Abstract
Arbuscular mycorrhizae (AM) are the most widespread symbioses on Earth, promoting nutrient supply of most terrestrial plant species. To unravel gene expression in defined stages of Medicago truncatula root colonization by AM fungi, we here combined genome-wide transcriptome profiling based on whole mycorrhizal roots with real-time reverse transcription-PCR experiments that relied on characteristic cell types obtained via laser microdissection. Our genome-wide approach delivered a core set of 512 genes significantly activated by the two mycorrhizal fungi Glomus intraradices and Glomus mossae. Focusing on 62 of these genes being related to membrane transport, signaling, and transcriptional regulation, we distinguished whether they are activated in arbuscule-containing or the neighboring cortical cells harboring fungal hyphae. In addition, cortical cells from nonmycorrhizal roots served as a reference for gene expression under noncolonized conditions. Our analysis identified 25 novel arbuscule-specific genes and 37 genes expressed both in the arbuscule-containing and the adjacent cortical cells colonized by fungal hyphae. Among the AM-induced genes specifying transcriptional regulators were two members encoding CAAT-box binding transcription factors (CBFs), designated MtCbf1 and MtCbf2. Promoter analyses demonstrated that both genes were already activated by the first physical contact between the symbionts. Subsequently, and corresponding to our cell-type expression patterns, they were progressively up-regulated in those cortical areas colonized by fungal hyphae, including the arbuscule-containing cells. The encoded CBFs thus represent excellent candidates for regulators that mediate a sequential reprogramming of root tissues during the establishment of an AM symbiosis.
Ecto- and endomycorrhizal symbioses between higher plants and soil fungi are the most widespread beneficial plant-microbe interactions on Earth. Mycorrhizae are characterized by the transfer of limiting nutrients, in particular phosphorus and nitrogen, from fungal hyphae to the plant. In return, plants deliver hexoses to the fungi (Nehls et al., 2007), leading to a strongly increased photosynthate allocation to mycorrhizal roots. Apart from direct advantageous effects resulting from improved nutrition, indirect benefits of mycorrhizal interactions are enhanced resistances against abiotic and biotic stress conditions (Smith and Read, 2008).
Some 80% of all terrestrial plants enter an arbuscular mycorrhiza (AM) symbiosis with Glomeromycota fungi (Schüssler et al., 2001). During AM, extraradical hyphae emerging from germinating spores penetrate the rhizodermis via hyphopodia, pass through outer cortical cells, and proliferate in the inner cortex (Parniske, 2008). In Arum-type AM, these intraradical hyphae form highly branched intracellular structures termed arbuscules (Harrison, 1999). It has been shown that arbuscules are transient structures that only operate for a couple of days, indicating a tight control of their development and function (Harrison, 2005). Transfer of phosphorus and other minerals from fungal hyphae to the plant cytoplasm occurs at the periarbuscular interface that comprehends the fungal arbuscular membrane, the periarbuscular matrix, and the plant periarbuscular membrane (Parniske, 2000, 2008). The uptake of phosphorus is energy dependent and requires plant and fungal H+-ATPases (Requena et al., 2003) for an acidification of the periarbuscular space. Based on the localization of reporter proteins, Pumplin and Harrison (2009) demonstrated that different proteins are apparently targeted to specific domains of the periarbuscular membrane, indicating the presence of functional compartments at the symbiotic interface. In addition to AM-specific phosphate transporters essential for symbiotic inorganic phosphate transfer (Harrison et al., 2002; Bucher, 2007; Javot et al., 2007), two half ATP-binding cassette (ABC) transporters have recently been shown to be required for a functional AM symbiosis (Zhang et al., 2010).
An important benefit of legumes as AM models derives from the fact that this genus is able to enter a second beneficial plant-microbe interaction that leads to the development of nitrogen-fixing root nodules (Brewin, 1991). To initiate nodulation, secreted flavonoids first induce rhizobial genes required for the synthesis of lipochitooligosaccharide (LCO) nodulation (Nod) factors. These Nod factors, after perception by plant LysM-domain receptor kinases (Arrighi et al., 2006), activate downstream responses; that way inducing the formation of nodule primordia and mediating bacterial infection (Oldroyd and Downie, 2008). During AM, a similar molecular dialogue is initiated by the host plant via strigolactones that promote the branching of fungal hyphae and activate fungal metabolism (Akiyama et al., 2005; Besserer et al., 2006). Subsequent recognition of AM fungi by the host involves the perception of diffusible Myc signals (Kosuta et al., 2003) including LCOs structurally related to rhizobial Nod factors (Maillet et al., 2011). Together, the diffusible Myc signals prepare root infection by communicating the symbiotic nature of mycorrhizal fungi (Oldroyd et al., 2005; Ercolin and Reinhardt, 2011). Following their entry via hyphopodia, fungal hyphae grow along a prepenetration apparatus (PPA), a cytoplasmic channel formed after the establishment of hyphopodia (Genre et al., 2005, 2008). This coordinated cytological response of epidermal cells indicates that in addition to diffusible Myc signals acting at a distance (Kuhn et al., 2010), there are others that require fungal contact (Kosuta et al., 2003; Kloppholz et al., 2011). It has to be pointed out that signaling also takes place in later stages of the symbiosis; where genes specifically expressed in arbuscule-containing cells have to be activated (Harrison, 2005). One such late signal triggering the induction of a phosphate transporter gene was identified as lyso-phosphatidylcholine (Drissner et al., 2007).
Targeted molecular research on AM suffers from two obstacles: an asynchronous development of the symbiosis leading to the concomitant presence of different stages, and an obligate biotrophy of AM fungi. In the last couple of years, untargeted high-throughput expression profiling was pursued to generate an inventory of AM-induced genes (Liu et al., 2003; Wulf et al., 2003; Frenzel et al., 2005; Hohnjec et al., 2005, 2006; Küster et al., 2007b). These approaches benefited from the identification of two legumes that proved to be excellent AM models: Medicago truncatula (Barker et al., 1990; Rose, 2008) and Lotus japonicus (Handberg and Stougaard, 1992). In case of M. truncatula, current research relies on an advanced genome project (Cannon et al., 2009; Young and Udvardi, 2009), more than 250,000 ESTs in the Dana-Farber Cancer Institute (DFCI) Medicago Gene Index (Quackenbush et al., 2001), and different microarray as well as GeneChip tools (Küster et al., 2004, 2007a; Hohnjec et al., 2005; Lohar et al., 2006; Benedito et al., 2008). Publicly available expression profiles based on EST, microarray, and GeneChip data can be queried using the DFCI Medicago Gene Index (Quackenbush et al., 2001), the Truncatulix (Henckel et al., 2009), and MediPlEx (Henckel et al., 2010) data warehouses as well as the M. truncatula Gene Expression Atlas, the latter exclusively integrating expression profiles generated via GeneChips (Benedito et al., 2008).
Although the transcriptome studies performed so far resulted in an identification of hundreds of AM-related genes (Balestrini and Lanfranco, 2006; Hohnjec et al., 2006), there is limited information on the signaling components activated during the formation of arbuscules and in particular on the transcriptional regulators involved in the reprogramming of root cortical cells toward an accommodation of symbiotic fungi. One obvious reason for this can be seen in the fact that most AM-related expression profiles were based on pooled tissue samples containing a mixture of different cell types and stages of arbuscule development. To overcome this problem, Balestrini et al. (2007) pioneered the use of laser microdissection for the identification of arbuscule-specific phosphate transporter genes in AM roots of tomato (Solanum lycopersicum) via reverse transcription (RT)-PCR. A similar approach was later used in the studies of Gomez et al. (2009) and Guether et al. (2009) to track genes up-regulated in arbuscule-containing cells of M. truncatula and L. japonicus, respectively. On the level of individual genes, detailed in situ expression analyses were performed for selected genes (Küster et al., 2007b), including among others the phosphate transporter gene MtPt4 (Liu et al., 2003), the MtBcp1 gene encoding a blue-copper-binding protein (Hohnjec et al., 2005), and members of the AM-induced lectin gene family (Frenzel et al., 2005). Nevertheless, to our knowledge no promoter of a transcription factor (TF) gene specifically activated in AM roots was investigated so far.
In this study, we intended to sharpen our view on AM-related gene expression by a two-step approach. First, we performed a global transcriptome analysis of M. truncatula roots inoculated with the two widely studied AM fungi Glomus intraradices and Glomus mosseae. By using these two different microsymbionts, we were able to make use of the overlap of gene activation in both AM interactions, that way minimizing strain- or inoculum-related effects. To achieve a genome-wide identification of AM-activated genes, we relied on Medicago GeneChips; reported to cover more than 80% of the gene space in the model legume M. truncatula (Benedito et al., 2008). This marked extension of gene-specific probes significantly advanced earlier microarray-based AM transcriptome studies (Liu et al., 2003; Manthey et al., 2004; Hohnjec et al., 2005), leading to the identification of a core set of 512 M. truncatula genes involved in both AM interactions. In a second step, we intended to shed light on the spatial expression of a subset of AM-related genes. To achieve this goal, we performed cellular expression studies via real-time RT-PCR, using RNA isolated from distinct pools of laser-microdissected cells. In our study, gene expression in M. truncatula arbuscule-containing cells was directly compared with transcription in the adjacent cortical cells colonized by fungal hyphae. As a control, cortical cells from nonmycorrhized roots were used to gain information on gene expression in the absence of a symbiotic interaction. With an emphasis on genes encoding membrane transporters, signaling-related proteins, and transcriptional regulators, this approach identified novel components of the cell-specific program orchestrating AM symbiosis. In total, we identified 25 arbuscule-specific genes, while 37 genes were activated both in arbuscule-containing and in the neighboring cells. Together, these results highlight general mechanisms underlying fungal colonization up to the formation of arbuscules. Among the transcriptional regulators, we identified two highly similar genes encoding CAAT-box binding TFs (CBFs), which we analyzed in more detail via the expression of promotor-GUS fusions in transgenic roots. Remarkably, both genes are already activated by the initial physical contact between fungal hyphae and the plant epidermis and are expressed concomitantly with fungal colonization of the root cortex up to the formation of arbuscules, making the encoded CBFs excellent candidates for regulators mediating the sequential reprogramming of root tissues during the establishment of an AM symbiosis.
RESULTS AND DISCUSSION
Genome-Wide Transcriptome Profiling Identifies a Core Set of 512 M. truncatula Genes Related to Root Colonization by AM Fungi
M. truncatula roots colonized with either G. intraradices or G. mosseae under conditions of phosphate limitation (20 μm phosphate) were stained for fungal structures 28 d post inoculation (dpi; McGonigle et al., 1990). To minimize dilution by noncolonized regions, we selected roots with a similarly high root length colonization (RLC; 60%–80%) and high arbuscule frequencies (60%–75%) for the isolation of total RNA. Samples were checked for AM (MtPt4) and nodule marker (MtEnod18) gene expression via RT-PCR, as described by Hohnjec et al. (2005). Since some AM-induced genes can be activated by phosphate, we incorporated a study of gene expression in nonmycorrhizal roots grown for 28 d in the presence of a high (2 mm) phosphate concentration. As a common control, nonmycorrhizal roots grown for 28 d under phosphate limitation (20 μm phosphate) were harvested.
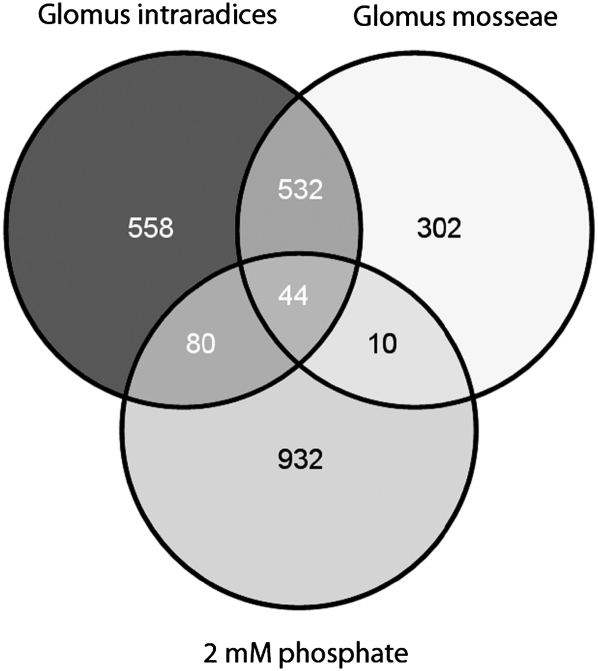
Total RNA isolated from three biological replicates of mycorrhizal and nonmycorrhizal roots was used for GeneChip hybridizations. In our study, each biological replicate is defined as a pool comprising five root systems. The complete dataset is available from the Gene Expression Omnibus (accession no. GSE32208) and is also included as Supplemental Table S1. Applying a 2-fold induction at a false discovery rate (FDR)-corrected P value of P < 0.05 as a cutoff, we found that 1,214 and 888 genes were up-regulated in G. intraradices- and G. mosseae-colonized roots, respectively, whereas 576 genes were coactivated in both AM interactions (Fig. 1; Supplemental Table S2).
