Abstract
Glutaredoxins (GRXs) are small, ubiquitous, glutathione-dependent oxidoreductases that participate in redox-regulated processes associated with stress responses. Recently, GRXs have been shown to exert crucial functions during flower developmental processes. GRXs modulate their target protein activities by the reduction of protein disulfide bonds or deglutathionylation reactions. The Arabidopsis (Arabidopsis thaliana) GRX ROXY1 participates in petal primordia initiation and further petal morphogenesis. ROXY1 belongs to a land plant-specific class of GRXs with a CC-type active site motif, deviating from the ubiquitously occurring CPYC and CGFS GRX classes. ROXY1 was previously shown to interact with floral TGA transcription factors in the nucleus, and this interaction is a prerequisite for ROXY1 to exert its activity required for Arabidopsis petal development. Deletion analysis further identified the importance of the ROXY1 C terminus for the ROXY1/TGA protein interactions and for the ROXY1 function in petal development. Here, by dissecting the ROXY1 C terminus, an α-helical L**LL motif immediately adjacent to the ROXY1 C-terminal eight amino acids was identified that is essential for the interaction with TGA transcription factors and crucial for the ROXY1 function in planta. Similar to the α-helical L**LL motifs binding to transcriptional coactivators with liganded nuclear receptors in animals, a hydrophobic face formed by the conserved leucines in the L**LL motif of ROXY1 possibly mediates the interaction with TGA transcription factors. Thus, the α-helical L**LL sequence is a conserved protein-protein interaction motif in both animals and plants. Furthermore, two separate TGA domains were identified by deletion experiments as being essential for mediating TGA protein interactions with ROXYs.
Glutaredoxins (GRXs) are small, ubiquitous, glutathione-dependent oxidoreductases that play an essential role in response to oxidative stress (Buchanan and Balmer, 2005). According to the amino acid sequences of active site motifs, three classes of GRXs are defined in plants, namely the CPYC, the CGFS, and the CC-type classes (Rouhier et al., 2004). The CPYC and CGFS classes are common to all prokaryotes and eukaryotes, while the CC-type class is specific for land plants (Lemaire, 2004; Rouhier et al., 2006; Xing et al., 2006). The Arabidopsis (Arabidopsis thaliana) genome encodes 31 GRX genes, and 21 belong to the CC-type class. The CC-type class expanded exclusively and strongly during the evolution of land plants, suggesting that this class of GRXs might have been recruited to participate in the evolution of more complex land plants (Xing et al., 2006; Li and Zachgo, 2009; Ziemann et al., 2009). Contrasting with wide-ranging roles of plant thioredoxins in chloroplast and mitochondrial processes, seed development and germination, and auxin signaling (Buchanan and Balmer, 2005; Reichheld et al., 2007; Arsova et al., 2010; Bashandy et al., 2010), the biological and physiological functions of plant GRXs have remained largely elusive. Recently, two CC-type GRX genes, ROXY1 and ROXY2, were functionally characterized, revealing their functions during Arabidopsis flower development (Xing et al., 2005; Xing and Zachgo, 2008; Li et al., 2009). The roxy1 mutant initiates only 2.5 petal primordia on average, instead of 4.0 observed in wild-type flowers, and exhibits abnormalities during further petal morphogenesis (Xing et al., 2005). ROXY1 and its closest homolog ROXY2 act redundantly during anther development and microspore formation (Xing and Zachgo, 2008). The other two CC-type GRX genes with known functions, ROXY19 (originally named GRX480) and ROXY4, participate in pathogen defense responses and GA signaling in Arabidopsis, respectively (Ndamukong et al., 2007; Hou et al., 2008).
TGA transcription factors have been recently identified as interaction proteins of ROXYs and thus are likely posttranslationally modified by ROXYs (Ndamukong et al., 2007; Li et al., 2009). TGA transcription factors belong to the basic leucine zipper (bZIP) protein superfamily and comprise 10 members in Arabidopsis: TGA1 to TGA7, PERIANTHIA (PAN), TGA9, and TGA10 (Jakoby et al., 2002; Murmu et al., 2010). TGA1 to TGA7 are implicated as regulators of PATHOGENESIS-RELATED (PR) genes and hence disease resistance in Arabidopsis (Zhang et al., 2003; Kesarwani et al., 2007). Contrarily, PAN is involved in flower development, regulating the number of floral organ primordia. The pan mutant reveals a pentamerous arrangement of floral organs in the first three whorls (Running and Meyerowitz, 1996; Chuang et al., 1999). Expression of a dominant-negative form of PAN (PAN-SRDX) uncovered additional redundant functions of TGA proteins during later petal morphogenesis (Li et al., 2009). Recently, TGA9 and TGA10 have also been shown to participate in flower development and to be required for microspore formation (Murmu et al., 2010).
Genetic analysis and protein interaction experiments indicate that the CC-type GRXs might modify TGA transcription factors by catalyzing thiol-disulfide exchange or deglutathionylation reactions through conserved Cys residues, thereby regulating their activity and related biological processes (Xing et al., 2005; Ndamukong et al., 2007; Li et al., 2009; Murmu et al., 2010). Upon pathogen infection and salicylic acid accumulation, NONEXPRESSOR OF PR GENES1 (NPR1) is translocated from the cytoplasm into the nucleus and interacts with TGA transcription factors. The NPR1/TGA1 protein interaction depends on the reduced state of two Cys residues in TGA1, as an intramolecular disulfide bridge formed between Cys-260 and Cys-266 prevents this interaction (Després et al., 2003). Salicylic acid-induced ROXY19 interacts with several TGA transcription factors and negatively modulates jasmonic acid-responsive PDF1.2 transcription in Arabidopsis, suggesting that this GRX is involved in the reduction of TGA1 (Ndamukong et al., 2007). The floral CC-type GRX ROXY1 protein interacts with PAN in the nucleus, which was shown to be a prerequisite to exert proper functions during floral organ development (Li et al., 2009). Unlike TGA1, PAN contains only one conserved Cys, Cys-340, corresponding to Cys-260 in TGA1 (Li and Zachgo, 2009). Mutagenesis of PAN Cys-340 into a Ser caused a failure of the mutagenized protein to rescue the pan mutant (Li et al., 2009). This demonstrates the importance of this single Cys in PAN and suggests that this Cys might represent a target for a posttranslational modification by ROXY1 (Li et al., 2009). Thus, the mechanisms by which TGA1 and PAN are posttranslationally modulated might differ.
Deletion experiments coupled with protein interaction analysis demonstrated the importance of the ROXY1 C terminus for the ROXY1/TGA protein interactions and for ROXY1 to exert its function in petal development (Li et al., 2009). Here, a comprehensive dissection analysis of the ROXY1 C terminus was conducted and a crucial α-helical L**LL motif immediately adjacent to the ROXY1 C-terminal eight amino acid residues was identified. This L**LL motif is shown to be essential for an interaction with TGA transcriptional factors and is indispensable for Arabidopsis petal development. Similar to the α-helical L**LL motifs of transcriptional coactivators that interact with liganded nuclear receptors as well as many other transcription factors in animals (Le Douarin et al., 1996; Heery et al., 1997; Voegel et al., 1998), a hydrophobic face formed by the conserved Leu residues in the L**LL motif of ROXY1 likely mediates the interaction with TGA transcription factors. Although crucial Cys residues of TGA proteins were identified as potential targets to be posttranslationally modified by ROXYs (Després et al., 2003; Li et al., 2009; Murmu et al., 2010), little is known about the TGA domains mediating the interaction with ROXYs. Domain truncation experiments demonstrate that the second Gln-rich domain (Q2) and an intervening region between bZIP and the first Gln-rich region (Q1) may form a ROXY1-interacting interface.
