Abstract
Understanding iron (Fe) sensing and regulation is important for targeting key genes for important nutritional traits like Fe content. The basic helix-loop-helix transcription factor FIT (for FER-LIKE FE DEFICIENCY-INDUCED TRANSCRIPTION FACTOR) controls Fe acquisition genes in dicot roots. Posttranscriptional regulation of transcription factors allows rapid adaptation to cellular changes and was also described for FIT. However, the mechanisms behind this regulation of FIT were for a long time not known. Here, we studied the posttranscriptional control mechanisms of FIT in Arabidopsis (Arabidopsis thaliana) and identified nitric oxide as a stabilizing stimulus for FIT protein abundance. Using cycloheximide, we confirmed that the level of FIT protein was regulated by way of protein turnover in wild-type and hemagglutinin-FIT plants. Upon cycloheximide treatment, FIT activity was hardly compromised, since Fe deficiency genes like IRON-REGULATED TRANSPORTER1 and FERRIC REDUCTASE OXIDASE2 were still inducible by Fe deficiency. A small pool of “active” FIT was sufficient for the induction of Fe deficiency downstream responses. Nitric oxide inhibitors caused a decrease of FIT protein abundance and, in the wild type, also a decrease in FIT activity. This decrease of FIT protein levels was reversed by the proteasomal inhibitor MG132, suggesting that in the presence of nitric oxide FIT protein was less likely to be a target of proteasomal degradation. Independent of FIT transcription, FIT protein stability and FIT protein activity, therefore, were targets of control mechanisms in response to Fe and nitric oxide. We summarize our results in a model that explains the different steps of FIT regulation integrating the plant signals that control FIT.
Iron (Fe) is an essential micronutrient for most organisms. Bioavailable Fe is only present in limited amounts, for example, in vegetarian diets for humans or in calcareous field conditions for crop plants. Understanding the molecular basis of the Fe homeostasis network in plants will help to breed higher nutritious quality food crops by enabling the targeting of the major key genes of the traits related to Fe content.
A prime controlling step for Fe content in aerial and subterranean plant parts is the efficiency by which the root takes up external Fe. Due to the low solubility of Fe in soil conditions, plants need to mobilize Fe for efficient acquisition. The Fe acquisition strategy I of nongraminaceous plants is based on soil acidification and Fe reduction, while the strategy II of grasses relies on phytosiderophore action (Römheld and Marschner, 1986). In Arabidopsis (Arabidopsis thaliana), reduction of ferric Fe occurs by the Fe reductase FRO2 (for FERRIC REDUCTASE OXIDASE2; Robinson et al., 1999). Uptake of the ferrous Fe into the root epidermis is carried out by the metal transporter IRT1 (for IRON-REGULATED TRANSPORTER1; Eide et al., 1996; Vert et al., 2002). The basic helix-loop-helix (bHLH) transcription factor FIT (for FER-LIKE FE DEFICIENCY-INDUCED TRANSCRIPTION FACTOR) is required for high-level expression of FRO2 and IRT1 (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Bauer et al., 2007). In contrast to many Fe deficiency mutants, fit loss-of-function mutants are Fe deficient but at the same time fail to highly induce FRO2 and IRT1 for compensation, suggesting that the FIT transcription factor up-regulates these important Fe deficiency genes (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). The FIT gene is expressed in a root-specific manner and is induced by Fe deficiency (Jakoby et al., 2004).
In addition to this transcriptional control, FIT is also regulated at the posttranscriptional level. First, ectopic overexpression of FIT only resulted in an activation of downstream targets like FRO2 and IRT1 upon –Fe but not +Fe conditions (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). Since FIT protein amounts were equally present in +Fe and –Fe conditions in these FIT-overexpressing (FIT Ox) plants, the effect can only be attributed to differential activity of FIT (Lingam et al., 2011). Second, FIT protein stability is regulated in plant cells via proteasomal degradation (Lingam et al., 2011; Sivitz et al., 2011). This process is counteracted by ethylene signaling (Lingam et al., 2011). Third, FIT protein interacts with other transcription factors in plant cells, and this interaction serves increased Fe acquisition responses. On the one hand, FIT interacts with EIN3/EIL1, transcription factors of the ethylene pathway, which augments FIT stability (Lingam et al., 2011). On the other hand, the HLH domain of bHLH proteins promotes protein-protein interaction, allowing the formation of homodimeric or heterodimeric complexes (Massari and Murre, 2000). BHLH genes of the subgroup Ib family, such as BHLH038 and BHLH039, are highly up-regulated upon Fe deficiency in roots and leaves independent of FIT (Heim et al., 2003; Vorwieger et al., 2007; Wang et al., 2007). Heterodimerization of FIT with BHLH038 or BHLH039 contributes to high Fe reduction levels in roots (Yuan et al., 2008).
In addition to FIT, at least three other proteins related to Fe homeostasis were found to be regulated at the posttranslational level. FERRITIN2 was found to be posttranslationally regulated in response to metal content, as shown by using different mutants impaired in internal Fe distribution and storage (Arnaud et al., 2006; Ravet et al., 2009a, 2009b). IRT1 underlies protein turnover, and site-directed mutagenesis of certain Lys residues of IRT1 resulted in altered protein turnover and increased metal contents (Kerkeb et al., 2008). Analysis of FRO2 overexpression lines revealed posttranscriptional control for FRO2 (Connolly et al., 2003).
Understanding transcription factor regulation at the protein level is highly important for plant breeding approaches that aim at gaining control of Fe acquisition. FIT is a conserved regulator for Fe uptake in dicotyledonous plants, and knowledge about its regulatory mechanisms can be transferable to crops that rely on a similar Fe uptake strategy (Ling et al., 2002; Bauer et al., 2004; Brumbarova and Bauer, 2005). bHLH transcription factors have also been identified as regulators of Fe acquisition in important graminaceous crops such as rice (Oryza sativa; Ogo et al., 2006; Zheng et al., 2010). Transcriptional regulation of Fe-regulated rice BHLH genes and their impact on the transcriptome have been quite extensively studied (Ogo et al., 2006, 2007, 2011; Zheng et al., 2010), while protein regulation has not been uncovered yet.
Posttranscriptional control of FIT may operate to integrate versatile external and internal signals. In the past, Fe deficiency gene regulation has been brought into the context of ethylene and nitric oxide (NO) signaling (Graziano et al., 2002; Lucena et al., 2006; Graziano and Lamattina, 2007; Besson-Bard et al., 2009; Chen et al., 2010; García et al., 2010; Lingam et al., 2011; Wu et al., 2011). Elucidating the regulation of FIT protein in response to such multipurpose signals may ultimately allow unraveling of the mechanisms by which plants adapt to Fe deficiency at the molecular level.
Since ethylene stabilizes FIT, and ethylene and NO may act in conjunction (García et al., 2010, 2011; Lingam et al., 2011; Romera et al., 2011), we inspected the effect of NO on the regulation of FIT protein levels. We identified NO as a signal that promoted not only the activation of FIT but also its protein stability, independent of the transcriptional control by ethylene and NO. We summarize our results in an integrative model.
