Abstract
Serine hydroxymethyltransferases (SHMs) are important enzymes of cellular one-carbon metabolism and are essential for the photorespiratory glycine-into-serine conversion in leaf mesophyll mitochondria. In Arabidopsis (Arabidopsis thaliana), SHM1 has been identified as the photorespiratory isozyme, but little is known about the very similar SHM2. Although the mitochondrial location of SHM2 can be predicted, some data suggest that this particular isozyme could be inactive or not targeted into mitochondria. We report that SHM2 is a functional mitochondrial SHM. In leaves, the presequence of SHM2 selectively hinders targeting of the enzyme into mesophyll mitochondria. For this reason, the enzyme is confined to the vascular tissue of wild-type Arabidopsis, likely the protoxylem and/or adjacent cells, where it occurs together with SHM1. The resulting exclusion of SHM2 from the photorespiratory environment of mesophyll mitochondria explains why this enzyme cannot substitute for SHM1 in photorespiratory metabolism. Unlike the individual shm1 and shm2 null mutants, which require CO2-enriched air to inhibit photorespiration (shm1) or do not show any visible impairment (shm2), double-null mutants cannot survive in CO2-enriched air. It seems that SHM1 and SHM2 operate in a redundant manner in one-carbon metabolism of nonphotorespiring cells with a high demand of one-carbon units; for example, during lignification of vascular cells. We hypothesize that yet unknown kinetic properties of SHM2 might render this enzyme unsuitable for the high-folate conditions of photorespiring mesophyll mitochondria.
Ser hydroxymethyltransferases (SHMs) are important for two central aspects of plant metabolism: generation of one-carbon units for a variety of biosynthetic pathways and the photorespiratory Gly-into-Ser conversion (Douce and Neuburger, 1999; Mouillon et al., 1999; Appaji Rao et al., 2003). Hence, it is not surprising that multiple isoforms of this pyridoxal 5′-P-dependent enzyme exist in the cytosol, the plastids, and the mitochondria (Besson et al., 1995). SHM operates in a reversible manner to produce N5,N10-methylene-tetrahydrofolate (CH2-THF) and Gly from Ser and THF (the one-carbon unit-generating reaction) or, in the reverse direction, Ser and THF from Gly and CH2-THF (the Ser-regenerating reaction in both one-carbon and photorespiratory metabolism). CH2-THF generation is the dominating function of SHM in all extramitochondrial cellular compartments, but, at least during the day, photorespiratory Ser synthesis dominates in leaf mesophyll mitochondria. Here, SHM closely collaborates with Gly decarboxylase (GDC), which provides CH2-THF for the reverse reaction of SHM.
The photorespiratory SHM reaction is mostly confined to leaf mesophyll mitochondria, where the enzyme is present in large amounts to match the high photorespiratory carbon flux (Douce et al., 2001). In silico analyses indicated the presence of two very similar SHMs, SHM1 (At4g37930) and SHM2 (At5g26780), in Arabidopsis (Arabidopsis thaliana) mitochondria (McClung et al., 2000; Bauwe and Kolukisaoglu, 2003). SHM1, which is the dominating isozyme in leaves, was identified as the photorespiratory SHM (Voll et al., 2006). Its inactivation or deletion is deleterious, and such mutants require elevated CO2 levels for normal development (Somerville and Ogren, 1981; Voll et al., 2006). The in vivo activity of SHM1 is affected by many factors (Collakova et al., 2008; Jamai et al., 2009). In addition, this particular SHM is supposed to play a critical role in controlling the cell damage provoked by abiotic stresses (Moreno et al., 2005).
In contrast with the well-known role of SHM1 in photorespiratory metabolism, the metabolic function of SHM2 is not known, and its relation to SHM1 is enigmatic. For example, Arabidopsis SHM1 knockout mutants could be complemented by overexpression of AtSHM1, but overexpression of AtSHM2, driven by its native promoter or the constitutive 35S promoter, did not cure their photorespiratory phenotype (Voll et al., 2006). This was surprising because the amino acid sequences of SHM1 and SHM2 are very similar (87% identity). Hence, the authors hypothesized that AtSHM2 does not encode a fully functional SHM protein; alternatively, the AtSHM2 gene product is not targeted to the mitochondrial matrix.
Of the other five predicted SHMs in Arabidopsis (McClung et al., 2000; Hanson and Roje, 2001; Bauwe and Kolukisaoglu, 2003), functional information is available only for the plastidic isoform, SHM3 (Zhang et al., 2010). This enzyme, as well as the cytosolic isoforms SHM4 and SHM5, is believed to be involved in general one-carbon metabolism. SHM6 and SHM7 likely reside in the nuclei of Arabidopsis, but their function is not yet known.
In this study, we report that Arabidopsis SHM2 encodes a functional mitochondrial SHM, which is constitutively expressed in most, if not all, organs. Although SHM1 is the dominant mitochondrial SHM in leaves, SHM2 is on par with or even dominates over SHM1 in roots. In leaves, the presequence of SHM2 selectively precludes targeting into mesophyll mitochondria. This is why the enzyme was detectable only within the vascular bundles, likely the protoxylem and adjacent cells, where it occurs together with SHM1. It is likely that SHM2 and SHM1 redundantly satisfy the high demand of one-carbon units for the synthesis of lignin precursors and related biosynthetic processes in this tissue. The restriction of SHM2 to the vasculature of leaves also explains why overexpression of SHM2 could not complement the shm1 allele in previous experiments. The mechanism of this unexpected import selectivity is not yet known.
RESULTS AND DISCUSSION
SHM2 Is a Functional Ser Hydroxymethyltransferase
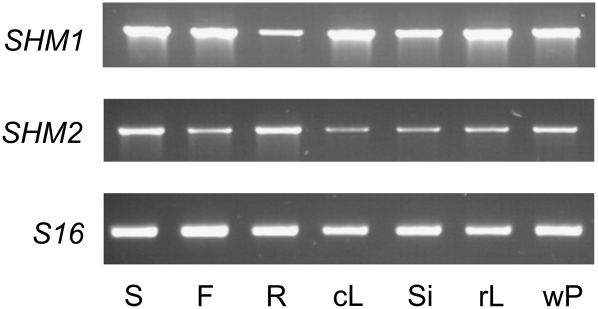
Similar to reported expression patterns (Voll et al., 2006), we found that the two paralogous genes, SHM1 and SHM2, are expressed in a variety of Arabidopsis organs (Fig. 1). Typically, SHM1 is expressed to distinctly higher levels in most organs other than roots, in which SHM2 transcripts slightly dominate. This pattern roughly corresponds with publicly available electronic northern data (Genevestigator; Zimmermann et al., 2004).
