of the TDC promoter (–583 to –559 relative to the TSS) were synthesized and ligated into the EcoRI and SacI sites of the pHIS2 vector, upstream of the HIS3 minimal promoter, to generate the reporter plasmid (pHIS2-TDC). A reporter plasmid with the mutated W box sequence (pHIS2-mTDC) was also made to serve as a control. The full-length CrWRKY1 cDNA was cloned into the yeast expression plasmid, pAD-GAL4-2.1 (Stratagene), to generate the effector plasmid. The reporter and effector plasmids were transformed into yeast strain Y187 (Clontech), and transformants were selected on synthetic dropout (SD) medium lacking Leu and Trp. Transformed colonies were then streaked on SD-His-Leu-Trp medium with 100 mm 3-AT to check TDC promoter activation.
Plasmid Construction for TDC Promoter Assay and CrWRKY1 Nuclear Localization
The two reporter plasmids used in the protoplast assay consist of the firefly Luciferase coding sequence under the control of a minimal CaMV 35S (−46 to +8) promoter with three tandem copies of the W box region of the TDC promoter (–583 to –559 relative to the TSS) upstream and an rbcS terminator. The W box core sequence was mutated in one reporter plasmid to serve as a control. The effector plasmids consist of the CaMV 35S promoter and the GAL4 activation domain (GALAD) fused to CrWRKY1 coding sequences and the rbcS terminator. A plasmid containing the GUS gene under the control of the CaMV 35S promoter and the rbcS terminator was used as an internal control.
For subcellular localization, two plasmids were constructed, one containing the enhanced GFP (eGFP) and the other containing a CrWRKY1-eGFP fusion. The expression of both genes was under the control of the CaMV 35S promoter and the rbcS terminator.
Protoplast Isolation and Electroporation
Protoplast isolation from C. roseus cell suspension cultures (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH no. 510) and electroporation with supercoiled plasmid DNA were performed using protocols previously developed for tobacco (Nicotiana tabacum) protoplasts (Pattanaik et al., 2010). The reporter, effector, and internal control plasmids were electroporated into C. roseus protoplasts; luciferase and GUS activities in transfected protoplasts were measured as described previously (Suttipanta et al., 2007). The luciferase activity was normalized against GUS activity and expressed as fold activation relative to the reporter-only control. For subcellular localization, the plasmids, containing either eGFP or CrWRKY1-eGFP, were individually electroporated into C. roseus protoplasts and visualized with a fluorescence microscope after 20 h of incubation at room temperature.
Enzyme Assay, Alkaloid Extraction, TLC, HPLC, and LC-MS
For TDC activity assay and alkaloid extraction, we followed the protocols described by Miranda-Ham et al. (2007). In routine assays, 20-μL aliquots of hairy root extracts or commercial alkaloid standards were applied onto TLC plates (silica G60; Merck). The TLC was run on solvent ethyl acetate:NH4OH:chloroform (75:2:23), and alkaloids were identified under UV (λ = 254 and 366 nm) and by color reaction with ceric ammonium sulfate.
The alkaloid extracts were further analyzed using LC-ESI-MS/MS with a Varian 1200 L Triple Quadruple MS/MS device equipped with a UV-diode array detection spectrophotometer. The LC was carried out on a 250-mm × 4.5-mm, 5-μm C-18 column (Zorbax Eclipse XDB; Agilent Technologies) with a flow rate of 0.2 mL min−1. The mobile phases were water-0.1% formic acid (A) and methanol-0.1% formic acid (B). A linear gradient from 90% A and 10% B to 10% A and 90%B was applied in 30 min. The MS/MS detector equipped with an ESI system was operated with a capillary voltage of 30 to 60 kV and heated at 300°C. Nitrogen was used as a flowing dry gas at a pressure of 20 ψ and as a nebulizing gas at a pressure of 50 ψ. Analysis was performed in scan mode ranging from mass-to-charge ratio 100 to 900. Collision-induced dissociation experiments were performed in the ion trap using helium as the collision gas. The collision energy varied from 6 to 27. The isolation width of the parent ion for following MS fragmentation events was set at ±0.5. Alkaloids were verified by their retention time (Rt), diode array profiles, and MS/MS spectra compared with authentic standards. Serpentine is detected at Rt of 12 min with λmax of 249 nm/306 nm/367 nm and +MS: 349[M+H]+, +MS2(349): 317; catharanthine at Rt of 17 min with λmax of 225 nm/283 nm and +MS: 337[M+H]+, +MS2(337): 173, 144; ajmalicine at Rt of 22 min with λmax of 226 nm/244 nm/283 nm and +MS: 353[M+H]+, +MS2(353): 321, 222, 210, 144; and tabersonine at Rt of 24 min with λmax of 227 nm/292 nm/267 nm and +MS: 337[M+H]+, +MS2(377): 305.