RESULTS
Analysis of HA-FIT Protein Levels Showed Posttranslational Control of HA-FIT Activity
To monitor the regulation of FIT protein in planta, we recently described the generation of a specific polyclonal affinity-purified antiserum directed against the C-terminal peptide of FIT to specifically monitor endogenous FIT protein in plants with the wild-type FIT background (Lingam et al., 2011; Fig. 1A). Generation of the specific FIT antiserum is very labor intensive (see “Materials and Methods”; Supplemental Protocol S1). In this work, we report a second tool to evaluate FIT regulation in planta. We generated hemagglutinin (HA)-tagged FIT overexpression transgenic lines. HA-FIT protein can be monitored through the immunogenic HA tag using commercial monoclonal antibodies (Lee et al., 2007). By using in our study the cauliflower mosaic virus (CaMV) 35S promoter, we intended to ensure that HA-FIT was constitutively produced. Thereby, we can attribute any differences in protein abundance to a regulation at the protein level and exclude the influence of transcriptional regulation. The functionality of the HA-FIT protein in planta was confirmed by functional complementation experiments of the fit loss-of-function mutant and by ectopic induction of IRT1 and FRO2 conferred by HA-FIT (Supplemental Results S1; Supplemental Figs. S1 and S2). Out of a total of 18 lines, we selected two different homozygous HA-FIT lines for further analysis. The line HA-FIT 9 (containing the construct Pro-2xCaMV35S:HA3-FIT) harbors a triple HA tag fused to FIT and shows weak ectopic HA-FIT mRNA overexpression (Supplemental Fig. S2). The line HA-FIT 8 (containing Pro-2xCaMV35S:HA7-FIT) harbors a septuple HA tag fused to FIT and has high ectopic HA-FIT mRNA overexpression (Supplemental Fig. S2). Detailed results about the generation, initial characterization, and selection of these lines are described in Supplemental Results S1 (Supplemental Fig. S2). Briefly, by selecting HA-FIT 8 and HA-FIT 9, we were able to monitor the regulation of the presence and activity of HA-FIT in roots and leaves upon strong and weak HA-FIT overexpression, respectively.
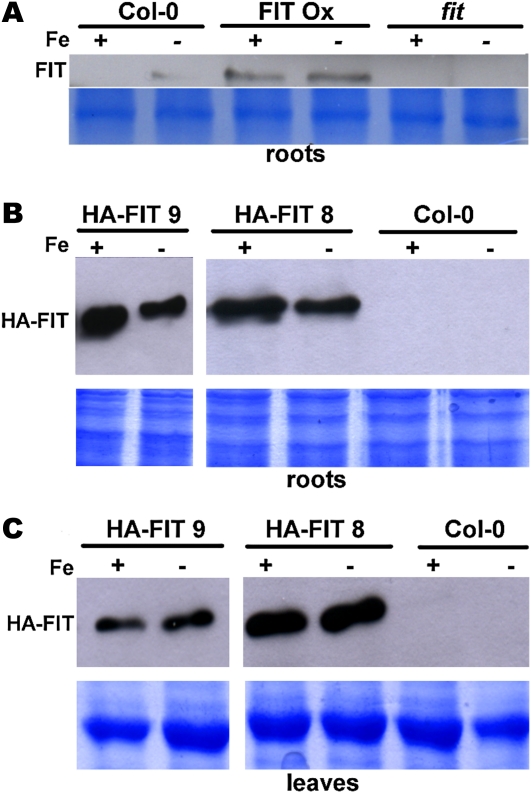
Figure 1.
FIT protein abundance at +Fe and –Fe in FIT overexpression plants. A, FIT protein in roots of wild-type Col-0, FIT Ox (positive control; Jakoby et al., 2004), and fit (negative control; note the specificity of the antiserum). Plants were grown in the 14-d agar growth system. FIT protein was detected by western blot using anti-FIT-C polyclonal antiserum (top panel); Coomassie blue staining represents the loading control (bottom panel). B, HA-FIT protein in roots of HA-FIT plants of the lines HA-FIT 9 and HA-FIT 8 and untransformed Col-0 as a negative control. Plants were grown in the 14-d agar growth system. HA-FIT protein was detected by western blot using anti-HA monoclonal antibodies (top panels); Coomassie blue staining represents the loading control (bottom panels). C, HA-FIT protein in leaves of HA-FIT plants, as described for B. [See online article for color version of this figure.]
As shown recently, in nontransgenic wild-type roots, FIT was detectable under –Fe conditions but not under +Fe conditions (Fig. 1A; Lingam et al., 2011). In the FIT Ox plants, strong FIT protein bands were detectable under both Fe supply conditions (Fig. 1A), demonstrating that FIT protein was produced at +Fe and –Fe in FIT Ox plants (Fig. 1A; Lingam et al., 2011). The differential activity of FIT at +Fe and –Fe in FIT Ox plants, resulting in the induction of FRO2 and IRT1 at –Fe but not at +Fe, therefore must be explained by differential activity of FIT. One of the first questions that arose was to determine whether HA-FIT levels were regulated similarly in the HA-FIT 8 and 9 lines as FIT in FIT Ox plants.
We found that in the two HA-FIT lines, HA-FIT protein was present under +Fe as well as –Fe conditions in roots and leaves (Fig. 1, B and C), in accordance with the results for the untagged FIT Ox line (Fig. 1A).
We conclude that HA-FIT protein behaved similar to FIT when overexpressed. HA-FIT protein was produced at +Fe and –Fe. In the strong HA-FIT overexpression line, HA-FIT 8, HA-FIT was active in –Fe leaves, while HA-FIT was inactive in –Fe leaves in the weak overexpression line, HA-FIT 9. The use of HA-FIT plants allows studying FIT protein regulation in a convenient manner, since a highly specific antiserum is commercially available. Hence, the two selected HA-FIT lines are useful tools for monitoring FIT regulation in plants. In addition, wild-type FIT can be evaluated using the FIT antiserum.
Investigation of FIT Protein Abundance in Response to Cycloheximide Demonstrated Turnover Control of FIT in Wild-Type and HA-FIT Plants
As just described, the activity of HA-FIT protein, measured by downstream FRO2 and IRT1 gene induction, was regulated at the posttranslational level with respect to Fe supply. It was recently shown that tagged FIT-GFP was the subject of to a turnover control in respective overexpression plants and that FIT-GFP was targeted by the proteasome (Sivitz et al., 2011). Therefore, we investigated whether FIT was indeed controlled by a turnover in wild-type plants as well as in the HA-FIT plants we had generated. Toward this end, we applied the protein translation inhibitor cycloheximide (CHX) to block FIT translation. Plants were grown under +Fe and –Fe in the hydroponic system. After 1 h of treatment with CHX, plant samples were harvested either immediately after the treatment (time point 0) or were retransferred to medium without CHX and harvested after several hours (as indicated) for protein detection and gene expression analysis. In non-CHX-treated wild-type control roots, FIT protein was detectable at –Fe at the two time points 0 and 4 h (Fig. 2A; in agreement with Fig. 1A). However, FIT protein was not detectable upon –Fe at time points 0 and 4 h if roots had been treated with CHX (Fig. 2A). In +Fe roots, FIT protein bands were generally not detectable in the wild type (Figs. 1A and 2A). Thus, inhibition of protein synthesis caused a down-regulation of FIT protein in the wild type, indicating that FIT protein produced before CHX treatment at –Fe had been degraded in the wild type. This observation suggests that FIT underlies a constant turnover control in wild-type plants.