Figure 1.
Nonquantitative RT-PCR analysis of SHM1 and SHM2 transcripts in different organs of Arabidopsis. The constitutively expressed gene At2g09990 encoding the 40S ribosomal protein S16 served as an internal control. cL, Cauline leaves; F, flowers; R, roots; rL, rosette leaves; S, stems; Si, siliques; wP, whole plant.
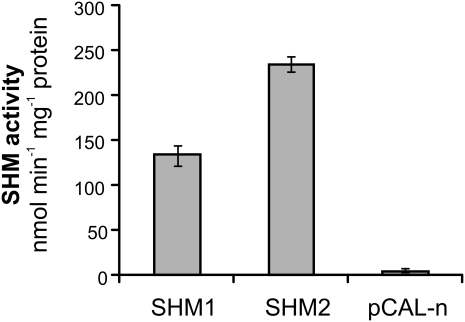
We next examined whether the SHM2-encoded protein is a functional SHM or, alternatively, nonfunctional as hypothesized by Voll et al. (2006). To this end, complementary DNA (cDNA) encoding the mature SHM2 was ligated into the expression vector pCal-n and overexpressed in Escherichia coli. SHM1-encoding cDNA was used as a positive control, and the nonrecombinant vector served as a negative control (Supplemental Fig. S1). This in vitro experiment showed distinct SHM activity for both recombinant enzymes (Fig. 2), and the measured activities were in the range of reported values for recombinant and native SHM from other organisms (Bourguignon et al., 1988; Jagath-Reddy et al., 1995; diSalvo et al., 1998; Vidal et al., 2005). The somewhat higher specific activity of recombinant SHM2 was likely because of a lower level of contaminating protein.
Figure 2.
Arabidopsis SHM2 is a functional SHM. Mature SHM2 was overexpressed in E. coli and tag-purified by affinity chromatography. SHM1 overexpressed from the same vector (pCal-n), and the empty vector served as positive and negative controls, respectively. SHM activity was determined using 14C-labeled Ser. Bars are mean ± sd from two measurements.
SHM2 Is Confined to the Vascular Tissue in Leaves
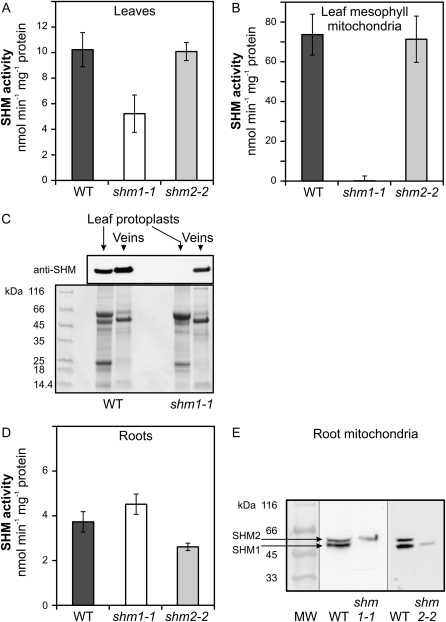
Evidence for the cellular distribution of SHM2 was obtained by the examination of individual null mutants for SHM1 (shm1-1) and SHM2 (shm2-2). The phenotypes of these two mutants were very similar to those reported by Voll et al. (2006): SHM1-deficient mutants required elevated CO2 for normal growth, whereas the SHM2-deficient mutant did not display any visible defect in normal air. This matched our observation that total SHM activities in leaves of shm2-2 were not significantly altered in comparison with the wild type (Fig. 3A), indicating that SHM2 likely represents only a very small fraction of total SHM activity in leaves. In the shm1-1 mutant, leaf SHM activity was distinctly reduced, although not as much as reported by Voll et al. (2006). Quantitatively, about 50% of the remaining total SHM activity in shm1-1 (Fig. 3A) fits well to the reported distribution of SHM isozymes in spinach (Spinacia oleracea) leaf protoplasts, in which the mitochondrial activity also represented approximately 50% of the total cellular SHM activity, whereas chloroplastic and cytosolic activities each represented approximately 20% to 25% (Besson et al., 1995).
Figure 3.
SHM activities, protein levels, and transcript levels in the individual shm1-1 and shm2-2 mutants in comparison with Arabidopsis wild type. A, B, and D, SHM activity in extracts from leaves, purified leaf mitochondria, and roots of the wild type (WT), shm1-1, and shm2-2, respectively. SHM activity for shm1-1 leaf mitochondria was not different from blank, indicating absence of SHM2 from leaf mesophyll mitochondria. C, SHM2 is undetectable in protoplasts but present in the vascular tissue of the SHM1-deficient mutant. Top, An immunoblot image of signals for mitochondrial SHM in extracts from leaf protoplasts and leaf veins prepared from wild-type and shm1-1 plants. Bottom, A protein-staining image of control gel run in parallel. E, Immunoblotting of SHM1 and SHM2 in root mitochondria purified from wild-type, shm1-1, and shm2-2 plants grown on MS medium. Both isoforms are present in root mitochondria. Alternative splicing produces a larger SHM2 in roots. Bars in A, B, and D are mean ± sd from three measurements (three biological replicates for leaf and root extracts; two replicates for mitochondria). For immunoblotting experiments, 10 μg of total protein was loaded per lane, except in the case of stroma protein prepared from mutant root mitochondria (approximately 3 μg). The primary antibody equally recognizes SHM1 and SHM2, but not SHM3 through SHM5.
Of note, residual SHM activity was undetectably low (at blank rate level) in purified leaf mesophyll mitochondria of the shm1 mutant (Fig. 3B). By contrast, and corresponding with the nearly unaltered whole-leaf SHM activity, the knockout of SHM2 did not significantly reduce total SHM activity in these organelles. This observation suggested that all SHM activity in Arabidopsis leaf mesophyll mitochondria could possibly represent SHM1 activity. Because SHM2 is expressed in leaves (Fig. 1), we therefore wanted to find out whether SHM2 protein can be detected in the leaf vasculature tissue. To this end, we used the Tape-Arabidopsis sandwich method described by Wu et al. (2009) to prepare protoplasts and a debris fraction highly enriched in veins from the wild-type and shm1-1 leaves. The extracted proteins were subjected to immunoblotting analysis (Fig. 3C) using an antibody that binds only to the mitochondrial SHMs and not to the extramitochondrial isoenzymes (Turner et al., 1992; Schjoerring et al., 2006). Although both fractions of the wild-type control leaves showed SHM1/2 signals, SHM2 was present only in the vascular bundle fraction but not in mesophyll protoplasts of shm1-1 leaves. This finding strongly substantiated the results obtained with purified mesophyll mitochondria and showed that most, if not all, leaf SHM2 is confined to the vasculature.