RESULTS
Analysis of the ROXY1 C-Terminal L**LL Motif
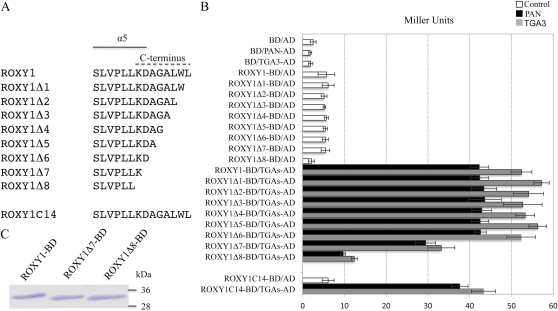
All GRXs possess a conserved thioredoxin fold comprising three α-helices and four β-sheets (β1-α2-β2-α3-β3-β4-α4 for ROXY1; Holmgren et al., 1975). In addition, all Arabidopsis CC-type GRXs share a predicted α-helix at the N terminus (α1), a predicted α-helix at the C terminus (α5), and an additional C-terminal extension (Li et al., 2009). NMR spectroscopy confirmed that a poplar (Populus spp.) GRX with a CGFC active site motif bears a similar secondary structure consisting of five α-helices and four β-sheets (Feng et al., 2006). In comparison with other CC-type GRXs, ROXY1 has a specific 28-amino acid N-terminal extension and an additional 15-amino acid intervening region between its α3 helix and β3 sheet, which are present only in its closest homolog, ROXY2 (Li et al., 2009). Deletion analysis has shown that the C-terminal eight amino acids of ROXY1 (hereafter termed the ROXY1 C terminus) are crucial for mediating the ROXY1/TGA protein interactions and for an in planta ROXY1 function, whereas the N-terminal extension and the intervening region of ROXY1 are dispensable (Li et al., 2009). As the α5 helix of ROXY1 has a two-amino acid overlap with the analyzed C terminus of ROXY1 (Fig. 1A), the removal of the C terminus may impair the formation of the α5 helix, thereby causing an inability of ROXY1 to interact with TGA transcription factors and consequently a failure to complement the roxy1 mutant (Li et al., 2009). To test this possibility, yeast two-hybrid experiments were performed to quantify interaction strengths for tested protein pairs by monitoring reporter gene expression using β-galactosidase assays. One to eight amino acids of the ROXY1 C terminus were removed successively, and eight corresponding mutant ROXY1 proteins were generated (ROXY1Δ1–ROXY1Δ8; Fig. 1A). Wild-type or mutagenized ROXY1 proteins were used as bait and expressed as a fusion to the GAL4 DNA-binding domain (GAL4-BD; hereafter BD), whereas two TGA transcription factors, PAN and TGA3 (Supplemental Fig. S1), were used as prey and expressed as fusion proteins to the GAL4 activation domain (GAL4-AD; hereafter AD; Fig. 1B). In agreement with our earlier report (Li et al., 2009), strong protein interactions were detected for ROXY1-BD/PAN-AD (42.30 ± 2.26 Miller units) and ROXY1-BD/TGA3-AD (52.40 ± 2.50 Miller units; Fig. 1B). As a negative control, cotransformation of empty bait and prey vectors (BD/AD) resulted in a background level of around 2.50 ± 0.70 Miller units (Fig. 1B). When BD was separately coexpressed with prey proteins PAN-AD and TGA3-AD, β-galactosidase activities recorded were comparable to that of BD/AD (Fig. 1B), indicating that PAN-AD or TGA3-AD alone is unable to activate reporter gene expression. With the exception of ROXY1Δ8-BD, ROXY1-BD and deletion mutants ROXY1Δ1-BD to ROXY1Δ7-BD exhibited very weak autoactivation in the absence of prey proteins (approximately 5–6 Miller units; Fig. 1B). Gradual removal of one to six amino acids from the ROXY1 C terminus (ROXY1Δ1–ROXY1Δ6) did not impair interaction affinities with TGA proteins (approximately 40 Miller units for PAN and approximately 50 Miller units for TGA3; Fig. 1B). However, further deletion caused an approximately 20% drop of the TGA interaction affinities for ROXY1Δ7 (29.47 ± 2.38 and 33.20 ± 3.24 Miller units for PAN and TGA3, respectively; Fig. 1B) and an almost complete loss for ROXY1Δ8 (9.70 ± 0.53 and 12.37 ± 0.70 Miller units for PAN and TGA3, respectively; Fig. 1B). Western-blot analysis of protein extracts from yeast cells harboring ROXY1Δ7-BD and ROXY1Δ8-BD demonstrated that the two truncated forms of ROXY1 proteins were still expressed in yeast at levels similar to wild-type ROXY1 (Fig. 1C). Together, these data indicate that the most N-terminal two amino acids (KD) of the ROXY1 C terminus (Fig. 1A) contribute significantly to the interaction with TGA proteins. As these two amino acids are part of the ROXY1 α5 helix (Fig. 1A), these findings imply that the α-helical domain may be required for binding to TGA transcription factors in yeast. To test this hypothesis, ROXY1C14-BD was engineered, which consists of the GAL4-BD fused solely to the 14 C-terminal ROXY1 amino acids (Fig. 1A). Compared with the strong interaction detected for wild-type ROXY1 and two TGA proteins (42.3 ± 2.3 Miller units for PAN and 52.4 ± 2.5 Miller units for TGA3), the interaction between ROXY1C14-BD and the two TGA proteins was only slightly reduced (37.6 ± 2.0 Miller units for PAN and 43.3 ± 2.9 Miller units for TGA3; Fig. 1B). Collectively, these data indicate that the ROXY1 α5 helix is required to mediate an interaction with PAN and TGA3 proteins in yeast.
Figure 1.
Analysis of protein interactions between deleted versions of ROXY1 and two TGA proteins. A, Schematic representation of deleted forms of ROXY1 proteins. In ROXY1Δ1 to ROXY1Δ8, the C-terminal one to eight amino acids were deleted, respectively; ROXY1C14 comprises the ROXY1 C-terminal 14 amino acids alone. Amino acids comprising the α5 helix and the C terminus are indicated by solid and dashed lines, respectively. B, Quantitative β-galactosidase activity assays measuring the interaction of the ROXY1 C-terminal deletion mutants and the ROXY1 C-terminal 14 amino acids with two TGA proteins. ROXY1Δ1 to ROXY1Δ8, ROXY1C14, as well as wild-type ROXY1 were used as bait and expressed as a fusion to the GAL4-BD, whereas PAN and TGA3 were used as prey and expressed as fusion proteins to the GAL4-AD. Black and gray bars indicate PAN-AD and TGA3-AD, respectively; white bars represent controls. Data are means ± sd of three independent experiments. C, Western-blot analysis of protein extracts from yeast cells expressing ROXY1Δ7 and ROXY1Δ8 fused to the GAL4-BD. ROXY1 and its two truncated forms fused to GAL4-BD have a molecular mass of about 33 to 34 kD (14 kD for ROXY1, 13 kD for ROXY1Δ7 and ROXY1Δ8, and 20 kD for GAL4-BD). Molecular mass markers (kD) are indicated at right. [See online article for color version of this figure.]