Figure 1.
Transcriptional response of M. truncatula roots to a colonization with different AM fungi and to a treatment with 2 mm phosphate. M. truncatula roots were inoculated with G. intraradices and G. mosseae for 28 d under conditions of phosphate limitation (20 μm phosphate). Alternatively, roots were grown for 28 d in the presence of 2 mm phosphate. Genes significantly up-regulated 2-fold at an FDR-corrected P value of P < 0.05 in relation to control roots grown under conditions of phosphate limitation were compared to identify coregulation of expression. Numbers indicate genes activated in different conditions. Diagrams were drawn using Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Of the 576 AM-induced genes, 44 were also activated at the cutoffs mentioned above in roots treated with 2 mm phosphate, leaving a core set of 532 genes significantly induced by two different AM fungi but not via elevated phosphate levels (Fig. 1; Supplemental Table S2). The low overlap between phosphate-induced gene expression and transcriptional activation by AM fungal colonization is in line with reports of Liu et al. (2003) and Hohnjec et al. (2005), illustrating that phosphate supply is unable to mimic AM-induced gene expression.
Our core set of 532 AM-related genes is supported by a search in the MediPlEx database (Henckel et al., 2010), combining in silico gene expression in AM EST collections from the DFCI Medicago Gene Index (libraries #ARE, #ARB, T1682, #GFS, 5520, #9CR) with the GeneChip hybridizations detailed above. This search returned 91 genes of the core set as AM induced with an R value larger than 2 (Stekel et al., 2000; Supplemental Table S2). Moreover, our core set contained 51 genes reported as coinduced in the same two AM interactions when relying on oligonucleotide microarrays (Hohnjec et al., 2005; Supplemental Table S2), whereas it encompassed 346 genes reported as activated at least 2-fold by G. intraradices under different experimental conditions on the basis of Medicago GeneChips (Gomez et al., 2009; Supplemental Table S2).
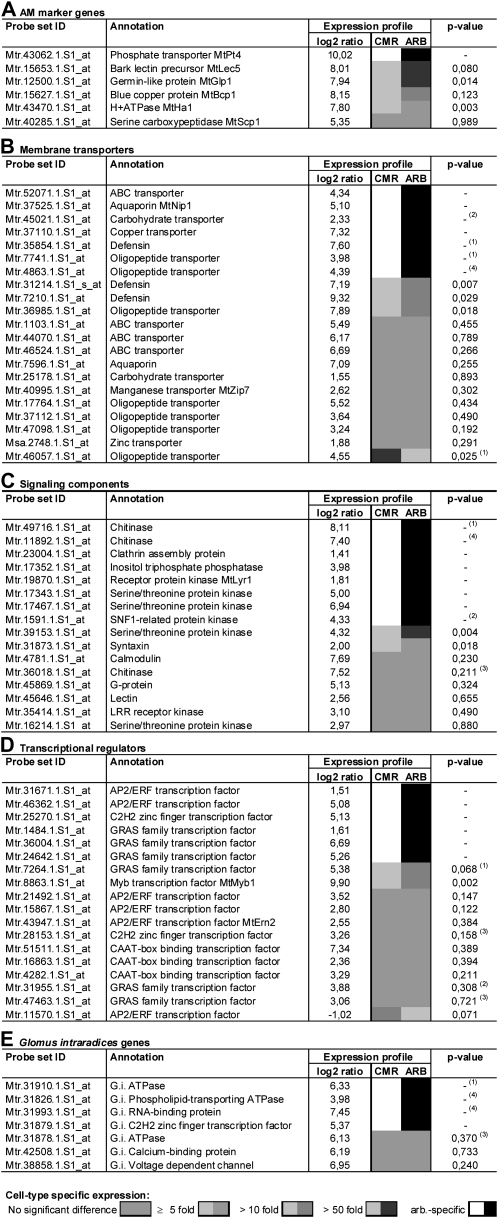
Among the core set of AM-related genes, 15 well-known AM expression markers (Hohnjec et al., 2006; Küster et al., 2007b) were identified as strongly induced in our conditions (Table I). As an example, the MtPt4 gene encoding a phosphate transporter known to be required for an efficient AM symbiosis (Javot et al., 2007) was activated more than 1,000 (log2 = 10.02)- and 400 (log2 = 8.65)-fold in G. intraradices- and G. mosseae-colonized roots, respectively. This very strong induction of AM-related marker genes (Table I) indicates that our approach should allow an identification also of those genes expressed either transiently or activated less strongly during arbuscule development.
Table I. M. truncatula AM marker genes activated in roots colonized with G. intraradices and G. mosseae.
Probe IDs of Medicago GeneChips are referenced to the corresponding DFCI M. truncatula Gene Index IDs (release 10) and to M. truncatula gene names from the literature. Log2 ratios of gene expression for G. intraradices-colonized (Gi-Myc), G. mosseae-colonized (Gm-Myc), and 2 mm phosphate-treated (2 mm-P) roots, all measured against roots grown at 20 μm phosphate, are given. Whereas all 15 AM marker genes are significantly up-regulated in the AM roots used for expression profiling, none of them is activated in roots treated with 2 mm phosphate. FDR-corrected P values (FDR-P) are indicated. References for the AM-induced genes identified are as follows: MtPt4 (Javot et al., 2007), MtMyb1 (Liu et al., 2003), MtTi1 (Grunwald et al., 2004), MtLec7 (Frenzel et al., 2005), MtGst1 (Wulf et al., 2003), MtBcp1 (Hohnjec et al., 2005), MtLec5 (Frenzel et al., 2005), MtGlp1 (Doll et al., 2003), MtHa1 (Krajinski et al., 2002), MtScp1 (Liu et al., 2003), MtVapyrin (Pumplin et al., 2010), MtSbtM1 (Takeda et al., 2011), MtZip7 (Burleigh et al., 2003), MtAnn2 (Manthey et al., 2004), and MtDxs2 (Floss et al., 2008).
| GeneChip Probe ID | DFCI 10 ID | Gene | Gi-Myc | FDR-P | Gm-Myc | FDR-P | 2 mm-P | FDR-P |
| Mtr.43062.1.S1_at | TC142142 | MtPt4 | 10.02 | 2.7E-10 | 8.65 | 1.0E-09 | −0.38 | 5.1E-01 |
| Mtr.8863.1.S1_at | TC146022 | MtMyb1 | 9.90 | 6.7E-10 | 8.99 | 2.3E-09 | 0.21 | 8.1E-01 |
| Mtr.16454.1.S1_at | TC152603 | MtTi1 | 9.71 | 5.0E-11 | 8.41 | 1.7E-10 | −0.04 | 9.6E-01 |
| Mtr.45648.1.S1_at | TC143767 | MtLec7 | 9.02 | 2.7E-10 | 8.30 | 8.7E-10 | 0.40 | 4.3E-01 |
| Mtr.15957.1.S1_at | TC166174 | MtGst1 | 8.98 | 1.7E-09 | 8.13 | 6.6E-09 | −0.14 | 8.9E-01 |
| Mtr.15627.1.S1_at | TC159695 | MtBcp1 | 8.15 | 5.1E-09 | 6.63 | 6.8E-08 | −0.19 | 8.5E-01 |
| Mtr.15653.1.S1_at | TC143161 | MtLec5 | 8.01 | 8.9E-10 | 7.64 | 2.0E-09 | −0.08 | 9.3E-01 |
| Mtr.12500.1.S1_at | TC153539 | MtGlp1 | 7.94 | 5.1E-10 | 7.80 | 8.8E-10 | −0.39 | 4.2E-01 |
| Mtr.43470.1.S1_at | TC141883 | MtHa1 | 7.80 | 8.3E-09 | 6.80 | 5.7E-08 | 0.11 | 9.2E-01 |
| Mtr.40285.1.S1_at | TC143816 | MtScp1 | 5.35 | 6.7E-08 | 5.16 | 1.9E-07 | −3.51 | 7.7E-06 |
| Mtr.39050.1.S1_at | TC149084 | MtVapyrin | 4.89 | 1.4E-06 | 3.77 | 3.2E-05 | −0.59 | 4.2E-01 |
| Mtr.32129.1.S1_at | AW584611 | MtSbtM1 | 4.80 | 2.2E-06 | 3.61 | 6.1E-05 | 0.38 | 6.7E-01 |
| Mtr.40995.1.S1_at | TC145398 | MtZip7 | 2.62 | 4.2E-06 | 2.29 | 2.7E-05 | 0.38 | 3.6E-01 |
| Mtr.20364.1.S1_at | TC153713 | MtAnn2 | 2.42 | 1.9E-02 | 1.53 | 3.2E-02 | 0.46 | 7.3E-01 |
| Mtr.43585.1.S1_at | TC141952 | MtDxs2 | 1.57 | 4.7E-04 | 1.87 | 2.1E-04 | −4.26 | 2.8E-07 |
Via comparisons of Medicago GeneChip probes to currently available G. intraradices sequences (Tisserant et al., 2011), 20 Glomus genes were identified among the set of 532 genes coinduced in both AM interactions (Supplemental Table S2). The remaining 512 AM-related M. truncatula genes were grouped into functional categories, based on automated annotations of the encoded gene products via the Sequence Analysis and Management System software (Bekel et al., 2009) and Gene Ontology classifications (http://www.medicago.org). In addition, MapMan (Usadel et al., 2005) was used to visualize gene expression profiles and to identify functional categories expressed most significantly different from others.
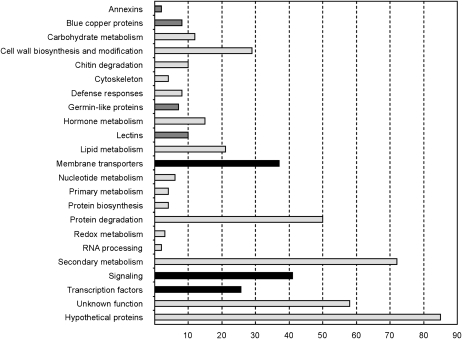
In Figure 2, the distribution of AM-coinduced genes into functional categories is shown. Typical AM-related gene families of unknown biological function encoding annexins (e.g. MtAnn2; Manthey et al., 2004), blue-copper proteins (e.g. MtBcp1; Hohnjec et al., 2005), germin-like proteins (e.g. MtGlp1; Doll et al., 2003), and lectins (e.g. MtLec5 and MtLec7; Frenzel et al., 2005) are classified separately. For all these gene families except the one encoding annexins, comparisons to the M. truncatula Gene Expression Atlas (He et al., 2009) revealed that their members were expressed either specifically or were almost exclusively activated in AM roots in comparison to all other tissues analyzed (data not shown), that way widening our knowledge on previously unknown AM-related family members.
Figure 2.
Cellular functions of M. truncatula genes activated in mycorrhizal roots. All 512 M. truncatula genes coinduced at least 2-fold at an FDR-corrected P value of P < 0.05 in response to the AM fungi G. intraradices and G. mosseae that were not induced by a treatment with 2 mm phosphate (Supplemental Table S2) were grouped into functional categories. The number of genes allocated to each functional category is indicated. The bars are colored as follows: black, functional categories studied by laser microdissection; dark gray, AM-related gene families; light gray, other functional categories.
In addition to these gene families, the following functional categories were most prominent (Fig. 2; Supplemental Table S2): (1) cell wall biosynthesis and modification, including several enzymes related to a remodeling of the extracellular matrix; (2) chitin degradation, including several chitinases that might be involved in the disassembly of fungal structures; (3) protein degradation, including a range of different proteinases and peptidases that can be connected to the dynamic turnover of mycorrhizal structures; (4) hormone metabolism, including several genes involved in gibberellin biosynthesis; and (5) secondary metabolism, including a high number of cytochromes, components of the carotenoid metabolism, and UDP-sugar transferases. The activation of these functions is in line with previous reports on gene expression in AM roots (Liu et al., 2003; Wulf et al., 2003; Hohnjec et al., 2006; Küster et al., 2007b; Guether et al., 2009).
Interestingly, the categories membrane transporters, signaling, and TFs were not only prominent but their members were also expressed most significantly different from others in AM roots (data not shown), based on statistical analyses implemented in MapMan (Usadel et al., 2005). In addition, many of these candidate genes either displayed a mycorrhiza-specific or mycorrhiza-enhanced expression in AM roots according to the Medicago Gene Expression Atlas (He et al., 2009), suggesting a relevance for AM interactions. Remarkably, these analyses also returned six TF genes coinduced in our conditions as being specifically expressed in AM tissues (Supplemental Fig. S1).
Since the three cellular functions mentioned are particularly relevant for coordinating the reprogramming of root cortical cells toward an accomodation of fungal structures, we investigated the cell-type expression of 71 candidate genes selected from these categories via laser microdissection.