In roots, the knockout of SHM1 did not reduce total SHM activity (Fig. 3D), whereas the knockout of SHM2 significantly lowered total SHM activity. This roughly corresponds to the dominance of the SHM2 over SHM1 transcripts in roots, as shown in Figure 1. Because SHM2 apparently was not present in mesophyll protoplasts and mesophyll mitochondria, it was important to examine root mitochondria for the presence of this enzyme. Indeed, the immunoblot analysis of matrix extracts prepared from purified root mitochondria showed that SHM1 and SHM2 are present in mitochondria of the wild-type roots but absent from the respective null mutants (Fig. 3E). This provides direct evidence for the mitochondrial location of SHM2 and is confirmed by the recent identification of SHM2 in the mitochondrial proteome (Tan et al., 2010).
In Figure 3E, it is interesting to note that root SHM2 is slightly larger than root SHM1, and the two proteins can be clearly separated. This difference in sizes does not correspond with the earlier gene models At5g26780.1 (SHM2) and At4g37930.1 (SHM1) and is most likely caused by the preferential use of an alternative splice acceptor site in intron 11 of the SHM2 pre-mRNA (SHM2 gene model At5g26780.2 in the most recent Arabidopsis genome release, The Arabidopsis Information Resource 10). Initial studies revealed that the two alternative transcripts are present in both leaves and roots (data not shown). One of them encodes 16 additional amino acids; however, we have no evidence for the presence of this larger SHM2 form in leaves or of the smaller SHM2 in roots. It is possible that alternative splicing of the SHM2 pre-mRNA is regulated organ specifically, with the larger form being specific for roots, but this will require future experiments and is outside the scope of our present report.
Cellular Distribution of SHM1 and SHM2 in Arabidopsis Leaves
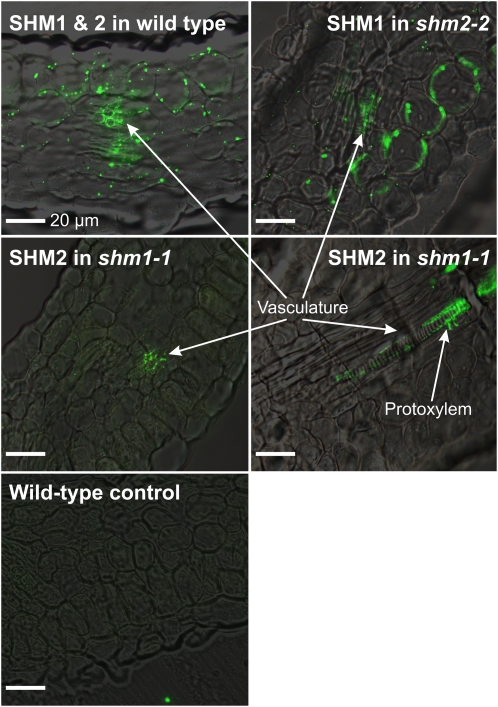
To independently confirm our finding that SHM2 is confined to the vasculature of leaves, we examined the cellular distribution of SHM1 and SHM2 in leaf cross sections of the wild type and the individual shm1 and shm2 mutants (Fig. 4). To reduce nonspecific fluorescence as much as possible, we used affinity-purified (with matrix-bound SHM2) anti-SHM1/2 antibodies in this experiment.
Figure 4.
Overlays of immunofluorescence signals of SHM1 and SHM2 and bright-field images of Arabidopsis leaf cross sections. Top left, SHM1 and SHM2 signals in mitochondria of the mesophyll and the vasculature of wild-type leaves. Top right, SHM1 signals in mitochondria of the mesophyll and the vasculature of shm2-2 leaves. Middle, SHM2 signals in mitochondria of the vasculature of shm1-1 leaves showing a likely location of SHM2 in the protoxylem. Bottom, Example of a control section subsequently treated with 5% bovine serum albumin instead of anti-SHM antibodies and the labeled secondary antibody. The primary antibody is specific for mitochondrial SHM and was affinity-purified against SHM2. The secondary antibody is conjugated with Alexa-Fluor 488. Bars = 20 μm.
The wild-type leaves showed signals for SHM1/2 protein in the mesophyll and in the leaf veins (Fig. 4, top left). A very similar distribution of fluorescent spots was observed in cross sections of shm2-2 leaves (Fig. 4, top right), indicating that SHM1 is present in both tissues. By contrast, SHM2 signals were exclusively associated with the vasculature and not observed in the mesophyll of shm1-1 leaves (Fig. 4, middle). This is best seen in the sectioned xylem vessel shown in the middle right of Figure 4 and suggests confinement of leaf SHM2 to the protoxylem and adjacent cell layers. This is an interesting finding because xylem differentiation involves lignification and, therefore, has a particularly high demand of one-carbon units to provide the required O-methylated phenylpropanoid precursors (Ye, 2002; Boerjan et al., 2003).
The Combined Knockout of SHM1 and SHM2 Is Lethal Even in Elevated CO2
In leaves, SHM2 cannot substitute for SHM1 in photorespiratory metabolism (Somerville and Ogren, 1981; Voll et al., 2006), and our results explain why such substitution is not possible. On the other hand, mitochondrial SHM activity is considered essential for one-carbon metabolism in all eukaryotic cells (Mouillon et al., 1999; Hanson and Roje, 2001). Similarly, there is a nonsubstitutable need for the collaborating enzyme GDC (Engel et al., 2007). In light of the viability of the SHM1 null mutant under nonphotorespiratory conditions, it was hence interesting to ask whether SHM1 is replaceable, at least on a low level, with respect to the requirements of one-carbon metabolism of mesophyll cells.