For quantitative determination of serpentine, catharanthine, ajmalicine, and tabersonine, reverse-phase HPLC was performed using a 250-mm × 4.5-mm, 5-μm C-18 column and a linear solvent gradient from 80% to 20% 5 mm phosphate buffer and 20% to 80% acetonitrile, at a flow rate of 1.0 mL min−1, and monitored by a Waters 2998 photodiode array detector. The authentic alkaloid standards were used to establish standard curves for quantification. Data were analyzed using the Masslynx software. Chromatographic peaks were integrated and compared with the respective standard curves to calculate the total amount of alkaloids in each sample.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number HQ646368.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Biosynthetic pathway of C. roseus TIAs.
Supplemental Figure S2. qRT-PCR analysis of CrWRKY1 and TDC expression levels in C. roseus seedlings upon induction by MJ.
Supplemental Figure S3. Plant expression vectors used for generating transgenic hairy roots and PCR confirmation of the transgenes.
Supplemental Figure S4. Relative expression levels of selected TIA pathway genes in CrWRKY1 and CrWRKY1-SRDX hairy roots.
Supplemental Figure S5. Alkaloid production in CrWRKY1, CrWRKY1-SRDX, and empty vector control hairy roots.
Supplemental Figure S6. UV spectra of serpentine, catharanthine, ajmalicine, and tabersonine.
Supplemental Figure S7. MS/MS spectra of serpentine, catharanthine, ajmalicine, and tabersonine.
Supplemental Figure S8. qRT-PCR analysis of Rps9 expression levels relative to Elongation Factor1a in C. roseus.
Supplemental Table S1. Oligonucleotides used in PCR and qPCR.
Acknowledgments
We thank J. May (Department of Civil Engineering and Environmental Research Training Laboratories, University of Kentucky) for assistance and advice on LC-MS/MS. We also thank S. O’Connor (John Innes Centre) for providing C. roseus cell culture. We appreciate K. Shen (University of Kentucky) for her helpful suggestions and critical reading of the manuscript.
