Abstract
In many species, the Y (or W) chromosome carries relatively few functional genes. This observation motivates the null hypothesis that the Y will be a minor contributor to genetic variation for fitness. Previous data and theory supported the null hypothesis, but evidence presented here shows that the Y of Drosophila melanogaster is a major determinant of a male's total fitness, with standing genetic variation estimated to be 68% of that of an entire X/autosome genomic haplotype. Most Y-linked genes are expressed during spermatogenesis, and correspondingly, we found that the Y influences fitness primarily through its effect on a male's reproductive success (sperm competition and/or mating success) rather than his egg-to-adult viability. But the fitness of a Y highly depended on the genetic makeup of its bearer, reverting from high to low in different genetic backgrounds. This pattern leads to large epistatic (inconsistent among backgrounds) but no additive (consistent among backgrounds) Y-linked genetic variance for fitness. On a microevolutionary scale, the observed large epistatic variation on the Y substantially reduces heritable variation for fitness among males, and on a macroevolutionary scale, the Y produces strong selection for genomic rearrangements that move interacting genes onto the nonrecombining region of the Y.
Comparative evidence indicates that the Y chromosome arises from an ordinary autosome when it stops recombining with the X and subsequently degenerates (1–3). The Y chromosome of Drosophila melanogaster constitutes about 12% of a male's genomic DNA but most of this is apparently noncoding heterochromatic DNA (4). The Y is known to code only for (i) six male fertility factors [which block sperm development when deleted (4)], (ii) a trans-acting enhancer of autosomal loci that is expressed in developing spermatocytes,† and (iii) the bobbed locus of ribosomal DNA repeats (5). Despite these important functions, the Y is not essential for viability in males (5, 6), has no conspicuous effect when placed in females (5, 6), has little or no influence on most quantitative traits (refs. 5 and 6, but see ref. 7), and contains proportionately few of thousands of mapped genes of Drosophila (8).
Given both its low gene content and minor influence on quantitative traits, the contribution of the Y to genetic variance in fitness might be expected to be small. This view was supported by theory indicating more stringent requisite conditions to maintain nonneutral polymorphism on the Y compared with the X and autosomes (9, 10). Yet, molecular variation at the Y-linked bobbed locus is substantial (11–13); polymorphism for suppressors of meiotic drive and for a ribosomal DNA deletion have been found on the Y of other species (14–21), and studies on the genetics of speciation (18, 22) and meiotic drive (14–17) suggest that the Y evolves rapidly in a nonneutral manner.
Data supporting the view that the Y chromosome of D. melanogaster contains little or no polymorphism for fitness come from studies in which no additive genetic variation was found for (i) total fitness, (ii) segregation distortion (i.e., meiotic drive), and (iii) noncompetitive male fertility (neither additive nor epistatic genetic variation; refs. 23–25). All previous studies, however, were unable to measure a critically important parameter: epistatic genetic variance for total fitness.
Epistatic fitness variation, with little or no additive variation, is the hallmark of stable nonneutral polymorphism on the Y that is maintained because of Y/X or Y/autosome genetic interactions. In this case, genetic variation is present but gene frequencies do not change in response to selection. An assay for the presence and relative magnitude of epistatic variation is critical to understanding the evolution of the Y chromosome. Here we report the results of such an assay.
Materials and Methods
Sampling Y Chromosomes.
Y chromosomes were sampled from a large outbred population (LHM). This base population was derived from a larger laboratory population (LH, named for its collector Larry Harshman, Davis, CA) that was produced from 400 inseminated females collected from central California in 1991. It was maintained subsequently on 2-week generations at high density in 20–26 half-pint vessels (mixed each generation) under a 12-h light/12-h dark diurnal cycle at 25°C with Ne > 5,000. LHM is a moderate-density population that was founded in November 1995 from 2,000 LH adults at generation 105 of their laboratory culture. All general features of the LH culture environment were preserved except that larvae were reared at moderate density (200–250 per 10-dram vial provisioned with 10 ml of cornmeal/molasses medium).
