Abstract
Postpollination nonrandom mating among compatible mates is a widespread phenomenon in plants and is genetically undefined. In this study, we used the recombinant inbred line (RIL) population between Landsberg erecta and Columbia (Col) accessions of Arabidopsis (Arabidopsis thaliana) to define the genetic architecture underlying both female- and male-mediated nonrandom mating traits. To map the genetic loci responsible for male-mediated nonrandom mating, we performed mixed pollinations with Col and RIL pollen on Col pistils. To map the genetic loci responsible for female-mediated nonrandom mating, we performed mixed pollinations with Col and Landsberg erecta pollen on RIL pistils. With these data, we performed composite interval mapping to identify two quantitative trait loci (QTLs) that control male-mediated nonrandom mating. We detected epistatic interactions between these two loci. We also explored female- and male-mediated traits involved in seed yield in mixed pollinations. We detected three female QTLs and one male QTL involved in directing seed number per fruit. To our knowledge, the results of these experiments represent the first time the female and male components of seed yield and nonrandom mating have been separately mapped.
The process of pollination and fertilization offers plants a wealth of opportunity to breed selectively. Plants exhibit a stunning variety of floral and phenological mechanisms to selectively attract and bind pollen (Richards, 1986; Zinkl et al., 1999; Zinkl and Preuss, 2000; Waser and Ollerton, 2006). Once bound, pollen tubes embark on an elaborate journey through the pistil to deliver their sperm to the unfertilized ovules of the ovary. The pistil provides everything the male gametophyte requires for success in this endeavor, including guidance cues and nutrients that allow pollen tubes to traverse through the pistil to deliver their cargo. Although the pistil is a great facilitator of male gametophyte function, it can also act as an arena for mate competition or as a screen for mate discrimination. Even when the male gametophyte is successful in fertilizing an ovule, the female tissue may yet exhibit preferences by selectively aborting unwanted seeds or fruits (Obeso, 2004).
Given these opportunities, it is not surprising that some pollen have greater mating success—a phenomenon called nonrandom mating. Nonrandom mating in plants occurs across a continuum of relatedness (Marshall and Folsom, 1991). On one end of the spectrum, pollen may fail because it is self in a self-incompatible species (for review, see Hiscock and Tabah, 2003; Charlesworth et al., 2005; Hua et al., 2008). On the other end of the spectrum, pollen may fail because it is from a different and genetically incompatible species (Hogenboom, 1973, 1975; de Nettancourt, 2001). Our focus, however, is on variation in mating success among compatible mates. Specifically, we focus on postpollination nonrandom mating mechanisms that operate to differentially distinguish between conspecific compatible pollen (for review, see Willson and Burley, 1983; Marshall and Folsom, 1991).
Experiments in postpollination nonrandom mating among compatible mates are not new. More than 90 years ago, geneticist D.F. Jones was demonstrating selective fertilization in mixtures of corn pollen to explain deviations from Mendelian ratios of traits in progeny (Jones, 1920). Jones exploited endosperm color morphs in strains of inbred maize (Zea mays) to determine whether equal amounts of pollen sire equal amounts of progeny. Iterations of Jones’ experimental strategy have been widely used to explore aspects of postpollination nonrandom mating in plants that span the physiological, ecological, and phylogenetic spectrum. Postpollination nonrandom mating occurs in cultivars and wild species (e.g., maize and Viola tricolor, respectively; Jones, 1920, 1922; Pfahler, 1965; Skogsmyr and Lankinen, 1999; Lankinen and Skogsmyr, 2002). It occurs in plants that display different pollination syndromes, from animal to wind pollination (e.g., Raphanus sativus and Betula pendula, respectively; Marshall and Ellstrand, 1986; Pasonen et al., 1999). It occurs in plants that are predominately outcrossing, predominately selfing, and that exhibit mixed mating systems (e.g., Lesquerella fendleri, Arabidopsis [Arabidopsis thaliana], and Erythronium grandiflorum, respectively; Rigney et al., 1993; Mitchell and Marshall, 1998; Carlson et al., 2009). It occurs in every major clade of eudicot, from basal tricolpates, core tricolpates, asterids, and rosids (e.g., Persoonia mollis, Silene latifolia, Campsis radicans, and Cucurbita pepo, respectively; Bertin and Sullivan, 1988; Bertin, 1990; Quesada et al., 1991; Taylor et al., 1999; Krauss, 2000). It occurs in monocots, including the commelinoid clade (e.g., Allium cepa and Eichhornia paniculata, respectively; Currah, 1981; Cruzan and Barrett, 1996). It even occurs in gymnosperms (e.g., Pseudotsuga menziesii; Apsit et al., 1989). Most empirical studies demonstrate that plants have the potential for postpollination nonrandom mating among compatible mates. Although the demonstration of this potential in predominately outcrossing plants, predominately selfing plants, and plants with mixed mating systems has led to genetic predictions based on the potential for intrasexual selection in these different mating systems (Mazer et al., 2010), very little work has been done to genetically dissect the process.
Part of the challenge of unraveling the genetic basis for nonrandom mating lies in the number of entities that may be involved. Physiologically, postpollination nonrandom mating can occur via competition among pollen for access to ovules (male-mediated nonrandom mating). Alternatively, nonrandom mating could be due to differences in the complementarities between female tissues and individual pollen grains: pistil favoritism toward some pollen donors or handicapping of some pollen donors (female-mediated nonrandom mating). The genetics of this process have the potential to extend beyond the gametophyte genomes. Heritable differences in pollen tube growth may be encoded by the paternal sporophyte, which preloads pollen grains with nutrients for the initial stages of pollen tube growth, as well as by the male gametophyte genome (Stephenson et al., 2003). If anything, things may be more complex on the female side of the equation. Female-mediated nonrandom mating may be influenced by the obvious female sporophyte tissue that pollen tubes grow through and seeds develop in as well as the female gametophyte, which plays a role in guiding the pollen tube during the last stages of fertilization (Higashiyama et al., 2001; Higashiyama, 2002; Okuda et al., 2009). This highlights the importance of assaying female and male controls separately when dissecting the physiology and genetics of this process.