References
- Aerts RJ, Gisi D, De Carolis E, De Luca V, Baumann TW. (1994) Methyl jasmonate vapor increases the developmentally controlled synthesis of alkaloids in Catharanthus and Cinchona seedlings. Plant J 5: 635–643 [Google Scholar]
- Choi PS, Kim YD, Choi KM, Chung HJ, Choi DW, Liu JR. (2004) Plant regeneration from hairy-root cultures transformed by infection with Agrobacterium rhizogenes in Catharanthus roseus. Plant Cell Rep 22: 828–831 [DOI] [PubMed] [Google Scholar]
- Costa MM, Hilliou F, Duarte P, Pereira LG, Almeida I, Leech M, Memelink J, Barceló AR, Sottomayor M. (2008) Molecular cloning and characterization of a vacuolar class III peroxidase involved in the metabolism of anticancer alkaloids in Catharanthus roseus. Plant Physiol 146: 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed M, Verpoorte R. (2004) Growth, metabolic profiling and enzymes activities of Catharanthus roseus seedlings treated with plant growth regulators. Plant Growth Regul 44: 53–58 [Google Scholar]
- El-Sayed M, Verpoorte R. (2007) Catharanthus terpenoid indole akaloids: biosynthesis and regulation. Phytochem Rev 6: 277–305 [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, De Luca V. (2008) Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J 54: 763–784 [DOI] [PubMed] [Google Scholar]
- Ferreres F, Pereira DM, Valentão P, Oliveira JM, Faria J, Gaspar L, Sottomayor M, Andrade PB. (2010) Simple and reproducible HPLC-DAD-ESI-MS/MS analysis of alkaloids in Catharanthus roseus roots. J Pharm Biomed Anal 51: 65–69 [DOI] [PubMed] [Google Scholar]
- Goddijn OJ, Pennings EJ, van der Helm P, Schilperoort RA, Verpoorte R, Hoge JH. (1995) Overexpression of a tryptophan decarboxylase cDNA in Catharanthus roseus crown gall calluses results in increased tryptamine levels but not in increased terpenoid indole alkaloid production. Transgenic Res 4: 315–323 [DOI] [PubMed] [Google Scholar]
- Goddijn OJ, van der Duyn Schouten PM, Schilperoort RA, Hoge JH. (1993) A chimaeric tryptophan decarboxylase gene as a novel selectable marker in plant cells. Plant Mol Biol 22: 907–912 [DOI] [PubMed] [Google Scholar]
- Goklany S, Loring RH, Glick J, Lee-Parsons CW. (2009) Assessing the limitations to terpenoid indole alkaloid biosynthesis in Catharanthus roseus hairy root cultures through gene expression profiling and precursor feeding. Biotechnol Prog 25: 1289–1296 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hong SB, Peebles CA, Shanks JV, San KY, Gibson SI. (2006) Expression of the Arabidopsis feedback-insensitive anthranilate synthase holoenzyme and tryptophan decarboxylase genes in Catharanthus roseus hairy roots. J Biotechnol 122: 28–38 [DOI] [PubMed] [Google Scholar]
- Hughes EH, Hong SB, Gibson SI, Shanks JV, San KY. (2004) Metabolic engineering of the indole pathway in Catharanthus roseus hairy roots and increased accumulation of tryptamine and serpentine. Metab Eng 6: 268–276 [DOI] [PubMed] [Google Scholar]
- Kato N, Dubouzet E, Kokabu Y, Yoshida S, Taniguchi Y, Dubouzet JG, Yazaki K, Sato F. (2007) Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol 48: 8–18 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Liu DH, Jin HB, Chen YH, Cui LJ, Ren WW, Gong YF, Tang KX. (2007) Terpenoid indole alkaloids biosynthesis and metabolic engineering in Catharanthus roseus. J Integr Plant Biol 49: 961–974 [Google Scholar]
- Memelink J, Gantet P. (2007) Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Phytochem Rev 6: 353–362 [Google Scholar]
- Menke FL, Champion A, Kijne JW, Memelink J. (1999) A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J 18: 4455–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Ham ML, Islas-Flores I, Vázquez-Flota AF. (2007) Accumulation of monoterpenoid indole alkaloids in periwinkle seedlings (Catharanthus roseus) as a model for the study of plant-environment interactions. Biochem Mol Biol Educ 35: 206–210 [DOI] [PubMed] [Google Scholar]
- Montiel G, Breton C, Thiersault M, Burlat V, Jay-Allemand C, Gantet P. (2007) Transcription factor Agamous-like 12 from Arabidopsis promotes tissue-like organization and alkaloid biosynthesis in Catharanthus roseus suspension cells. Metab Eng 9: 125–132 [DOI] [PubMed] [Google Scholar]
- Murata J, Bienzle D, Brandle JE, Sensen CW, De Luca V. (2006) Expressed sequence tags from Madagascar periwinkle (Catharanthus roseus). FEBS Lett 580: 4501–4507 [DOI] [PubMed] [Google Scholar]