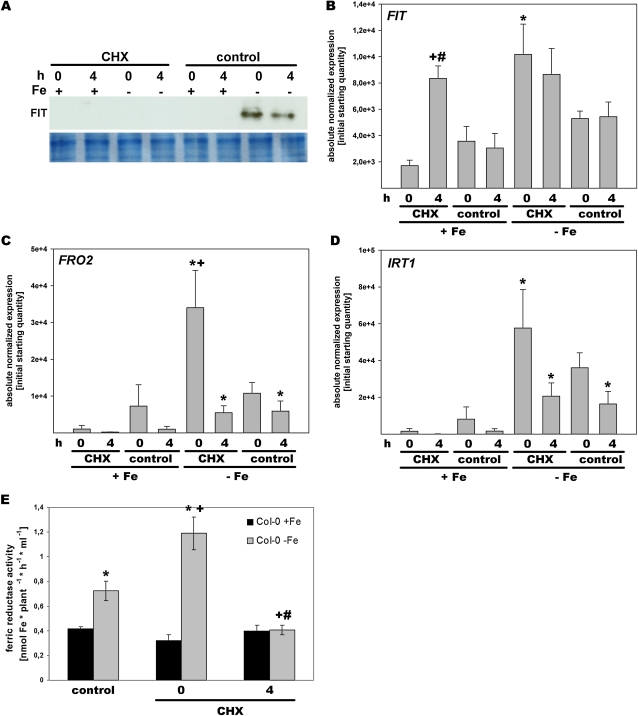
Figure 2.
CHX treatment reduced FIT protein abundance and FIT gene expression in wild-type plants. A, FIT protein in roots of the wild type treated for 1 h with 50 μm CHX or without (control). Plants were grown in the hydroponic system at +Fe or –Fe, and samples were harvested directly after the treatment (0-h time point) and 4 h after retransfer to +Fe or –Fe growth medium without CHX (4-h time point). FIT protein was detected by western blot using anti-FIT-C polyclonal antiserum (top panel); Coomassie blue staining represents the loading control (bottom panel). B to D, Reverse transcription-quantitative PCR analysis of FIT (B), FRO2 (C), and IRT1 (D) in roots at +Fe and –Fe. E, Fe reductase activity of wild-type plants (Col-0). * Significant change (P < 0.05) versus +Fe; + significant change (P < 0.05) versus the non-CHX control; # significant change (P < 0.05) of the 4-h time point versus the 0-h time point. n = 2 (B–D) and n = 5 (E). [See online article for color version of this figure.]
To evaluate FIT activity, we examined the effect of CHX treatment on the expression of Fe deficiency genes (Fig. 2, B–D). Very interestingly, we observed that FIT gene expression was up-regulated upon +Fe by CHX treatment at the 4-h time point versus the untreated control, so that at the 4-h time point, FIT expression levels were similar at +Fe and –Fe (Fig. 2B). Only at the 0-h time point was FIT expression low at +Fe upon CHX treatment as in the untreated control, while an induction of FIT was seen in both CHX-treated and untreated plant samples at –Fe at the 0-h time point compared with +Fe. One explanation for this finding is that a repressor protein might suppress FIT transcription at +Fe. This repressor protein might have been susceptible to CHX treatment (and presumably to –Fe). This would explain why CHX treatment resulted in a derepression of the FIT gene at the 4-h time point. This effect conferred by CHX at +Fe was specific for FIT, since IRT1 and FRO2 expression were not affected by CHX at +Fe (Fig. 2, compare B with C and D). Expression of FRO2 and IRT1 was found to be up-regulated by –Fe compared with +Fe, and the level of expression of FRO2 was even higher at the 0-h time point upon CHX treatment than in the control (Fig. 2, C and D). To test whether the increased FRO2 gene expression at the 0-h CHX time point would also result in increased protein activity, we performed an Fe reductase assay. We found that Fe reductase activity was at a constant low level at +Fe and 2-fold increased at –Fe, as expected. At the 0-h time point of CHX treatment, Fe reductase activity was 3-fold increased at –Fe, while at the 4-h time point of CHX treatment, Fe reductase activity was at the low level of +Fe (Fig. 2E). Therefore, Fe reductase activity paralleled FRO2 gene expression. Perhaps a negative regulatory mechanism was present at –Fe to prevent excessive reduction of Fe, and this mechanism might have been transiently eliminated by CHX treatment. Four hours after the CHX treatment, this negative control mechanism might have been restored.
Taken together, we could show that FIT was the subject of a protein turnover control in the wild-type condition. Moreover, when comparing the abundance of FIT (Fig. 1A) with gene expression (Fig. 2, B–D) in CHX-treated and untreated samples, it can be concluded that a high amount of FIT was not required for FRO2 and IRT1 induction.
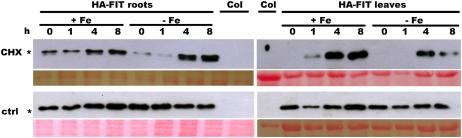
A similar experiment was conducted with HA-FIT plants (HA-FIT 8). Protein samples were harvested at the 0-h time point as well as 1, 4, and 8 h after retransfer to +Fe or –Fe medium (time points 0, 1, 4, and 8 in Fig. 3). By western-blot analysis, we observed that up to 1 h after the CHX treatment (time points 0 and 1), HA-FIT protein bands were less abundant than at 4 and 8 h after the treatment (time points 4 and 8) in both roots and leaves (Fig. 3). Very interestingly, in the –Fe root samples, protein bands were less abundant than in the +Fe root samples, by a factor of 3 at time point 0 and by a factor of 6 at time point 1 (Fig. 3). In the non-CHX-treated controls, HA-FIT protein levels varied only slightly. In leaves, no such striking difference of HA-FIT protein bands could be observed upon CHX treatment between +Fe and –Fe samples (Fig. 3). Thus, HA-FIT was the subject of a turnover control in the overexpression situation, which was more pronounced in –Fe roots than in +Fe roots.
Figure 3.
CHX treatment reduced HA-FIT protein abundance in HA-FIT plants. HA-FIT protein in roots (left) and leaves (right) of HA-FIT 8 plants grown at +Fe or –Fe and treated for 1 h with 50 μm CHX or without (control [ctrl]). Plants were grown in the hydroponic system, and samples were harvested directly after the treatment (0-h time point) or 1, 4, and 8 h after retransfer to +Fe or –Fe growth medium without CHX (1-, 4-, and 8-h time points). HA-FIT protein was detected by western blot using anti-HA monoclonal antibodies. * The HA-FIT band (top panels; Col is the negative untransformed plant sample control); Ponceau S staining represents the loading control (bottom panels). [See online article for color version of this figure.]
It was interesting, then, to determine whether CHX treatment had affected Fe deficiency gene expression in HA-FIT plants. We analyzed gene expression directly after the 1-h CHX incubation (time point 0), where HA-FIT protein was reduced or not detectable, and 4 h after the CHX treatment, where HA-FIT had again accumulated similar to the control (time point 4). FIT gene expression was at a constant high level in roots and leaves in both Fe supply conditions, irrespective of CHX treatment, as expected due to overexpression (Supplemental Figs. S3A and S4A). FRO2 and IRT1 expression levels were not significantly altered by CHX treatment in this case (Supplemental Figs. S3, B and C, and S4, B and C). These results suggest that Fe deficiency gene expression downstream of FIT did not rely on full FIT protein abundance after CHX treatment compared with the control. Presumably, a small pool of active FIT protein was sufficient to induce FRO2 and IRT1.
We can summarize three major points from these experiments. First, FIT protein levels were negatively affected by CHX treatment in wild-type and HA-FIT Ox plants, confirming turnover control of FIT. In roots, the effect was stronger at –Fe than at +Fe, suggesting that –Fe led to an enhanced degradation of HA-FIT. Second, reduction of FIT abundance, conferred by CHX, did not result in significantly lowered expression of FRO2 and IRT1. Third, FIT gene expression was up-regulated in +Fe wild-type roots upon CHX treatment. An explanation could be that FIT gene expression might be repressed at +Fe by a repressor that is susceptible to CHX.