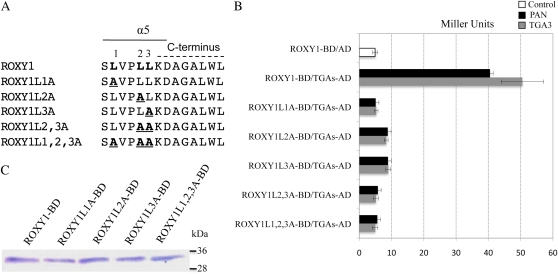
Intriguingly, the α5 helix of ROXY1 contains a short pentapeptide, LVPLL, immediately adjacent to the ROXY1 C terminus (Fig. 2A). This peptide resembles the α-helical L**LL motif known to mediate the interaction between transcriptional coactivators and liganded nuclear receptors as well as many other transcription factors in animals through a hydrophobic face formed by its Leu residues (Le Douarin et al., 1996; Heery et al., 1997; Voegel et al., 1998). To test by yeast two-hybrid interaction screens whether this α-helical L**LL motif of ROXY1 is essential for binding to plant TGA transcription factors, five mutagenized ROXY1 proteins were engineered by replacing one, two, or all three strongly hydrophobic Leu residues located in this motif by an Ala, which possesses a shorter and thus only weakly hydrophobic side chain (Fig. 2A). As observed for ROXY1 deletion proteins (Fig. 1, A and B), each of the five substitution mutants (ROXY1L1A-BD, ROXY1L2A-BD, ROXY1L3A-BD, ROXY1L2,3A-BD, and ROXY1L1,2,3A-BD) showed very weak autoactivation of the reporter gene (4–6 Miller units; Fig. 2B; Supplemental Fig. S2). When all the Leu residues in the L**LL motif were substituted with an Ala (ROXY1L1,2,3A-BD), interaction affinities with both TGA transcription factors were severely reduced to autoactivation levels, indicating a complete loss of the interaction with TGA transcription factors (Fig. 2B; Supplemental Fig. S2). Next, relative contributions of the three Leu residues to the interaction with TGA proteins were examined. Replacement of the first Leu with an Ala (ROXY1L1A-BD) led to a reduction of β-galactosidase activity by 87.4% and 89.6% for PAN and TGA3, respectively, similar to the triple substitution mutant (ROXY1L1,2,3A-BD; Fig. 2B). Mutations of the second or the third Leu to an Ala (ROXY1L2A-BD or ROXY1L3A-BD) still retained very weak interactions with TGA transcription factors (8–9 Miller units; Fig. 2B). The double substitution mutant ROXY1L2,3A-BD completely abrogated the residual weak interactions observed for ROXY1L2A-BD or ROXY1L3A-BD, resembling the triple substitution mutant ROXYL1,2,3A-BD (Fig. 2B). Immunoblot analysis of protein extracts from yeast clones expressing the three single and one triple substitution ROXY1 mutants showed that the Leu-to-Ala exchanges did not interfere with protein expression and stability (Fig. 2C).
Figure 2.
Effects of Leu-to-Ala substitutions in the L**LL motif on the capacity of ROXY1 to interact with TGA proteins. A, Schematic representation of mutant ROXY1 proteins with Leu-to-Ala substitutions for the L**LL motif. Amino acids comprising the α5 helix and the C terminus are indicated by solid and dashed lines, respectively. Numbers 1, 2, and 3 mark the first, second, and third Leu residues in the L**LL motif, respectively. For ROXY1L1A, ROXY1L2A, and ROXY1L3A, the first, second, and third Leu of the L**LL motif were substituted with an Ala, respectively. ROXY1L2,3A indicates a double and ROXY1L1,2,3A a triple Leu-to-Ala substitution. The three Leu residues of the L**LL motif are depicted by boldface letters, and their substituting Ala residues are depicted by boldface and underlined letters. B, Quantification of the effect of the ROXY1 Ala-to-Leu substitutions on the interaction with PAN and TGA3. Wild-type and mutagenized ROXY1 proteins were used as bait and fused to the GAL4-BD, whereas PAN and TGA3 were used as prey and fused to the GAL4-AD. Mating and cotransformation were performed for combinations containing PAN and TGA3, respectively. Data are means ± sd of three independent experiments. C, Immunoblot analysis of protein extracts from yeast clones harboring the ROXY1 Leu-to-Ala substitutions fused to the GAL4-BD. ROXY1 and the substituted versions fused to GAL4-BD have a molecular mass of about 34 kD. Molecular weight markers (kD) are indicated at right. [See online article for color version of this figure.]
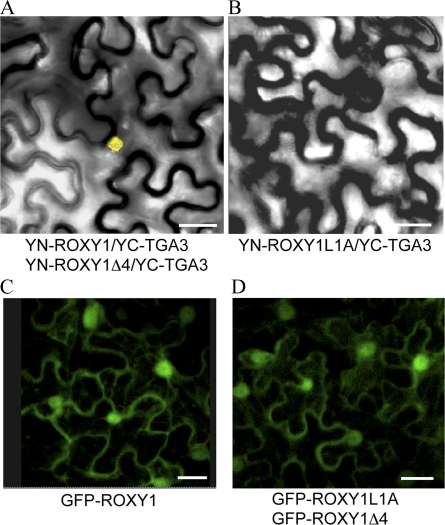
To explore whether the α5 L**LL motif of ROXY1 is also essential for ROXY1/TGA protein interactions in planta, the bimolecular fluorescence complementation (BiFC) technique was employed. For the BiFC experiments, the N terminus of yellow fluorescent protein (YFP; YN) was cloned in frame upstream of the wild-type ROXY1 and each of the five single, double, and triple ROXY1 substitution mutants, whereas the C terminus of YFP (YC) was fused N terminally to PAN or TGA3. By performing Agrobacterium tumefaciens coinfiltration, constructs were pairwise transiently expressed in abaxial epidermal cells of tobacco (Nicotiana benthamiana) leaves (Voinnet et al., 2003). Reconstitution of YFP fluorescence was examined by confocal microscopy 3 to 5 d after transient coexpression of protein pairs. Consistent with our recent report (Li et al., 2009), nuclear yellow fluorescence was observed for coexpression of wild-type ROXY1 with PAN and TGA3 (Fig. 3A). However, no yellow fluorescence was observed in the nucleus when TGA proteins were coexpressed with each of the five substituted ROXY1 proteins (Fig. 3B). As negative controls, coexpression of nonfused YN with nonfused YC or one of the fusion proteins with nonfused YN or nonfused YC failed to reconstitute a fluorescent YFP chromophore (data not shown). Similar to a nucleocytoplasmic expression pattern for the GFP-ROXY1 fusion protein (Fig. 3C), all the examined Leu-to-Ala substitution mutants of ROXY1 fused to GFP were still localized to both the nucleus and the cytoplasm (Fig. 3D). These findings demonstrate that Leu-to-Ala substitutions in the α5 helix of ROXY1 do not affect the intracellular localization of ROXY1 and that ROXY1 proteins cannot interact any more with TGA proteins in planta if any of the three Leu residues of the α-helical L**LL motif is replaced by an Ala.
Figure 3.
In planta interactions of mutagenized ROXY1 with TGA proteins and subcellular localization of ROXY1 variants. A, Wild-type ROXY1 and the truncated form of ROXY1 lacking the C-terminal four amino acids (ROXY1Δ4) interact with TGA3 in the nucleus. The same nuclear interaction was also observed for PAN. B, All investigated ROXY1 substitution variants (ROXY1L1A, ROXY1L2A, ROXY1L3A, ROXY1L2/3A, and ROXY1L1/2/3A) failed to interact in planta, which is shown exemplarily for ROXY1L1A. C, Nucleocytoplasmic distribution of the GFP/ROXY1 fusion protein. D, Replacement of the first Leu (L1) with an Ala in the ROXY1 L**LL and removal of the ROXY1 C-terminal four amino acids showed a wild-type-like nucleocytoplasmic localization. All other mutagenized ROXY1 proteins also accumulated in the nucleus and the cytoplasm. Bars = 50 μm.
Contribution of the L**LL Motif to Governing Petal Development
The roxy1 mutant (Fig. 4B) initiates a reduced number of petal primordia and forms only 2.5 petals on average instead of 4.0 petals developing in wild-type flowers (Xing et al., 2005; Fig. 4A). The combination of intracellular ROXY1 protein localization studies along with complementation experiments of the roxy1 mutant showed that a nuclear interaction between ROXY1 and TGA proteins is a prerequisite for ROXY1 to exert its function during petal development (Li et al., 2009). To examine the functional relevance of the L**LL motif in planta, each of the five mutagenized L**LL versions of ROXY1 genes was expressed in the roxy1 mutant under the control of ROXY1 regulatory sequences known to confer endogenous protein expression (Xing et al., 2005). Transgenic plants were scored to determine if mutagenized ROXY1 proteins are capable of rescuing the abnormal petal development of the roxy1 mutant. The wild-type ROXY1 gene served as a positive control and complemented petal phenotypes of the roxy1 mutant in all 56 analyzed ROXY1pro::ROXY1 T1 transgenic plants (Fig. 4C). In contrast, none of the examined T1 transformants expressing ROXY1L1A (64 T1 plants), ROXY1L2A (60 T1 plants), ROXY1L3A (70 T1 plants), ROXY1L2,3A (66 T1 plants), and ROXY1L1,2,3A (58 T1 plants) produced wild-type petals (Fig. 4D). These results prove that the three conserved Leu residues of the L**LL motif in the α5 helix of ROXY1 are crucial for a proper ROXY1 function during petal development.