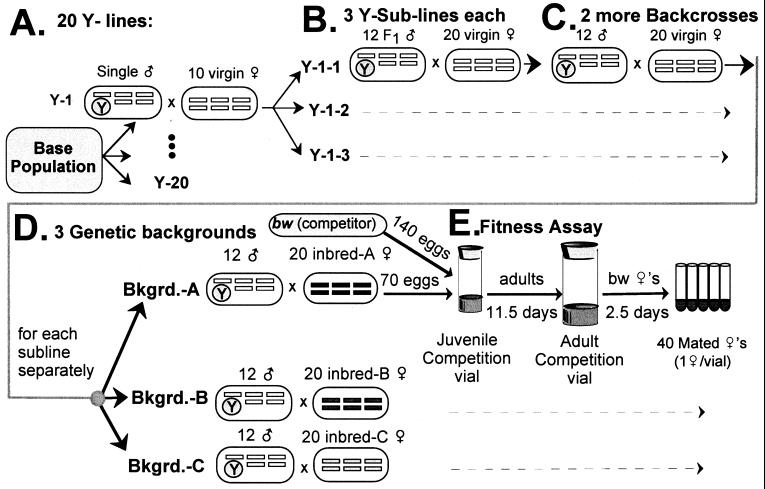
To begin the assay, 20 Y chromosomes were sampled randomly by collecting 20 males from the LHM base population (Fig. 1A). Each male was used to found a separate line (Y line) that was fixed for his Y chromosome. The 20 Y lines were split immediately into three replicate lineages (Y sublines) that were all fixed for the same Y chromosome (Fig. 1B). In sum, 60 lines were established—20 different Y-chromosome lines, each replicated with 3 sublines.
Figure 1.
Protocol to establish replicated iso-male lines and assay the fitness of their Y chromosomes. Karyotypes are depicted within ellipses with chromosome I (X/Y) (left), autosome II (center), and autosome III (right; dot chromosome IV is not shown). Rectangles depict wild-type chromosomes from the base population and dashed arrows indicate replication of the protocol steps immediately above. Except where noted, all females were taken from the LHM base population. (A) A single wild-type male from the base population initiated an iso-male line (Y-chromosome line) that carried a single, clonally propagated Y chromosome (20 different Y-chromosome lines were made in this way). (B) Each Y chromosome line was replicated three times (Y sublines). (C) Each Y subline was backcrossed to the base population to further remove the association between the Y and its original genetic background. (D) Males from each Y subline were crossed to females from three different inbred lines (A–C) immediately before the fitness assay. (E) Fitness of the target Y chromosomes was measured (see text).
Genetic Background of Assayed Y Chromosomes.
To accurately measure the fitness variation of Y chromosomes, each Y needed to be disassociated from its original genetic background. This disassociation was done by independently backcrossing each Y subline to the base population three times (Fig. 1 B and C) and then crossing each backcrossed Y subline to three different inbred lines (Fig. 1C).
Path analysis was used to calculate how much of the genetic background from the original male (or his 10 mates) remained identical by descent (ibd) among different sublines of the same Y at the time of the fitness assay [e.g., among sublines Y-1-1 {background-A} vs. Y-1-2 {background-A} vs. Y-1-3 {background-A}]. At this time, males expressing the same Y chromosome in different sublines shared [by descent from the same founding sire (or one of the 10 founding dams)] (i) 0% of their X chromosomes, (ii) 0% of their autosomes that were derived from the inbred line, and (iii) only 0.1% of the remaining autosomes that were potentially ibd from the founding cross (1 male × 10 females) that was used to initiate each Y-line (Fig. 1A), i.e., prob[ibd] = 0.001 for the uncontrolled autosomes. These calculations of ibd for the uncontrolled autosomes assume neutrality. The small effective population size used during the backcross procedure (Ne < Nmales/2 = 6), however, would have obviated most of the influence of selection, making the prob[ibd] calculations reasonable approximations.
In sum, the proportion of the total genome shared by males from the three different sublines of each Y line (which could potentially inflate the estimated genetic variation among Y chromosomes) is only 0.0002. So although the replicated Y sublines may share some genes from the original cross, the amount is expected to be small. This low genetic similarity for the uncontrolled autosomes guaranteed that ibd from the parents founding each Y-chromosome line did not substantially contribute to the estimate of Y-linked fitness variation.
To increase experimental power in detecting Y-linked fitness variation, each Y subline was crossed to an inbred line immediately before the fitness assay (Fig. 1D). This cross caused the genome of a male expressing a target Y to have two parts: (i) an experimentally controlled X/autosome genomic haplotype that was derived from one of three inbred lines (A, B, or C), and (ii) the remaining homologous part of the autosomal genome that was derived randomly from the LHM base population. The inbred lines were derived from the LHM base population by brother–sister mating for 8 generations. This protocol (i) permitted potential Y/X or Y/autosome genetic interaction to be expressed consistently among offspring that were derived from the same cross to an inbred line, (ii) produced a genetic background that had normal levels of heterozygosity and genetic diversity, and (iii) increased genetic uniformity among offspring and thereby increased experimental power in detecting subtle effects of the Y on fitness.
Assaying Fitness of the Y.
Fitness variation among the Y chromosomes was measured by competing males expressing a target Y against males expressing a visible genetic marker, bw, which produced brown eye color (the wild-type eye color of target males is red). The bw competitor stock was derived from a bw stock that was backcrossed repeatedly (8–9 times) through the LHM base population so that its genetic background closely matched that of the LHM base population.