- Naoumkina MA, He X, Dixon RA. (2008) Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor SE, Maresh JJ. (2006) Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep 23: 532–547 [DOI] [PubMed] [Google Scholar]
- Ouwerkerk PBF, Memelink J. (1999) Elicitor-responsive promoter regions in the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol Biol 39: 129–136 [DOI] [PubMed] [Google Scholar]
- Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, Patra B, Yuan L. (2010) Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231: 1061–1076 [DOI] [PubMed] [Google Scholar]
- Pauw B, Hilliou FA, Martin VS, Chatel G, de Wolf CJ, Champion A, Pré M, van Duijn B, Kijne JW, van der Fits L, et al. (2004) Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J Biol Chem 279: 52940–52948 [DOI] [PubMed] [Google Scholar]
- Peebles CA, Hughes EH, Shanks JV, San KY. (2009) Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metab Eng 11: 76–86 [DOI] [PubMed] [Google Scholar]
- Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S. (2008) A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol 49: 865–879 [DOI] [PubMed] [Google Scholar]
- Roepke J, Salim V, Wu M, Thamm AM, Murata J, Ploss K, Boland W, De Luca V. (2010) Vinca drug components accumulate exclusively in leaf exudates of Madagascar periwinkle. Proc Natl Acad Sci USA 107: 15287–15292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runguphan W, Maresh JJ, O’Connor SE. (2009) Silencing of tryptamine biosynthesis for production of nonnatural alkaloids in plant culture. Proc Natl Acad Sci USA 106: 13673–13678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Sibéril Y, Benhamron S, Memelink J, Giglioli-Guivarc’h N, Thiersault M, Boisson B, Doireau P, Gantet P. (2001) Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol Biol 45: 477–488 [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Vazquez-Flota FA, De Luca V. (1999) Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11: 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttipanta N, Pattanaik S, Gunjan S, Xie CH, Littleton J, Yuan L. (2007) Promoter analysis of the Catharanthus roseus geraniol 10-hydroxylase gene involved in terpenoid indole alkaloid biosynthesis. Biochim Biophys Acta 1769: 139–148 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Zhang H, Menke FLH, Deneka M, Memelink J. (2000) A Catharanthus roseus BPF-1 homologue interacts with an elicitor-responsive region of the secondary metabolite biosynthetic gene Str and is induced by elicitor via a JA-independent signal transduction pathway. Plant Mol Biol 44: 675–685 [DOI] [PubMed] [Google Scholar]
- Vázquez-Flota F, Hernández-Domínguez E, de Lourdes Miranda-Ham M, Monforte-González M. (2009) A differential response to chemical elicitors in Catharanthus roseus in vitro cultures. Biotechnol Lett 31: 591–595 [DOI] [PubMed] [Google Scholar]
- Vom Endt D, Soares e Silva M, Kijne JW, Pasquali G, Memelink J. (2007) Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol 144: 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CT, Liu H, Gao XS, Zhang HX. (2010a) Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Rep 29: 887–894 [DOI] [PubMed] [Google Scholar]
- Wang Q, Yuan F, Pan Q, Li M, Wang G, Zhao J, Tang K. (2010b) Isolation and functional analysis of the Catharanthus roseus deacetylvindoline-4-O-acetyltransferase gene promoter. Plant Cell Rep 29: 185–192 [DOI] [PubMed] [Google Scholar]
- Wei S. (2010) Methyl jasmonic acid induced expression pattern of terpenoid indole alkaloid pathway genes in Catharanthus roseus seedlings. Plant Growth Regul 61: 243–251 [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, et al. (2005) Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17: 944–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pre M, Gantet P, Memelink J. (2011) The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J 67: 61–71 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang L. (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ML, Shao JR, Tang YX. (2009) Production and metabolic engineering of terpenoid indole alkaloids in cell cultures of the medicinal plant Catharanthus roseus (L.) G. Don (Madagascar periwinkle). Biotechnol Appl Biochem 52: 313–323 [DOI] [PubMed] [Google Scholar]