Figure 4.
Complementation experiments of mutagenized ROXY1 proteins. Wild-type and mutant ROXY genes driven by the ROXY1 promoter were transformed into the roxy1 mutant. Petal phenotypes of transgenic T1 plants were scored, and representative phenotypes are shown. The roxy1 mutant flowers (B) develop only 2.5 petals per flower instead of 4.0 petals as observed in wild-type (WT) flowers (A). The wild-type ROXY1 gene is able to complement the roxy1 mutant (C). In contrast, replacement of the first Leu (L1) with an Ala in the L**LL motif of ROXY1 (ROXY1L1A) or removal of the ALWL motif from the ROXY1 C terminus (ROXY1Δ4) failed to rescue abnormal petal phenotypes of the roxy1 mutant (D). All other mutagenized ROXY1 proteins (ROXY1L2A, ROXY1L3A, ROXY1L2,3A, and ROXY1L1,2,3A) also lost their capacities to complement the roxy1 mutant. At least 50 T1 transformants were analyzed for each construct.
Functional Analysis of the ROXY1 C-Terminal ALWL Motif
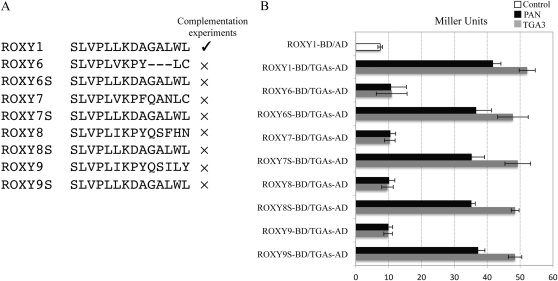
Arabidopsis contains 21 CC-type ROXYs, encompassing ROXY1 to ROXY21 (Li et al., 2009; Ziemann et al., 2009). Sequence alignments and complementation experiments of the roxy1 mutant using different Arabidopsis CC-type GRXs revealed that all complementing GRXs (seven out of 12 investigated) share a conserved C-terminal motif, AL/IWL, whereas noncomplementing GRXs do not possess this conserved sequence at the C terminus (Li et al., 2009; Fig. 5A; Supplemental Fig. S1). Yeast two-hybrid analysis has shown that deletion of only the ALWL motif from the ROXY1 C terminus (ROXY1Δ4) did not impair interaction affinities with TGA transcription factors (Fig. 1, A and B). Further BiFC experiments also revealed the dispensability of the ALWL motif for an in planta interaction with the investigated TGA3 and PAN proteins (Fig. 3A). Similar to the GFP-ROXY1 fusion protein (Fig. 3C), ROXY1Δ4 fused C terminally to GFP also exhibited a nucleocytoplasmic localization (Fig. 3D). To test whether the ROXY1 C-terminal four amino acids are required for petal development, ROXY1Δ4, lacking only the conserved AL/IWL motif at the C terminus, was expressed in the roxy1 mutant under the control of ROXY1 regulatory sequences. None of the tested T1 transformants harboring ROXY1pro::ROXY1Δ4 (55 T1 plants) developed wild-type petals (Fig. 4D). This result indicates that the C-terminal four amino acids of ROXY1 are dispensable for an interaction with PAN and TGA3 proteins but are required for ROXYs to function properly during petal development.
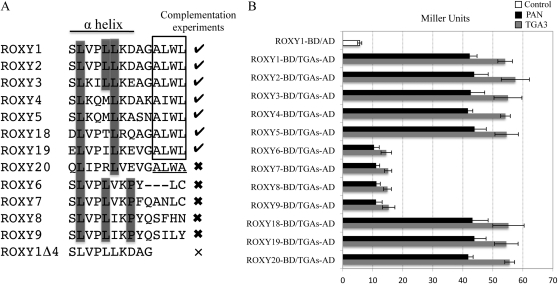
Figure 5.
Comparison of L**LL motifs for different ROXYs and analyses of their competence to interact with TGA proteins. A, Alignment of C-terminal sequences comprising the L**LL motif of the ROXY1 α5 helix for representative ROXYs. Check marks indicate ROXYs capable of complementing the roxy1 mutant, thick crosses denote previously tested ROXYs that failed to complement the ROXY1 mutant, and the thin cross represents a mutagenized ROXY1 lacking the C-terminal four amino acids. The C-terminal four amino acids A(L/I)WL conserved for all the ROXYs complementing the roxy1 mutant are boxed, and the C-terminal four amino acids ALWA of the noncomplementing ROXY20 are underlined. Leu residues (corresponding to L1–L3 in ROXY1) are indicated for all the tested ROXYs, and conserved Pro residues for four of the five noncomplementing ROXYs are highlighted by a gray box. Amino acid sequences corresponding to the ROXY1 α5 helix are shown below a solid line. B, Quantitative yeast protein interaction data of representative ROXYs tested in complementation experiments. Data are means ± sd of three independent experiments.
Dissection of the ROXY L**LL Motif
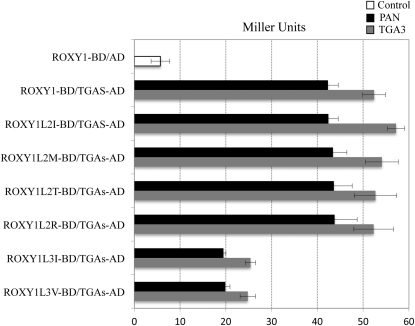
The inability of the five noncomplementing ROXYs (ROXY6–ROXY9 and ROXY20) to govern normal petal development could be due to their failure to interact with TGA proteins. To clarify this issue, the interaction of ROXY1 to ROXY9 and ROXY18 to ROXY20 with PAN and TGA3 was analyzed by yeast two-hybrid experiments (Fig. 5). All 12 tested ROXYs displayed a very weak autoactivation in the absence of prey proteins (5–6 Miller units; Fig. 5B; Supplemental Fig. S2). Strong protein interactions were observed when each of the complementing ROXYs (ROXY1–ROXY5, ROXY18, and ROXY19) was coexpressed with PAN (about 40 Miller units) and TGA3 (about 50 Miller units; Fig. 5). Unlike all the other noncomplementing ROXYs, ROXY20 possesses a C-terminal ALWA sequence similar to the AL/IWL motif but failed to complement the roxy1 mutant (Li et al., 2009; Fig. 5A). However, this CC-type GRX still showed a strong interaction with TGA transcription factors in yeast (41.8 ± 1.64 Miller units for PAN and 55.6 ± 4.07 Miller units for TGA3; Fig. 5B). Significantly compromised interactions with TGA transcription factors (10–15 Miller units) were detected for each of the other noncomplementing ROXYs (Fig. 5B).