To begin the fitness assay, eggs carrying the target Y were combined at a 1:2 ratio with those from the bw competitor stock (Fig. 1E). Flies were reared under the same culturing protocol to which the base population had adapted for over 200 generations. To duplicate these culturing conditions, 210 eggs first were added to a juvenile competition vial (containing fresh medium), where larvae, pupae, and newly emerged adults were reared (Fig. 1E). After 11 days, all adult flies were transferred, with brief CO2 anesthesia, from their juvenile rearing vial to an adult competition vial (containing fresh medium seeded with live yeast; Fig. 1E). Juvenile fitness was scored by counting the number of adult target males alive on day 14 of the 2-week generation cycle. Mortality between days 11 and 14 was rare, as ascertained by searching for males that had died during this interval (dead males were visible on the surface of the adult competition vials).
Adult fitness was measured by the number females fertilized by target vs. bw competitor males. To ensure that this measure was not confounded by juvenile fitness (egg-to-adult survival), we compared the number of females mated by target males to the number of target vs. competitor adult males that were available to mate these females. Paternity analysis of a sample of bw females was made on the last day of the 14-day generation cycle. Because the bw females were homozygous for the recessive bw marker, the identity of her mate(s) (red-eyed target male vs. brown-eyed competitor male) could be determined by the eye color of her offspring. A total of 40 homozygous bw females was taken from each adult competition vial and then individually cultured at 1 female per vial. Families from the individually cultured females were binned into five categories, based on a scan using a dissecting microscope, of the anesthetized progeny. The bin categories were all brown, strong-majority brown, many brown and red, strong-majority red, and all red. Only the many-brown-and-red category was counted individually. Females were assigned dichotomously to be mated to a target, red-eyed male or a brown-eyed competitor when >66% of her offspring expressed the respective eye color.
Families not expressing a strong eye-color majority (33–65% red-eyed progeny) were rare (4.4%) and scored as uninformative. The binning procedure has the pragmatic advantage of reducing labor and thereby permits far more females to be scored, compared with the case where all progeny were counted. More importantly, the binning procedure does not confound the viability of progeny with the reproductive success of sires, because the last male to mate a female generally sires most of her offspring. Any fitness variation that was missed because of the binning procedure can bias the estimated variation among Y chromosomes only downward.
In sum, juvenile fitness of target males was measured by the number of adult target males observed on day 14 of the culturing cycle compared with the expected number of 35 target male eggs present at the start of the assay. Comparison of the percentage of females mated to red- vs. brown-eyed males to the percentages of these males in the pool of available adult males measured the adult fitness of target males. The same comparison to a value of 33% (starting frequency of target-male zygotes) measured the total fitness of target males.
Genomewide Fitness Variation.
To assess the magnitude of the variation in fitness among Y chromosomes on a relative scale, we needed to establish a benchmark for comparison. We chose the additive genetic variation in total fitness of genomic haplotypes to represent such a benchmark. This is the genetic variation associated with a genomewide set of genes, equivalent to the genome of an egg or sperm. The genomic haplotypes contained all genes located on chromosomes I (X), II, and III, excepting for pragmatic reasons the small number (<1%) located on the dot chromosome IV. Comparing the genetic variation on the Y to that of a nearly full set of genes allowed us to express the genetic variation on the Y as a percentage of genomewide fitness variation.
To measure the fitness of genomic haplotypes, we used several cytogenetic tools that permitted an entire genomic haplotype to be cloned and then made to cosegregate in a manner similar to a single giant Y chromosome. As a consequence, the fitness assay of genomic haplotypes was functionally equivalent to that of Y chromosomes described in Fig. 1. A detailed description of the protocol used to sample, clone, and measure the fitness of genomic haplotypes can be found in ref. 26.
Fitness Measures and Statistical Procedures.
Net fitness of a Y chromosome, and its juvenile and adult components, was expressed as relative fitness. Each raw data point (e.g., percent survival) was standardized by dividing the observed measure by the highest average value among the 20 sampled Y chromosomes. This procedure causes the highest fitness Y chromosome to have an average relative fitness of 1.0, with the fitness of all other Y chromosomes scaled proportionately. The same scaling method was used in analyzing the benchmark of 69 genomic haplotypes to standardize measurements and to permit quantitative comparison to the assay of Y-linked fitness variation. Nonstandardized raw data are provided in the Appendix.
Additive genetic variance for net fitness, and its juvenile and adult components, was estimated with a random-effects ANOVA by using the jmp (SAS Institute, Cary, NC) statistical package. The random effect of primary interest was genotype (i.e., the 20 Y chromosomes or 69 genomic haplotypes). Additional random effects were (i) blocks [fitness assays were replicated independently over three consecutive days (blocks) with all genetic units (i.e., the 20 Y chromosomes or 69 genomic haplotypes) represented within each block] and, in the case of the Y chromosomes, (ii) genetic backgrounds (A, B, or C). Epistatic variance for the Y chromosomes was estimated by combining the data from all three genetic backgrounds (A, B, and C) and estimating the epistatic variance by using the “variance components estimates” routine within the jmp statistical package.