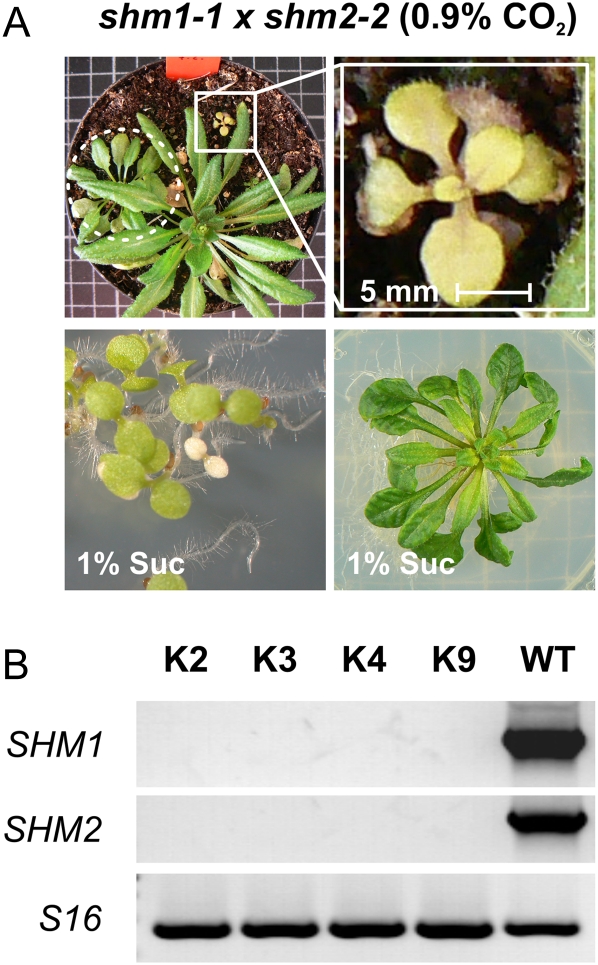
Because SHM2 represents the only known alternative to SHM1 in Arabidopsis, we crossed shm2-2 with shm1-1 and isolated several lines that were homozygous for both mutant alleles. If germinated and grown in dim light with 0.9% CO2 on soil, these double-null mutants developed very few small and yellowish leaves before they eventually died (Fig. 5A, top). This was clearly different from the wild-type-like growth of the parental mutants under these conditions and showed that the presence of at least one of the two isozymes, either SHM1 or SHM2, is essential for nonphotorespiratory one-carbon metabolism. Under similar environmental conditions, but using Suc-supplemented Murashige and Skoog (MS) medium instead of soil, the double mutants also first developed pale seedlings (Fig. 5A, bottom). Unlike the fully lethal GDC-deficient mutant (Engel et al., 2007), however, these seedlings turned green a few weeks later, and the plants eventually even flowered. Flowering occurred approximately 3 to 4 months after germination, but the produced seeds were not fertile.
Figure 5.
The combined deletion of SHM1 and SHM2 is detrimental. A, Top, shm1-1 × shm2-2 double mutant (white square on left image; enlarged in the right image) together with an shm1-1 homozygous plant (dotted circle) and a double-heterozygous plant after growth for 10 weeks in soil with 0.9% CO2-enriched air. Bottom, A shm1-1 × shm2-2 double-homozygous individual after germination (left) and after growth for 8 weeks in 0.9% CO2-containing air on MS medium with 1% Suc (right). B, RT-PCR confirms absence of SHM1 and SHM2 transcripts in leaves of four double-knockout lines (K2, K3, K4, and K9) in comparison with their presence in wild-type (WT) plants and the constitutively expressed S16 (At2g09990) transcripts used for internal calibration.
Despite these highly artificial growth conditions, the survival of the shm1 × shm2 double mutant is nevertheless remarkable because it is generally accepted that polyglutamylated folates do not equilibrate between mitochondria and other subcellular compartments (Mouillon et al., 1999). One could speculate that, under the artificial conditions of highly elevated CO2 and Suc feeding, the P-Ser pathway provides Ser from Suc degradation (Ho and Saito, 2001) for the synthesis of CH2-THF and Gly from Ser by the extramitochondrial SHMs. GDC could then produce CH2-THF from Gly for mitochondrial metabolism. A similar route could allow the shm1 mutant, which has neither SHM1 (because of the mutation) nor mesophyll SHM2 (because import into mitochondria is not possible), adequate mesophyll one-carbon metabolism in nonphotorespiratory conditions.
Complementation of the shm1 Mutant by SHM2 Requires the SHM1 Presequence
These data consistently show that SHM2 is a functional mitochondrial SHM that does not occur in the leaf mesophyll but is instead present in mitochondria of nonphotorespiring tissues. They do not explain, however, why overexpression of SHM2 under control of the 35S promoter is unable to complement the SHM1 null mutant as reported by Voll et al. (2006).
The inspection of the predicted SHM1 and SHM2 presequences gave no clue to functional differences (Huang et al., 2009). To experimentally test whether the SHM2 presequence selectively prevents import of the enzyme into mesophyll mitochondria, we designed a complementation construct in which the SHM1 promoter drives expression of a fusion protein comprising the SHM1 presequence and the SHM2 mature protein (Fig. 6A). The respective cleavage sites were derived from sequence similarity with the known N terminus of mature pea (Pisum sativum) SHM1 (Turner et al., 1992). To identify stably transformed, homozygous shm1-1 derivatives, T1 seedlings were grown in elevated CO2 and selected by their resistance to the herbicide BASTA. Transgenic plants were then transferred to ambient air and selected for the wild-type-like individuals. Four lines were selected from this two-step screening and selfed over several generations.
Figure 6.
SHM1pro-driven expression of SHM2 fused to the SHM1 presequence complements the shm1-1 allele. A, Schematic structure of the SHM1:SHM2 (S1/2) chimerical transgene. B, Eight-week-old wild-type (WT), shm1-1, and shm2-2 plants and two stably complemented shm1-1 mutant lines P10 and P20 transformed with the S1/2 chimerical transgene. Plants were grown for 6 weeks in air enriched with 0.15% CO2 and then in normal air for another 2 weeks to allow direct comparison of growth with the shm1-1 mutant. C, Example of genotype verification in the transgenic lines, showing that line P20 is homozygous for the shm1-1 allele and harbors the SHM1:SHM2 chimerical transgene. [See online article for color version of this figure.]