In previous work, we developed a system in Arabidopsis to investigate the genetics of postpollination nonrandom mating among compatible mates (Carlson et al., 2009). We demonstrated that genetically and geographically distinct accessions of Arabidopsis display both female- and male-mediated nonrandom mating. Of the nonrandom phenotypes we identified, we chose Columbia (Col) and Landsberg erecta (Ler) accessions to investigate the genetics of nonrandom mating because of the large nonrandom mating signal they display. When Col-neomycin phosphotransferase II (NPTII) (see “Materials and Methods”) and Ler pollen compete on a Col female, Col-NPTII sires 89% of the progeny, whereas Ler sires only 11% of the progeny. When Col-NPTII and Ler pollen compete on a Ler female, Col-NPTII again sires the majority of the progeny (92%), whereas Ler sires only 8%. We also chose these accessions for genetic analysis because of the large population of densely mapped recombinant inbred lines (RILs) available (Lister and Dean, 1993). RILs are powerful tools that allow high-resolution genetic mapping of loci that direct complex traits. Each RIL contains chromosomes that are a different, defined homozygous patchwork of Col and Ler DNA. By assaying nonrandom mating phenotypes with these RILs, we can statistically associate nonrandom mating phenotypes with chromosomal regions.
In this paper, we use this system to perform quantitative trait locus (QTL) mapping to construct genetic maps of loci responsible for both female- and male-mediated nonrandom mating and seed yield traits in a pair of accessions. To our knowledge, this is the first time that female and male loci involved in nonrandom mating have been separately mapped; this strategy may be one of the surest ways around the challenges of revealing separate roles of the female and male tissue in postpollination nonrandom mating among compatible mates.
RESULTS
Male-Mediated Nonrandom Mating and Seed Yield Phenotypes in the Ler/Col RIL Collection
The nonrandom sorting of pollen grains can be male mediated, female mediated, or a combination of the two. Our system allows us to genetically map these separate entities. To map male-mediated nonrandom mating loci, we perform mixed pollinations of RIL pollen and a common marker pollen (Col-NPTII, Col pollen with an integrated kanamycin resistance marker; see “Materials and Methods”) on virgin Col pistils. Thus, variations in the results of these pollinations can be statistically associated with Col and Ler chromosomal regions in the RIL pollen.
We performed 983 mixed pollinations using pollen from 99 different RILs. From these data, we excluded any pollination that did not yield live seed from our analysis. We also excluded any RIL that failed to yield at least five successful pollinations. We define a successful pollination as one that yields live seed. This resulted in our analysis of 928 pollinations with 98 different RILs. We performed between six and 20 pollinations with each RIL, with the mean number of pollinations with each RIL of 9.45 ± 1.99. We assayed the paternity of the 38,267 resulting seeds from these pollinations as described in “Materials and Methods.” Because of the nature of the data, we also had the opportunity to investigate seed yield per fruit. To do so, we counted the total live seed yield for each fruit that resulted from mixed pollinations. The average seed yield in these pollinations was 41.82 ± 8.78 seeds per fruit.
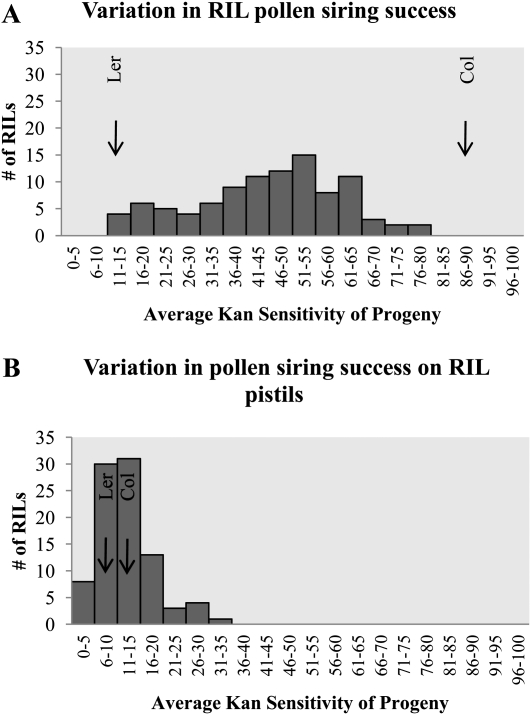
The RIL pollen display a wide array of phenotypic variation consistent with postpollination nonrandom mating among compatible mates being a quantitative trait (Fig. 1A). Given the proportional nature of the data and the large discrepancy between the Col and Ler pollen siring abilities, it is unsurprising that we see very little transgressive variation. The variation we see between pollinations could be due to genuine genetic differences in the RILs or random error. We partitioned variance in siring ability into genetic (VG) and error (VE) components in a random-effects ANOVA using the mean squared (MS) values using the SPSS statistical package (SPSS Inc.; Table I). We used these data to estimate broad-sense heritability for each trait: H2 = VG/(VG + VE). The broad-sense heritability for pollen siring ability in the RILs is 0.45. The broad-sense heritability for seed yield per fruit in mixed pollinations is 0.22. A Pearson correlation coefficient was computed to assess the relationship between seed yield per fruit and pollen siring ability phenotypes. There was no correlation between these two variables (r2 = −0.058, n = 928, P = 0.075).
Figure 1.