Results
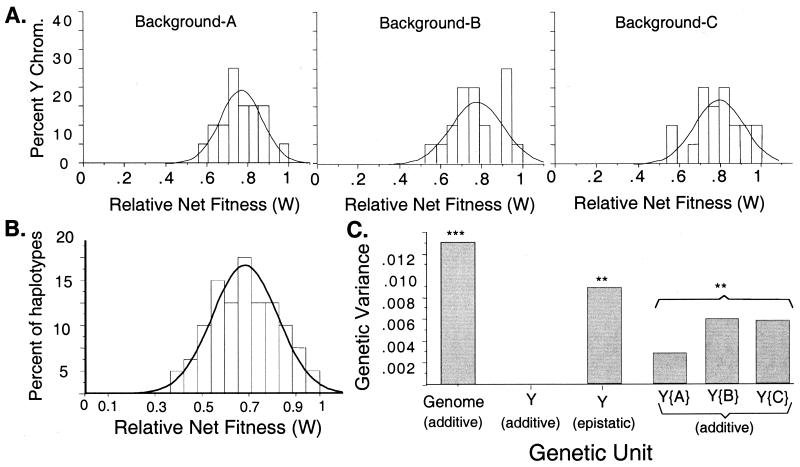
The benchmark assay of 69 genomic haplotypes revealed high levels of additive genetic variation for net fitness within the LHM base population (VG(add) = 0.013, SDG(add) = 0.114, and genetic coefficient of variation = 17%; Fig. 2B). The best genomic haplotypes produced offspring with average fitness almost 200% higher than the worst. The estimated VG(add) represents the standing genetic variance for the average fitness of outbred offspring that would be produced by different randomly selected sperm or egg genotypes (genomic haplotypes). The observed high magnitude of VG(add) has important implications for many evolutionary phenomena, e.g., adaptive female choice of mates, background selection, background trapping, selective sweeps, and the adaptive significance of recombination.
Figure 2.
The phenotypic distribution of relative net fitness (W) for the 20 sampled Y chromosomes in three different genetic backgrounds (A, B, and C) in A, and the benchmark sample of 69 genomic haplotypes in B. Each fitness measure is the average from 3 replicates, and in each replicate, the genomic haplotype was expressed in an average of 35 different genetic backgrounds (chromosomes Y, II, and III). (C) Comparison of the genetic variances of the 20 Y chromosomes (Table 1) to that of the 69 benchmark genomic haplotypes. The designation Y{i} represents the additive genetic variance among Y chromosomes within genetic backgrounds i = A, B, or C. ***, P < 0.001; **, P < 0.01, for :Ho. Genetic variance = 0.
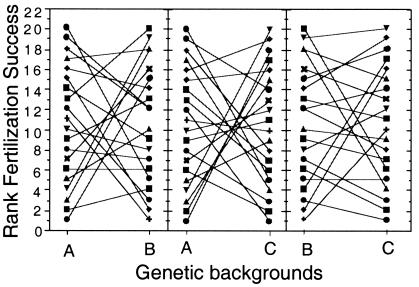
Within each of the three tested genetic backgrounds, there was substantial additive genetic variance among the sample of 20 Y chromosomes [Fig. 2A and entries Y{A},Y{B}, and Y{C} in Fig. 2C]. When combined across genetic backgrounds, however, VG(add) of the Y chromosomes was not significantly different from zero (its point estimate was negative and set to 0, Table 1; Fig. 2C). This pattern of high additive genetic variation within genetic backgrounds, but not between, is diagnostic of epistatic genetic variation (i.e., genetic variation that is inconsistent in its expression among different genetic backgrounds). The epistatic genetic variance of the Y is statistically significant (ANOVA, P = 0.0088, Table 1) and its biological significance is indicated by its estimated magnitude, i.e., 68% of that of the benchmark of entire sets of genes (Fig. 2C). The high epistatic variance in total fitness was associated with strong reversals in the rank order of reproductive success of the same Y chromosome in different genetic backgrounds (Fig. 3). Collectively, these data demonstrate that the selective value of a particular Y chromosome varies over a large range, with the fitness of a Y depending strongly on the genetic background in which it is expressed.
Table 1.