Longitudinal Sections Allow an Accurate Separation of Specific Cell Types from AM Roots via Laser Microdissection
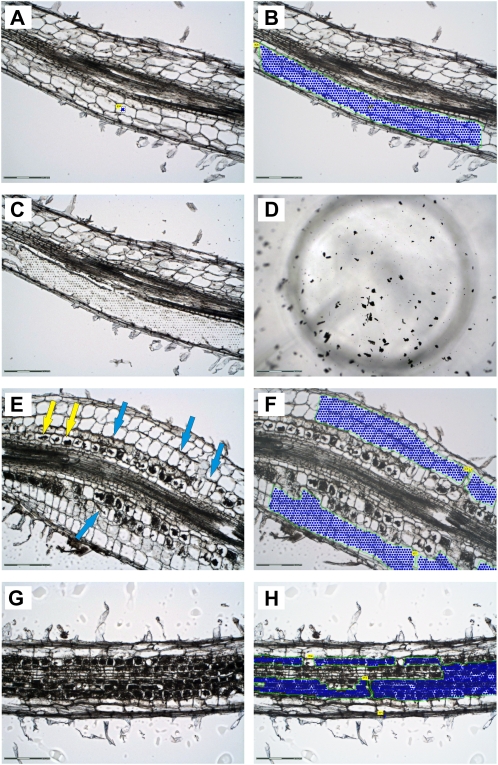
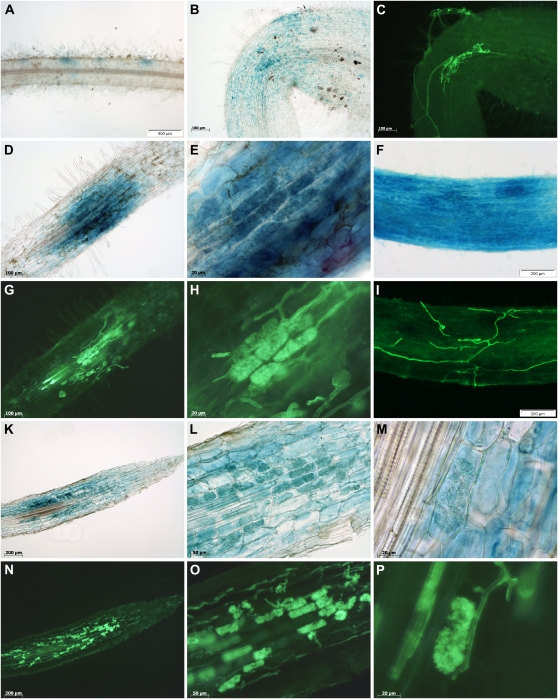
We used laser microdissection to obtain specific pools of three different cell types in four biological replicates: cortical cells from noncolonized control roots (CCR), cortical cells from mycorrhizal roots (CMR), and cortical cells containing arbuscules (ARB; Fig. 3). We found that embedding of roots in Steedman’s wax (Gomez et al., 2009) preserves root morphology, allowing an identification of vascular tissues, cortical cells, and the epidermal cell layer including root hairs (Fig. 3, A–C). In contrast to transverse sections (Balestrini et al., 2007; Gomez et al., 2009), our longitudinal sections offered the possibility to evaluate the colonization status of complete root regions. Chains of arbuscules in different developmental stages and fungal hyphae growing in the extracellular space of adjacent cortical cells were clearly visible (Fig. 3, E–H). This facilitated cell harvest and allowed us to focus not only on mature arbuscules filling the complete cell lumen (ARB samples; Fig. 3, D, G, H), but also on cortical cells interspersed with fungal hyphae in the immediate neighborhood of arbuscule-containing cells (CMR samples; Fig. 3, E and F). Since each cell pool from the CMR cell type differs in the density and growth of fungal hyphae, the CMR samples are expected to display the strongest variation of transcriptional changes.
Figure 3.
Laser microdissection of three specific cell types from M. trunctula roots. Root areas designated for cell harvest are marked with a green line and blue dots. Along the line the laser dissects the cells from the surrounding tissue, while dots represent single catapulting events. A to C, Longitudinal section of a nonmycorrhized root used for the collection of cortical cells from control roots (CCR). A and B, Section before and after selection of CCR for laser microdissection. C, Section after laser microdissection of CCR. D, View into the collection tube showing typical flakes of harvested cells (in this case arbuscule-containing cells). E to H, Longitudinal sections of mycorrhized roots displaying chains of arbuscules at different developmental stages and fungal hyphae growing in the apoplast of outer cortical cells. E and F, Section before and after selection of cortical cells from mycorrhized roots (CMR). The harvested area was extended to inner cortical cells in case no arbuscules were visible in these cells. Fungal hyphae are present in the apoplast (blue arrows). G and H, Section before and after selection of arbuscule-containing cells (ARB). Only cells harboring mature arbuscules filling up the whole lumen were harvested. These cells could be easily distinguished from those containing young or severely degraded arbuscules (yellow arrows in E). Scale bars represent 300 μm for D and 150 μm for all other sections.
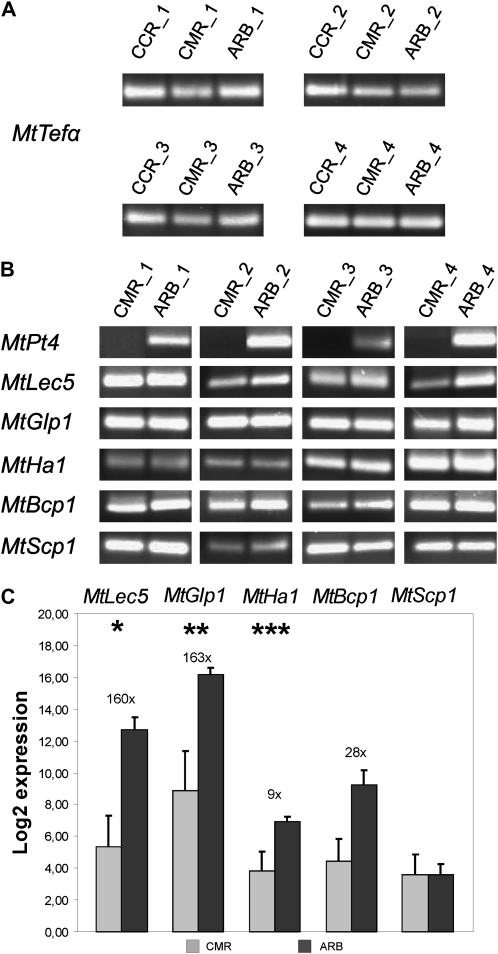
To assess the suitability of the collected samples and to check in particular for cross-contamination between CMR and ARB samples, the expression of the six AM marker genes MtPt4, MtBcp1, MtScp1, MtLec5, MtGlp1, and MtHa1 (Harrison et al., 2002; Krajinski et al., 2002; Doll et al., 2003; Liu et al., 2003; Frenzel et al., 2005; Hohnjec et al., 2005) was measured in all four biological replicates. Whereas transcripts of the MtTefα gene used as a constitutive control were amplified from all three cell types (Fig. 4A), transcripts of the six AM-specific genes tested were not detected in CCR samples, as could be expected (data not shown).
Figure 4.
Detection of AM marker gene transcripts in laser-microdissected cell types. Marker gene expression was measured by real-time RT-PCR in four biological replicates of three different cell types: cortical cells from nonmycorrhizal control roots (CCR), cortical cells from mycorrhizal roots containing fungal hyphae (CMR), and arbuscule-containing cells (ARB). A and B, Gel electrophoresis of the final real-time RT-PCR amplification products representing the control gene MtTefα and six AM marker genes. All amplified fragments had the correct sizes. Note that MtPT4 transcripts were not detected in CMR. C, Real-time RT-PCR measurement of five AM marker genes induced in ARB in comparison to CMR. Expression values are displayed as log2 mean values of all four biological replicates. Numbers and bars represent fold induction and ses, respectively. Asterisks indicate significance levels of a Student’s t test on the expression values in the two different cell types: * = P < 0.1; ** = P < 0.05; *** = P < 0.005. MtTefα, Transcriptional elongation factor α; MtPT4, phosphate transporter 4; MtBcp1, blue copper protein 1; MtScp1, Ser carboxypeptidase 1; MtLec5, lectin 5; MtGlp1, germin-like protein 1; MtHa1, H+-ATPase 1.
The phosphate transporter gene MtPt4 was used as an expression marker for cross-contamination between the CMR and ARB samples, since this gene is exclusively transcribed in cells containing arbuscules (Harrison et al., 2002). The detection of MtPt4 transcripts in all ARB in contrast to their absence in all CMR samples (Fig. 4B) indicates that no significant cross-contamination of CMR samples with material from arbuscule-containing cells occurred. In contrast to MtPt4, transcripts of the other five genes tested could be amplified both in CMR and ARB samples (Fig. 4B). Of these, the Ser carboxypeptidase gene MtScp1 was the only one expressed at similar levels in both cell types, whereas the MtBcp1 gene encoding a blue-copper protein, the lectin gene MtLec5, the MtGlp1 gene specifying a germin-like protein, and the H+-ATPase gene MtHa1 displayed an up to 160-fold induction in ARB cells (Fig. 4C). While the results obtained for MtScp1 and MtBcp1 are in accordance with promoter studies indicating expression outside of arbuscule-containing cells (Liu et al., 2003; Hohnjec et al., 2005), MtLec5, MtGlp1, and MtHa1 have so far been reported to be arbuscule specific (Krajinski et al., 2002; Doll et al., 2003; Frenzel et al., 2005). This deviance might be due to a higher sensitivity of our PCR-based method in comparison to the in situ expression methods used in the cited studies.
Together, our results demonstrate that the total RNA prepared from all three cell types was suitable for identifying arbuscule-specific as well as arbuscule-enhanced expression patterns, even if the harvested cell types were collected in close proximity.
Laser Microdissection Identifies Novel Arbuscule-Specific Genes and Genes Being Generally Expressed in Cortical Cells Colonized by AM Fungi
Expression of 71 candidate genes was measured by one-step real-time RT-PCR in at least three biological replicates. In these experiments, nine genes could not be amplified reproducibly from any cell type and were not considered further. Figure 5 gives an overview of the remaining 62 genes and their expression patterns in the laser-microdissected samples. Except for three genes encoding an ABC transporter (Mtr.44070.1.S1_at), a calcium-binding protein (Mtr.4781.1.S1_at), and an AP2/ERF TF (Mtr.11570.1.S1_at), none of these genes could be amplified reproducibly from CCR cells (data not shown), suggesting that they are either truly mycorrhiza specific or are expressed in other cell types than the ones investigated here, e.g. the vascular tissue.
Figure 5.
Cell-type-specific expression of genes activated in mycorrhizal roots. Real-time RT-PCR measurement of gene expression in three biological replicates of the laser-microdissected cell types CMR (cortical cells from mycorrhized roots) and ARB (arbuscule-containing cells). Differences in transcription are indicated by different shades of gray (legend see below). P values represent significance levels of a Student’s t test on the expression values in the two different cell types. In addition, the log2 expression ratios of gene expression in roots mycorrhized with G. intraradices versus nonmycorrhizal roots (Supplemental Table S1) are shown. Footnotes: (1): One of the three biological replicates was replaced by replicate four, to obtain three gene-specific PCR products of the correct size or a consistent expression pattern. (2): A gene-specific PCR product could only be obtained for two out of three biological replicates of ARB. (3): A gene-specific PCR product could only be obtained for two out of three biological replicates of CMR. (4): A gene-specific PCR product was obtained for only one out of three biological replicates of CMR.
Among the 62 genes analyzed for cell-specific expression, 21 displayed a specific activation in the arbuscule-containing cells, including six membrane transporter genes, seven signaling-related genes, six genes encoding TFs, and two fungal genes (Fig. 5; Supplemental Fig. S2). In addition, four genes were detected in all ARB, but in only one out of three CMR samples, indicating that they are expressed at a very low level in the latter cell type. Six genes specifically expressed in arbuscule-containing cells were described to be activated in these cells before (Gomez et al., 2009), but none of them was so-far known to be restricted to this cell type within mycorrhizal roots.
Interestingly, the majority of the genes investigated was found to be expressed both in arbuscule-containing cells (ARB) and in the adjacent cortical cells being colonized by fungal hyphae at different levels (CMR). Here, transcripts of 37 genes, including 14 membrane transporter genes, eight signaling-related genes, 12 TF genes, and three fungal genes were detected. Of these 37 genes, 28 were expressed at comparable levels in both cell types, whereas nine were significantly induced in either CMR or ARB (Fig. 6). So far, only four genes of this group were known to be expressed in arbuscule-containing cells (Gomez et al., 2009), but no information was available on their activity in the adjacent cells being connected to hyphal growth.
Figure 6.
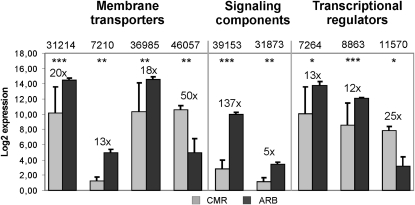
Genes differentially expressed in cortical and arbuscule-containing cells. Real-time RT-PCR measurement of the expression of selected genes classified as CMR or ARB induced in Figure 5. Gene expression is displayed as the log2 mean value of three biological replicates. Numbers and bars represent fold-induction and ses, respectively. Different genes are labeled with abbreviated GeneChip probe IDs (compare Fig. 5). Asterisks indicate significance levels of a Student’s t test on expression values in the two different cell types: * = P < 0.1; ** = P < 0.05; *** = P < 0.005.