Frequency distributions of average kanamycin sensitivity of progeny in mixed pollinations using the Ler/Col RIL population. Arrows depict mean of parental lines (Col parental arrow indicates average kanamycin resistance of progeny). A, Variation in pollen siring success in mixed pollinations of Col-NPTII pollen and RIL pollen on Col pistils. B, Variation in pollen siring success in mixed pollinations between Col-NPTII pollen and Ler pollen on RIL pistils.
Table I. Variance components for mixed pollination traits.
KanS refers to the proportion of kanamycin-sensitive seeds resulting from mixed pollinations. Components of variance due to genetic differences among RILs (VG) and error measures within RILs (VE) were estimated from an ANOVA of MS values, where the dependent variable was either kanamycin sensitivity or seed set, and the random factor was RILs. Heritability (H2) was calculated as VG/(VG + VE). The F ratio (MSTrait/MSError) and resulting P value are also indicated.
| Trait | N | Mean | sd | VG | VE | H2 | F Ratio | P |
| RIL pollen | 931 | |||||||
| Proportion KanS progeny | 0.457 | 0.223 | 0.030 | 0.036 | 0.45 | 8.775 | <0.000 | |
| Seeds per fruit | 41.82 | 8.78 | 55.02 | 193.91 | 0.22 | 3.65 | <0.000 | |
| RIL pistil | 843 | |||||||
| Proportion KanS progeny | 0.134 | 0.153 | 0.006 | 0.052 | 0.10 | 2.092 | <0.000 | |
| Seeds per fruit | 32.65 | 15.17 | 41.76 | 188.98 | 0.18 | 3.05 | <0.000 |
Female-Mediated Nonrandom Mating and Seed Yield Phenotypes in the Ler/Col RIL Collection
Our previous results suggest that there is not a strong female-mediated nonrandom mating signal in mixed pollinations of Col-NPTII and Ler pollen on Col and Ler females (Carlson et al., 2009). Nevertheless, it is possible that a female signal could be revealed in the RILs and may be mapped via transgressive phenotypes. To map female-mediated nonrandom mating loci, we performed mixed pollinations of Ler pollen and Col-NPTII pollen on virgin RIL pistils. Thus, variations in the results of these pollinations can be statistically associated with Col and Ler chromosomal regions in the RIL females.
We performed 993 mixed pollinations on pistils from 99 different RILs. From these data, we excluded any pollination that did not yield live seed from our analysis. We also excluded any RIL that failed to yield at least five successful pollinations. This resulted in our analysis of 843 pollinations with pistils from 90 different RILs. We performed between six and 18 pollinations with each RIL, with the mean number of pollinations with each RIL of 9.40 ± 2.43. We assayed the paternity of the 27,523 resulting seeds from these pollinations as described in “Materials and Methods.” We also counted the total live seed yield for each fruit that resulted from mixed pollinations. The average seed yield in these pollinations was 32.65 ± 15.17 seeds per fruit.
Unsurprisingly, given the parental phenotypes, the RIL pistils show little variability in the proportion of progeny sired by Col-NPTII as compared with the variability when we use RIL pollen (Fig. 1B). Nevertheless, some transgressive segregation is seen in the proportion of progeny sensitive to kanamycin when RIL pistils are varied (Fig. 1B). We partitioned variance in siring ability into VG and VE components in a random-effects ANOVA using the MS values using the SPSS statistical package (SPSS Inc.; Table I). We used these data to estimate broad-sense heritability for each trait: H2 = VG/(VG + VE). The broad-sense heritability for changes in pollen siring success on RIL females is 0.10. The broad-sense heritability for seed yield per fruit in mixed pollinations is 0.18. A Pearson correlation coefficient was computed to assess the relationship between seed yield per fruit and pollen siring success phenotypes on RIL females. There was no correlation between these two variables (r2 = −0.047, n = 843, P = 0.176).
Mapping of Male and Female Nonrandom Mating QTLs and Analysis of Epistatic Interactions
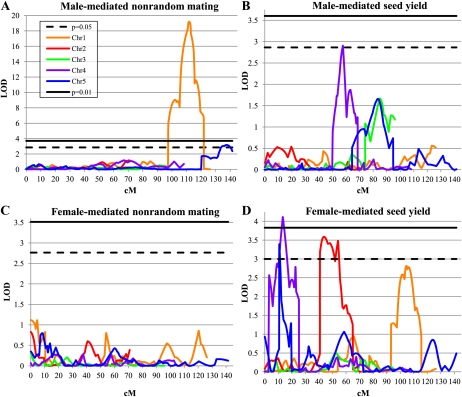
We used the above data to perform composite interval mapping (Fig. 2). Unsurprisingly, for the female- and male-mediated nonrandom mating traits, only the RIL pollen data yielded QTLs of appreciable significance.
Figure 2.
QTL map of loci for nonrandom mating and seed yield traits. The dashed black line and solid black line indicate the LOD scores for P = 0.05 and 0.01 thresholds, respectively. Orange, red, green, purple, and blue lines indicate the LOD scores for chromosomes 1, 2, 3, 4, and 5, respectively. A, Profile for male-mediated nonrandom mating phenotype. B, Profile for male-mediated seed yield. C, Profile for female-mediated nonrandom mating phenotype. D, Profile for female-mediated seed yield.