Random-effects ANOVA for the 20 Y chromosomes
| Source | MS | df | F ratio | Prob > F |
|---|---|---|---|---|
| Net fitness | ||||
| Y chromosome | 0.0408 | 19 | 0.616 | 0.8662 |
| Block | 0.1517 | 2 | 3.341 | 0.1102 |
| Y chromo. × Block | 0.0396 | 38 | 1.342 | 0.1383 |
| Gen. Back. | 0.8395 | 2 | 13.556 | 0.0017 |
| Y chromo. × Gen. Back. | 0.0507 | 38 | 1.902 | 0.0088 |
| Block × Gen. Back. | 0.0353 | 4 | 1.198 | 0.3186 |
| Juvenile fitness | ||||
| Y chromosome | 0.0218 | 19 | 1.577 | 0.2320 |
| Block | 0.1638 | 2 | 2.612 | 0.2030 |
| Y chromo. × Block | 0.0148 | 38 | 0.778 | 0.8012 |
| Gen. Back. | 0.3450 | 2 | 5.230 | 0.0801 |
| Y chromo. × Gen. Back. | 0.0181 | 38 | 0.950 | 0.5592 |
| Block × Gen. Back. | 0.0669 | 4 | 3.520 | 0.0109 |
| Adult fitness | ||||
| Y chromosome | 0.0418 | 19 | 0.796 | 0.6938 |
| Block | 0.4692 | 2 | 10.517 | 0.0231 |
| Y chromo. × Block | 0.0293 | 38 | 1.097 | 0.3596 |
| Gen. Back. | 0.4806 | 2 | 7.356 | 0.0146 |
| Y chromo. × Gen. Back. | 0.0500 | 38 | 1.873 | 0.0103 |
| Block × Gen. Back. | 0.0420 | 4 | 1.574 | 0.1897 |
| Denominator MS Synthesis | |
|---|---|
| Test denominator synthesis | |
| Y chromosome (Y) | Y × Block + Y × Gen. Back. − 1 × Residual |
| Block | Y × Block + Block × Gen. Back. − 1 × Residual |
| Y chromo. × Block | Residual |
| Gen. Back. | Y × Gen. Back. + Block × Gen. Back. − 1 × Residual |
| Y chromo. × Gen. Back. | Residual |
| Block × Gen. Back. | Residual |
See text for details of the model's main effects and design. MS, mean square.
Figure 3.
An interaction plot of the rank order of adult fitness of the 20 Y chromosomes. Only adult fitness, as opposed to juvenile fitness, contributed significantly to the total genetic variation for fitness of Y chromosomes (Table 1). The rank order of fitness reverts from high to low in different genetic backgrounds.
Decomposition of total fitness into its two major components (Table 1) demonstrates that the Y has no measurable effect on juvenile fitness but has a strong effect on adult fitness. Regression of total fitness on juvenile and adult fitness components indicates that juvenile and adult fitness account for r2 = 7% (juvenile) and r2 = 82% (adult) of the variation in total fitness.
As a further check on the potential confound of a genetic association between the Y chromosomes and their original autosomal genetic background, we have repeated the assay described in Fig. 1 in a new genetic background (inbred line D). This procedure was done in the context of a new experiment designed to resolve whether the Y-linked fitness variation was caused by male mating success vs. sperm competition. In this experiment, 28 new Y chromosomes were assayed for fitness, the number of backcrosses in step C of Fig. 1 was increased from 2 to 8, and the number of sublines (Fig. 1B) was increased from 3 to 9. The observed additive genetic variation among Y chromosomes in genetic background D was estimated to be 0.0086 (H0: VG(add) = 0.0; P = 0.0006), i.e., a value corroborating the high genetic variance observed previously in backgrounds A, B, and C.
Discussion
The data presented here indicate high variation among iso-male lines (Y lines). Although we cannot rule out the existence of some unknown sexually transmitted disease or other such factor influencing the fitness of progeny, Y-linked polymorphism is likely to be responsible for the statistically significant variation in fitness that we observed among the 20 Y-chromosome lines.
With the present data we cannot resolve whether the observed Y-linked variation in fertilization success is caused primarily by a male's mating success or his competence in sperm competition; the disproportionate role of Y in spermatogenesis suggests the latter candidate function. The strong role of Y in spermatogenesis, however, may also influence fitness through segregation distortion (meiotic drive).
Segregation distortion of sex chromosomes occurs when XY males produce Y-bearing sperm at an expected frequency different from 50%. Segregation distortion influences the sex ratio of offspring caused by the mortality of Y- (or X-) bearing sperm. Prior studies of the Y chromosome's influence on segregation distortion (23–25) found subtle additive genetic effects but no epistasis between the Y and the rest of the genome. We observed high epistatic variation among Y chromosomes when using an assay protocol that was relatively insensitive to segregation distortion (the fitness assay pooled sons and daughters during the scoring of paternity), suggesting that segregation distortion was not a major factor contributing to our observed Y-linked genetic variation. Nonetheless, recent data suggest that segregation distortion may be important in the fitness variation of Y chromosomes in other Drosophila species and may have been important in the past in D. melanogaster (14–17).