In total, cell-specific expression patterns were identified for 62 AM-related genes. In the following, our results are discussed in detail with respect to the functional classification of the genes.
Glomus Intraradices Genes
Seven AM-related genes of fungal origin were selected for cell-specific transcriptome analysis. These genes encoded five membrane transporters, one calcium-binding protein, and two TFs (Fig. 5). Interestingly, our analysis of the expression patterns in the specific cell pools revealed that four genes seemed to be specifically expressed in arbuscules, indicating that the distribution of fungal transcripts can be restricted to these, irrespective of the coenocytic nature of the microsymbiont.
The genes investigated included three members specifying ATPases, two of which represented cytoplasmatic ATPases probably associated with proteasomes and thus being involved in protein degradation, whereas the third one was predicted to be membrane localized and involved in the transport of phospholipids or cations (Fig. 5). While two of the ATPase genes showed a specific expression in arbuscules, one was induced at comparable levels in both CMR and ARB samples. In addition, a gene specifying a voltage-dependent channel protein was also activated in both cell types, similar to a signaling-related gene encoding a calcium-binding protein (Fig. 5).
Interestingly, the two fungal genes related to transcriptional regulation, encoding a zinc finger and an RNA-binding protein, showed specific expression in ARB (Fig. 5). These genes might therefore control the transcription of fungal genes specifically required during arbuscule formation or function.
M. truncatula Genes Encoding Membrane Transporters
A functional AM symbiosis is characterized by the exchange of nutrients between the micro- and macrosymbionts. It is thus not surprising that 37 M. truncatula genes encoding membrane transporters were coactivated in the two AM interactions studied (Supplemental Table S3), including 13 genes that Benedito et al. (2010) identified previously on the basis of expression data reported by Gomez et al. (2009). Apart from the AM marker genes MtPt4 and MtHa1 already discussed above as well as the mycorrhiza-induced aquaporin gene MtNip1 (Uehlein et al., 2007), cellular expression patterns were determined for 20 genes of this functional category (Fig. 5).
The largest group comprised genes encoding oligopeptide transporters of the H+/oligopeptide symporter type. Eleven members of this class were coinduced in both AM interactions and seven of them were examined in the laser-microdissected cell pools (Fig. 5). Proton-dependent oligopeptide transporters (POTs) are reported to be involved in the uptake of small peptides into eukaryotic cells (Paulsen and Skurray, 1994). Possible functions are thus connected to an intake of degraded fungal proteins subsequent to the action of AM-activated proteases (Supplemental Table S2), alternatively the uptake of fungal effector peptides, such as the recently reported SP7 protein (Kloppholz et al., 2011; Plett et al., 2011), is an intriguing possibility. The wide range of possible POT functions is mirrored by cellular expression patterns, since we found two arbuscule-specific POT genes (Fig. 5), three POT genes equally expressed in CMR and ARB cells (Fig. 5), and one POT gene being induced in each cell type (Figs. 5 and 6).
A second prominent group of AM-related genes encoded five ABC transporters and three aquaporins. From this collection, we identified one ABC-transporter gene as ARB specific (Fig. 5) and three more as being expressed with no significant difference between CMR and ARB (Fig. 5). With MtStr and MtStr2, two M. truncatula ABC transporter genes were recently reported to be important for AM (Zhang et al., 2010). Both genes belong to the G subfamily of ABC transporters and are specifically expressed in arbuscule-containing cells. The ABC transporter identified as arbuscule specific in our study belongs to the same subfamily, suggesting a similar function. Zhang et al. (2010) speculated that strigolactones might be a substrate for these transporters, inducing the strong ramification of fungal hyphae that leads to arbuscule formation, which would explain the cell-specific expression. In contrast, the three ABC-transporter genes we found expressed in arbuscule-containing and adjacent cortical cells specify P-glycoprotein multidrug resistance transporters of the ABCB subfamily. Since several members of this group are known to transport auxins (Geisler and Murphy, 2006), a role for these might be the fine tuning of auxin distribution in colonized root cells.
In the group of aquaporins, we investigated the expression pattern of two Nod 26-like intrinsic proteins (NIP) genes including MtNip1, which was described as AM induced by Uehlein et al. (2007) and was also strongly AM induced here. Due to the fact that this gene is slightly induced by phosphate, it was not included in our core set of 512 AM-related genes. In line with the results of Gomez et al. (2009), we found MtNip1 to be activated in ARB cells. In addition, we could show here that this gene is expressed exclusively in this cell type, whereas the second NIP gene was expressed in the surrounding, hyphae-containing cortical cells as well (Fig. 5). Interestingly, MtNip1 and other NIPs did not facilitate water uptake in heterologous expression systems, but acted as low-affinity transport system for ammonium (Uehlein et al., 2007), which is the main form in which nitrogen is supplied to the plant by AM fungi (Govindarajulu et al., 2005). These observations support the idea that nitrogen is provided by the microsymbiont via arbuscules as well as fungal hyphae growing in the apoplast of cortical cells, and the two NIPs might be involved in uptake of these nutrients into host cells.
Strikingly, the most strongly induced transporter genes in our experiment encoded three defensins. These seem to be of special importance for arbuscule-containing cells, since one defensin gene was exclusively expressed (Fig. 5) and the other two were significantly up-regulated in ARB (Figs. 5 and 6). Many members of the defensin gene family are activated in plants during defense reactions in response to diverse pathogens (Thomma et al., 2002), including harmful fungi. Although the activation of plant defense responses has been reported for the initial stages of mycorrhizal colonization, they seem to be effectively controlled and down-regulated when the symbiosis is completely established (Harrison and Dixon, 1993). Interestingly, some defensins are known to reduce the elongation of fungal hyphae and to cause strong hyphal branching (Broekaert et al., 1995). Therefore, those defensins specifically activated during a mature mycorrhiza may in fact influence hyphal ramification during early arbuscule development or control arbuscule lifespan.
The remaining membrane transporter genes we found to be up-regulated during AM are mainly involved in the transfer of micronutrients, macronutrients such as nitrate, or carbohydrates. Concerning ion transporters of the MtZIP family (Burleigh et al., 2003), we identified a gene encoding the manganese transporter MtZip7 (López-Millán et al., 2004). We found the MtZip7 gene and a zinc transporter gene to be expressed in CMR and ARB cells alike (Fig. 5), whereas a copper transporter gene was found to be ARB specific (Fig. 5). Since AM fungi probably improve micronutrient supply of host plants (Clark and Zeto, 2000; Parniske, 2008), the three transporter genes might be activated by the plant to enhance uptake of these elements from the microsymbiont. The fact that we found two of the transporter genes mentioned to be expressed in both arbuscule-containing and surrounding hyphae-containing cortical cells supports the assumption that an exchange of nutrients by the symbiotic partners or at least an uptake by the plant is not restricted to arbuscules.
With respect to sugar allocation in AM roots, we analyzed the expression patterns of two carbohydrate transporters. Due to the fact that hexoses derived from the plant metabolism are supplied to mycorrhizal fungi, AM roots represent strong carbon sinks. Whereas hexoses generated from Suc via the activity of apoplastic invertases are directly available to the fungus (Schaarschmidt et al., 2007), hexoses provided by cytoplasmic invertases or Suc synthases (Hohnjec et al., 1999) first have to be exported to the extracellular space (Baier et al., 2010). Up to now, little is known about the carbohydrate transporters involved in the translocation of symplastic hexoses to the apoplastic interface. The two genes investigated here are candidates for this function, but showed no consistent expression. Since only one was ARB specific (Fig. 5), Suc transfer is probably not limited to arbuscules, which is consistent with the expression of genes encoding the cytoplasmatic Suc-cleaving enzyme MtSucS1 (Hohnjec et al., 2003) and the hexose transporter Mtst1 (Harrison, 1996).
M. truncatula Genes Related to Signaling
In Supplemental Table S4, the expression characteristics of 41 genes encoding signaling-related components induced in both AM interactions are summarized. A large group consists of genes probably involved in intracellular signal transduction, including 13 genes encoding protein kinases, a phosphatase, and a protein phosphatase inhibitor. Furthermore, genes specifying two inositol polyphosphate phosphatases, a phosphatidylinositol transfer protein, a calmodulin-binding protein, two Rop guanine nucleotide exchange factors, and a G-protein deserve attention. Together, they might represent those members of large gene families that mediate calcium, phosphoinosite, or G-protein signaling in AM roots. Seven genes from this group and an additional calmodulin gene only induced by G. intraradices were investigated for their cellular expression pattern (Fig. 5). Interestingly, we found genes encoding three protein kinases (two Ser/Thr kinases and a SNF1-related kinase) and one inositol triphosphate phosphatase to be exclusively expressed in ARB (Fig. 5). To our knowledge, these are the first genes reported to be involved in internal signaling processes that are specifically up-regulated in arbuscule-containing in comparison to the neighboring cells. Another Ser/Thr protein kinase gene was strongly induced in ARB (Figs. 5 and 6), whereas a calmodulin gene, a G-protein gene, and one additional Ser/Thr protein kinase gene were expressed at equal levels in ARB and CMR (Fig. 5), hinting that they are involved in signal processes occurring in cortical cells of colonized root areas in general.
A second group of AM-activated, signaling-related genes encodes components associated with vesicle-mediated transport, including a syntaxin, a clathrin assembly protein, a GTP-binding protein, and a basic secretory protein. It is tempting to speculate that these proteins are involved in the membrane biogenesis associated with an intracellular accomodation of fungal structures. The expression pattern of the two genes we analyzed in the specific cell pools is in line with the assumption that in particular the arbuscule-containing cells are places of intensive membrane buildup and turnover, since a gene encoding a clathrin assembly protein was exclusively detected in ARB (Fig. 5) and a syntaxin gene was found to be ARB induced (Figs. 5 and 6).
Three chitinase genes strongly induced in both AM interactions were included in the analysis of signaling-related genes (Fig. 5). Chitinases hydrolyze β-1,4-glycosidic bonds (Salzer et al., 2000) and are mostly regarded as part of plant defense mechanisms (Arlorio et al., 1992). Although several chitinase genes are up-regulated during early phases of nodulation and mycorrhization, this activation is transient and they are in general not regarded to play a role during later stages. Nevertheless, several members of the class III chitinase gene family are activated specifically during AM (Salzer et al., 2000), and for one of these genes an arbuscule-specific localization was found by in situ hybridization (Bonanomi et al., 2001). In this study, we investigated three members of the chitinase gene family and found two of them only in ARB (Fig. 5), whereas the third one was active in CMR and ARB (Fig. 5). The predominant ARB expression of chitinase genes might thus support the formation of functional symbiotic interfaces by reducing the amount of chitinous elicitors.
The largest group of signaling-related genes encompassed 15 genes encoding receptor kinases with a predicted membrane localization that presumably perceive extracellular AM-related signals. From this group, we investigated genes encoding a leucin-rich repeat receptor kinase and the LysM receptor kinase MtLyr1. In addition, a gene encoding a membrane-bound lectin with predicted kinase activity was analyzed (Fig. 5). Whereas the lectin gene and the leucin-rich repeat receptor kinase gene were equally expressed in both cell types studied, MtLyr1 was specifically expressed in ARB cells (Fig. 5). In M. truncatula, the two LysM receptor kinases MtNfp, representing the Lyr type, and MtLyk3, representing the Lyk type of this family, were identified as Nod-factor receptors (Gough and Cullimore, 2011). MtNfp also plays a role during early AM interactions, since it is essential for the perception of the recently characterized Myc-LCO signals (Maillet et al., 2011). Since MtNfp is not essential for the establishment of an AM (Amor et al., 2003), Maillet et al. (2011) speculated that another receptor active at higher Myc-LCO concentrations must exist. These could be achieved during arbuscule formation, as a result of a tighter contact between the two symbiotic partners during infection of cortical cells. Interestingly, it was shown recently that in the nonlegume Parasponia, an ortholog of MtNfp named PaNfp is essential for the symbioses with nitrogen-fixing bacteria as well as AM fungi (Op den Camp et al., 2011). In contrast to legume plants, the Parasponia genome only contains a single Lyr-type receptor kinase gene, whereas legume plants contain two copies (Op den Camp et al., 2011). It was speculated that during evolution of legume plants and their ability to form nodules, duplication of the initial Lyr gene occurred and one copy became the receptor for rhizobial LCOs, whereas the other one is targeted by Myc LCOs. The closest relative of MtNfp is MtLyr1, which could thus represent a potential receptor for Myc LCOs (Op den Camp et al., 2011). We could show here that MtLyr1 is exclusively expressed in ARB and not in the surrounding cortical cells from mycorrhized roots. This fits well to the phenotype of RNAi knockdowns of the more ancient PaNfp gene, since here the AM symbiosis is aborted at the point of arbuscule formation. Our finding that MtLyr1 is exclusively expressed in arbuscule-containing cells supports the fact that the encoded receptor is needed for this step, possibly to perceive Myc LCOs secreted during later stages of AM.