Analysis of sequence conservation in the L**LL motif revealed that both the L1 and the L3 of the ROXY1 L**LL motif are conserved for all the ROXYs interacting strongly with TGA proteins, including all complementing ROXYs and the noncomplementing ROXY20 (Fig. 5A). By contrast, the L2 of the L**LL motif for all interacting ROXYs is either unchanged (ROXY1–ROXY3) or replaced by a hydrophobic Met (ROXY4 and ROXY5), a weakly hydrophilic Thr (ROXY18), a hydrophobic Ile (ROXY19), or a hydrophilic Arg (ROXY20; Fig. 5A). Although L1 and L2 are conserved for all the other noncomplementing ROXYs that showed weak interactions with TGA transcription factors, L3 is replaced by either a hydrophobic Val (ROXY6 and ROXY7) or a hydrophobic Ile (ROXY8 and ROXY9; Fig. 5A). This indicates that the presence of a conserved L2 seems less important for the interaction with TGA proteins. Substitution experiments were further performed to dissect the contribution of different amino acids in the L2 and L3 positions of the ROXY1 L**LL motif to the interaction with TGA proteins. Mutant ROXY1 proteins with L2 substituted by a Met (ROXY1L2M), a Thr (ROXY1L2T), an Ile (ROXY1L2I), or an Arg (ROXY1L2R) were engineered, and mutational effects on protein interaction capacities were examined using yeast two-hybrid assays (Fig. 6; Supplemental Fig. S2). Quantification of β-galactosidase activity revealed that the replacement of L2 with each of these amino acids did not impair interaction strengths significantly (Fig. 6). The necessity of a conserved L3 to meditate the interaction with TGA proteins was also analyzed in yeast-two hybrid assays. Two mutagenized ROXY1 proteins with L3 substituted by either a Val (ROXY1L3V) or an Ile (ROXY1L3I) were engineered (Fig. 6; Supplemental Fig. S2). Compared with wild-type ROXY1, both mutants displayed severely reduced β-galactosidase activity (20–25 Miller units; Fig. 6). However, this interaction was still slightly stronger than that shown for noncomplementing ROXYs interacting weakly with TGA transcription factors (15 Miller units; Figs. 5B and 6). This could be attributed to the presence of a Pro at the last position of the α5 helix for the four noncomplementing ROXYs (Fig. 5A), as this amino acid might impair the formation of the α5 helix and thus interaction strengths with TGA proteins. Collectively, these results further strengthen the observation that the interaction with TGA proteins depends on the L**LL motif. The L1 and L3 of the L**LL motif are crucial for mediating the interaction with TGA proteins, and the lack of L3 for ROXY6 to ROXY9 seems to abrogate an interaction capacity. The exchange of the L2 in the ROXY1 L**LL motif with a Met, a Thr, an Ile, or an Arg does not affect the interaction with TGA proteins, whereas the substitution of this Leu by an Ala is detrimental for this interaction. Although ROXY20 contains the crucial L3 and interacts with TGA proteins in yeast, its C terminus terminates with an ALWA motif (Fig. 5A). Deviation from the conserved AL/IWL motif, which is required for an in planta ROXY activity (Fig. 4D), may render ROXY20 unable to exert a full ROXY1-like activity.
Figure 6.
Effects of exchanging the second and third Leu residues of the ROXY1 L**LL motif on an interaction with TGA proteins. Quantitative β-galactosidase assays were performed to measure interaction strengths in the yeast two-hybrid system. ROXY1L2I, ROXY1L2M, ROXY1L2T, and ROXY1L2R indicate that the second Leu of the ROXY1 L**LL motif is replaced by an Ile, a Met, a Thr, and an Arg, respectively. ROXY1L3V and ROXY1L3I denote that the third Leu of the ROXY1 L**LL motif is mutated into a Val and an Ile, respectively. Data are means ± sd of three independent experiments.
Interaction Analysis of ROXYs with Swapped C Termini
Next, we tested whether the transfer of the ROXY1 C-terminal L**LL motif to noncomplementing ROXYs is sufficient to mediate a ROXY1-like activity. ROXY6 to ROXY9, which showed severely compromised interactions with TGA proteins (Fig. 5B), were selected for domain-swapping experiments. According to sequence alignments of ROXY1 and ROXY6 to ROXY9 (Fig. 7A), the C-terminal 11 amino acids of ROXY6, as well as the C-terminal 14 amino acids of ROXY7, ROXY8, and ROXY9, were separately replaced by the ROXY1 C-terminal 14 amino acids (ROXY6S, ROXY7S, ROXY8S, and ROXY9S; Fig. 7A). The capacities of swapped ROXYs containing both the ROXY1 α-helical L**LL motif and the ROXY1 C-terminal ALWL motif to interact with TGA proteins were analyzed in yeast (Fig. 7B; Supplemental Fig. S2). As negative controls, all swapped ROXYs displayed a very weak autoactivation in the absence of prey proteins (5–6 Miller units; Supplemental Fig. S2). Compared with their wild-type proteins, strongly increased interactions with TGA proteins were observed for all swapped mutants (30–40 Miller units; Fig. 7B), further supporting the notion that the ROXY1 C-terminal L**LL motif is necessary for binding to TGA proteins in yeast. BiFC experiments revealed a reconstituted nuclear yellow fluorescence for coexpression of ROXY8S and PAN or TGA3, whereas no yellow fluorescence was detectable when ROXY8 was cotransformed with TGA proteins (data not shown). This indicates that swapped ROXYs acquired a capacity to interact in planta with TGA proteins. To examine whether the swapped ROXY proteins can also exert a ROXY1-like activity in flower development, each of the mutant versions of the four ROXY genes was expressed in the roxy1 mutant and petal phenotypes were scored (Supplemental Table S1). The roxy1 mutant transformed with each of the swapped ROXY genes and their wild-type versions developed on average only 2.5 petals per flower (Fig. 7A; Supplemental Table S1), demonstrating that they are unable to replace ROXY1 functionally in planta and thus fail to complement the roxy1 mutant. To exclude the possibility that the lack of roxy1 complementation for all the swapped ROXYs is caused by altered subcellular protein localization, swapped ROXYs and their wild-type versions were C-terminally fused in frame with GFP. All analyzed ROXY proteins were detected in both the nucleus and the cytoplasm and thus were not affected in their intracellular localization (data not shown). Together, these results imply that the transfer of the ROXY1 C terminus to noninteracting ROXYs is sufficient to mediate an interaction with TGA proteins in yeast and in planta. However, additional overall sequence differences of the four swapped ROXYs likely hinder the chimeric proteins from exerting a full ROXY1-like activity, including posttranslational target protein modifications in planta.
Figure 7.
Interaction strengths between swapped mutants of noncomplementing ROXYs containing the ROXY1 C-terminal sequences and two TGA factors. A, Alignment of C-terminal sequences of ROXY1, four noncomplementing ROXYs (ROXY6–ROXY9), as well as their swapped mutants (ROXY6S–ROXY9S). According to the sequence alignment, the C-terminal 11 amino acids of ROXY6 and the C-terminal 14 amino acids of ROXY7 to ROXY9 were exchanged with the ROXY1 C-terminal 14 amino acids, generating ROXY6S to ROXY9S, respectively. As indicated by the check mark, ROXY1 is capable of complementing the roxy1 mutant, whereas ROXY6 to ROXY9 as well as their swapped mutants (ROXY6S–ROXY9S) failed to rescue the roxy1 mutant (denoted by crosses). B, Quantitative β-galactosidase activity assays measuring the interaction between two TGA proteins and four swapped mutants as well as their wild-type proteins. Data are means ± sd of three independent experiments.