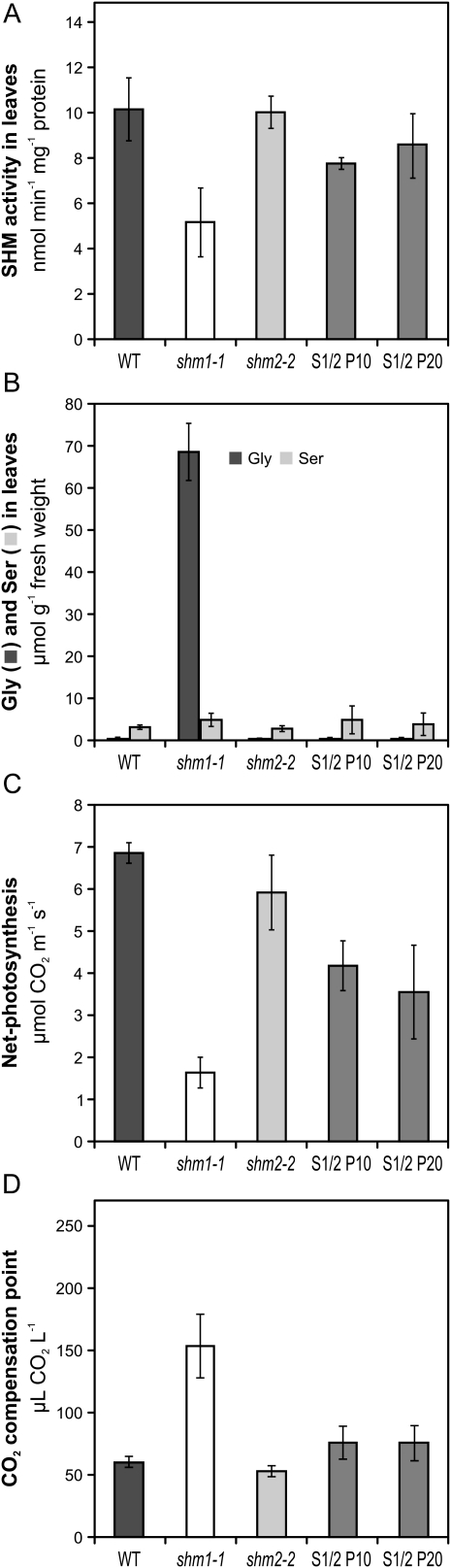
Although the air-grown progeny of two of these lines showed some segregation of the air-tolerant phenotype, possibly by inactivation of the transgene, the progenies of lines P10 and P20 were perfectly stable and developed very similar to the wild-type plants (Fig. 6B). Genotype analysis of the T3 generation reconfirmed homozygosity of the shm1-1 allele and the presence of the SHM1:SHM2 chimerical transgene (Fig. 6C). In addition to the restored wild-type phenotype, leaf SHM activity was distinctly greater than shm1 level in individuals of both lines and came close to wild-type level in individuals of line P20 (Fig. 7A). The leaf contents of Gly and Ser were also very much reduced and close to wild-type levels in both lines. The Gly-to-Ser ratio was about 0.20, which is very similar to the wild-type ratio of about 0.15 (Fig. 7B). Photosynthetic rates were distinctly elevated in both transgenic lines although still somewhat lower than that of the wild type (Fig. 7C), and CO2 compensation points were also close to the wild type (Fig. 7D).
Figure 7.
Biochemical and gas-exchange parameters of complemented shm1-1 plants in comparison with the wild-type (WT) and parent plants. A, SHM activity in extracts from leaves harvested in the middle of the light period. Note that some data from Figure 3 (wild type, shm1-1, and shm2-2) were included for easier comparison. B, Gly and Ser contents of rosette leaves. C, Photosynthetic rates of rosette leaves at 380 μL L−1 CO2. D, CO2 compensation points at 21% O2. Plants were grown in 0.15% CO2 (A and C) followed by 5 d in normal air (B). Bars represent means ± sd from three (A and B, three biological replicates) and five (C, five biological replicates) measurements, respectively.
These quantitative data confirmed that the SHM1:SHM2 fusion protein was efficiently expressed and properly imported into leaf mesophyll mitochondria. We speculate that specific features of the SHM2 presequence, which harbors the mitochondria-targeting sequence, selectively prevent the import of SHM2 into mesophyll mitochondria. Such restrictions apparently do not exist for the import of SHM1 into mitochondria of these different tissues.
CONCLUSION
The aim of this study was to examine whether SHM2 is a functional SHM, whether it is targeted to the mitochondria, and what its function for cellular metabolism could possibly be. These questions resulted from the previous finding in another group (Voll et al., 2006) that overexpression of SHM2 cDNA was unable to complement a SHM1-deficient mutant, neither under control of the SHM1 promoter (which should provide adequate tissue specificity and expression level) nor with the strong constitutive cauliflower mosaic virus 35S promoter.
Our results provided several levels of evidence that SHM2, which is structurally very similar to SHM1, is a functional mitochondrial SHM. With the exception of roots, SHM1 is generally somewhat more strongly expressed than SHM2, but the two isozymes are probably present in all organs of Arabidopsis, although in varying amounts.
With respect to their operation in general one-carbon metabolism, SHM1 and SHM2 seem to be more or less redundant enzymes. This follows from the lack of any visible phenotype of the shm2 mutation in normal air and the high-CO2–curable photorespiratory phenotype of the shm1 mutation (Voll et al., 2006). It is further supported by our observation that the combined knockout of SHM1 and SHM2 is detrimental even in high-CO2 conditions. Because an elevated CO2 concentration efficiently suppresses photorespiratory metabolism, it is also likely that extramitochondrial SHMs cannot compensate for the simultaneous absence of both mitochondrial isozymes, except under the artificial condition of Suc feeding. This is similar to but, in light of this exception, also different from the operation of GDC. GDC is absolutely confined to mitochondria, and null mutants cannot survive under any tested condition, including the combination of highly elevated CO2 and Suc feeding (Engel et al., 2007).
With respect to photorespiration, however, SHM1 and SHM2 are not redundant (Voll et al., 2006). This is not simply due to low-level expression of SHM2 in leaves but to an intriguing feature of SHM2: The presequence of this particular enzyme selectively does not allow import into mesophyll mitochondria. After exchange of the SHM2 presequence against that of SHM1, SHM2 could fully replace SHM1 in photorespiratory metabolism. On the other hand, as we have shown with purified root mitochondria and by immunolocalization in leaves, SHM2 is normally imported into mitochondria of other tissues.
At present, it remains speculative what the molecular mechanism of the observed cell type selectivity of the SHM2's presequence may be. Tissue-specific subcellular localization of enzymes is known for many enzymes and organisms, such as Gln synthetase, which is dual-targeted to chloroplasts and mitochondria in Arabidopsis (Taira et al., 2004). In chicken and other uricotelic vertebrates, Gln synthetase is directed to the mitochondria in liver and to the cytoplasm in brain by a sorting mechanism that is most likely toggled by differences in the mitochondrial membrane potential (Matthews et al., 2010). A cell type-dependent selection of a member of an isoenzyme family for import into an organelle, however, has not yet been reported. Our observation of tissue-specific restrictions for the organellar import of one of a pair of isoenzymes provokes speculation about the existence of a yet unknown layer of cellular control.