FIT Protein Accumulation Is Counteracted by NO Inhibitors and Restored by Inhibitors of Proteasomal Degradation
The above results confirmed that FIT was the subject of a turnover control at the protein level in wild-type and HA-FIT plants. Recently, we showed that FIT stability was increased by ethylene signaling (Lingam et al., 2011). Here, we asked the question whether NO would also affect FIT protein abundance and activity. Previous reports had shown that NO positively affects Fe deficiency responses in tomato (Solanum lycopersicum) and Arabidopsis (Graziano et al., 2002; Graziano and Lamattina, 2007; Besson-Bard et al., 2009; Chen et al., 2010) and that NO may act at a similar level to ethylene to promote the Fe deficiency responses (Lucena et al., 2006; García et al., 2010; Wu et al., 2011).
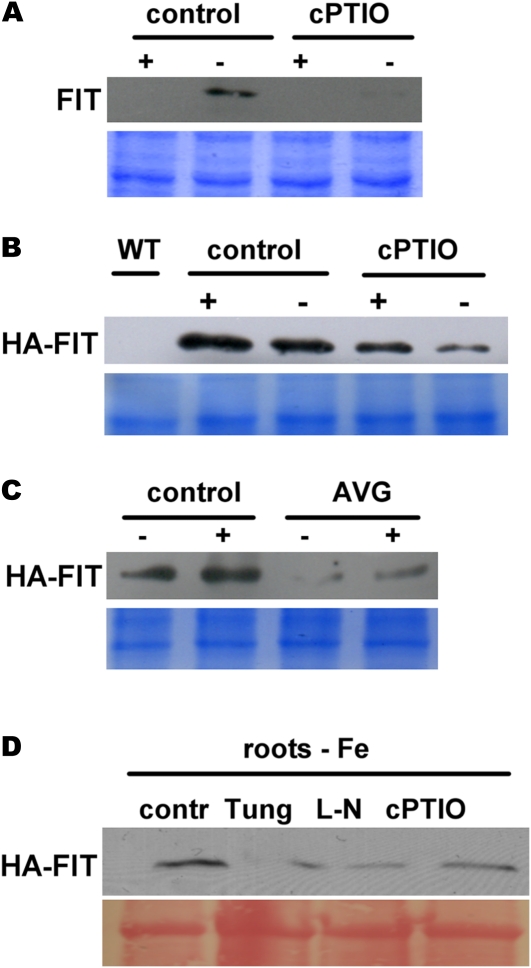
To test the effect of NO, we grew wild-type and HA-FIT 9 plants in the 6-d growth system that we found most suitable for the NO pharmacological treatments with the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO). We selected cPTIO because it was described as a common plant inhibitor for NO in the literature (Graziano and Lamattina, 2007; Chen et al., 2010). In these conditions, we were able to detect FIT protein in wild-type control roots at –Fe (Fig. 4A), which was in accordance with our previous results (Figs. 1A and 2A). cPTIO treatment caused a strong down-regulation of FIT protein to 2% at –Fe compared with control roots, suggesting that inhibition of NO signaling resulted in reduced FIT protein abundance (Fig. 4A).
Figure 4.
NO inhibition caused a reduction of FIT and HA-FIT protein levels. A, FIT protein in roots of the wild-type untreated (control) and plants treated for 24 h with 1 mm cPTIO. Plants were grown in the 6-d agar system at +Fe and –Fe. FIT protein was detected by western blot using anti-FIT-C polyclonal antiserum (top panel); Coomassie blue staining represents the loading control (bottom panel). B, HA-FIT in roots of HA-FIT 9 plants, treated and grown as in A. WT, Wild type. C, HA-FIT in roots of HA-FIT 9 plants, treated with 10 μm AVG and grown as in A. D, HA-FIT protein abundance in roots of –Fe HA-FIT 9 plants, untreated (control [contr]) or treated with 1 mm tungstate (Tung), 1 mm l-NAME (L-N), and 1 mm cPTIO, showing that several NO inhibitors caused reduction of HA-FIT protein amounts. Plants were grown as in A. In B to D, HA-FIT protein was detected by western blot using anti-HA monoclonal antibodies (top panels). In B and C, Coomassie blue staining represents the loading control (bottom panels); in D, Ponceau S was used as the loading control (bottom panel). [See online article for color version of this figure.]
In the HA-FIT plants, cPTIO treatment also resulted in a decrease of HA-FIT protein (to 30% at –Fe and to 50% at +Fe versus controls; Fig. 4B). As a control, we also confirmed the ethylene effect on HA-FIT protein stability by testing HA-FIT protein abundance upon treatment with the ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG), as reported previously for endogenous FIT (Lingam et al., 2011). As expected (Lingam et al., 2011), AVG caused a reduction of HA-FIT protein levels at +Fe and –Fe (Fig. 4C).
We further confirmed the effect of NO inhibition on FIT protein by testing additional NO inhibitors, namely tungstate and N-nitro-l-arginine methylester hydrochloride (l-NAME). We found that all three NO inhibitors reduced HA-FIT protein at –Fe, namely to 40% (tungstate), 30% (l-NAME), and 50% (cPTIO) versus the controls (Fig. 4D). Thus, inhibition of NO indeed decreased FIT protein accumulation.
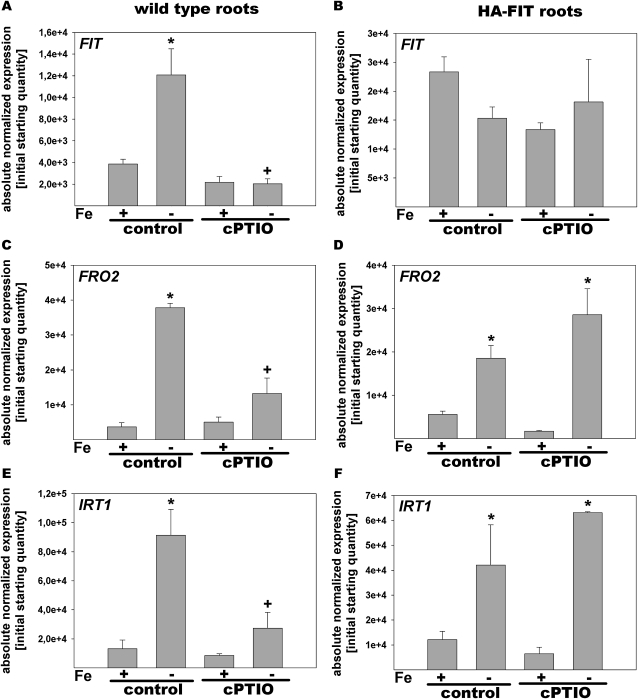
We then investigated whether cPTIO treatments had affected the expression of the Fe deficiency genes in wild-type plants and HA-FIT plants grown as just described. In wild-type control roots exposed to +Fe or –Fe, gene expression was as expected and corresponded to the results described in the previous paragraphs: FIT was induced 3-fold, whereas IRT1 and FRO2 were at least 8-fold induced by –Fe (Fig. 5, A, C, and E). The same was observed for HA-FIT plants, except that FIT was overexpressed compared with the wild type (Fig. 5, B, D, and F). cPTIO application resulted in a decreased expression of FIT, FRO2, and IRT1 gene expression in –Fe wild-type roots compared with the –Fe control (Fig. 5, A, C, and E). This was as expected according to the literature (Graziano et al., 2002; Graziano and Lamattina, 2007; Besson-Bard et al., 2009; Chen et al., 2010). On the other hand, cPTIO treatment had no effect on gene expression in HA-FIT plants (Fig. 5, B, D, and F). Presumably, the remaining pool of HA-FIT protein in the transgenic overexpression plants was sufficient to trigger FRO2 and IRT1 induction. The decrease of HA-FIT by cPTIO shows that HA-FIT protein regulation cannot be explained merely by a reduced transcriptional activation due to cPTIO but that reduced NO affected HA-FIT also at the protein level.