Identification of TGA Protein Domains Interacting with ROXY1
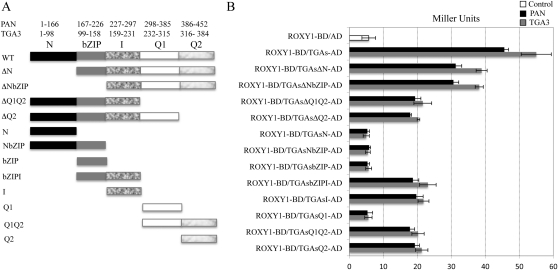
TGA transcription factors share three highly conserved domains of known functions, including a bZIP involved in DNA binding, nuclear localization, and homodimerization or heterodimerization as well as two Gln-rich domains (Q1 and Q2) involved in transcriptional activation (Katagiri et al., 1989, 1990; Fig. 8A; Supplemental Fig. S1). Two additional regions of as yet undefined functions exist in all TGA proteins, namely a nonconserved N-terminal region (N) before the bZIP domain and a conserved portion, localized between the bZIP and Q1 domains, which was named the intervening (I) region (Fig. 8A; Supplemental Fig. S1). The C-terminal Gln-rich domain of TGA3 is known to be crucial for the interaction with ROXY1 and TaROXY-α3, a ROXY1 ortholog in Triticum aestivum (Ziemann et al., 2010). Given the observation that the ROXY1 α-helical L**LL motif mediates binding to TGA proteins, we also wanted to determine the TGA protein domains required for an interaction with ROXYs. Four deletion mutants for both PAN and TGA3 were engineered: ΔN, N terminus removed; ΔNbZIP, N terminus plus bZIP deleted; ΔQ1Q2, two Gln-rich domains removed; and ΔQ2, lacking the second Gln-rich region (Fig. 8A). These deleted forms of TGA transcription factors were cloned into the prey vector and examined for their capability to interact with ROXY1 in the yeast two-hybrid system using the full length of PAN and TGA3 as positive controls (Fig. 8B; Supplemental Fig. S2). Deletion of the N terminus (ΔN) reduced β-galactosidase activity levels by about 30%. Further removal of the bZIP (ΔNbZIP) did not result in a further reduction of β-galactosidase activity. Deletion of the second Gln-rich region (ΔQ2) or further removal of the first Gln-rich region (ΔQ1Q2) caused an approximately 60% reduction of the β-galactosidase activity. Thus, the first Gln-rich region (Q1) and the bZIP are unlikely to be required for the interaction between ROXY1 and TGA proteins. However, the N terminus and the second Gln-rich domain (Q2) are indispensable for a strong interaction between the two TGA proteins and ROXY1. Given the observation that the N terminus of TGA transcription factors destabilizes TGA proteins in yeast (Zhou et al., 2000), reduced protein interactions as observed for TGA transcription factors lacking the N terminus are unlikely to result from a lower abundance of truncated proteins. As the N terminus of TGA transcription factors is nonconserved (Supplemental Fig. S1), it might not participate in the formation of a ROXY1-binding interface but possibly affects the structural stability of TGA transcription factors and thereby facilitates an interaction with ROXY1. To test this hypothesis, a second set of deletion mutants for two TGA transcription factors was constructed: N, N terminus; NbZIP, N terminus plus bZIP; bZIP; bZIPI, bZIP plus intervening region; I, intervening region; Q1, first Gln-rich region; Q1Q2, two Gln-rich regions; Q2, second Gln-rich region (Fig. 8A). Coexpression of ROXY1 with N, NbZIP, bZIP, or Q1 gave only background levels of the β-galactosidase activity, whereas cotransformation of ROXY1 with bZIPI, I, Q1Q2, or Q2 retained about 35% to 43% of the β-galactosidase activity observed for wild-type TGA transcription factors (Fig. 8B; Supplemental Fig. S2). Collectively, these findings indicate that the intervening region and the second Gln-rich domain of the investigated TGA proteins are essential for the interaction with ROXY1, while the nonconserved N terminus likely facilitates a strong interaction with ROXY1.
Figure 8.
Protein interactions between ROXY1 and truncated versions of two TGA proteins. A, Schematic representation of wild-type and deleted forms of PAN and TGA3 used in yeast two-hybrid experiments. TGA transcription factors share five regions, the N terminus (N; amino acids 1–166 [PAN] and 1–98 [TGA3]), the bZIP (amino acids 167–226 [PAN] and 99–158 [TGA3]), the intervening region (I; amino acids 227–297 [PAN] and 159–231 [TGA3]), the first Gln-rich region (Q1; amino acids 298–385 [PAN] and 232–315 [TGA3]), and the second Gln-rich region (Q2; amino acids 386–452 [PAN] and 316–384 [TGA3]). ΔN, ΔNbZIP, ΔQ1Q2, and ΔQ2 represent four deleted versions of TGA proteins. B, Quantitative β-galactosidase activity assays measuring the interaction of ROXY1 and truncated versions of PAN and TGA3. ΔN, ΔNbZIP, ΔQ1Q2, ΔQ2, N, NbZIP, bZIPI, bZIP, I, Q1, Q1Q2, and Q2 represent truncated forms of TGA proteins. Mating and cotransformation were performed for combinations containing PAN and TGA3, respectively. Data are means ± sd of three independent experiments.
DISCUSSION
GRXs are known to participate in redox-related processes associated with stress responses. Recently, ROXY1, an Arabidopsis CC-type GRX, has been shown to be involved in flower development, participating in the initiation and morphogenesis of petals (Xing et al., 2005). Genetic and intracellular localization experiments revealed that a nuclear interaction between ROXY1 and TGA transcription factors requires the C terminus of ROXY1 (Li et al., 2009). Here, dissection of the ROXY1 C terminus and its flanking sequences by a combination of different complementary approaches led to the identification of an L**LL motif located in the α5 helix of ROXY1. This Leu-rich motif of ROXY1 is essential for the interaction with PAN and TGA3 proteins and is together with the AL/IWL motif required for governing Arabidopsis petal development. Domain-deletion experiments of these TGA proteins demonstrate that the second Gln-rich domain and an intervening region between the bZIP domain and the first Gln-rich region are necessary for the interaction with ROXY1.
The ROXY1 C-Terminal L**LL Motif Is Essential for the Interaction with TGA Transcription Factors
The C-terminal eight amino acids of ROXY1 were previously shown to be required for the interaction with TGA proteins and for ROXY1 function in petal development, whereas the N terminus and the intervening region of ROXY1 are dispensable (Li et al., 2009). Here, yeast two-hybrid assays coupled with the ROXY1 C-terminal deletion experiments show that the ROXY1 C-terminal six amino acids are not required for the interaction with TGA proteins. However, the most C-terminal two amino acids of the ROXY1 α5 helix contribute significantly to the interaction with TGA proteins. Furthermore, when only the ROXY1 C-terminal 14 amino acids were tested in yeast two-hybrid experiments, these 14 amino acids were able to mediate an interaction with PAN and TGA3 proteins. Together, these data indicate that the ROXY1 α5 helix is required for the interaction with TGA proteins. Closer inspection of the ROXY1 α5 helix revealed the presence of an L**LL motif. In animals, this short L**LL motif exists in a large variety of nuclear proteins functioning as transcriptional coactivators, such as SRC-1/p160, TIF-2/GRIP-1, CBP/p300, and RIP-140. The animal L**LL motif mediates binding to liganded nuclear receptors and other transcription factors, such as the cAMP response element-binding (CREB) protein, which is one of the best characterized bZIP proteins in animals (Ribeiro et al., 1994; Le Douarin et al., 1996; Heery et al., 1997; Voegel et al., 1998).
To analyze the function and sequence conservation of a plant L**LL motif, substitution experiments were conducted. This showed that substitution of each of the three Leu residues by an Ala abrogates an interaction with PAN and TGA3 proteins in yeast. These substitutions also disable a nuclear in planta interaction with the two analyzed TGA proteins. The mutagenized ROXY1 proteins can no longer exert a full ROXY1 activity and fail to complement the roxy1 mutant. For the second Leu of the ROXY1 L**LL motif, changes to an Ile, a Met, a Thr, and an Arg did not affect the interaction with the two analyzed TGA proteins in yeast. The L**LL motif is thus indispensable for the interaction of ROXY1 with the two analyzed TGA proteins. TGA transcription factors belong to a group of bZIP proteins that are categorized into 10 groups (A–I and S) in Arabidopsis. TGA transcription factors belong to group D, and all members of this group have been investigated (Running and Meyerowitz, 1996; Chuang et al., 1999; Jakoby et al., 2002; Zhang et al., 2003; Kesarwani et al., 2007; Murmu et al., 2010). Nuclear transcriptional coactivators of animals can contain one or more copies of the L**LL motif. CBP/p300 acts as a transcriptional coactivator in animals and interacts with the bZIP protein CREB through two copies of the L**LL motif. The ability of the Leu-rich motif to mediate binding to a nuclear receptor or a transcription factor likely relies not solely on the α-helicity but also on the surface accessibility of this short motif (Heery et al., 1997). A second L**LL motif also exists in the α3 helix of ROXY1 (L84IRL87L88; Supplemental Fig. S1). However, replacement of the three Leu residues by Ala in this motif neither impaired the interaction of ROXY1 with TGA proteins nor the capability of the mutagenized ROXY1 to rescue abnormal petal phenotypes of the roxy1 mutant (S. Li and S. Zachgo, unpublished data). Only the L**LL motif in the ROXY1 α5 helix, therefore, is required and responsible for mediating the interaction with TGA proteins.