Although most genes known to reside on the Y influence spermatogenesis, and hence potentially contribute to sperm competition, the large magnitude of the observed Y-linked fitness variation, in combination with strong sperm displacement in D. melanogaster, suggests to us that mating behavior also may be a contributor. Because the Y is male-limited, theory predicts it to be a hot spot for the recruitment (by means of translocation or transposition, see refs. 27 and 28) of male-benefit/female-detriment alleles (3). Because behavior is selected in fundamentally different ways between the sexes, genes mediating behavioral traits would be feasible candidates for recruitment to the Y.
Our finding of substantial epistatic fitness variation on the Y, with little or no additive variation, suggests major Y-linked balanced polymorphism for fitness. Superficially, this result seems incongruent with previous theories and experiments that indicated little or no fitness variation on the Y of D. melanogaster. The epistatic fitness variation for total fitness that we observed, however, was not measured in previous experimental work. The earlier modeling work demonstrated that frequency-independent selection was unlikely to support substantial Y-linked polymorphism (9, 10). Newer studies (14–17, 29–31), however, suggest that frequency-dependent selection—or more complex selection based on meiotic drive, intransitive fitness, and/or genotype × genotype interactions between males and females—may provide the requisite theoretical framework for Y-linked polymorphism.
A recent molecular analysis of a 1738-bp coding region of a Y-linked structural gene in D. melanogaster found no polymorphism among 10 alleles that were collected from around the world (32). This study suggests that the Y is not polymorphic. The small Y-linked DNA region that was sequenced, however, may not accurately reflect the level of chromosomewide polymorphism on the Y. Heterogeneity in molecular variation among Y-linked regions was illustrated recently by a large-scale analysis of four Y-linked genes in humans (33), which found one gene to have no polymorphism within its coding region whereas the other three had substantial polymorphism (about 20% as high as a sample of autosomal genes).
The strong reversals in the rank order of the fitness of Y chromosomes in different genetic backgrounds (Fig. 3) have important evolutionary consequences. Although comparative evidence indicates extensive degeneration of the nonrecombining region of the Y over geological time, it also is known to accumulate new genes by means of translocations and transpositions (1, 27, 28, 34–36). Strong epistasis between Y-linked and X- or autosome-linked genes would favor any process that brought these interacting genes into tight linkage. This linkage could occur by extending the region of the X and Y that do not recombine (e.g., by an inversion in species with recombination occurring in males), or through transfer of genes on the X or autosomes to the nonrecombining region of the Y by means of transposition or translocation. All of these process are known to have occurred during the evolution of Y chromosomes in many different taxa (see for examples refs. 27 and 28 and 34–38). Our data demonstrate that the requisite epistatic selection and polymorphism are currently present in D. melanogaster to strongly favor genetic rearrangements that move new genetic material to the nonrecombining Y.
The major finding from this research is the large influence of the Y chromosome on genetic variation for male fitness. The Y was estimated to have as much polymorphic influence on the total fitness of males as 2/3 of an entire set of X/autosomal genes. This large, epistatic component of genomewide fitness may substantially reduce the heritability of male fitness, and thereby reduce the efficiency of selection on other loci. Determining the mechanisms that produce this extensive fitness polymorphism, and the contributing genes, presents a major challenge in our understanding of the evolutionary genetics of Y chromosomes.
Acknowledgments
We thank A. Clark, J. Gibson, A. B. Carvalho, and B. Lyon for comments on the manuscript. This work was supported by the National Science Foundation Grants DEB9623479 and DEB986917.
Abbreviation
- ibd
identical by descent
Appendix
Raw data from the adult and juvenile fitness assays are presented in Table 2. Adult fitness is a deviation—proportion of bw females inseminated by target males minus the expected proportion when mating occurred at random among the pool of available adult males (Fig. 1E). For example, if the available male pool was 35 target males and 70 bw competitor males, and if target males inseminated 40% of bw females, then the mating deviation would be 0.40 − 0.33 = 0.067. Juvenile fitness is the number of surviving male offspring carrying a target Y that was derived from the 70 target eggs (35 of which are expected to be males) used to begin each fitness assay (Fig. 1E). The sample size is 3 for each table entry. W, fitness.
Table 2.