Two QTLs were identified that affect male-mediated nonrandom mating, qMNRM1-2 (for QTL for Male-mediated Nonrandom Mating; Fig. 2A). The 2-log of the odds (LOD) interval of qMNRM1 maps between markers g4552-mi103 on chromosome 1, with an LOD peak near marker TAG1 (111.0 centimorgans [cM] from the top of the chromosome). This locus shows an average decrease of 14% in the siring ability of pollen (as measured in the percentage of kanamycin-sensitive seeds sired in mixed pollinations) when the Ler allele is present and explains 66.9% of the variance of the trait (Table II). The 2-LOD interval of qMNRM2 maps between markers emb514-CATHANK, with an LOD peak near marker g2368 (137.5 cM from the top of the chromosome). This locus shows an average decrease of 4.6% in the siring ability of pollen when the Ler allele is present and explains 5.3% of the variance of the trait (Table II). It is not likely that qMNRM1-2 is a male sterility factor because it does not overlap with fertility (seed yield per fruit) QTLs mapped from single-pollen pollinations of this RIL population (J.N. Fitz Gerald, unpublished data). It is interesting to note that these QTLs colocalize with chromosomal areas that displayed segregation distortion in the construction of the RILs (see “Discussion”).
Table II. QTL detected for male-mediated nonrandom mating (MNRM) and female and male seed yield (FSY, MSY) traits.
Associated Marker, Position refers to the closest marker associated with the QTL, as well as the QTL peak. CI refers to the 2-LOD confidence interval (in cM), as well as the associated markers. For additive effect, when numbers are positive, then Col alleles increase the trait. When numbers are negative, then Ler alleles increase the trait. Variance refers to the percentage of variance explained by the QTL. Asterisk, Not significant by GLM.
| Trait | Chromosome | Associated Marker, Position (cM) | CI (cM) | LOD | Additive Effect (P = ) | Variance |
| qFSY1 | 2 | er, 43.7 | 40.7-61.2 (GPA1-LTP) | 3.59 | −2.21 (0.01) | 12.1 |
| qFSY2 | 4 | app, 13.3 | 6.88-23.7 (g3843- mi167) | 4.11 | −2.65 (0.0015) | 16.9 |
| qFSY3 | 5 | 06569, 10.7 | 10.6-14.3 (mi121-mi97) | 3.36 | −1.21 (0.138)* | 2.1 |
| qMSY1 | 4 | AG, 57.7 | 50.2-68.6 (mi260-mi123) | 2.9 | 2.52 (0.0031) | 6.8 |
| qMNRM1 | 1 | Tag1, 111.9 | 109.3-114.0 g4552-mi103 | 19.2 | 0.14 (<0.000) | 66.9 |
| qMNRM2 | 5 | g2368, 137.5 | 120.4-141.5 (emb514-CATHHANK) | 3.17 | 0.046 (<0.000) | 5.3 |
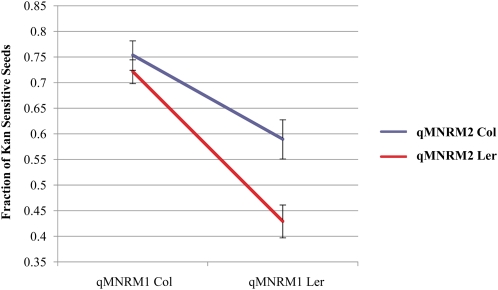
We detected significant epistatic interaction between qMNRM1 and qMNRM2 in the male-mediated nonrandom mating data (Table III; Fig. 3). When qMNRM1 contains a Col allele, qMNRM2 has little effect on pollen siring ability. By contrast, when qMNRM1 contains a Ler allele, a Col allele at qMNRM2 increases the siring ability of pollen (Table III; Fig. 3).
Table III. QTL epistatic effects for male-mediated nonrandom mating.
QTL_i and QTL_j are the two QTLs involved in epistatic interactions. aa, Estimated additive by additive effect. Variance refers to the percentage of variance explained by the epistasis.
| QTL_i | P | QTL_j | Position | aa (P = ) | Variance |
| cM | |||||
| qMNRM1 | 111.9 | qMNRM2 | 137.5 | −0.035 (0.0051) | 6.8 |
Figure 3.
Epistasis effect between qMNRM1 and qMNRM2. Graph shows the average pollen siring success (and se), as denoted by the percentage of progeny that are kanamycin sensitive, when the Col or Ler allele is at qMNRM1 and qMNRM2. When qMNRM1 is Col, qMNRM2 has very little effect on pollen siring success. When qMNRM1 is Ler, however, the Col allele of qMNRM2 increases the siring success of the pollen grain.
When we analyzed the data on seed yield per fruit, we detected both female- and male-mediated seed yield QTLs, qFSY1-3 (for QTL for Female-mediated Seed Yield) and qMSY1 (for QTL for Male-mediated Seed Yield). We detected three female-mediated seed yield QTLs. The 2-LOD interval of qFSY1 maps between markers GPA-LTP on chromosome 2, with an LOD peak near marker er (43.7 cM from the top of the chromosome). This locus shows an average increase of 2.21 seeds when the Ler allele is present and explains 12.1% of the variance of the trait (Table II). This same locus was identified in an earlier mapping experiment for seed yield per fruit between the Arabidopsis accessions Ler and Cape Verde Islands (CVI; see “Discussion”; Alonso-Blanco et al., 1999). The 2-LOD interval of qFSY2 maps between markers g3843-mi167 on chromosome 4, with an LOD peak near marker app (13.3 cM from the top of the chromosome). This locus shows an average increase of 2.65 seeds when the Ler allele is present and explains 16.9% of the variance of the trait (Table II). Finally, the 2-LOD interval of qFSY3 maps between markers mi121-mi97 on chromosome 5, with an LOD peak near marker 06569 (10.7 cM from the top of the chromosome). This locus shows an average increase of 1.21 seeds when the Ler allele is present and explains 2.1% of the variance of the trait (Table II). Although this last peak was detected in our composite interval mapping, it was not significant by general linear model (GLM) and is thus highly suspect.