M. truncatula Genes Encoding Transcriptional Regulators
In total, 25 genes encoding TFs were identified as coinduced in AM roots (Table II), comprising the families AP2/ERF, Z-C2H2, CAAT-box binding, GRAS, MYB, WRKY, and NAC. Of these, only five GRAS-TF genes and one gene encoding a Myb-TF (designated MtMyb1 in Table I) were previously reported to be specifically activated in roots colonized with Glomus spp. (Liu et al., 2003; Gomez et al., 2009), while a few members of other gene families were reported to be of relevance in the root nodule symbiosis.
Table II. Overview of 25 M. truncatula AM-induced genes encoding transcriptional regulators.
Probe IDs of Medicago GeneChips are referenced to M. truncatula gene names, where applicable. Log2 ratios of gene expression for G. intraradices-colonized (Gi-Myc), G. mosseae-colonized (Gm-Myc), and 2 mm phosphate-treated (2 mm-P) roots, all measured against roots grown at 20 μm phosphate, are given. FDR-corrected P values (FDR-P) are indicated. References for the AM-induced TF genes identified are as follows: MtErn1 (Middleton et al., 2007), MtErn2 (Andriankaja et al., 2007), MtMyb1 (Liu et al., 2003), MtCbf1 (this work), and MtCbf2 (this work).
| GeneChip Probe ID | Gene | Annotation | Gi-Myc | FDR-P | Gm-Myc | FDR-P | 2 mm-P | FDR-P |
| Mtr.46362.1.S1_at | AP2/ERF TF | 5.08 | 2.4E-05 | 4.07 | 3.3E-04 | 0.05 | 9.8E-01 | |
| Mtr.21492.1.S1_at | AP2/ERF TF | 3.52 | 1.2E-06 | 3.49 | 2.6E-06 | −4.05 | 9.0E-07 | |
| Mtr.15867.1.S1_at | AP2/ERF TF | 2.80 | 3.6E-04 | 2.39 | 2.4E-03 | 0.61 | 4.3E-01 | |
| Mtr.31671.1.S1_s_at | AP2/ERF TF | 2.77 | 1.2E-04 | 2.31 | 1.1E-03 | 0.65 | 2.9E-01 | |
| Mtr.43947.1.S1_at | MtErn2 | AP2/ERF TF | 2.55 | 4.3E-05 | 2.32 | 1.9E-04 | −1.38 | 4.4E-03 |
| Mtr.7556.1.S1_at | MtErn1 | AP2/ERF TF | 1.61 | 5.0E-03 | 1.27 | 3.1E-02 | −0.04 | 9.7E-01 |
| Mtr.25270.1.S1_at | C2H2 zinc finger TF | 5.13 | 5.9E-06 | 3.99 | 1.1E-04 | 0.01 | 9.9E-01 | |
| Mtr.28153.1.S1_at | C2H2 zinc finger TF | 3.26 | 3.2E-06 | 2.18 | 2.4E-04 | −0.78 | 8.3E-02 | |
| Mtr.41957.1.S1_at | C2H2 zinc finger TF | 1.06 | 1.5E-03 | 1.12 | 2.0E-03 | 0.78 | 1.0E-02 | |
| Mtr.51511.1.S1_at | MtCbf1 | CAAT-box TF | 7.34 | 1.8E-09 | 6.15 | 1.5E-08 | −0.67 | 1.5E-01 |
| Mtr.51511.1.S1_s_at | MtCbf1 | CAAT-box TF | 6.45 | 1.0E-08 | 5.58 | 7.7E-08 | −0.82 | 9.2E-02 |
| Mtr.4282.1.S1_at | CAAT-box TF | 3.29 | 4.8E-07 | 3.32 | 8.1E-07 | −0.45 | 2.7E-01 | |
| Mtr.16863.1.S1_at | MtCbf2 | CAAT-box TF | 2.36 | 1.8E-04 | 1.56 | 7.3E-03 | −0.34 | 6.1E-01 |
| Mtr.36004.1.S1_at | GRAS TF | 6.69 | 1.4E-08 | 6.08 | 6.9E-08 | −0.78 | 1.5E-01 | |
| Mtr.7264.1.S1_at | GRAS TF | 5.38 | 1.9E-08 | 3.05 | 9.3E-06 | −0.66 | 1.4E-01 | |
| Mtr.31954.1.S1_at | GRAS TF | 4.05 | 1.9E-05 | 3.14 | 3.5E-04 | −1.18 | 7.9E-02 | |
| Mtr.31955.1.S1_at | GRAS TF | 3.88 | 2.2E-06 | 3.05 | 4.1E-05 | −2.39 | 1.8E-04 | |
| Mtr.47463.1.S1_at | GRAS TF | 3.06 | 7.8E-06 | 2.85 | 2.8E-05 | −1.33 | 5.6E-03 | |
| Mtr.1484.1.S1_at | GRAS TF | 1.61 | 1.6E-03 | 1.21 | 1.7E-02 | 0.04 | 9.7E-01 | |
| Mtr.8863.1.S1_at | MtMyb1 | Myb TF | 9.90 | 6.7E-10 | 8.99 | 2.3E-09 | 0.21 | 8.1E-01 |
| Mtr.10894.1.S1_at | Myb TF | 2.59 | 6.2E-04 | 1.47 | 3.8E-02 | 1.01 | 1.2E-01 | |
| Mtr.33210.1.S1_at | Myb TF | 1.60 | 3.1E-02 | 1.71 | 3.2E-02 | 1.06 | 1.6E-01 | |
| Mtr.49044.1.S1_at | NAC-domain TF | 5.42 | 1.7E-07 | 2.72 | 2.2E-04 | −0.23 | 8.0E-01 | |
| Mtr.51555.1.S1_at | WRKY TF | 1.63 | 2.2E-02 | 1.82 | 1.8E-02 | 0.06 | 9.7E-01 | |
| Mtr.23616.1.S1_at | WRKY TF | 1.59 | 4.2E-02 | 1.96 | 2.1E-02 | −1.54 | 4.2E-02 |
With six members each, genes encoding GRAS and AP2/ERF transcriptional regulators were most prominent among the TFs identified. This is particularly interesting, since analogous proteins are involved in early signaling in the root nodule symbiosis. Here, the GRAS-TFs MtNsp1 and MtNsp2 (Kaló et al., 2005; Smit et al., 2005) as well as the AP2/ERF-TF MtErn1 (Middleton et al., 2007) are essential for the activation of symbiosis-related genes via Nod-factor signaling. Detailed studies on these TFs led to the identification of two further AP2/ERFs (MtErn2, MtErn3) and revealed that GRAS and AP2/ERF proteins interact with promoter sequences of early nodulin genes (Andriankaja et al., 2007; Hirsch et al., 2009). Since the TF proteins mentioned obviously represent an important control system in the regulation of symbiosis-specific genes, it is interesting that with MtErn1 and MtErn2, two of these were found to be induced in AM roots in our study.
Additional prominent TF genes induced in AM roots specified Cbfs of the HAP3 and HAP5 type, C2H2 zinc-finger proteins, and Myb proteins, with three members each (Table II). In the root nodule symbiosis, the MtHap2-1 CBF was identified as a key developmental regulator by Combier et al. (2006), whereas the zinc-finger protein Mszpt2-1 was shown to be essential for the differentiation of the nitrogen-fixing zone of root nodules (Frugier et al., 2000). So far, both gene families were not reported to be related to Glomus-colonized roots.
Since we regarded transcriptional regulators as particularly interesting for our study, we investigated the cellular expression pattern of a large subset of 17 TF genes, including members of the five most prominent families. Additionally, an AP2/ERF induced by diffusible factors from AM fungi (N. Hohnjec, unpublished data) was included in the analysis (Fig. 5).
Interestingly, we found arbuscule-specific or ARB-induced genes in four of the families investigated. Whereas a C2H2 zinc finger, two AP2/ERF, and three GRAS TF genes were ARB specific (Fig. 5), MtMyb1 and a further GRAS gene were significantly activated in ARB cells (Fig. 5). The ensconced TFs thus represent candidates for regulators that control the expression of genes required for proper arbuscule development and function. On the other hand, all gene families containing ARB-specific members also included genes expressed at equal levels in CMR and ARB (Figs. 5 and 6), indicating that different members of a gene family control different steps of the symbiosis.
In contrast to the TF genes mentioned so far, all three Cbf genes were expressed at similar levels in CMR and ARB (Fig. 5), indicating a more general role in the coordination of fungal colonization. Cbf proteins are known to regulate the expression of genes containing CCAAT motifs in their promoter sequences by forming heterotrimeric complexes that bind to the CAAT box (Combier et al., 2008). Since CCAAT motifs are present in about 30% of eukaryotic promotors (Mantovani, 1998), Cbf proteins represent global regulators of gene expression that probably gain specificity via interactions with other TFs (Maity and de Crombrugghe, 1998). That way, Cbf activation during an AM symbiosis can mediate an expression of whole sets of AM-related genes via the recognition of their promoter regions and a subsequent interaction with other transcriptional regulators. We therefore analyzed the two genes Mtr.51511.1.S1_at (designated MtCbf1) and Mtr.16863.1.S1_at (designated MtCbf2), both encoding CBFs of the HAP5 type, in more detail.
Expression of the MtCbf1 and MtCbf2 Genes Encoding CBFs Correlates with Fungal Contact and Spread
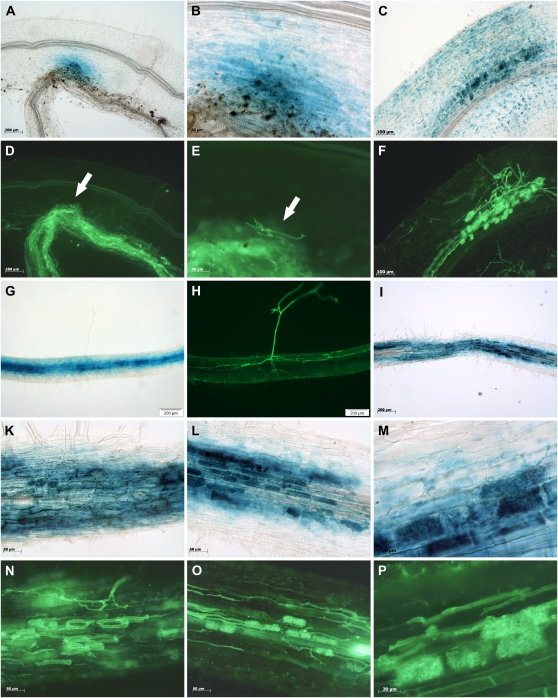
The two genes MtCbf1 and MtCbf2 are highly similar (96.3% identity on the level of nucleic acids; Supplemental Fig. S3), indicating that they might be derived from a duplication event. They are located in close proximity on M. truncatula chromosome 2 (http://www.medicagohapmap.org/, bacterial artificial chromosome clone AC136138), being separated by two different M. truncatula genes. Both MtCbf promoter sequences display no marked similarities, except of the region immediately upstream of the start codons (Supplemental Fig. S4). To obtain a comprehensive insight into the up-regulation of the MtCbf1 and MtCbf2 genes during successive stages of fungal colonization, their activity was analyzed both via real-time RT-PCR and via the expression of promoter-GUS fusions in transgenic roots, using a 4-week time course of mycorrhization. The results obtained confirmed the expression patterns of MtCbf1 and MtCbf2 detected in ARB and CMR cell pools, and in addition revealed a striking activation of these genes already during very early stages of the AM interaction.
The analysis of reporter gene expression in transgenic roots showed that both promoters displayed no activity in roots grown in the absence of mycorrhizal fungi (data not shown). In mycorrhizal transgenic roots, first spots of blue staining in epidermal cell layers were observed as early as 5 dpi for MtCbf1 and MtCbf2 in those places where fungal hyphae just attached to the plant epidermis, but had not yet entered the host cells (Figs. 7, A–C and 8, A, B, D, and E). This activation was always dependent on direct physical contact between the two symbiotic partners and is therefore most probably not induced by diffusible signals from AM fungi. Once the fungus had entered the root cortex, promotor activity expanded to cortical and arbuscule-containing cells, always related to the progression of fungal hyphae (Figs. 7, D, E, G, and H and 8, C and F). Whereas epidermal staining became even more pronounced for MtCbf1 during these stages and was so intense that staining of underlying cell layers could hardly be distinguished (Fig. 7, F and I), epidermal staining for MtCbf2 remained at a lower level, and the activity of this promoter appeared stronger in the cortex (Fig. 8, G and H). During later AM stages, when fungal progress is mostly achieved by an intraradical spread of the hyphae, no activity in epidermal cells was observed any more for both promoters, and reporter gene activity became restricted to those cortical cells that were either in contact with fungal hyphae or that contained arbuscules (Figs. 7, K–P and 8, I–P). No GUS staining was observed in root regions that did not contain fungal infection units (data not shown).