Raw fitness data
| Line | Adult W
|
Juvenile
W
|
Line | Adult
W
|
Juvenile W
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| Y1{A} | 0.141 | 0.050 | 30.0 | 2.000 | Y11{A} | 0.070 | 0.139 | 25.3 | 1.155 |
| Y1{B} | −0.021 | 0.048 | 25.3 | 4.041 | Y11{B} | 0.132 | 0.105 | 24.7 | 2.309 |
| Y1{C} | −0.013 | 0.065 | 32.0 | 7.000 | Y11{C} | 0.173 | 0.055 | 31.7 | 2.887 |
| Y2{A} | 0.144 | 0.089 | 24.7 | 1.155 | Y12{A} | 0.188 | 0.053 | 24.0 | 1.732 |
| Y2{B} | 0.136 | 0.012 | 22.7 | 2.082 | Y12{B} | 0.052 | 0.031 | 23.3 | 4.933 |
| Y2{C} | 0.100 | 0.083 | 25.3 | 1.155 | Y12{C} | 0.170 | 0.044 | 28.7 | 3.215 |
| Y3{A} | 0.088 | 0.042 | 28.0 | 3.000 | Y13{A} | 0.156 | 0.030 | 28.0 | 4.583 |
| Y3{B} | 0.122 | 0.052 | 24.7 | 5.686 | Y13{B} | 0.052 | 0.116 | 25.0 | 6.083 |
| Y3{C} | 0.162 | 0.117 | 34.0 | 2.646 | Y13{C} | 0.130 | 0.179 | 29.0 | 2.000 |
| Y4{A} | 0.105 | 0.070 | 27.0 | 3.606 | Y14{A} | 0.115 | 0.060 | 28.7 | 1.155 |
| Y4{B} | 0.067 | 0.038 | 22.0 | 6.928 | Y14{B} | −0.035 | 0.057 | 25.0 | 1.000 |
| Y4{C} | 0.151 | 0.162 | 28.0 | 3.000 | Y14{C} | 0.138 | 0.055 | 26.7 | 7.234 |
| Y5{A} | 0.084 | 0.041 | 28.7 | 4.163 | Y15{A} | 0.074 | 0.023 | 28.3 | 1.528 |
| Y5{B} | 0.048 | 0.133 | 22.0 | 3.606 | Y15{B} | 0.134 | 0.068 | 22.0 | 1.000 |
| Y5{C} | 0.137 | 0.073 | 28.0 | 2.000 | Y15{C} | 0.258 | 0.064 | 26.0 | 3.464 |
| Y6{A} | 0.093 | 0.113 | 24.0 | 2.646 | Y16{A} | 0.112 | 0.110 | 29.7 | 2.309 |
| Y6{B} | 0.031 | 0.051 | 27.7 | 4.726 | Y16{B} | 0.034 | 0.075 | 21.0 | 7.000 |
| Y6{C} | 0.066 | 0.164 | 33.0 | 3.464 | Y16{C} | 0.157 | 0.148 | 29.0 | 5.568 |
| Y7{A} | 0.146 | 0.069 | 24.0 | 2.646 | Y17{A} | 0.063 | 0.033 | 27.7 | 7.371 |
| Y7{B} | 0.037 | 0.148 | 24.7 | 6.807 | Y17{B} | −0.014 | 0.076 | 25.3 | 4.933 |
| Y7{C} | 0.107 | 0.120 | 26.3 | 4.509 | Y17{C} | 0.205 | 0.089 | 25.0 | 2.000 |
| Y8{A} | 0.155 | 0.081 | 26.0 | 3.606 | Y18{A} | 0.147 | 0.107 | 28.3 | 6.110 |
| Y8{B} | 0.132 | 0.087 | 22.7 | 8.505 | Y18{B} | −0.028 | 0.028 | 20.3 | 2.082 |
| Y8{C} | 0.081 | 0.128 | 28.0 | 4.000 | Y18{C} | 0.203 | 0.082 | 23.3 | 2.887 |
| Y9{A} | 0.213 | 0.135 | 28.0 | 3.606 | Y19{A} | 0.002 | 0.059 | 29.3 | 6.658 |
| Y9{B} | 0.017 | 0.054 | 23.7 | 4.041 | Y19{B} | 0.119 | 0.138 | 27.0 | 9.539 |
| Y9{C} | 0.092 | 0.096 | 26.3 | 1.528 | Y19{C} | 0.252 | 0.124 | 25.0 | 5.568 |
| Y10{A} | 0.085 | 0.057 | 28.3 | 2.082 | Y20{A} | 0.149 | 0.124 | 30.3 | 3.786 |
| Y10{B} | 0.028 | 0.075 | 22.3 | 1.155 | Y20{B} | 0.070 | 0.100 | 26.7 | 3.055 |
| Y10{C} | −0.002 | 0.090 | 26.3 | 4.933 | Y20{C} | 0.253 | 0.094 | 27.7 | 1.528 |
W, fitness.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Stankewicz, R. & Zhang, P. A. (1998) Conf. Drosoph. Res. 39, 826 (abstr.).