Not only the how but also the why of this sorting remains to be investigated. What is the advantage of strictly separating SHM2 from photorespiratory metabolism? For mitochondrial SHM and for the H-protein, which is one of the GDC protein components, it has been suggested that some isoforms could be specialized to either photorespiratory or one-carbon metabolism, respectively. In aspen (Populus tremuloides), for example, each particular SHM and H-protein isoform abundantly occur in the xylem, where they presumably contribute to lignin formation during xylogenesis (Vander Mijnsbrugge et al., 2000; Rajinikanth et al., 2007). This is an important process; moreover, lignification in woody stems of perennial species is one of the most one-carbon unit-demanding metabolic processes in plants (Hanson and Roje, 2001). Lignification is also a distinctive feature of the tracheary elements of herbaceous plants, such as Arabidopsis. The close association of leaf SHM2 with these structures suggests participation in this metabolism. If adopted to our study, SHM1 seems to be a “Jack of all trades,” whereas the (yet unknown) kinetic properties of SHM2 are possibly optimized for operation under the low-folate conditions in nonphotorespiratory cells with a high demand of one-carbon units. Vice versa, such specialization might render SHM2 unsuitable for the high-folate conditions of photorespiring mesophyll mitochondria.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type and mutant lines for the isolation of the homozygous transferred (T)-DNA insertion mutants (SALK 0083735 and SALK 096265), all of the Columbia-0 ecotype, were obtained from the Nottingham Arabidopsis Stock Centre and grown on soil (Type Mini Tray; Einheitserdewerk) and vermiculite (4:1 mixture) in Percival growth chambers (12-/12-h light/dark cycle, 22°C/18°C, photosynthetically active photon flux density of approximately 150 μmol m−2 s−1) and watered with 0.1% Wuxal liquid fertilizer (Aglukon Spezialdünger GmbH). For some experiments, the air CO2 concentration was increased to 0.15% or 0.9% (low photorespiratory condition) and continuously monitored.
The shm1 × shm2 double mutant was produced by crossing homozygous Atshm1-1 and Atshm2-2, both grown at 0.9% CO2, and selfing the T1 progeny. Of 30 T2 individuals, 12 were homozygous with respect to the shm1-1 allele and heterozygous with respect to shm2-2. After selfing these lines, double-homozygous mutants were identified in the T3 generation grown in 0.9% CO2, both on soil:vermiculite and on solidified MS (Murashige and Skoog, 1962) basal medium (Duchefa Biochemie) containing 1% Suc and 1% agar.
Transcript Analyses and Genotype Verification
RNA was isolated (Nucleospin RNA Plant Kit; Macherey-Nagel) from Arabidopsis organs harvested in the middle of the light period and used for cDNA synthesis (RevertAid H minus cDNA synthesis kit; MBI Fermentas). Primers for PCR amplification were SHM1-RT-S, 5′-CAT TCG TCC TCT TAT TCG ATC CAC-3′ (SHM1 sense), SHM1-RT-A, 5′-GTT CTT GTA CTT CAT GGT TTC TTT CTC-3′ (SHM1 antisense), SHM2-RT-S, 5′-CAC CCA ACT CCA ATG CTC ACT TAT ACA GAA G-3′ (SHM2 sense), and SHM2-RT-A, 5′-CTC TTT GTA TCT CAT CGT CTC TTT CTC G-3′ (SHM2 antisense). PCR analysis was performed with 28 cycles, and the amounts of cDNA were calibrated according to signal intensities of a reverse transcription (RT)-PCR fragment of the constitutively expressed At2g09990 mRNA encoding the 40S ribosomal protein S16 with primers S16S, 5′-GGC GAC ACA ACC AGC TAC TGA-3′ (sense); and S16A, 5′-CGG TAA CTC TTC TGG TAA CGA-3′ (antisense).
Genomic DNA of T-DNA lines SALK 0083735 (Atshm1-1) and SALK 096265 (Atshm2-2) was subjected to standard PCR (Master Mix; Qiagen) with primers specific for the left border (LB1, 5′-AAT CAG CTG TTG CCC GTC TCA CTG GTG AA-3′) and gene-specific primers for SHM1 (SHM1-S, 5′-GCC TCA TGA AAG AAT CAT GGC ACT TG-3′) and SHM2 (SHM2-S, 5′-GGA CAT CTT TCT CAT GGT TAT CAG-3′), respectively. Homozygosity of the T-DNA insertion was verified using the sense primers SHM1-S and SHM2-S, respectively, in combination with the reverse primers SHM1-A (5′-GTT CTT GTA CTT CAT GGT TTC TTT CTC-3′) and SHM2-A (5′-CCA CAC GAT GAA AAC GGA TCC TTG TAT C-3′). By PCR analysis as described above, no SHM1 or SHM2 transcripts were detectable in the respective individual and double mutants.
Overexpression in Escherichia coli
The coding sequences for the mature SHM1 and SHM2 (in analogy to pea [Pisum sativum] SHM1; Turner et al., 1992) were amplified from leaf cDNA prepared as above using the following oligonucleotides (introduced BamHI and SalI restriction sites underlined): SHM1-5, 5′-AA GGA TCC CTT TCT TCT TCA ATT GAC AAA CCC ATT CG-3′; SHM1-3, AAG TCG ACC GTT CTT GTA CTT CAT GGT TTC TTT CTC; SHM2-5′, AA GGA TCC ATG TCG TCT TTA TCA ACC GCA GCT ATG-3′; and SHM2-3, AAG TCG ACC CTC TTT GTA TCT CAT CGT CTC TTT CTC G-3′. BamHI-SalI fragments were ligated in frame downstream of the small calmodulin-binding protein affinity tag of expression vector pCal-n (Stratagene). Correctness of constructs was verified by sequencing. Expression was induced in E. coli strain BL21 by adding 1 mm isopropyl-β-d-thiogalactopyranosid for 4 h at 37°C. The proteins were then affinity-purified following the batch method of the manufacturer's protocol.
SHM Activity
Leaf and root extracts were individually prepared from three plants per line grown in 0.15% CO2 as previously described (Ewald et al., 2007). Leaf mitochondria were prepared according to Method B in Keech et al. (2005) from two independent sets of plants grown in normal air (wild type and shm2-2) and air enriched with 0.15% CO2 (shm1-1), respectively. SHM activity was measured in these extracts, with mitochondrial stroma extracts, and with the recombinant proteins using 14C-labeled Ser as also described before (Eisenhut et al., 2006).
Immunoblotting
Leaf and root extracts were prepared in 25 mm HEPES, pH 7.0, 0.5 mm EDTA, 8 mm dithiothreitol, and 1 mm phenylmethanesulfonyl fluoride. Next, to determine of protein concentration (Bradford, 1976), proteins were separated on 12% denaturing polyacrylamide gels (Laemmli, 1970) and electrotransferred onto a polyvinylidene difluoride membrane. SHM1 and SHM2 were identified with an anti-SHM1/2 polyclonal antiserum raised in rabbits against recombinant potato SHM1 (Schjoerring et al., 2006) and using an anti-rabbit IgG-horseradish peroxidase conjugate in combination with the Amersham ECL Advance Western Blotting Detection Kit (GE Healthcare Europe GmbH). Specificity testing is shown in Supplemental Figure S1.