Figure 7.
Activity of the MtCbf1 promoter in M. truncatula mycorrhizal roots. A, B, D to F, and K to M, Light micrographs of M. truncatula mycorrhizal roots expressing the gusAint gene under the control of the MtCbf1 promoter. C, G to I, and N to P, Corresponding fluorescence micrographs showing counterstaining of fungal structures with Alexa Fluor 488 WGA conjugate at exactly the same root position. In A to C, F, and I, whole roots are shown; whereas D, E, G, H, and K to P show 60-μm-thin sections. E, H, L, and O represent enlarged regions of the roots shown in D, G, K, and N, respectively. A to C, Promoter activity during early AM stages, with fungal hyphae being just attached to the root epidermis. D, E, G, and H, Promoter activity in a young infection unit. F and I, Strong epidermal promoter activity in a region with an expanding infection unit. K, L, N, and O, Promoter activity in a densely colonized root. M and P, Enlargement of a single arbuscule. Scale bars represent 500 μm for A; 200 μm for F, I, K, and N; 100 μm for B, C, D, and G; 50 μm for L and O; 20 μm for E, H, M, and P.
Figure 8.
Activity of the MtCbf2 promoter in M. truncatula mycorrhizal roots. A to C, G, I, and K to M, Light micrographs of M. truncatula mycorrhizal roots expressing the gusAint gene under the control of the MtCbf2 promoter. D to F, H, N to P, Corresponding fluorescence micrographs showing counterstaining of fungal structures with Alexa Fluor 488 WGA conjugate at exactly the same root position. In A to H, whole roots are shown, whereas I to P show 60-μm-thin sections. B and E represent enlarged regions of the root shown in A and D. K, L, N, and O represent enlarged regions of the root shown in I. A, B, D, and E, Promoter activity during early stages, with fungal hyphae being just attached to the root epidermis. In this case, the hyphae in contact to the root surface (indicated by arrows) emerged from a highly mycorrhized leek root attached to the M. truncatula root. C and F, Promoter activity in a young infection unit. G and H, Promoter activity in a region with an expanding infection unit. In contrast to MtCbf1, no strong MtCbf2 activity can be observed in epidermal cell layers in this stage, hence the strong GUS staining in the cortex is visible from the outside. I, K, L, N, and O, Promoter activity in a densely colonized root. M and P, Enlargement of a group of arbuscules. Scale bars represent 200 μm for A, D, G, H, and I; 100 μm for C and F; 50 μm for B, E, K, L, N, and O; and 20 μm for M and P.
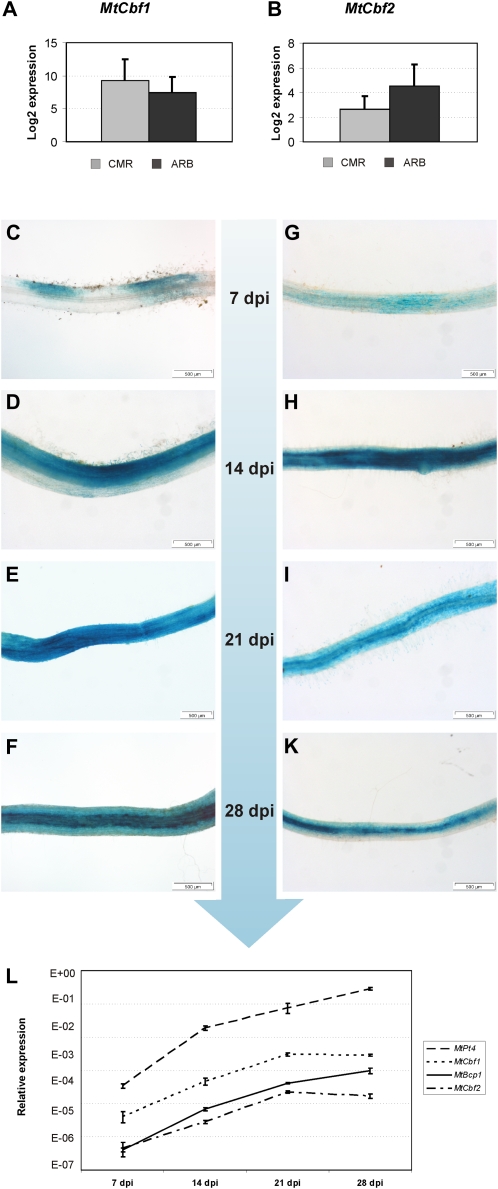
In our laser-microdissection experiments, both genes were identified as expressed with no significant difference between CMR and ARB (Fig. 9, A and B), although MtCbf1 tended to be more strongly expressed in CMR (3.5-fold induction) and MtCbf2 appeared activated in ARB (3.8-fold induction). This tendency was also mirrored in the reporter gene expression patterns driven by the two promoters, since GUS staining mediated by the MtCbf2 promoter seemed to be more intense in the arbuscule-containing cells (Fig. 8M), whereas GUS staining mediated by the MtCbf1 promoter was equal in arbuscule-containing and surrounding cortical cells interspersed with fungal hyphae (Fig. 7M). Taking into account that GUS staining appears more intense in the densely filled cells containing arbuscules, these expression patterns largely confirm our results obtained via laser microdissection.
Figure 9.
Expression of MtCbf1 and MtCbf2 in specific cell types and during a time course of mycorrhization. A and B, Real-time RT-PCR measurement of MtCbf1 and MtCbf2 expression in arbuscule-containing cells (ARB) and adjacent cortical cells colonized by fungal hyphae (CMR). Gene expression is displayed as the log2 mean value of three biological replicates. Bars represent ses. C to F, Promoter activity of MtCbf1 during a time course covering 7 to 28 dpi of mycorrhization, represented by roots displaying typical GUS staining patterns for each time point. G to K, Promoter activity of MtCbf2 during a time course covering 7 to 28 dpi of mycorrhization, represented by roots displaying typical GUS staining patterns for each time point. L, Real-time RT-PCR measurement of MtCbf1, MtCbf2, MtPt4, and MtBcp1 in M. truncatula roots during the 7 to 28 dpi time course of mycorrhization. Bars represent ses.
During the mycorrhizal time course performed from 7 to 28 dpi, the MtCbf1 and MtCbf2 promoters displayed a constantly rising activity, visualized by representative GUS staining patterns from different time points (Fig. 9, C–K). This increase closely correlated to the expression of the AM marker genes MtPt4, representing an arbuscule-specific gene, and MtBcp1, representing a gene expressed in arbuscule-containing as well as in the surrounding, hyphae-containing cortical cells (Fig. 9L). In our time course, MtPt4 and MtBcp1 from 7 to 28 dpi showed a constantly increased transcription, leading to an up-regulation of 836- and 341-fold, respectively, at 28 dpi in comparison to 7 dpi. Interestingly, the expression of both MtCbf1 and MtCbf2 was strongest already at 21 dpi, where 70- and 37-fold inductions in comparison to 7 dpi was reached. At 28 dpi, the activity of both TF genes already declined, probably due to a reduced number of epidermal infection events at this time point. Together with our promoter studies (Fig. 9, F and K), this observation underlines the importance of both genes already for early infection stages.
It can be concluded that MtCbf1 and MtCbf2 seem to be relevant during all stages of the AM symbiosis being characterized by a direct physical contact between the two partners. Therefore, the encoded CAAT-box TFs represent excellent candidates for novel regulators not only during later AM stages, but especially for the first steps of the interaction, where knowledge both on regulators mediating infection as well as infection-related expression markers is scarce. Interestingly, after the first activation in the epidermal cell layer, activity of both promoters seemed to precede the actual fungal colonization, since GUS staining did not only spread into the surrounding epidermal cells, but was several cells ahead of the proceeding fungal hyphae in the cortex. It is tempting to speculate that this gene expression pattern is promoted by a short-distance signal that prepares cells for an arrival of the microsymbiont, possibly facilitating fungal entry in the proximity of the first infection site or the hyphal spread in the cortex. In this context, the ability of CBFs to interact with a large range of promoters becomes particularly interesting, since they might thus be able to activate large parts of the symbiotic program in colonized root cortical cells, ultimately leading to the reprogramming of host cells toward an accomodation of symbiotic fungi.
CONCLUSION
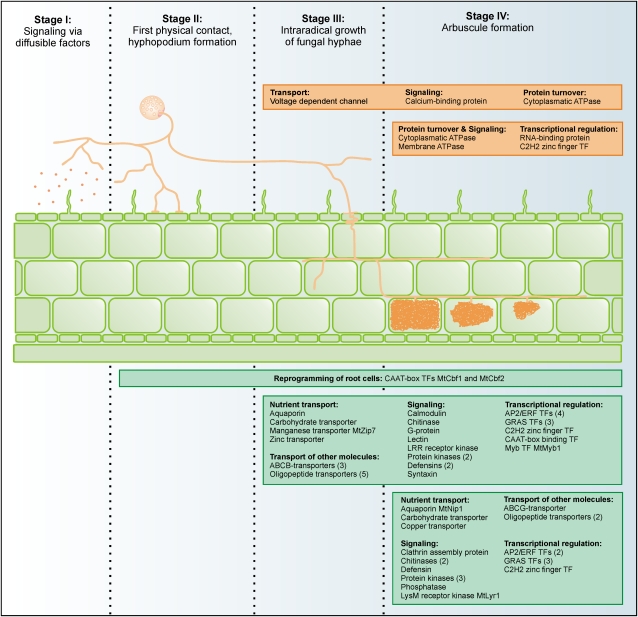
An attribution of gene activity to defined developmental stages is an important step toward the understanding of complex biological processes. That way, our identification of specific cellular expression patterns for a large subset of AM-related genes in mycorrhizal roots allows an association of these genes with the different developmental stages of an AM symbiosis, as shown in Figure 10. The colonization of plant roots by AM fungi is characterized by at least four distinct steps. While the first step is characterized by an exchange of diffusible signals between the two partners, causing the activation of signal cascades in epidermal and cortical cells, the second step involves direct physical contact, leading to hyphopodium formation by the fungus and PPA formation on the plant side, followed by penetration of rhizodermal cells by fungal hyphae. In the third step, fungal hyphae spread in the apoplast, resulting in the fourth and most intimate step, the formation of arbuscules. It has to be noted that steps two, three, and four are achieved in a short time period, where AM development is characterized by a spread of infection units from initial entry points, being accompanied by a sequential buildup and decay of arbuscules.
Figure 10.
Schematic summary of fungal and plant gene expression patterns during four different stages of the AM symbiosis. Proteins encoded by the genes identified are grouped according to functional categories. Fungal gene products are listed in orange, plant gene products in green boxes. The total number of genes with identical annotations is indicated in brackets. Note that it remains to be elucidated to what extent genes identified as expressed in arbuscule-containing as well as in the adjacent cortical cells colonized by fungal hyphae are already active during stages I and II.
Due to the fact that our AM expression profiles were recorded on the basis of pooled tissue samples of strongly mycorrhized roots, it is likely that processes occurring during later, functional stages of AM (steps three and four) were preferentially identified, and these are also represented by our specific cell pools derived by laser microdissection. We propose that the genes we identified to be active in cortical cells colonized by fungal hyphae and arbuscule-containing cells are related to the general progression of fungal hyphae in the root cortex, whereas genes only active in arbuscule-containing cells account for specific functions of this highly specialized symbiotic interface. Our identification of transporters, signaling-related genes, and TFs with distinct expression patterns provide insight into the genetic processes accompanying and allowing progression toward a functional symbiosis (Fig. 10). Interestingly, only two genes with enhanced expression in cortical cells colonized by fungal hyphae in comparison to arbuscule-containing cells were identified, suggesting that processes exclusively related to the growth of fungal hyphae in the cortex, which are not needed for arbuscule formation and function, are rare. It remains unclear, whether the mechanisms allowing fungal penetration of roots also guide intraradical growth of fungal hyphae. Regarding the fact that the primary infection is guided intracellularly via the PPA, whereas the growth of fungal hyphae in the cortex is mostly apoplastic and keeping in mind that fungal hyphae in contact to cortical cells may also be involved in nutrient transfer to the plant, there are differences between these two steps. It therefore seems feasible that there will be a set of genes whose activity is required during all stages, like the two CAAT-box TF genes identified in this study (Fig. 10). In addition, some genes specifically needed for the initial infection likely exist, analogous to the arbsucule-specific genes we identified here. Both steps are tightly controlled by the plant, mirrored in two types of mutants identified for the AM symbiosis (Harrison, 2005). These either mediate no entry of fungal hyphae at all, or allow entry and spread but no arbsucule formation, suggesting the existence of specific fungal signal molecules and transduction pathways required in these stages.
The identification of two CBF genes specifically activated during all stages of an AM infection delivers two candidates mediating a high-level control of gene expression during the colonization of roots by AM fungi. It will be interesting to see, whether other CAAT-box TF genes identified as AM induced show a similar expression pattern and how many genes active during apoplastic growth of fungal hyphae are already activated during earlier stages. These questions can only be solved via an analysis of cell types from early stages of AM interaction such as epidermal regions challenged with AM fungal signals or cortical regions harboring PPAs.