We detected one male-mediated seed yield QTL, qMSY1 (Fig. 2B). The 2-LOD interval of qMSY1 maps between markers mi260-mi123 on chromosome 4, with an LOD peak near marker AG (57.7 cM from the top of the chromosome). This locus shows an average increase of 2.52 seeds when the Col allele is present and explains 6.8% of the variance of the trait (Table II). The identification of a male-mediated seed yield trait in Col/Ler RILs is consistent with experiments that examined paternal effects in seed size and number in reciprocal crosses between different Arabidopsis accessions (see “Discussion”; House et al., 2010).
DISCUSSION
Although there has been more than a century of work on nonrandom mating among compatible mates in plants, genetic studies of this process are rare. Studies have explored whether there is a genetic component to male competitive ability and established heritability and evolvability values for pollen competition traits (Snow and Mazer, 1988; Sari-Gorla et al., 1992; Havens, 1994; Skogsmyr and Lankinen, 2000; Lankinen et al., 2009). However, very little information exists defining the genetic architecture of nonrandom mating among compatible mates. In previous work, we surveyed the nonrandom mating phenotypes in lab and geographical accessions of Arabidopsis (Carlson et al., 2009). In almost all cases, we found statistically significant differences in paternity. In some cases, such as mixed pollinations with Col-NPTII and Ler pollen, the differences between the observed seed paternities are extreme (89% and 11%, respectively, favored toward Col-NPTII-fathered progeny; Carlson et al., 2009). In this study, we used the Ler/Col RILs to dissect the female and male genetic architecture that underlies this nonrandom mating. Because of the nature of the data we collected, we also had the opportunity to study female- and male-mediated controls of seed yield in mixed pollinations.
qMNRM1 and qMNRM2 Account for 72.2% of the Variance in Male-Mediated Nonrandom Mating
We identified two QTLs implicated in male-mediated nonrandom mating (qMNRM1 and qMNRM2). qMNRM1 accounts for 66.9% of the variance in male-mediated nonrandom mating and is located on chromosome 1. The 2-LOD interval for this QTL maps to the region bounded by markers g4552 and mi103. Using a higher-density map (see “Materials and Methods”) narrowed the 2-LOD interval to an area between markers Athb13 and Mi462 that contains roughly 50 genes, ranging between At1G69980 to At1G70250. Several of these genes are viable candidates for the causative gene. We examined these candidates for tissue-specific expression using the Electronic Fluorescent Pictograph browser (Winter et al., 2007) available at the Bio-Array Resource for Plant Biology (http://bar.utoronto.ca/welcome.htm). We focused on genes that show expression in either developing pollen (Honys and Twell, 2004) or mature pollen/pollen tubes (dry, in vitro germinated, or after growth through styler tissue; Qin et al., 2009). Of the 39 genes available in the database, only three showed expression levels markedly above background in mature pollen/pollen tubes: At1G69840, At1G69940, and At1G70170. Near the Athb13 side of the interval, At1G69840 encodes a putative hypersensitive response protein that contains a stomatin/prohibitin/flotillin/HflK/C domain (Qi and Katagiri, 2009). Near the center of the interval, At1G69940 encodes a pollen-specific pectin methylesterase involved in pollen tube growth (Tian et al., 2006; Röckel et al., 2008). Near the mi462 side of the interval, At1G69940 encodes a protein that contains a matrix metalloproteinase motif. Although only three genes showed expression in mature pollen/pollen tubes, 17 of the 39 genes showed expression levels markedly above background in developing pollen (a list that includes At1G69940 and At1G70170). Finally, the causative gene may act in the sporophytic anther tissue, rather than in the developing or mature gametophyte. We will likely be able to narrow candidates using cell biology to identify whether the Col allele provides an advantage over the Ler allele in germinating pollen, early pollen tube growth, or late pollen tube growth.
Identifying candidate genes for qMNRM2 will be more difficult. qMNRM2 accounts for only 5.3% of the variance in male-mediated nonrandom mating and is located on chromosome 5. The 2-LOD interval for this QTL maps to the region bounded by markers emb514 and CATHHANK. This is a much larger interval than what we see in qMNRM1, encompassing 21.1 cM compared with the 4.7-cM distance for qMNRM1. This region contains more than 500 genes. The small effect this locus has, coupled with the epistatic interaction qMNRM2 displays with qMNRM1, makes the prospect of cloning the causative gene in this region via the candidate gene approach much more challenging.
Both male-mediated nonrandom mating QTLs localized at chromosomal regions that were overrepresented in the construction of the RILs, the likely result of segregation distortion (Lister and Dean, 1993). This overrepresentation could be the result of the Col alleles in these regions providing a competitive advantage over Ler alleles during the self-fertilizations that led to the construction of these lines (Lister and Dean, 1993). Lister and Dean identified a bias for Col alleles at five markers between 88 and 118 cM on the lower arm of chromosome 1. qMNRM1 maps to this region, with a confidence interval that spans 109.3 to 114.0 cM. The only other region that Lister and Dean identified as having a bias for Col alleles in the RILs is on the lower arm of chromosome 5 at 132 cM. qMNRM2 maps to this region, with a confidence interval that spans 120.4 to 141.5 cM. Lister and Dean also identified three regions that had Ler bias in the RILs on chromosome 2, the upper arm of chromosome 4, and the upper arm of chromosome 5. It is possible that Ler alleles in these regions give pollen a competitive advantage, but our assay is not sensitive enough to detect and map these signals. Alternatively, the allelic bias seen in the RILs may be due to Ler DNA in these regions affecting seed yield (qFSY1 is present on chromosome 2, and qFSY2 and qFSY3 map very close to the biased regions on chromosome 4 and 5), seedling viability, chance, or other mechanisms.
qFSY1-3 and qMSY1 Account for 31.1% and 6.8% of the Variance in Female- and Male-Mediated Control of Seed Yield, Respectively
Seed yield is usually determined by three components: fruits per plant, seeds per fruit, and seed weight. Although seed yield has been the subject of genetic analysis in Arabidopsis, few studies have separated the different components (with seed weight/size most often separately analyzed), and no study has quantitatively mapped the differing female and male contributions to these traits (Alonso-Blanco et al., 1999; Barth et al., 2003; House et al., 2010; Li et al., 2010). From an applied perspective, the genetic dissection of the contributions of male and female tissue to seed yield components may be quite useful in developing strategies to improve agricultural yield.