Figure 5.
Altered gene expression in response to cPTIO. Reverse transcription-quantitative PCR analysis is shown in wild-type (A, C, and E) and HA-FIT (B, D, and F) roots treated with or without cPTIO. Plants were grown in the 6-d agar growth system with FIT (A and B), FRO2 (C and D), and IRT1 (E and F). * Significant change versus +Fe of each treatment (P < 0.05); + significant change versus the control at each Fe supply (P < 0.05). n = 2.
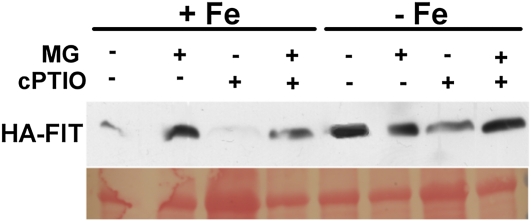
It was interesting, then, to further investigate the mechanism by which NO could prevent FIT protein degradation. Toward this end, we incubated cPTIO-treated HA-FIT plants with the common proteasome inhibitor MG132. In this experiment, HA-FIT was reduced to 50% at –Fe and to 6% at +Fe upon cPTIO treatment compared with the controls (Fig. 6; in agreement with Fig. 4B). When cPTIO-grown seedlings were treated with MG132, FIT protein levels were restored at –Fe and +Fe (Fig. 6).
Figure 6.
MG132 reversed the cPTIO-mediated repression of HA-FIT. HA-FIT abundance is shown in roots of HA-FIT 9 plants grown at –Fe or +Fe and treated as indicated for 24 h with 1 mm cPTIO and for 4 h with 100 μm MG132 (MG). Plants were grown in the 6-d agar growth system. HA-FIT protein was detected by western blot using anti-HA monoclonal antibodies (top panel); Ponceau S was used as the loading control (bottom panel). [See online article for color version of this figure.]
Hence, we conclude that, upon inhibition of NO signaling, FIT protein was more susceptible to degradation by the proteasome. Therefore, application of proteasome inhibitors could result in the restoration of FIT protein levels after cPTIO treatment. We propose that NO promotes FIT protein stability at a similar level to ethylene by inhibiting proteasomal degradation of FIT.
DISCUSSION
Here, we investigated regulatory mechanisms acting at the protein level upon a key transcription factor of the Fe deficiency response. FIT protein was the subject of a turnover control under both +Fe and –Fe conditions and was susceptible to proteasomal degradation. The turnover control took place in a stronger manner at –Fe than at +Fe. NO was identified as an internal signal for achieving full-level FIT protein accumulation. NO counteracted the proteasomal degradation of FIT and presumably acted in a similar manner to ethylene. Irrespective of the amount of FIT protein at +Fe, downstream activation of FRO2 and IRT1 did not take place at +Fe. A –Fe signal was required to trigger FIT activation. We suggest that FIT exists in both “active” and “inactive” forms, being more prone to proteasomal degradation at –Fe. We discuss the mechanisms by which FIT is regulated.
FIT Activity Is Controlled at Multiple Steps from Transcription to Active FIT Protein
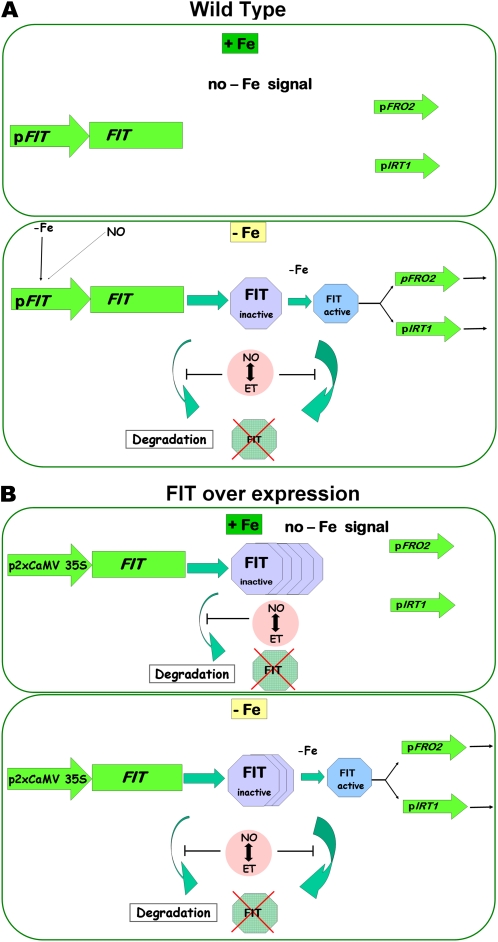
Due to our combined analysis of FIT protein regulation in wild-type plants on one side and in HA-FIT overexpression plants on the other side, we could discriminate multiple regulatory mechanisms acting upon FIT protein. The wild-type situation allowed uncovering transcriptional and posttranslational regulation. The overexpression plants allowed us to confirm that, indeed, posttranslational effects took place, since the regulation of FIT protein abundance could be uncoupled from the transcriptional control. Our findings about the multiple mechanisms that confer FIT activation are summarized in Figure 7.
Figure 7.
Model summarizing the regulation of FIT activity in wild-type and HA-FIT Ox plants by multiple control steps. A, The wild type. In +Fe wild-type roots, FIT induction does not take place. It might be repressed by a negative regulator. Downstream targets of FIT, like FRO2 and IRT1, are not induced. In –Fe wild-type roots, FIT transcription is induced. The presumptive FIT repressor protein might be removed by –Fe. Subsequently, FIT protein is produced. Due to a –Fe signal, FIT is activated and promotes the induction of FRO2 and IRT1. The transition from inactive to active FIT could be achieved through FIT protein modification by the addition or removal of covalent modifications, or by a differential interaction with proteins present only at –Fe, such as bHLH038 and bHLH039 (Yuan et al., 2008), or perhaps both. FIT itself is degraded due to protein turnover (compare this work with Lingam et al., 2011; Sivitz et al., 2011). NO and ethylene (ET) increase the accumulation of FIT by counteraction of proteasomal FIT degradation (compare this work and Lingam et al., 2011). B, HA-FIT overexpression. Transcriptional control of HA-FIT is not relevant due to the 2xCaMV 35S promoter. In +Fe HA-FIT roots, HA-FIT is targeted by protein turnover. NO and ethylene increase the accumulation of FIT, probably by counteraction of proteasomal FIT degradation. In the absence of a –Fe activating signal, FRO2 and IRT1 targets are not induced. In –Fe HA-FIT roots, HA-FIT is activated by a –Fe signal and promotes the expression of FRO2 and IRT1. Further details in B are as in A. [See online article for color version of this figure.]