Arabidopsis protoplasts were prepared by the Tape-Sandwich method (Wu et al., 2009) from leaves (approximately 2-cm wide, 4-cm long) harvested from 4-week-old Arabidopsis wild-type and shm1-1 plants grown as described above (0.9% CO2). As the only modification of the original method, we fixed the adaxial epidermis to a strip of Tesa Extra Power Tape (Tesa Hamburg) and the lower epidermis to Leukopor tape (BSN Medical). After the complete removal of protoplasts, the remaining leaf veins and debris bound to the Tesa tape were carefully detached and frozen in liquid nitrogen. Soluble proteins were extracted, separated by SDS-PAGE (10 μg per lane), and immunoblotted as described above.
For the isolation of Arabidopsis root mitochondria, the wild-type and mutant seedlings were grown in a 12-/12-h light/dark cycle (160 μE m−2 s−1) at 22°C on vertical plates (one-half-strength MS medium, 1% Suc, 8% agar). After 3 weeks, roots (8–10 g fresh weight) were harvested from 30 plates and used for the isolation of mitochondria as described before (Gupta et al., 2005). Matrix proteins were extracted, separated by SDS-PAGE (10 μg per lane), and immunoblotted as described above.
Immunolocalization
To minimize unspecific signals, anti-SHM1/2 antibodies were affinity-purified from the high-titer anti-SHM1/2 polyclonal antiserum mentioned above using recombinant Arabidopsis SHM2. In short, overexpressed and tag-purified SHM2 was separated on a preparative denaturing polyacrylamide gel and blotted onto nitrocellulose. The SHM1-containing portion of the membrane was then identified by staining with Ponceau S, excised, and incubated with the antiserum. Bound antibodies were eluted with an acidic Gly buffer, immediately brought to pH 7, and stored at 4°C (http://medicine.yale.edu/labs/koelle/Site/Home.html).
Sections of 4- to 6-week-old leaves of Arabidopsis wild-type and shm mutants were vacuum-infiltrated with 4% (w/v) paraformaldehyde and 0.1% Triton X-100 in phosphate-buffed saline (PBS), fixed for 2 h, dehydrated in a standard ethanol series, embedded into Technovit 8100 (Kulzer Heraeus), and blocked after polymerization. Cross sections of 3- to 5-μm thickness were cut with a rotation microtome (Leica), attached to adhesive-treated microscope slides, and equilibrated in PBS for 20 min. They were then incubated in 0.1 m ammonium chloride in PBS to quench background autofluorescence and enhance antigenicity (5 min), washed with PBS (5 min), and blocked with 5% (w/v) bovine serum albumin in PBS (30 min). Sections were incubated with the affinity-purified anti-SHM1/2 antibody (1:200 dilution in PBS, 5% bovine serum albumin) at 4°C for 16 h. After washing with PBS (four times for 10 min), we used Alexa-Fluor 488-conjugated anti-rabbit IgG (1:500 dilution in PBS, 5% [w/v] bovine serum albumin; Molecular Probes) to detect anti-SHM1/2 IgG at 37°C for 1 h). Sections were covered with 75% glycerol in PBS and analyzed by conventional fluorescence microscopy (Zeiss Axioskop equipped with a Zeiss Imager A.1 camera; Software AxioVision Rel. 4.6).
Construction of the Chimeric SHM1/SHM2 Gene
The SHM1 promoter sequence (approximately 950 bp) including the 5′-nontranslated region and the SHM1 presequence was amplified from Arabidopsis chromosomal DNA using the primers SHM1-Pro-5-BamHI, 5′ AA GGA TCC CCA CCA AGA GCA AAC GAA TAA CAG CAG-3′ (sense), and SHM1-Pro-3-EcoRI, 5′-AA GAA TTC GTA ACA TGA AGT GGA TCG AAT AAG AGG-3′ (antisense). The resulting fragment was first ligated into pGEM-T Easy (Promega), excised via BamHI and EcoRI, and inserted into the corresponding sites of the lacZ gene in plant vector pGreenII 0229 (http://www.pgreen.ac.uk). Next, a genomic fragment of the SHM2 gene beginning with the mature SHM2-encoding part was amplified using primers SHM2-CDS-5-EcoRI, 5′-AA GAA TTC ATG TCG TCT TTA TCA ACC GCA GCT ATG-3′ (sense) and SHM2-CDS-3-XhoI, 5′-AA CTC GAG CTC TTT GTA TCT CAT CGT CTC TTT CTC-3′ (antisense), cloned into pGEM-T Easy, excised via EcoRI and XhoI, and ligated into corresponding sites of the recombinant pGreenII 0229 vector, in frame with the SHM1 sequence. The resulting plasmid was verified by partial sequencing, transformed into Agrobacterium tumefaciens strain GV3102 pSoup (Hellens et al., 2000), and used for the stable transformation of the shm1-1 mutant (Clough and Bent, 1998). T1 seeds were grown on soil in elevated CO2. BASTA-selected transformants were transferred to normal air as a second test of successful complementation. Four selected lines were then repeatedly selfed. Genotypes, including the absence of native SHM1 and SHM2 transcripts, were verified by PCR and RT-PCR using primers that did not recognize the transgene's transcript and were reexamined in all following generations (T2–T3).
Leaf Amino Acids
For amino acid determination, 100 mg of leaf material was ground in liquid nitrogen and extracted in 1.8 mL of 80% ethanol for 30 min. After centrifugation, the supernatant was vacuum dried, and the residue was dissolved in 8 mm sodium phosphate (pH 6.8) containing 2.5% tetrahydrofurane. Individual amino acids were separated by HPLC and quantified as described earlier (Hagemann et al., 2005).
Gas Exchange Measurements
Photosynthetic parameters were measured with the portable gas exchange system LI-6400 (Li-Cor Biosciences) equipped with an Arabidopsis leaf chamber. Fully developed rosette leaves were first adapted in the leaf chamber for 20 min. Measurements were performed at 25°C at a photosynthetic photon flux density of 250 μmol m−2 s−1, 210 ml L−1 O2, and variable CO2. Carbon assimilation rates (A) at 380 μL L−1 CO2 were calculated by the LI-6400 software. Apparent CO2 compensation points were calculated from A/Ci curves by linear regression of the data in the low-CO2 range.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_119954 for SHM1 and NP_851081.1 for SHM2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Specificity of the anti-potato SHM1 antibody for SHM1 and SHM2.