ROXYs Sharing the L**LL and AL/IWL Motifs Exert Similar Biochemical Activities
A subclass of ROXYs, comprising ROXY1 to ROXY5, ROXY18, and ROXY19, exhibits strong interactions with TGA transcription factors and is able to replace the ROXY1 function in petal development if expressed under the control of the ROXY1 promoter (Li et al., 2009). Several of these ROXYs have been shown to be involved in different biological processes. ROXY2, which interacts with TGA9 and TGA10 proteins in yeast and in planta, functions redundantly together with ROXY1 in anther development and microspore formation (Xing and Zachgo, 2008; Murmu et al., 2010). ROXY4 is a target gene up-regulated by DELLA and participates in GA signaling and floral organ development (Hou et al., 2008). ROXY19 is known to function in pathogen defense (Ndamukong et al., 2007). The capability of this subclass of ROXYs to exhibit ROXY1-like activities in flower development depends on the presence of an L**LL motif in their α5 helix. Here, we show that the presence of L1 and L3 is required, whereas L2 is dispensable for a ROXY1-like activity, as L2 can be replaced by a Met (ROXY4 and ROXY5), a Thr (ROXY18), or an Ile (ROXY19). This suggests that the functional properties of these ROXYs required for interaction and modification of TGA proteins are conserved. However, as these ROXYs are involved in different biological processes, such as stress and hormone responses as well as flower development, the recruitment to different biological processes likely occurred via modifying their expression domains. Given their conserved biochemical activity, subfunctionalization and neofunctionalization processes of ROXYs were likely realized by evolving differences in their cis-regulatory mechanisms and thereby contributed to generating functional diversification when the ROXY class expanded during land plant evolution.
Analysis of ROXYs that failed to complement the roxy1 mutant revealed further requirements for exerting an in planta ROXY1 activity. ROXY20 also possesses a conserved L1 and L3 in the α5 helix and shows an interaction with PAN and TGA3 proteins in yeast, but it cannot exert a ROXY1-like activity in planta. The failure of noncomplementing ROXYs to rescue the roxy1 mutant is likely caused by altered amino aid sequences conferring specificity to the protein activity in planta. The most C-terminal Leu of the AL/IWL motif is replaced by an Ala in ROXY20, and this motif thus terminates with a less hydrophobic side chain. The importance of this motif is further emphasized by the finding that the deletion of the last four amino acids impairs ROXY1 activity in planta. ROXY6 to ROXY9 lack the C terminus with the α-helical L**LL and AL/IWL motifs and neither interact with PAN and TGA3 nor complement the roxy1 mutant. The transfer of the ROXY1 C terminus to ROXY6 to ROXY9 suffices to restore the interaction with TGA proteins in yeast. However, these swapped proteins cannot exert a full ROXY1-like activity in Arabidopsis. ROXYs lacking this C terminus likely accumulated further overall amino acid deviations that contributed to establishing different biochemical activities during the expansion of the CC-type GRXs.
Together, the combination of protein interaction studies in yeast and functional analyses in planta allowed us to determine the minimal protein interaction domains in yeast and to ascertain whether these sequences are sufficient for mediating a full protein activity in planta or whether additional protein parts contribute to further protein activities, such as posttranslational modifications of target proteins.
ROXY1 Might Affect PAN Protein Activities by Posttranslational Cys Modifications
Deletion experiments of PAN and TGA3 demonstrate that the second Gln-rich domain and an intervening region participate in the formation of a ROXY1-interacting interface, whereas the nonconserved N terminus of TGA transcription factors possibly affects the structural stability of TGA transcription factors, thereby facilitating an interaction with ROXY1. Of the four Cys residues in TGA1 (Cys-172, Cys-260, Cys-266, and Cys-287), Cys-260, Cys-266, and Cys-287 are located in the first Gln-rich domain. Cys-260 and Cys-266 were previously shown to form an intramolecular disulfide bridge under oxidative conditions, which is reduced in a redox-dependent manner (Després et al., 2003). Most recently, additional complex posttranslational modifications have been reported for TGA1 Cys residues, including the formation of an intramolecular disulfide bridge between Cys-172 and Cys-287 and S-nitrosylation or S-glutathionylation of Cys-260 and Cys-266 (Lindermayr et al., 2010). In PAN, five Cys residues exist in the N terminus, and only one conserved Cys (Cys-340), corresponding to the TGA1 Cys-260, was found to be present in the first Gln-rich region of PAN (Li and Zachgo, 2009). Mutagenesis analysis of the PAN Cys-340 into Ser-340 followed by complementation experiments of the pan mutant showed that this mimicked reduced status of the single conserved Cys, Cys-340, resulted in a nonfunctional PAN protein that fails to complement pan mutants (Li et al., 2009). Given the observed interaction of ROXY1 and PAN required for petal development and the identification of only one crucial Cys in PAN, Cys-340 represents a potential target Cys for ROXY1 to modify posttranslationally. Current data available from Cys-to-Ser substitution experiments of the ROXY1 CCMC motif combined with roxy1 complementation analyses demonstrate that only the N-terminal Cys of the CCMC motif is crucial, indicating that ROXY1 exerts its activity via the monothiol mechanism (Xing et al., 2005; Li et al., 2009). Similar to Cys-260 and Cys-266 of TGA1, Cys-340 of PAN could also be S-nitrosylated or S-glutathionylated in vivo. Conceivably, ROXY1 may dock on the interaction interface formed by the second Gln-rich domain and the intervening region and catalyze deglutathionylation or denitrosylation reactions through target sites located in the first Gln-rich region. In animals, liganded nuclear receptors are known to recognize the L**LL motif of transcriptional coactivators by two main interactions, hydrophobic interactions and hydrogen bonding between specific charged amino acids (Savkur and Burris, 2004). Two highly conserved charged residues in liganded nuclear receptors (i.e. a Lys and a Glu) form a “charge clamp” that positions the α-helical L**LL motif by forming hydrogen bonds with the polypeptide backbone of the L**LL motif and allows the Leu side chains to pack into the intervening hydrophobic groove (Savkur and Burris, 2004).
Having identified the respective interaction domains of ROXY1 and PAN proteins, it will now be interesting to conduct structural analyses of TGA and ROXY proteins to resolve and model their interaction. This will ultimately advance our knowledge of how DNA-binding and/or transcriptional activities of TGA transcription factors are regulated by GRXs in developmental and stress-related processes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) mutants and wild-type plants were all Columbia ecotypes. Arabidopsis seeds were sown in soil and treated at 4°C for 6 d. Both tobacco (Nicotiana benthamiana) and Arabidopsis plants were grown in the greenhouse under controlled environmental conditions (22°C–23°C, 100 μmol photons m−2 s−1, 60% relative humidity, 16 h of light/8 h of dark).
Yeast Two-Hybrid Assays
Mutagenized versions of ROXYs were generated by a PCR-based mutagenesis approach. Wild-type ROXYs as well as their mutagenized forms were fused in frame as bait to the GAL4-BD in the bait vector pGBKT7 by EcoRI and BamHI restriction sites. To examine whether ROXYs as well as their corresponding mutant versions interact with TGA transcription factors, full-length cDNAs encoding TGA3 and PAN were PCR amplified and cloned into pGADT7 by EcoRI/BamHI restriction sites and pGADT7rec by a recombination strategy as described previously (Li et al., 2009), respectively. To test whether truncated forms of TGA3 and PAN affect the interaction with ROXY1, deleted TGA genes were created by a PCR-based approach. Both TGA proteins were used as prey and expressed as fusion proteins to the GAL4-AD. After cotransformation using Y2HGold or mating using both Y187 and Y2HGold (www.clontech.com), yeast cells were plated on selection medium containing SD/-Trp-His-Leu-Ade-aureobasidin A and then incubated in a growth chamber at 30°C for 2 to 5 d. All primers used for cloning are listed in Supplemental Table S2.