The first control step in FIT activation takes place at the transcriptional level. FIT gene expression is induced at –Fe (Fig. 7A shows the wild-type situation; compare with Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). FIT induction requires transcription factors acting upstream of FIT. Positive regulators must be themselves activated by –Fe. Alternatively, a negative regulator might suppress FIT transcription at +Fe. Elimination of the repressor at –Fe would equally result in an induction of FIT. In this respect, it is interesting that the application of CHX at +Fe resulted in increased FIT gene expression. This effect was previously not observed, because CHX effects were not shown in wild-type plants at +Fe (Sivitz et al., 2011). An explanation for this observation is that CHX destroyed the repression at +Fe. The presumptive repressor of FIT at +Fe might be the subject of a protein turnover control. It is appealing to speculate that the turnover control of the FIT repressor responds to Fe, so that the repressor may be effectively diminished upon –Fe. In future experiments, it would be interesting to confirm the repression effect by identifying promoter-binding sites and the repressor itself.
As a second regulatory step, FIT was controlled by posttranslational turnover control (Fig. 7; Lingam et al., 2011; Sivitz et al., 2011). Inhibition of protein translation led to a decrease of HA-FIT protein abundance irrespective of transcription. In wild-type plants, turnover of FIT was tractable only at –Fe, because at +Fe, FIT was not detected. FIT protein, therefore, must undergo cycles of degradation and resynthesis so that FIT/HA-FIT levels appear constant in the absence of CHX. From our studies using HA-FIT plants, we can conclude that turnover of FIT took place at +Fe and –Fe in roots and leaves, whereby it was most pronounced upon –Fe in roots. The wild-type results that we present here show that the regulation is relevant at –Fe only, since at +Fe, FIT was not detectable. In this respect, our results agree with those of Sivitz et al. (2011). These authors had observed a turnover of FIT-GFP in overexpression plants mainly at –Fe, but they did not perform any experiments with the wild type in this respect. Sivitz et al. (2011) found, by using a GFP-tagged FIT overexpression line, that in these plants FIT-GFP was present at a lower level at –Fe than at +Fe. However, when Sivitz et al. (2011) used an untagged FIT line, FIT protein was detected equally at +Fe and –Fe, more similar to our results shown here. Perhaps technical reasons or the GFP tag itself accounted for this altered protein abundance at +Fe and –Fe in FIT-GFP plants. We could show that NO inhibitors had a negative effect on FIT abundance, which was reversed by MG132. Therefore, all the evidence points to the possibility that FIT is degraded by the proteasome and that NO may act to prevent it. A further discussion of the involvement of NO is presented below.
A third level of regulation took place at the level of protein activity (Fig. 7). The output of FIT activity was measured as the induction of IRT1 and FRO2. The abundance of FIT protein was not found proportional to the level of the activity of FIT protein. To this point, our findings agree with those of Sivitz et al. (2011) and Lingam et al. (2011). An elevated level of FIT/HA-FIT protein does not imply a general increase of FIT activity. On the other hand, a reduction in FIT levels does not necessarily cause a reduction in downstream responses.
What are the reasons and mechanisms for the turnover and activation control of FIT? Sivitz et al. (2011) proposed that the activity of FIT was related to its constant turnover and that ubiquitination and proteasomal degradation of FIT, stimulated by –Fe, might be needed to maintain a turnover of FIT for its transcriptional activity at its target binding sites. Lingam et al. (2011), on the other hand, proposed that the differential FIT activity was due to the activation of FIT from a large inactive pool to a small active pool, both of which might be targeted by the proteasome. Here, we showed that the activity of FIT was not compromised by CHX treatment. Obviously, low amounts of FIT protein were sufficient to trigger IRT1 and FRO2 induction, so we assume that these low amounts contained sufficient active FIT that the synthesis of “fresh” FIT (Sivitz et al., 2011) was not immediately needed. By comparing the amounts of protein at +Fe and –Fe upon CHX and cPTIO treatment and the untreated controls, we suggest that a large pool of FIT that was targeted by the proteasome must have been inactive FIT. Since the proteasome did not appear to select between active and inactive FIT, the proteasomal degradation may not play an important role for increasing the pool of active FIT. This leads to the question of what other mechanism could activate FIT. One possibility is that the active and inactive states differ by specific covalent modifications. If the transfer from the inactive state to the active state has a bottleneck, this could be achieved through limitation of the enzymes that may confer or remove covalent modifications to “activate” FIT. On the other hand, regulation of FIT activity may also be conferred by an interaction with another protein that is needed for downstream responses. This other protein should be present at –Fe only, and it should be regulated by different upstream mechanisms in a FIT-independent manner. Limitation in the availability of this binding partner of FIT could also represent a bottleneck at –Fe, explaining why there are only low amounts of active FIT. Interestingly, bHLH038 and bHLH039 fulfill the criteria of such an interaction partner (Yuan et al., 2008), since they are expressed only at –Fe in a FIT-independent manner (Wang et al., 2007), much better than the other interaction partners, EIN3/EIL1 (Lingam et al., 2011). Although such an interaction could be the bottleneck, posttranslational modification of FIT could be mandatory to initiate subsequent interaction with other proteins.
What are the reasons for the turnover of FIT if the activation of FIT would occur at a different level? It is very common for transcription factors that their degradation occurs after or even before they have been modified and activated (Shen et al., 2007, 2009).
FIT also controls its own induction (Jakoby et al., 2004) and could be regulated by a feedback loop. The physiological reason for switching off key transcription factors is to allow cells to continuously remain responsive to incoming signals. FIT could be inside a negative feedback loop that restricts its own abundance. In the context of Fe regulation, the reason for rapidly switching off FIT may lie in preventing Fe toxicity symptoms that would be expected if FIT caused excessive uptake of Fe. Excessive cellular Fe would augment the pool of free metals, resulting in radical production through the Fenton reaction and cellular damage.
How far the three mechanisms of covalent modification, protein interaction, and protein degradation are intermingled and contribute to the regulation of FIT activation has to be studied in the future by analyzing the genes for the upstream regulation of FIT. In some well-studied cases, numerous regulatory mechanisms were found to be involved in activating and deactivating a single nuclear factor (Perkins, 2006), and such examples can provide a framework for the mechanisms that control FIT.
NO Reduced the Proteasomal Degradation of FIT
As shown previously, cPTIO caused a decrease of Fe deficiency gene expression (Graziano et al., 2002; Graziano and Lamattina, 2007; Besson-Bard et al., 2009; Chen et al., 2010). At the transcriptional level, therefore, NO can induce FIT, IRT1, and FRO2. In addition, we demonstrated here that cPTIO also caused a reduction of FIT protein. cPTIO, a scavenger of NO, caused a reduction of FIT protein levels at +Fe and –Fe. Thus, it can be inferred that NO promoted FIT protein levels. The observed inhibitory effect of cPTIO on FIT protein levels was not merely the result of reduced transcriptional activation of FIT, for two reasons. First, reduced protein accumulation caused by cPTIO was also apparent in HA-FIT overexpression plants, where HA-FIT transcription was not regulated by NO and thus not affected by cPTIO. Second, MG132, which acts to inhibit the proteasome, restored FIT protein levels upon cPTIO treatment. These observations suggest that inhibition of NO provoked a stronger proteasomal degradation of FIT. Hence, NO may act to prevent the proteasomal degradation of FIT. To our knowledge, this is a new link between FIT activity and NO that could be concluded from our work.
Although principally low amounts of FIT protein are sufficient to trigger FIT downstream responses to the full level, this was not the case upon cPTIO treatment in the wild type. Perhaps the remaining levels of active FIT were too low in the wild type treated with cPTIO to cause downstream gene induction. In HA-FIT plants treated with cPTIO, the levels of the remaining FIT protein were higher than in the wild type, and presumably, sufficient amounts of active FIT were among it. This could be the reason why in HA-FIT plants, cPTIO application did not affect downstream gene expression.