Acknowledgments
We thank Klaudia Michl (HPLC analyses), Kathrin Jahnke (preparation of mitochondria), and Ursula Bauwe (growth facilities) for technical assistance. Seeds provided by the Nottingham Arabidopsis Stock Center and the information available through The Arabidopsis Information Resource were essential prerequisites.
References
- Appaji Rao N, Ambili M, Jala VR, Subramanya HS, Savithri HS. (2003) Structure-function relationship in serine hydroxymethyltransferase. Biochim Biophys Acta 1647: 24–29 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Kolukisaoglu Ü. (2003) Genetic manipulation of glycine decarboxylation. J Exp Bot 54: 1523–1535 [DOI] [PubMed] [Google Scholar]
- Besson V, Neuburger M, Rebeille F, Douce R. (1995) Evidence for three serine hydroxymethyltransferases in green leaf cells: purification and characterization of the mitochondrial and chloroplastic isoforms. Plant Physiol Biochem 33: 665–673 [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bourguignon J, Neuburger M, Douce R. (1988) Resolution and characterization of the glycine-cleavage reaction in pea leaf mitochondria. Properties of the forward reaction catalysed by glycine decarboxylase and serine hydroxymethyltransferase. Biochem J 255: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collakova E, Goyer A, Naponelli V, Krassovskaya I, Gregory JF, III, Hanson AD, Shachar-Hill Y. (2008) Arabidopsis 10-formyl tetrahydrofolate deformylases are essential for photorespiration. Plant Cell 20: 1818–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Salvo ML, Delle Fratte S, De Biase D, Bossa F, Schirch V. (1998) Purification and characterization of recombinant rabbit cytosolic serine hydroxymethyltransferase. Protein Expr Purif 13: 177–183 [DOI] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rébeillé F. (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Douce R, Neuburger M. (1999) Biochemical dissection of photorespiration. Curr Opin Plant Biol 2: 214–222 [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Kahlon S, Hasse D, Ewald R, Lieman-Hurwitz J, Ogawa T, Ruth W, Bauwe H, Kaplan A, Hagemann M. (2006) The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol 142: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu Ü, Morgenthal K, Weckwerth W, Pärnik T, Keerberg O, Bauwe H. (2007) Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol 144: 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald R, Kolukisaoglu Ü, Bauwe U, Mikkat S, Bauwe H. (2007) Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant Physiol 145: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ, Stoimenova M, Kaiser WM. (2005) In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J Exp Bot 56: 2601–2609 [DOI] [PubMed] [Google Scholar]
- Hagemann M, Vinnemeier J, Oberpichler I, Boldt R, Bauwe H. (2005) The glycine decarboxylase complex is not essential for the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Biol (Stuttg) 7: 15–22 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Roje S. (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52: 119–137 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Ho CL, Saito K. (2001) Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids 20: 243–259 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Whelan J, Millar AH. (2009) Refining the definition of plant mitochondrial presequences through analysis of sorting signals, N-terminal modifications, and cleavage motifs. Plant Physiol 150: 1272–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagath-Reddy J, Ganesan K, Savithri HS, Datta A, Rao NA. (1995) cDNA cloning, overexpression in Escherichia coli, purification and characterization of sheep liver cytosolic serine hydroxymethyltransferase. Eur J Biochem 230: 533–537 [DOI] [PubMed] [Google Scholar]
- Jamai A, Salomé PA, Schilling SH, Weber APM, McClung CR. (2009) Arabidopsis photorespiratory serine hydroxymethyltransferase activity requires the mitochondrial accumulation of ferredoxin-dependent glutamate synthase. Plant Cell 21: 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardeström P. (2005) Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant 124: 403–409 [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Matthews GD, Gur N, Koopman WJH, Pines O, Vardimon L. (2010) Weak mitochondrial targeting sequence determines tissue-specific subcellular localization of glutamine synthetase in liver and brain cells. J Cell Sci 123: 351–359 [DOI] [PubMed] [Google Scholar]
- McClung CR, Hsu M, Painter JE, Gagne JM, Karlsberg SD, Salomé PA. (2000) Integrated temporal regulation of the photorespiratory pathway. Circadian regulation of two Arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiol 123: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JI, Martín R, Castresana C. (2005) Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J 41: 451–463 [DOI] [PubMed] [Google Scholar]
- Mouillon JM, Aubert S, Bourguignon J, Gout E, Douce R, Rébeillé F. (1999) Glycine and serine catabolism in non-photosynthetic higher plant cells: their role in C1 metabolism. Plant J 20: 197–205 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Rajinikanth M, Harding SA, Tsai CJ. (2007) The glycine decarboxylase complex multienzyme family in Populus. J Exp Bot 58: 1761–1770 [DOI] [PubMed] [Google Scholar]
- Schjoerring JK, Mäck G, Nielsen KH, Husted S, Suzuki A, Driscoll S, Boldt R, Bauwe H. (2006) Antisense reduction of serine hydroxymethyltransferase results in diurnal displacement of NH4+ assimilation in leaves of Solanum tuberosum. Plant J 45: 71–82 [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. (1981) Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol 67: 666–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira M, Valtersson U, Burkhardt B, Ludwig RA. (2004) Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 16: 2048–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YF, O’Toole N, Taylor NL, Millar AH. (2010) Divalent metal ions in plant mitochondria and their role in interactions with proteins and oxidative stress-induced damage to respiratory function. Plant Physiol 152: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Ireland R, Morgan CL, Rawsthorne S. (1992) Identification and localization of multiple forms of serine hydroxymethyltransferase in pea (Pisum sativum) and characterization of a cDNA encoding a mitochondrial isoform. J Biol Chem 267: 13528–13534 [PubMed] [Google Scholar]
- Vander Mijnsbrugge K, Meyermans H, Van Montagu M, Bauw G, Boerjan W. (2000) Wood formation in poplar: identification, characterization, and seasonal variation of xylem proteins. Planta 210: 589–598 [DOI] [PubMed] [Google Scholar]
- Vidal L, Calveras J, Clapés P, Ferrer P, Caminal G. (2005) Recombinant production of serine hydroxymethyl transferase from Streptococcus thermophilus and its preliminary evaluation as a biocatalyst. Appl Microbiol Biotechnol 68: 489–497 [DOI] [PubMed] [Google Scholar]
- Voll LM, Jamai A, Renné P, Voll H, McClung CR, Weber APM. (2006) The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol 140: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH. (2002) Vascular tissue differentiation and pattern formation in plants. Annu Rev Plant Biol 53: 183–202 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun K, Sandoval FJ, Santiago K, Roje S. (2010) One-carbon metabolism in plants: characterization of a plastid serine hydroxymethyltransferase. Biochem J 430: 97–105 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]