MATERIALS AND METHODS
Plant Growth, AM Fungal Inoculation, and Visualization of AM Fungal Structures
Medicago truncatula Gaertn ‘Jemalong’ genotype A17 seeds were surface sterilized and scarified as reported by Hohnjec et al. (2003). Plants were grown in the climate chamber (humidity: 70%; photosynthetic photon flux: 150 μmol m−2 s−1) at a 16-h light (23°C) and 8-h dark (18°C) regime. For subsequent Medicago GeneChip hybridizations using whole roots, plants were mycorrhized with AM fungi under conditions of phosphate limitation (20 μm phosphate). In addition, nonmycorrhizal roots grown at 20 μM phosphate as well as nonmycorrhizal roots grown at 2 mm phosphate were generated as described previously (Hohnjec et al., 2005). Two different AM fungal inocula were used: Glomus mosseae granular AM fungus inoculum BEG 12 (Biorize R&D), and Glomus intraradices Schenck and Smith DAOM197198 inoculum (Premier Tech Biotechnologies), the latter having recently been reassigned to Rhizophagus irregularis (Błaszk., Wubet, Renker, and Buscot) C. Walker and A. Schüβler comb. nov. (Stockinger et al., 2009). At 28 dpi with AM fungi or at 28 d growth under nonmycorrhizal conditions, roots were harvested and frozen in liquid nitrogen. Randomly selected areas of mycorrhizal roots were stained for fungal colonization using the gridline intersection method according to McGonigle et al. (1990). Here, the percentage of RLC (scoring hyphae, spores, vesicles, or arbuscules) ranged from 60% to 80%, while the relative arbuscule frequency in colonized fragments varied between 60% and 75%.
To obtain mycorrhizal roots for embedding in Steedman’s wax, 2-week-old seedlings were mycorrhized by adding 15% (v/v) inoculum G. intraradices isolate 49 (Maier et al., 1995) produced in leek (Allium porrum ‘Elefant’) cultures to the substrate. Mycorrhizal and nonmycorrhizal plants were fertilized with half-strength Hoagland solution containing 20 μm phosphate and an additional 2 mm NH4NO3. RLC was checked regularly via ink staining according to the protocol of Vierheilig et al. (1998) and gridline intersection counting according to McGonigle et al. (1990). Seventy percent to 80% RLC turned out to be most convenient for the selection of arbuscule-containing and adjacent cortical cells. This level of colonization was usually reached at 21 dpi.
Tissue Embedding, Tissue Sectioning, and Laser Microdissection
Roots were embedded using the Steedman’s wax protocol (Gomez et al., 2009) with the following modifications: Eosin was already added in the first step of the ethanol series (75% [v/v] ethanol with 0.1% [v/v] Eosin y), the overnight fixation step in Farmer’s fixative, and the overnight incubation step in 1:1 ethanol:wax were extended from 12 to 14 h, and root pieces were embedded in TurbOflowII molds as well as cassettes (McCormick Scientific). Blocks were stored in vacuum-sealed plastic bags containing desiccant bags at 4°C.
Longitudinal root sections of 12 μm were obtained using a Hyrax M55 rotary microtome (Zeiss). Ribbons were spread on heat-sterilized glass slides and were stretched with diethylpyrocarbonate-treated autoclaved water. Slides were dried for 1 h in a hybridization oven at 32°C. Slides with sections were used on the same or the following 2 d and stored in vacuum-sealed plastic bags with desiccation bags at 4°C if necessary. Sections were dewaxed immediately before cell harvest by washing the slides with absolute ethanol several times at 38°C on a heating plate, until the wax was not visible anymore. Subsequently, slides were dried on the heating plate.
The P.A.L.M. microbeam system with a Capmover (Zeiss) was used for laser microdissection and pressure catapulting. To collect cells, the CloseCut and Auto-LPC function was used according to the manufacturer’s instructions. Cells were collected into 500-μL adhesive caps (Zeiss) and stored directly at −80°C after the harvest was completed.
For each cell type, four biological replicates were produced, based on distinct rounds of plant cultivation and root embedding. Biological replicates consisted of three technical replicates of approximately 1,000 cells each, which were pooled after RNA isolation and amplification.
RNA Isolation and Amplification
Whole-root mycorrhizal samples were taken from frozen stocks, pooled, and ground using lysing matrix D tubes (MP Biomedicals) in a FastPrep (MP Biomedicals) prior to RNA extractions. Total RNA isolation and DNase I on-column digestion was performed via RNeasy kits (Qiagen) according to the manufacturer’s instructions. RNA preparations were quality checked both via spectrophotometry (NanoDrop ND-1000, Peqlab) and via capillary electrophoresis in RNA nano chips (Agilent Bioanalyzer, Agilent), as recommended by the manufacturers.
Total RNA was isolated from laser-microdissected cells using the RNeasy micro kit (Qiagen). Three hundred and fifty microliters of RLT buffer containing β-mercaptoethanol were added to each sample followed by a 30-min incubation at room temperature. The lysate was spun down for 5 min at 13,400g, mixed 1:1 with ethanol absolute, and transferred to the clean-up column. On-column DNAse I digestion was performed according to the manufacturer’s instructions. RNA from laser-microdissected cells was amplified using the TargetAmp 2-round aRNA amplification kit (Epicentre Biotechnologies), as specified by the manufacturer. Quantity and quality of total RNA as well as T7-amplified aRNA was checked via capillary electrophoresis in RNA pico and nano chips, respectively, using an Agilent 2100 bioanalyzer (Agilent).
Medicago GeneChip Hybridizations
RNA was processed for use on Affymetrix GeneChip Medicago genome arrays, according to the manufacturer’s GeneChip 3′ in vitro transcription (IVT) express kit user manual. Briefly, 100 ng of total RNA with an RNA integrity number (Agilent 2100 bioanalyzer, Agilent) of at least 8.5 containing spiked-in poly-A+ RNA controls was used in a RT reaction (GeneChip 3′ IVT express kit; Affymetrix) to generate first-strand cDNA. After second-strand synthesis, double-stranded cDNA was used in a 16-h IVT reaction to generate aRNA (GeneChip 3′ IVT express kit; Affymetrix). Size distribution of in vitro transcribed aRNA and fragmented aRNA, respectively, was assessed via an Agilent 2100 bioanalyzer (Agilent), using an RNA 6000 nano assay. Ten micrograms of fragmented aRNA was added to a 300-μL hybridization cocktail also containing hybridization controls. Two hundred microliters of the mixture was hybridized on GeneChips for 16 h at 45°C. Standard post-hybridization wash and double-stain protocols (FS450_0001; GeneChip HWS kit; Affymetrix) were used on an Affymetrix GeneChip fluidics station 450. GeneChips were scanned on an Affymetrix GeneChip scanner 3000 7G.
Evaluation of Data from Medicago GeneChip Hybridizations
Cel files obtained from Medicago GeneChip hybridizations were analyzed using the Robin software (http://mapman.gabipd.org/web/guest/robin). Normalization was performed across all GeneChips using the robust multichip average algorithm. Intensity values calculated for each probe set were log2 transformed and averaged across all three biological replicates. Log2 differences between the conditions studied were evaluated statistically by applying a FDR correction for P values implemented in Robin. Original annotations of probes from Medicago GeneChips were replaced by automated annotations as well as functional classifications generated via Sequence Analysis and Management System (Bekel et al., 2009) and Gene Ontology classifications (http://www.medicago.org/GeneChip). To visualize gene expression profiles, MapMan (Usadel et al., 2005) was used. Data from Medicago GeneChip hybridizations were related to in silico expression profiles using MediPlEx (Henckel et al., 2010), applying the AM root libraries preselection. Since Medicago GeneChips are based on gene models from EST and genomic sequences, the number of probe sets exceeds the number of genes represented to a certain extent. Nevertheless, we refer to genes instead of probe sets in this work for reasons of simplicity.
Real-Time RT-PCR
Primers were designed using the Primer3 Web interface (Rozen and Skaletsky, 2000) according to the following criteria: product size range 100 to 150 bp; primer size min: 18 bp, opt: 21 bp, max: 24 bp; primer Tm min: 52°C, opt: 53°C, max: 55°C. If the position of the coding region was known, the amplicon was preferentially positioned in the 3′ region of the gene or the 3′ untranslated region. The primer pairs suggested by the primer3 program were blasted against the DFCI Medicago Gene Index (Quackenbush et al., 2001) to check for mispriming in known M. truncatula transcript sequences. The number of matching bp in off-target genes and especially matches at the 3′ end of the primer were taken into account and the most suitable primer pair according to these parameters was chosen. All primer pairs were tested for performance and specificity with RNA from whole-root tissue prior to using them for RNA from laser-microdissected samples. For some of the genes, primer pairs published by Gomez et al. (2009) were used. These are indicated in Supplemental Table S5, where all primer pairs are listed.
For the laser-microdissected samples, 50 ng of T7-amplified RNA (aRNA) were used for real-time RT-PCR, using the SensiMix SYBR one-step kit (Bioline). RT-PCR conditions (Realplex cycler, Eppendorf) were as follows: 10 min 42°C, 10 min 95°C, 50 cycles (15 s 95°C, 30 s 55°C, 30 s 72°C), 15 s 95°C, melting curve from 40°C to 95°C. In addition to a melting curve analysis, all real-time PCR products were separated on 2% (w/v) agarose gels to check both for specificity and a correct amplification size. Resequencing of selected PCR products was used to additionally confirm the specific amplification of the target gene. Expression results were averaged over three biological replicates. In case one replicate delivered a significantly deviant result from the other two, the measurement was repeated. If the difference persisted, the fourth replicate was included into the analysis and replaced the replicate in question, if this resulted in a more consistent expression pattern. The constitutive translation elongation factor gene MtTef1α (TC178258 in the DFCI Medicago Gene Index) was used for normalization across different conditions. MtTef1α expression was analyzed in four technical replicates, and the average value was used to calculate relative gene expression levels using the 2−ΔCT value with ΔCT = CTgene − CTMtTef1α. Expression differences were analyzed for significance using the Student’s t test incorporated in MS Excel 2007 (Microsoft Corp.).
For the whole-root samples, 50 ng of total RNA were used for real-time RT-PCR, using the SensiMix SYBR one-step kit (Bioline) according to the manufacturer’s instructions. The constitutive translation elongation factor gene MtTef1α (TC178258 in the DFCI Medicago Gene Index) was used for normalization across different conditions. All expression results were averaged over three biological replicates. Relative gene expression levels were calculated as described above.
Construction and Histological Analysis of Transgenic Hairy Roots
The promoters of two CAAT-box TF genes represented by probe sets Mtr.51511.1.S1_at (referred to as MtCbf1) and Mtr.16863.1.S1_at (referred to as MtCbf2) were PCR amplified with phusion hot start DNA polymerase (Finnzymes, Biozym) from M. truncatula Gaertn ‘Jemalong’ genotype A17 genomic DNA, using gene-specific primers containing appropriate restriction sites. The amplified products covered the −1,513/−3 region for MtCbf1 and the −1,484/−6 region for MtCbf2 relative to the start codon. The promoter regions were cloned in front of the gusAint gene of pGUSint (Hohnjec et al., 2003) using SphI/SmaI restriction sites for MtCbf1 and SphI/EcoRI restriction sites for MtCbf2. Whereas the promoter-GUS fusion of MtCbf1 was excised using SpeI, the promoter-GUS fusion of MtCbf2 was excised using SalI/StuI. Both fragments were filled in with Klenow polymerase, and cloned into the SmaI site of the binary plasmid pRedRoot (Limpens et al., 2004). The plasmids obtained were electroporated into Agrobacterium rhizogenes ARqua1, and the resulting strains were used to generate hairy roots on M. truncatula ‘Jemalong A17’ according to Vieweg et al. (2004). Transgenic roots were identified using dsRed fluorescence (Leica MZ 10F, Leica Microsytems) and were mycorrhized by adding 15% (v/v) inoculum G. intraradices isolate 49 (Maier et al., 1995) produced in leek cultures (cv Elefant) to the substrate. GUS assays were performed as described by Hohnjec et al. (2003), without preheating of the samples. To obtain thin sections, GUS-stained roots were embedded in 5% agarose and cut using a Leica VT1000S vibratome (Leica). Counterstaining of fungal structures was performed with Alexa Fluor 488 wheat germ agglutinin conjugate (Invitrogen). Roots were bleached for 3 min in 10% KOH at 95°C, washed three times with water, and stained in phosphate-buffered saline buffer containing 20 μg/mL Alexa Fluor 488 WGA conjugate overnight. For staining of fungal structures in thin sections and staining of extraradical hyphae on the root surface, samples were directly transferred into phosphate-buffered saline buffer containing 20 μg/mL Alexa Fluor 488 WGA conjugate and stained overnight. Photo documentation was performed with a Leica MZ 10F stereomicroscope (Leica Microsytems), equipped with an Olympus XC50 camera (Olympus), and with a Zeiss Axio Observer Z1 microscope equipped with an AxioCam ICc1 (Zeiss).