NO could be involved in activating and stabilizing FIT directly or indirectly. Direct effects could be exerted by way of the nitrosylation of Cys residues, which are also present in FIT (Tada et al., 2008; Lindermayr and Durner, 2009), while indirect modifications may occur through alterations of enzyme activities occurring as a response to NO. If modifications of FIT occur in response to NO, this can be the explanation why the physiological output differed between untreated controls, CHX, and cPTIO application.
Interestingly, the NO effect on gene expression and FIT protein regulation paralleled that of ethylene (Graziano et al., 2002; Lucena et al., 2006; Graziano and Lamattina, 2007; Besson-Bard et al., 2009; Chen et al., 2010; García et al., 2010; Lingam et al., 2011; Wu et al., 2011). Ethylene, like NO, is required for full-level up-regulation of Fe deficiency gene expression and FIT protein abundance. This observation suggests that NO and ethylene act in the same way and perhaps in sequential order. It was recently proposed that a strictly linear relationship between NO and ethylene action may not exist and that they may promote each other (García et al., 2010, 2011; Romera et al., 2011).
CONCLUSION
Protein turnover of transcription factors is prevalent in plants. Posttranslational modifications affect the protein interaction capacities of bHLH proteins and their functions (Bracken et al., 2003; Barnes and Firulli, 2009). In plants, functional modifications of bHLH proteins were reported to occur during light perception and development (Shen et al., 2007, 2009; Lampard et al., 2008; Park et al., 2008; Kang et al., 2009; Han et al., 2010). Auxin signaling and other hormone response transcription factors are also targets for proteasome action, for example of the ubiquitin-26S proteasome system (Schwager et al., 2007; Vierstra, 2009). The interpretation is that in this way, cells remain continuously responsive to the incoming signals and that the reorientation of the activity of these transcription factors can be modulated in a flexible manner. Once activated, transcription factors can be rapidly removed from the cell by protein degradation to prevent their excessive action.
Our study is among the first to address the control of a key Fe deficiency transcription factor in response to NO. Controlled turnover and activity of FIT strengthens the importance of FIT. FIT may be the key regulator in processing the versatile incoming signals from hormonal and intracellular triggers. Due to FIT turnover and activity control, plant roots remain responsive to changing Fe availability in the soil as well as changing demands for Fe nutrition during the day and throughout the plant’s life cycle.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0 ) was used as the wild type in the experiments. The fit mutant is represented by the fit-3 allele (Jakoby et al., 2004; Bauer et al., 2007). For physiological assays, seeds were surface sterilized as described by Jakoby et al. (2004).
In the 2-week growth system, plants were grown and exposed for 3 d to –Fe as described (Jakoby et al., 2004). In the 6-d growth system, seeds were directly germinated on 50 μm Fe (+Fe) or 0 μm Fe (–Fe) Hoagland agar medium. In the hydroponic growth system, plants were grown to the age of 4 weeks in hydroponic quarter-strength Hoagland medium containing 10 μm Fe (+Fe) and then transferred for 1 week to 10 μm Fe (+Fe) or 0 μm Fe (–Fe) as described (Klatte et al., 2009).
Generation of Transgenic HA-FIT Overexpression Plants
Two different HA-FIT overexpression constructs were generated. First, FIT cDNA was introduced by Gateway cloning (Invitrogen) into pDONR207 and then into the binary vector pAlligator2 (http://www.isv.cnrs-gif.fr/jg/alligator/vectors.html) to obtain the Pro-2xCaMV35S:HA3-FIT fusion construct. Second, FIT cDNA was amplified by PCR flanked by SalI and PstI restriction sites and inserted by restriction-ligation into pPILY (Ferrando et al., 2000). The obtained HA7-FIT construct was then transferred by Gateway cloning into pDONR207 and subsequently into the binary vector pMDC32 (Curtis and Grossniklaus, 2003) to finally obtain the Pro-2xCaMV35S:HA7-FIT fusion construct. Both destination vectors were transferred into Agrobacterium tumefaciens strain GV2260 (containing pGV2260). The transformation of Arabidopsis ecotype Col-0 was performed following the “floral dip” method (Clough and Bent, 1998). Selection of transgenic seeds was based on GFP fluorescence in the case of pAlligator and hygromycin resistance for the pMDC transformants. Homozygous transgenic lines termed HA3-FIT Ox and HA7-FIT Ox were selected by PCR and multiplied to the T3 generation (Supplemental Results S1). The fit mutant was transformed with these constructs in a similar manner to show functional complementation by HA-FIT (Supplemental Results S1). In addition, the functionality of HA-FIT was confirmed by analyzing downstream responses in plants (Supplemental Results S1).
Pharmacological Treatments of Plants
For protein translation inhibition experiments using CHX, plants were grown in the hydroponic growth system. Plants were transferred to liquid Hoagland medium containing 50 μm CHX (Sigma-Aldrich; using a 50 mm stock solution dissolved in dimethyl sulfoxide) and incubated for 1 h. Roots and leaves were harvested either directly after treatment (0-h time point) or roots were washed and transferred to fresh Hoagland medium for 1 to 8 h after treatment as indicated. Roots and leaves were harvested separately and frozen in liquid nitrogen until further processing.
NO experiments were conducted using the 6-d growth assay. Five-day-old seedlings were transferred to fresh 50 μm or 0 μm Fe Hoagland agar medium, containing as treatments 1 mm cell-permeating NO scavenger cPTIO (Sigma-Aldrich), 1 mm sodium tungstate (hereafter named tungstate), or 1 mm l-NAME. After 24-h treatments, roots were harvested and further processed. If indicated, 10 μm AVG (Sigma-Aldrich) was added to the growth medium. For MG132 treatment, 6-d-old seedlings were treated for 4 h in liquid Hoagland medium containing 100 μm MG132 (Calbiochem) and harvested for analysis.
FIT Antibody Preparation and Immunological Detection
A polyclonal mouse FIT antiserum was produced that was directed against the C-terminal part of FIT excluding the bHLH domain. For use on western blots with plant protein extracts, anti-FIT-C antiserum was purified (Supplemental Protocol S1).
Total protein extracts were prepared from roots of 6-d-old seedlings (Supplemental Protocol S1; Scharf et al., 1998). Ten micrograms of protein was loaded per lane on a 10% SDS-PAGE device and subsequently blotted to a nitrocellulose membrane.
For the detection of FIT protein, freshly purified undiluted anti-FIT-C mouse antiserum was applied in western-blot experiments according to standard procedures. These primary antibodies were detected with anti-mouse IgG conjugated with horseradish peroxidase (1:8,000 dilution; Sigma-Aldrich). HA-FIT protein was detected by using the SNAP identifier system (Merck-Millipore). Membranes were incubated with anti-HA high-affinity monoclonal rat antibody (1:1,000; clone 3F10; Roche) and as secondary antibody anti-rat IgG (whole molecule) conjugated with horseradish peroxidase (1:10,000; Sigma-Aldrich). Detection signals were developed by using chemiluminescence. Relative quantification of protein bands was calculated using ImageJ software (Abramoff et al., 2004) and normalization to the Coomassie blue- or Ponceau S-stained bands.
Gene Expression Analysis
Gene expression analysis was performed by reverse transcription-quantitative real-time PCR as described previously (Klatte and Bauer, 2009). Briefly, DNase-treated RNA was used for cDNA synthesis. SYBR Green I-based real-time PCR analysis was performed using ExTaq R-PCR (TaKaRa) in the My IQ single-color real-time PCR detection system (Bio-Rad). Each PCR was performed with two technical replicates, from which an average was calculated (Klatte and Bauer, 2009). For each gene, the absolute quantity of the initial transcript was determined by standard curve analysis. Absolute expression data were normalized against the averaged expression values of the internal control gene EF1BALPHA2. Primer sequences have been published (Wang et al., 2007). Two biological replicate RNA/cDNA samples were generated. Statistical evaluation was performed by t test using the values of biological replicates.