Because of the nature of the data we collected, we were able to investigate genetic traits involved in female and male control of seeds per fruit in mixed pollinations. We identified three QTLs implicated in female-mediated seed yield (qFSY1–qFSY3). qFSY1 accounts for 12.1% of the variance in female-mediated seed yield and is located on chromosome 2. The 2-LOD interval for this QTL maps to the region bounded by markers GPA1 and LTP, with a peak at marker er. This is consistent with earlier mapping studies. In mapping allelic variation in life history traits in the Arabidopsis accessions Ler and CVI, Alonso-Blanco et al. (1999) identified four QTLs that affected seed number per fruit, one of which centered on marker er on chromosome 2. Interestingly, in their study, the CVI allele at er increases the phenotypic value of seed yield per fruit over that of the Ler allele. In our study, the Ler allele at er increases the phenotypic value of seed yield per fruit over the Col allele. Despite the differences in pollination method (Alonso-Blanco et al. [1999] used self-pollination, whereas we emasculated pistils and performed mixed pollinations) and the use of different accessions, the localization of a QTL peak at er on chromosome 2 verifies the importance of this locus for the seed yield per fruit trait.
qFSY2 accounts for 16.9% of the variance in female-mediated seed yield and is located on chromosome 4. Like qFSY1, the Ler allele at this locus increases the phenotypic value of seed yield per fruit. This QTL, like qMNRM2, encompasses a large, 16.8-cM region that contains more than 600 genes. Identification of the causative gene at this locus will require fine mapping to narrow the candidates. Finally, qFSY3 accounts for a paltry 2.1% of the variance in female-mediated seed yield. This, and the failure of GLM to identify this QTL, makes this locus for female-mediated seed yield highly suspect.
We identified one QTL implicated in male-mediated seed yield (qMSY1). qMSY1 accounts for 6.8% of the variance in male-mediated seed yield and is located on chromosome 4. The 2-LOD interval for this QTL maps to the region bounded by markers mi260 and mi123 and encompasses a 18.4-cM region that contains more than 1000 genes. The Col allele at this locus increases the phenotypic value of seed yield per fruit, consistent with data from reciprocal crosses between different accessions of Arabidopsis (House et al., 2010). To study the role of conflict between parents and between parents and offspring, House et al. performed a series of reciprocal crosses between four accessions of Arabidopsis, including Col and Ler. They concluded that the number of viable seeds per fruit was influenced by the identity of the pollen donor, with Ler fathers producing significantly less seeds than Col fathers (House et al., 2010).
CONCLUSION
Nonrandom mating among compatible mates has long captured the interest of researchers because of its potential significance for processes, such as inbreeding depression, sexual selection, reproductive barrier reinforcement, and speciation. In flowering plants, it has been shown to operate in every major taxa, and yet its genetics remain a mystery. In this study, for the first time to our knowledge, we have separately mapped female and male nonrandom mating traits as well as seed yield traits. Given the sheer number of tissues that may be involved in directing nonrandom mating, the ability to unambiguously separate female and male roles is an important first step toward disentangling the influence different tissues have on this process. The data presented here provide insight into nonrandom mating in predominately selfing plants and are an important first step in identifying and defining the genic architecture that directs this widespread phenomenon.
MATERIALS AND METHODS
Arabidopsis (Arabidopsis thaliana) Ecotypes
The RIL set used for this study was derived from a cross between ecotypes Ler and Col (Lister and Dean, 1993). Residual heterozygosity in this population is calculated to be 0.42% (slightly less than the theoretical expectation of 0.78%; Lister and Dean, 1993). For nonrandom mating assays, we substituted the wild-type Col accession (Col-4) for a Col accession (Col-0) containing an integrated, intergenic kanamycin antibiotic resistance marker (Col-NPTII). This allowed us to quantify the progeny of mixed pollinations via a simple plate germination assay. Col-NPTII is an F2 homozygous transfer DNA insertion mutant obtained from the SIGnAL project (GenBank accession no. BZ377762; Alonso et al., 2003). The intergenic transfer DNA lies between At1g28440 and At1g28450. This strain displays a 4:0 segregation of NPTII-mediated kanamycin resistance, demonstrating homozygosity (Carlson et al., 2009). We obtained all strains from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/).
Plant Growth
Seeds were imbibed and cold treated at 4°C for 3 d in the dark to break dormancy and to promote uniform germination. Plants are grown in 4.5-inch pots with generally 10 plants per pot in Percival growth chambers (Percival Scientific). Plants were grown in Shultz premium potting soil, watered every second day, and fertilized (18-18-21) twice per week. Plants are subjected to 12 h of 130-μE fluorescent lighting at 22°C.
Mixed Pollinations
Mixed pollinations were performed as described on primary inflorescences of maternal plants (Carlson et al., 2009). We emasculated buds during stages 11 to 12 of development (Smyth et al., 1990). We allowed pistils to develop to stage 14 before we performed pollinations. We harvested anthers from stage 14 flowers and visually inspected them for levels of dehiscence. From these anthers, we chose two from each potential father and readied them on forceps. We used a stereomicroscope (Leica ZOOM2000) to better visualize the stigma when we applied pollen from Col-NPTII on one-half of the available surface area of the virgin stigma. We then applied pollen from the competing father on the remaining surface area. We completed each assay within 1 min. Mature siliques were collected, and seed paternity was assayed by growing seeds on standard Murashige and Skoog media with Suc containing 50 μg/mL kanamycin (Murashige and Skoog, 1962).