Quantification of β-Galactosidase Activity
β-Galactosidase activity from cotransformed or mated yeast was measured using the o-nitrophenyl β-d-galactopyranoside assay, where o-nitrophenyl produced from cleavage of o-nitrophenyl β-d-galactopyranoside by β-galactosidase was detected by spectrophotometry (Miller, 1972; Möckli and Auerbach, 2004).
Transient Expression Experiments
Leaves from 3- to 4-week-old tobacco plants were transformed by infiltration using a 1-mL syringe of Agrobacterium tumefaciens cells harboring appropriate plasmids. A. tumefaciens strain GV3101::pMP90 (RG) was used to examine the intracellular localization of GFP fusion proteins. GFP was fused to the N terminus of wild-type or mutant forms of ROXYs. All fusion genes encoding these fusion proteins were introduced into the pBAR-35S expression vector by XmaI and XbaI restriction sites, transiently expressed in tobacco leaf epidermal cells under the control of the cauliflower mosaic virus 35S promoter, and then monitored by capturing GFP fluorescence. To further confirm whether wild-type or mutagenized ROXYs (ROXY1L1A, ROXY1L2A, ROXY1L3A, ROXY1L2,3A, ROXY1L1,2,3A, ROXY1Δ4, ROXY8, and ROXY8S) interact in planta with TGA transcription factors, the BiFC technique was applied. Gateway-compatible vectors, pE-SPYNE and pE-SPYCE, were used to generate expression vectors using a Gateway cloning strategy (Walter et al., 2004; www.invitrogen.com), where fusion genes are under the control of the cauliflower mosaic virus 35S promoter. The YN was cloned upstream of mutagenized ROXY1 proteins in the pE-SPYNE vector. The YC was in frame fused N terminally for pE-SPYCE to TGA3 or PAN. The wild-type ROXY1 protein served as a control. Constructs were transformed into A. tumefaciens strain GV3101::pMP90 (RK). To avoid cosuppression, A. tumefaciens strain GV3101::pMP90 (RG) carrying the p19 viral silencing suppressor gene was always included in the infiltration mixture (Voinnet et al., 2003). Infiltrated tobacco leaves stayed attached to the plants, were examined after 3 to 5 d for the reconstitution of YFP fluorescence, and images were captured with a Leica TCS SP2 AOBS confocal laser scanning microscope.
Western-Blot Analysis
Preparation of yeast protein extracts was performed as described by Cabrera et al. (2009). Equal amounts of protein extracts were loaded onto a 15% SDS-PAGE gel. Fractionated proteins were transferred to nitrocellulose membranes, western blotted using the anti-Myc primary antibody (Santa Cruz Biotechnology), then the alkaline phosphatase-conjugated IgG secondary antibody, followed by immunodetection using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium staining.
Complementation Experiments
To test whether wild-type or mutagenized versions of ROXYs exert a ROXY1-like activity and complement the roxy1 mutant, respective cDNAs were cloned in pGSA1252 (www.chromdb.org) by XbaI restriction sites upstream of a NOS terminator and downstream of a 3.6-kb ROXY1 promoter fragment known to confer its endogenous expression (Xing et al., 2005). All these constructs were introduced into A. tumefaciens strain GV3101::pMP90 (RG) and transformed into the roxy1 mutant using the floral dip method (Clough and Bent, 1998). T1 transgenic plants were obtained by spraying with a 0.2% (v/v) BASTA solution. For complementation analysis of the roxy1 mutants, petal phenotypes of at least 50 T1 transgenic plants were examined.
Sequence Analysis
Protein sequence alignment of TGA factors and ROXYs was performed using the ClustalW program with default settings (http://www.ebi.ac.uk/clustalw) and adjusted manually.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein sequence alignment of all CC-type GRXs (A) and all TGA proteins (B) in Arabidopsis.
Supplemental Figure S2. Data of control experiments for yeast two-hybrid analyses.
Supplemental Table S1. Petal number of roxy1 mutants transformed with ROXY1, ROXY6 to ROXY9, and the respective swapped ROXYs.
Supplemental Table S2. Oligonucleotide primers used in this study.
References
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F. (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP. (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Cabrera M, Ostrowicz CW, Mari M, LaGrassa TJ, Reggiori F, Ungermann C. (2009) Vps41 phosphorylation and the Rab Ypt7 control the targeting of the HOPS complex to endosome-vacuole fusion sites. Mol Biol Cell 20: 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Running MP, Williams RW, Meyerowitz EM. (1999) The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev 13: 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YG, Zhong N, Rouhier N, Hase T, Kusunoki M, Jacquot JP, Jin CW, Xia B. (2006) Structural insight into poplar glutaredoxin C1 with a bridging iron-sulfur cluster at the active site. Biochemistry 45: 7998–8008 [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387: 733–736 [DOI] [PubMed] [Google Scholar]
- Holmgren A, Söderberg BO, Eklund H, Brändén CI. (1975) Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 A resolution. Proc Natl Acad Sci USA 72: 2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XL, Hu WW, Shen LS, Lee LYC, Tao Z, Han JH, Yu H. (2008) Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiol 147: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F; bZIP Research Group (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua NH. (1989) Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340: 727–730 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Yamazaki K, Horikoshi M, Roeder RG, Chua NH. (1990) A plant DNA-binding protein increases the number of active preinitiation complexes in a human in vitro transcription system. Genes Dev 4: 1899–1909 [DOI] [PubMed] [Google Scholar]
- Kesarwani M, Yoo J, Dong X. (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. (1996) A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J 15: 6701–6715 [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD. (2004) The glutaredoxin family in oxygenic photosynthetic organisms. Photosynth Res 79: 305–318 [DOI] [PubMed] [Google Scholar]
- Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S. (2009) Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21: 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zachgo S. (2009) Glutaredoxins in development and stress responses of plants. Adv Bot Res 52: 333–361 [Google Scholar]
- Lindermayr C, Sell S, Müller B, Leister D, Durner J. (2010) Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22: 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. (1972) Assay of β-galactosidase. Miller JH, , Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 352–355 [Google Scholar]
- Möckli N, Auerbach D. (2004) Quantitative β-galactosidase assay suitable for high-throughput applications in the yeast two-hybrid system. Biotechniques 36: 872–876 [DOI] [PubMed] [Google Scholar]
- Murmu J, Bush MJ, DeLong C, Li S, Xu M, Khan M, Malcolmson C, Fobert PR, Zachgo S, Hepworth SR. (2010) Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol 154: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. (2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Khafif M, Riondet C, Droux M, Bonnard G, Meyer Y. (2007) Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19: 1851–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A, Brown A, Lee KAW. (1994) An in vivo assay for members of the cAMP response element-binding protein family of transcription factors. J Biol Chem 269: 31124–31128 [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Jacquot JP. (2006) Genome-wide analysis of plant glutaredoxin systems. J Exp Bot 57: 1685–1696 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. (2004) Plant glutaredoxins: still mysterious reducing systems. Cell Mol Life Sci 61: 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running MP, Meyerowitz EM. (1996) Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122: 1261–1269 [DOI] [PubMed] [Google Scholar]
- Savkur RS, Burris TP. (2004) The coactivator LXXLL nuclear receptor recognition motif. J Pept Res 63: 207–212 [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJS, Tini M, Vivat V, Chambon P, Gronemeyer H. (1998) The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J 17: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Xing S, Lauri A, Zachgo S. (2006) Redox regulation and flower development: a novel function for glutaredoxins. Plant Biol (Stuttg) 8: 547–555 [DOI] [PubMed] [Google Scholar]
- Xing S, Rosso MG, Zachgo S. (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S. (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Zhang YL, Tessaro MJ, Lassner M, Li X. (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 13: 191–202 [DOI] [PubMed] [Google Scholar]
- Ziemann M, Bhave M, Zachgo S. (2009) Origin and diversification of land plant CC-type glutaredoxins. Genome Biol Evol 1: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann M, Bhave M, Zachgo S. (2010) Bioinformatic studies of the wheat glutaredoxin gene family and functional analysis of ROXY1 orthologues. Funct Plant Biol 38: 28–34 [DOI] [PubMed] [Google Scholar]