Fe Reductase Activity Assay
Fe reductase activity was determined according to Jakoby et al. (2004) using 6-d-old seedlings of similar size, grown on Hoagland agar plates, and treated as described.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Functional complementation of fit mutants by a HA3-FIT Ox transgene.
Supplemental Figure S2. Gene expression analysis of different HA-FIT Ox lines used in the selection of lines HA-FIT 8 and HA-FIT 9.
Supplemental Figure S3. Gene expression was not affected in HA-FIT roots in response to CHX.
Supplemental Figure S4. Gene expression was not affected in HA-FIT leaves in response to CHX.
Supplemental Results S1. Generation, characterization, and selection of HA-tagged FIT overexpression lines.
Supplemental Protocol S1. FIT antibody preparation and immunological detection.
Acknowledgments
We thank F. Parcy (CNRS Grenoble) for providing pAlligator2. We thank T. Potuschak (CNRS Strasbourg) for pPILY. We are grateful to I. Fuchs and U. Müller (Zoology Department, Saarland University) for FIT-C antiserum production. We are thankful for discussions with M. Graziano and L. Lamattina (National University of Mar del Plata) about NO experiments. We thank R. Ivanov and T. Brumbarova (Saarland University) for fruitful discussions.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophot Int 11: 36–42 [Google Scholar]
- Arnaud N, Murgia I, Boucherez J, Briat JF, Cellier F, Gaymard F. (2006) An iron-induced nitric oxide burst precedes ubiquitin-dependent protein degradation for Arabidopsis AtFer1 ferritin gene expression. J Biol Chem 281: 23579–23588 [DOI] [PubMed] [Google Scholar]
- Barnes RM, Firulli AB. (2009) A twist of insight: the role of Twist-family bHLH factors in development. Int J Dev Biol 53: 909–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Ling HQ, Guerinot ML. (2007) FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem 45: 260–261 [DOI] [PubMed] [Google Scholar]
- Bauer P, Thiel T, Klatte M, Bereczky Z, Brumbarova T, Hell R, Grosse I. (2004) Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiol 136: 4169–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Gravot A, Richaud P, Auroy P, Duc C, Gaymard F, Taconnat L, Renou JP, Pugin A, Wendehenne D. (2009) Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol 149: 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Whitelaw ML, Peet DJ. (2003) The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cell Mol Life Sci 60: 1376–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P. (2005) Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol 137: 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Yang JL, Qin C, Jin CW, Mo JH, Ye T, Zheng SJ. (2010) Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis thaliana. Plant Physiol 154: 810–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML. (2003) Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol 133: 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A, Farras R, Jasik J, Schell J, Koncz C. (2000) Intron-tagged epitope: a tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J 22: 553–560 [DOI] [PubMed] [Google Scholar]
- García MJ, Lucena C, Romera FJ, Alcántara E, Pérez-Vicente R. (2010) Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J Exp Bot 61: 3885–3899 [DOI] [PubMed] [Google Scholar]
- García MJ, Suárez V, Romera FJ, Alcántara E, Pérez-Vicente R. (2011) A new model involving ethylene, nitric oxide and Fe to explain the regulation of Fe-acquisition genes in strategy I plants. Plant Physiol Biochem 49: 537–544 [DOI] [PubMed] [Google Scholar]
- Graziano M, Beligni MV, Lamattina L. (2002) Nitric oxide improves internal iron availability in plants. Plant Physiol 130: 1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano M, Lamattina L. (2007) Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant J 52: 949–960 [DOI] [PubMed] [Google Scholar]
- Han YJ, Kim HS, Kim YM, Shin AY, Lee SS, Bhoo SH, Song PS, Kim JI. (2010) Functional characterization of phytochrome autophosphorylation in plant light signaling. Plant Cell Physiol 51: 596–609 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ. (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkeb L, Mukherjee I, Chatterjee I, Lahner B, Salt DE, Connolly EL. (2008) Iron-induced turnover of the Arabidopsis IRON-REGULATED TRANSPORTER1 metal transporter requires lysine residues. Plant Physiol 146: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte M, Bauer P. (2009) Accurate real-time reverse transcription quantitative PCR. Methods Mol Biol 479: 61–77 [DOI] [PubMed] [Google Scholar]
- Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P. (2009) The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol 150: 257–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC. (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Durner J. (2009) S-Nitrosylation in plants: pattern and function. J Proteomics 73: 1–9 [DOI] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P. (2011) Interaction between the bHLH transcription factor FIT with ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between iron acquisition regulation and ethylene signaling in Arabidopsis. Plant Cell 23: 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena C, Waters BM, Romera FJ, García MJ, Morales M, Alcántara E, Pérez-Vicente R. (2006) Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J Exp Bot 57: 4145–4154 [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Kobayashi T, Aung MS, Nakanishi H, Nishizawa NK. (2011) OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol Biol 75: 593–605 [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishizawa NK. (2006) Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot 57: 2867–2878 [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itail RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, Nishizawal NK. (2007) The rice bHLH protein OslRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J 51: 366–377 [DOI] [PubMed] [Google Scholar]
- Park HJ, Ding L, Dai M, Lin R, Wang H. (2008) Multisite phosphorylation of Arabidopsis HFR1 by casein kinase II and a plausible role in regulating its degradation rate. J Biol Chem 283: 23264–23273 [DOI] [PubMed] [Google Scholar]
- Perkins ND. (2006) Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene 25: 6717–6730 [DOI] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. (2009a) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57: 400–412 [DOI] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Kim SA, Cellier F, Thomine S, Guerinot ML, Briat JF, Gaymard F. (2009b) Post-translational regulation of AtFER2 ferritin in response to intracellular iron trafficking during fruit development in Arabidopsis. Mol Plant 2: 1095–1106 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Romera FJ, García MJ, Alcántara E, Pérez-Vicente R. (2011) Latest findings about the interplay of auxin, ethylene and nitric oxide in the regulation of Fe deficiency responses by strategy I plants. Plant Signal Behav 6: 167–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Marschner H. (1986) Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr 9: 695–713 [Google Scholar]
- Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. (1998) The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol 18: 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager KM, Calderon-Villalobos LI, Dohmann EM, Willige BC, Knierer S, Nill C, Schwechheimer C. (2007) Characterization of the VIER F-BOX PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell 19: 1163–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhou Z, Feng S, Li J, Tan-Wilson A, Qu LJ, Wang H, Deng XW. (2009) Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21: 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A, Grinvalds C, Barberon M, Curie C, Vert G. (2011) Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J 66: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vorwieger A, Gryczka C, Czihal A, Douchkov D, Tiedemann J, Mock HP, Jakoby M, Weisshaar B, Saalbach I, Bäumlein H. (2007) Iron assimilation and transcription factor controlled synthesis of riboflavin in plants. Planta 226: 147–158 [DOI] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Baumlein H, Weisshaar B, Bauer P. (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Wu J, Wang C, Zheng L, Wang L, Chen Y, Whelan J, Shou H. (2011) Ethylene is involved in the regulation of iron homeostasis by regulating the expression of iron-acquisition-related genes in Oryza sativa. J Exp Bot 62: 667–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ. (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zheng L, Ying Y, Wang L, Wang F, Whelan J, Shou H. (2010) Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol 10: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]