As noted previously, the presence of the NPTII gene does not change the competitive ability of Col-NPTII pollen. This was demonstrated in two ways. First, when Col-NPTII was crossed to Col-0, and the F1 was allowed to self-fertilize, the resulting progeny displayed 3:1 segregation of NPTII-mediated kanamycin resistance (Carlson et al., 2009). Because Col-NPTII shows the expected 3:1 ratio of segregation, this is fairly strong evidence that the presence of the antibiotic resistance marker insertion does not change the siring success of the pollen. Also, as noted previously, we delivered consistently equal amounts of pollen in mixed pollinations, demonstrated by our mixed pollinations of Col-NPTII and Col-4 pollen on virgin Col-4 pistils. These control pollinations show no statistical difference in the siring success of the two pollen types and do not differ substantially from the expected 1:1 ratio in progeny (Carlson et al., 2009).
Statistical Analysis and QTL Mapping
Because the data we collect from mixed pollinations are proportional data (the proportion of kanamycin-sensitive seeds) and, thus, are not free to vary, for statistical testing, we transformed the proportion of seeds sensitive to kanamycin using an arcsine square root transformation to normalize the distribution of data. We partitioned variance in siring ability into VG and VE components in a random-effects ANOVA using the SPSS statistical package (SPSS Inc.). We used these data to calculate broad-sense heritability for each trait based on variance within genotype [H2 = VG/(VG + VE)]. Phenotypic correlation analysis was performed by calculating the Pearson correlation coefficients using this software package.
The genetic linkage map for the Ler/Col RILs was constructed using public data for the genotypes of the RILs obtained from the Nottingham Arabidopsis Stock Centre (http://www.arabidopsis.info/) using Mapmaker/EXP version 3.0b (Lander et al., 2009). Maps were constructed using 222 genetic markers, with 44 ± 9.6 markers per chromosome at an intermarker distance of 2.5 ± 1.6 cM using the Kosambi mapping function. For candidate gene identification for male-mediated nonrandom mating QTLs, mapping was further refined using a set of 1288 markers with 257 ± 63 markers per chromosome at an intermarker distance of 1.4 ± 1.8 cM. Confidence interval mapping of major-effect QTL was conducted with Zmapqtl (QTL Cartographer v1.17j; Basten et al., 2002). The reported P value thresholds were obtained from the maximum LOD scores found in 10,000 permutations of the trait data. Reported QTL confidence intervals were derived from the markers at which the LOD score for a trait dropped 2-LOD below the peak maximum. QTL additive values were estimated from a generalized linear model including all detected QTL peaks (trait = μ + βmrk1 × Mrk1 + … + βmrkn × MrkN +ε), with a crossed term added when epistasis was detected (JMP version 8.0.1; SAS Institute). Variance components were estimated with a restricted maximum likelihood method (JMP). Although all computations for mixed pollination traits were performed with arcsine transformed data, results were subsequently untransformed back to trait values to report additive scores and to construct Figure 3.
Acknowledgments
We thank two anonymous reviewers for comments that greatly improved a previous version of this article.
References
- Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M. (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 4710–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Apsit VJ, Nakamura RR, Wheeler NC. (1989) Differential male reproductive success in Douglas fir. Theor Appl Genet 77: 681–684 [DOI] [PubMed] [Google Scholar]
- Barth S, Busimi AK, Friedrich Utz H, Melchinger AE. (2003) Heterosis for biomass yield and related traits in five hybrids of Arabidopsis thaliana L. Heynh. Heredity 91: 36–42 [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B. (2002) QTL Cartographer, Version 1.17. Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Bertin RI. (1990) Paternal success following mixed pollinations of Campsis radicans. Am Midl Nat 124: 153–163 [Google Scholar]
- Bertin RI, Sullivan M. (1988) Pollen interference and cryptic self-fertility in Campsis radicans. Am J Bot 75: 1140–1147 [Google Scholar]
- Carlson AL, Telligman M, Swanson RJ. (2009) Incidence and post-pollination mechanisms of nonrandom mating in Arabidopsis thaliana. Sex Plant Reprod 22: 257–262 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Vekemans X, Castric V, Glémin S. (2005) Plant self-incompatibility systems: a molecular evolutionary perspective. New Phytol 168: 61–69 [DOI] [PubMed] [Google Scholar]
- Cruzan MB, Barrett SCH. (1996) Postpollination mechanisms influencing mating patterns and fecundity: an example from Eichhornia paniculata. Am Nat 147: 576–598 [Google Scholar]
- Currah L. (1981) Pollen competition in onion (Allium cepa L). Euphytica 30: 687–696 [Google Scholar]
- de Nettancourt D. (2001) Incompatibility and incongruity in wild and cultivated plants. Springer, Berlin [Google Scholar]
- Havens K. (1994) Clonal repeatability of in vitro pollen tube growth rates in Oenothera organensis (Onagraceae). Am J Bot 81: 161–165 [Google Scholar]
- Higashiyama T. (2002) The synergid cell: attractor and acceptor of the pollen tube for double fertilization. J Plant Res 115: 149–160 [DOI] [PubMed] [Google Scholar]
- Higashiyama T, Yabe S, Sasaki N, Nishimura Y, Miyagishima S, Kuroiwa H, Kuroiwa T. (2001) Pollen tube attraction by the synergid cell. Science 293: 1480–1483 [DOI] [PubMed] [Google Scholar]
- Hiscock SJ, Tabah DA. (2003) The different mechanisms of sporophytic self-incompatibility. Philos Trans R Soc Lond B Biol Sci 358: 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenboom NG. (1973) Model for incongruity in intimate partner relationships. Euphytica 22: 219–233 [Google Scholar]