References
- 1.Bull J J. Evolution of Sex Determination Mechanisms. Menlo Park, CA: Benjamin-Cummings; 1983. [Google Scholar]
- 2.Charlesworth B. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 3.Rice W R. Bioscience. 1996;46:331–343. [Google Scholar]
- 4.Gatti M, Pimpinelli S. Chromosoma. 1983;88:349–373. [Google Scholar]
- 5.Williamson J H. In: The Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 1B. New York: Academic; 1976. pp. 667–699. [Google Scholar]
- 6.Ashburner M. Drosophila: A Laboratory Handbook. Plainview, New York: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 7.Stoltenberg S F. Anim Behav. 1997;53:853–864. doi: 10.1006/anbe.1996.0351. [DOI] [PubMed] [Google Scholar]
- 8.Lindsley D L, Zimm G G. The Genome of Drosophila. New York: Academic; 1992. [Google Scholar]
- 9.Clark A G. Genetics. 1987;115:569–577. doi: 10.1093/genetics/115.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark A G. Genetics. 1990;125:527–534. doi: 10.1093/genetics/125.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams S M, Furnier G R, Fuog E, Strobeck C. Genetics. 1987;116:225–232. doi: 10.1093/genetics/116.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyckegaard E M S, Clark A G. Proc Natl Acad Sci USA. 1989;86:1944–1948. doi: 10.1073/pnas.86.6.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark A G, Szumski F M, Lyckegaard E M S. Genet Res. 1990;58:7–13. doi: 10.1017/s0016672300029554. [DOI] [PubMed] [Google Scholar]
- 14.Jaenike J. Evolution (Lawrence, Kans) 1999;53:164–174. doi: 10.1111/j.1558-5646.1999.tb05342.x. [DOI] [PubMed] [Google Scholar]
- 15.Hurst L D. Genetics. 1992;130:229–230. doi: 10.1093/genetics/130.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst L D. Genetics. 1996;142:641–643. doi: 10.1093/genetics/142.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvalho A B, Vaz S C, Klaczko L B. Genetics. 1997;146:891–902. doi: 10.1093/genetics/146.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coyne J A. Nature (London) 1992;355:511–515. doi: 10.1038/355511a0. [DOI] [PubMed] [Google Scholar]
- 19.Wu C-I, Palopoli M F. Annu Rev Genet. 1994;28:283–308. doi: 10.1146/annurev.ge.28.120194.001435. [DOI] [PubMed] [Google Scholar]
- 20.Lamnissou K, Loukas M, Zouros E. Heredity. 1996;76:603–609. doi: 10.1038/hdy.1996.86. [DOI] [PubMed] [Google Scholar]
- 21.Hollocher H, Templeton A R, Desalle R, Johnston J S. Genetics. 1992;130:355–366. doi: 10.1093/genetics/130.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson N A, Hollocher H, Noonburg E, Wu C-I. Genetics. 1993;135:443–453. doi: 10.1093/genetics/135.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toro M A, Charlesworth B. Heredity. 1982;49:199–209. [Google Scholar]
- 24.Clark A G. Genetics. 1987;115:143–151. doi: 10.1093/genetics/115.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark A G, Lyckegaard E M S. Evolution (Lawrence, Kans) 1990;44:2106–2112. doi: 10.1111/j.1558-5646.1990.tb04315.x. [DOI] [PubMed] [Google Scholar]
- 26.Chippindale A K, Gibson J R, Rice W R. Proc Natl Acad Sci USA. 2001;98:1671–1675. doi: 10.1073/pnas.041378098. . (First Published January 30, 2001; 10.1073/pnas.041378098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahn B T, Page D C. Nat Genet. 1999;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- 28.Saxena R, Brown L G, Hawkins T, Alagappan R K, Skaletsky H, Reeve M P, Reijo R, Rozen S, Dinulos M B, Disteche C M, Page D C. Nat Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 29.Clark A G, Begun D J. Genetics. 1998;149:1487–1493. doi: 10.1093/genetics/149.3.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark A G, Begun D J, Prout T J. Science. 1999;283:217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- 31.Clark A G, Dermitzakis E T, Civetta A. Evolution. 2000;54:1030–1035. doi: 10.1111/j.0014-3820.2000.tb00102.x. [DOI] [PubMed] [Google Scholar]
- 32.Zurovcova M, Eanes W F. Genetics. 1999;153:1709–1715. doi: 10.1093/genetics/153.4.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen P, Wang F, Underhill P A, Franco C, Yang W-H, Roxas A, Sung R, Lin A A, Hyman R W, Vollrath D W, et al. Proc Natl Acad Sci USA. 2000;97:7354–7359. doi: 10.1073/pnas.97.13.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobzhansky T. Genetics. 1935;20:377–391. doi: 10.1093/genetics/20.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macknight R H. Genetics. 1939;24:180–201. doi: 10.1093/genetics/24.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinemann M, Steinemann S. Genetica (The Hague) 1998;103:409–420. [PubMed] [Google Scholar]
- 37.Graves J A M. J Exp Zool. 1998;281:472–481. [PubMed] [Google Scholar]
- 38.Lahn B T, Page D C. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]