- Hogenboom NG, Mather K. (1975) Incompatibility and incongruity. 2. Different mechanisms for non-functioning of intimate partner relationships. Proc R Soc Lond B Biol Sci 188: 361–374 [Google Scholar]
- Honys D, Twell D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House C, Roth C, Hunt J, Kover PX. (2010) Paternal effects in Arabidopsis indicate that offspring can influence their own size. Proc Biol Sci 277: 2885–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua ZH, Fields A, Kao TH. (2008) Biochemical models for S-RNase-based self-incompatibility. Mol Plant 1: 575–585 [DOI] [PubMed] [Google Scholar]
- Jones DF. (1920) Selective fertilization in pollen mixtures. Proc Natl Acad Sci USA 6: 66–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DF. (1922) Selective fertilization and the rate of pollen-tube growth. Biol Bull 43: 167–174 [Google Scholar]
- Krauss SL. (2000) The realized effect of postpollination sexual selection in a natural plant population. Proc Biol Sci 267: 1925–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. (2009) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Lankinen A, Maad J, Armbruster WS. (2009) Pollen-tube growth rates in Collinsia heterophylla (Plantaginaceae): One-donor crosses reveal heritability but no effect on sporophytic-offspring fitness. Ann Bot (Lond) 103: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankinen Å, Skogsmyr I. (2002) Pollen competitive ability: the effect of proportion in two-donor crosses. Evol Ecol Res 4: 687–700 [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. (2010) Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. (1993) Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J 4: 745–750 [DOI] [PubMed] [Google Scholar]
- Marshall DL, Ellstrand NC. (1986) Sexual selection in Raphanus sativus: Experimental-data on nonrandom fertilization, maternal choice, and consequences of multiple paternity. Am Nat 127: 446–461 [Google Scholar]
- Marshall DL, Folsom MW. (1991) Mate choice in plants - An anatomical to population perspective. Annu Rev Ecol Syst 22: 37–63 [Google Scholar]
- Mazer SJ, Hove AA, Miller BS, Barbet-Massin M. (2010) The joint evolution of mating system and pollen performance: predictions regarding male gametophytic evolution in selfers vs. outcrossers. Perspect Plant Ecol Evol Syst 12: 31–41 [Google Scholar]
- Mitchell RJ, Marshall DL. (1998) Nonrandom mating and sexual selection in a desert mustard: an experimental approach. Am J Bot 85: 48–55 [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Obeso JR. (2004) A hierarchical perspective in allocation to reproduction from whole plant to fruit and seed level. Perspect Plant Ecol Evol Syst 6: 217–225 [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Pasonen HL, Pulkkinen P, Kapyla M, Blom A. (1999) Pollen-tube growth rate and seed-siring success among Betula pendula clones. New Phytol 143: 243–251 [Google Scholar]
- Pfahler PL. (1965) Fertilization ability of maize pollen grains. I. Pollen sources. Genetics 52: 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi YP, Katagiri F. (2009) Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J 57: 932–944 [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R. (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada M, Schlichting CD, Winsor JA, Stephenson AG. (1991) Effects of genotype on pollen performance in Cucurbita pepo. Sex Plant Reprod 4: 208–214 [Google Scholar]
- Richards AJ. (1986) Plant Breeding Systems. Allen and Unwin, London [Google Scholar]
- Rigney LP, Thomson JD, Cruzan MB, Brunet J. (1993) Differential success of pollen donors in a self-compatible lily. Evolution 47: 915–924 [DOI] [PubMed] [Google Scholar]
- Röckel N, Wolf S, Kost B, Rausch T, Greiner S. (2008) Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J 53: 133–143 [DOI] [PubMed] [Google Scholar]
- Sari-Gorla M, Pé ME, Mulcahy DL, Ottaviano E. (1992) Genetic dissection of pollen competitive ability in maize. Heredity 69: 423–430 [Google Scholar]
- Skogsmyr I, Lankinen Å. (1999) Selection on pollen competitive ability in relation to stochastic factors influencing pollen deposition. Evol Ecol Res 1: 971–985 [Google Scholar]
- Skogsmyr I, Lankinen Å. (2000) Potential selection for female choice in Viola tricolor. Evol Ecol Res 2: 965–979 [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow AA, Mazer SJ. (1988) Gametophytic selection in Raphanus raphanistrum: a test for heritable variation in pollen competitive ability. Evolution 42: 1065–1075 [DOI] [PubMed] [Google Scholar]
- Stephenson AG, Travers SE, Mena-Ali JI, Winsor JA. (2003) Pollen performance before and during the autotrophic-heterotrophic transition of pollen tube growth. Philos Trans R Soc Lond B Biol Sci 358: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Saur MJ, Adams E. (1999) Pollen performance and sex-ratio evolution in a dioecious plant. Evolution 53: 1028–1036 [DOI] [PubMed] [Google Scholar]
- Tian GW, Chen MH, Zaltsman A, Citovsky V. (2006) Pollen-specific pectin methylesterase involved in pollen tube growth. Dev Biol 294: 83–91 [DOI] [PubMed] [Google Scholar]
- Waser NM, Ollerton J, (2006) Plant-Pollinator Interactions: From Specialization to Generalization. University of Chicago Press, Chicago, IL [Google Scholar]
- Willson MF, Burley N. (1983) Mate Choice in Plants. Princeton University Press, Princeton [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl GM, Preuss D. (2000) Dissecting Arabidopsis pollen-stigma interactions reveals novel mechanisms that confer mating specificity. Ann Bot (Lond) 85: 15–21 [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. (1999) Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440 [DOI] [PubMed] [Google Scholar]