GeneChip data are available from the Gene Expression Omnibus accession number GSE32208.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of AM-specific and AM-enhanced TF genes.
Supplemental Figure S2. Genes exclusively expressed in arbuscule-containing cells.
Supplemental Figure S3. Alignment of the coding sequences of the two CBF genes MtCbf1 and MtCbf2.
Supplemental Figure S4. Alignment of promoter sequences of the two CBF genes MtCbf1 and MtCbf2.
Supplemental Table S1. Gene expression in M. truncatula roots in response to G. intraradices colonization (28 dpi, at 20-μm phosphate), G. mosseae colonization (28 dpi, at 20-μm phosphate), and a 28-d treatment with 2-mm phosphate.
Supplemental Table S2. M. truncatula genes coactivated in response to G. intraradices and G. mosseae colonization.
Supplemental Table S3. AM-activated M. truncatula genes encoding membrane transporters.
Supplemental Table S4. AM-activated M. truncatula genes encoding signaling-related proteins.
Supplemental Table S5. Real-time RT-PCR primers used in this study and size of predicted PCR products.
Acknowledgments
We are grateful to Karen S. Gomez and Maria Harrison (Boyce Thompson Institute for Plant Research) for sharing protocols and for helpful discussions on laser microdissection. Excellent bioinformatics support was provided by Kolja Henckel (Bioinformatics Resource Facility, Center for Biotechnology, Bielefeld University).
References
- Akiyama K, Matsuzaki K, Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C. (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F. (2007) AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlorio M, Ludwig A, Boller T, Bonfante P. (1992) Inhibition of fungalm growth by plant chitinases and β-1,3-glucanases. Protoplasma 171: 34–43 [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier MC, Keck M, Gödde V, Niehaus K, Küster H, Hohnjec N. (2010) Knockdown of the symbiotic sucrose synthase MtSucS1 affects arbuscule maturation and maintenance in mycorrhizal roots of Medicago truncatula. Plant Physiol 152: 1000–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini R, Gómez-Ariza J, Lanfranco L, Bonfante P. (2007) Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol Plant Microbe Interact 20: 1055–1062 [DOI] [PubMed] [Google Scholar]
- Balestrini R, Lanfranco L. (2006) Fungal and plant gene expression in arbuscular mycorrhizal symbiosis. Mycorrhiza 16: 509–524 [DOI] [PubMed] [Google Scholar]
- Barker DG, Bianchi S, Blondon F, Dattée Y, Duc G, Essad S, Flament P, Gallusci P, Génier G, Guy P, et al. (1990) Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep 8: 40–49 [Google Scholar]
- Bekel T, Henckel K, Küster H, Meyer F, Mittard Runte V, Neuweger H, Paarmann D, Rupp O, Zakrzewski M, Pühler A, et al. (2009) The Sequence Analysis and Management System—SAMS-2.0: data management and sequence analysis adapted to changing requirements from traditional sanger sequencing to ultrafast sequencing technologies. J Biotechnol 140: 3–12 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Li H, Dai X, Wandrey M, He J, Kaundal R, Torres-Jerez I, Gomez SK, Harrison MJ, Tang Y, et al. (2010) Genomic inventory and transcriptional analysis of Medicago truncatula transporters. Plant Physiol 152: 1716–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N. (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanomi A, Wiemken A, Boller T, Salzer P. (2001) Local induction of a mycorrhiza-specific class III chitinase gene in cortical root cells of Medicago truncatula containing developing or mature arbuscules. Plant Biol 3: 194–200 [Google Scholar]
- Brewin NJ. (1991) Development of the legume root nodule. Annu Rev Cell Biol 7: 191–226 [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Terras FR, Cammue BP, Osborn RW. (1995) Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol 108: 1353–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M. (2007) Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol 173: 11–26 [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Kristensen BK, Bechmann IE. (2003) A plasma membrane zinc transporter from Medicago truncatula is up-regulated in roots by Zn fertilization, yet down-regulated by arbuscular mycorrhizal colonization. Plant Mol Biol 52: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Cannon SB, May GD, Jackson SA. (2009) Three sequenced legume genomes and many crop species: rich opportunities for translational genomics. Plant Physiol 151: 970–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RB, Zeto SK. (2000) Mineral acquisition by arbuscular mycorrhizal plants. J Plant Nutr 23: 867–902 [Google Scholar]
- Combier JP, de Billy F, Gamas P, Niebel A, Rivas S. (2008) Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev 22: 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll J, Hause B, Demchenko K, Pawlowski K, Krajinski F. (2003) A member of the germin-like protein family is a highly conserved mycorrhiza-specific induced gene. Plant Cell Physiol 44: 1208–1214 [DOI] [PubMed] [Google Scholar]
- Drissner D, Kunze G, Callewaert N, Gehrig P, Tamasloukht M, Boller T, Felix G, Amrhein N, Bucher M. (2007) Lyso-phosphatidylcholine is a signal in the arbuscular mycorrhizal symbiosis. Science 318: 265–268 [DOI] [PubMed] [Google Scholar]
- Ercolin F, Reinhardt D. (2011) Successful joint ventures of plants: arbuscular mycorrhiza and beyond. Trends Plant Sci 16: 356–362 [DOI] [PubMed] [Google Scholar]
- Floss DS, Hause B, Lange PR, Küster H, Strack D, Walter MH. (2008) Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J 56: 86–100 [DOI] [PubMed] [Google Scholar]
- Frenzel A, Manthey K, Perlick AM, Meyer F, Pühler A, Küster H, Krajinski F. (2005) Combined transcriptome profiling reveals a novel family of arbuscular mycorrhizal-specific Medicago truncatula lectin genes. Mol Plant Microbe Interact 18: 771–782 [DOI] [PubMed] [Google Scholar]
- Frugier F, Poirier S, Satiat-Jeunemaître B, Kondorosi A, Crespi M. (2000) A Krüppel-like zinc finger protein is involved in nitrogen-fixing root nodule organogenesis. Genes Dev 14: 475–482 [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. (2006) The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett 580: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P. (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ. (2009) Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough C, Cullimore J. (2011) Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol Plant Microbe Interact 24: 867–878 [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bücking H, Lammers PJ, Shachar-Hill Y. (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435: 819–823 [DOI] [PubMed] [Google Scholar]
- Grunwald U, Nyamsuren O, Tamasloukht M, Lapopin L, Becker A, Mann P, Gianinazzi-Pearson V, Krajinski F, Franken P. (2004) Identification of mycorrhiza-regulated genes with arbuscule development-related expression profile. Plant Mol Biol 55: 553–566 [DOI] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P. (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2: 487–496 [Google Scholar]
- Harrison MJ. (1996) A sugar transporter from Medicago truncatula: altered expression pattern in roots during vesicular-arbuscular (VA) mycorrhizal associations. Plant J 9: 491–503 [DOI] [PubMed] [Google Scholar]
- Harrison MJ. (1999) Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol 50: 361–389 [DOI] [PubMed] [Google Scholar]
- Harrison MJ. (2005) Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol 59: 19–42 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Dixon RA. (1993) Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vesicular-arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol Plant Microbe Interact 6: 643–654 [Google Scholar]
- He J, Benedito VA, Wang M, Murray JD, Zhao PX, Tang Y, Udvardi MK. (2009) The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henckel K, Küster H, Stutz LJ, Goesmann A. (2010) MediPlEx—a tool to combine in silico & experimental gene expression profiles of the model legume Medicago truncatula. BMC Res Notes 3: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henckel K, Runte KJ, Bekel T, Dondrup M, Jakobi T, Küster H, Goesmann A. (2009) TRUNCATULIX—a data warehouse for the legume community. BMC Plant Biol 9: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GE. (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N, Becker JD, Pühler A, Perlick AM, Küster H. (1999) Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Mol Gen Genet 261: 514–522 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Henckel K, Bekel T, Gouzy J, Dondrup M, Goesmann A, Küster H. (2006) Transcriptional snapshots provide insights into the molecular basis of arbuscular mycorrhiza in the model legume Medicago truncatula. Funct Plant Biol 33: 737–748 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Pühler A, Küster H. (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant Microbe Interact 16: 903–915 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Vieweg MF, Pühler A, Becker A, Küster H. (2005) Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 137: 1283–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kloppholz S, Kuhn H, Requena N. (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21: 1204–1209 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Chabaud M, Lougnon G, Gough C, Dénarié J, Barker DG, Bécard G. (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajinski F, Hause B, Gianinazzi-Pearson V, Franken P. (2002) Mtha1, a plasma membrane H+-ATPase gene from Medicago truncatula, shows arbuscule-specific induced expression in mycorrhizal tissue. Plant Biol 4: 754–761 [Google Scholar]
- Kuhn H, Küster H, Requena N. (2010) Membrane steroid-binding protein 1 induced by a diffusible fungal signal is critical for mycorrhization in Medicago truncatula. New Phytol 185: 716–733 [DOI] [PubMed] [Google Scholar]
- Küster H, Becker A, Firnhaber C, Hohnjec N, Manthey K, Perlick AM, Bekel T, Dondrup M, Henckel K, Goesmann A, et al. (2007a) Development of bioinformatic tools to support EST-sequencing, in silico- and microarray-based transcriptome profiling in mycorrhizal symbioses. Phytochemistry 68: 19–32 [DOI] [PubMed] [Google Scholar]
- Küster H, Hohnjec N, Krajinski F, El YF, Manthey K, Gouzy J, Dondrup M, Meyer F, Kalinowski J, Brechenmacher L, et al. (2004) Construction and validation of cDNA-based Mt6k-RIT macro- and microarrays to explore root endosymbioses in the model legume Medicago truncatula. J Biotechnol 108: 95–113 [DOI] [PubMed] [Google Scholar]
- Küster H, Vieweg MF, Manthey K, Baier MC, Hohnjec N, Perlick AM. (2007b) Identification and expression regulation of symbiotically activated legume genes. Phytochemistry 68: 8–18 [DOI] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ. (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar DP, Sharopova N, Endre G, Peñuela S, Samac D, Town C, Silverstein KA, VandenBosch KA. (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140: 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán AF, Ellis DR, Grusak MA. (2004) Identification and characterization of several new members of the ZIP family of metal ion transporters in Medicago truncatula. Plant Mol Biol 54: 583–596 [DOI] [PubMed] [Google Scholar]
- Maier W, Peipp H, Schmidt J, Wray V, Strack D. (1995) Levels of a terpenoid glycoside (blumenin) and cell wall-bound phenolics in some cereal mycorrhizas. Plant Physiol 109: 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Maity SN, de Crombrugghe B. (1998) Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci 23: 174–178 [DOI] [PubMed] [Google Scholar]
- Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Pühler A, Perlick AM, Küster H. (2004) Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant Microbe Interact 17: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1998) A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res 26: 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle T, Miller M, Evans D, Fairchild D, Swan J. (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls U, Grunze N, Willmann M, Reich M, Küster H. (2007) Sugar for my honey: carbohydrate partitioning in ectomycorrhizal symbiosis. Phytochemistry 68: 82–91 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Harrison MJ, Udvardi M. (2005) Peace talks and trade deals: keys to long-term harmony in legume-microbe symbioses. Plant Physiol 137: 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JS, Kudrna D, Wing R, Untergasser A, et al. (2011) LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331: 909–912 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2000) Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr Opin Plant Biol 3: 320–328 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Skurray RA. (1994) The POT family of transport proteins. Trends Biochem Sci 19: 404. [DOI] [PubMed] [Google Scholar]
- Plett JM, Kemppainen M, Kale SD, Kohler A, Legué V, Brun A, Tyler BM, Pardo AG, Martin F. (2011) A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol 21: 1197–1203 [DOI] [PubMed] [Google Scholar]
- Pumplin N, Harrison MJ. (2009) Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol 151: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ. (2010) Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J 61: 482–494 [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena N, Breuninger M, Franken P, Ocón A. (2003) Symbiotic status, phosphate, and sucrose regulate the expression of two plasma membrane H+-ATPase genes from the mycorrhizal fungus Glomus mosseae. Plant Physiol 132: 1540–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R. (2008) Medicago truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: past, present and future. Funct Plant Biol 35: 253–264 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Salzer P, Bonanomi A, Beyer K, Vögeli-Lange R, Aeschbacher RA, Lange J, Wiemken A, Kim D, Cook DR, Boller T. (2000) Differential expression of eight chitinase genes in Medicago truncatula roots during mycorrhiza formation, nodulation, and pathogen infection. Mol Plant Microbe Interact 13: 763–777 [DOI] [PubMed] [Google Scholar]
- Schaarschmidt S, González MC, Roitsch T, Strack D, Sonnewald U, Hause B. (2007) Regulation of arbuscular mycorrhization by carbon: the symbiotic interaction cannot be improved by increased carbon availability accomplished by root-specifically enhanced invertase activity. Plant Physiol 143: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüssler A, Schwarzott D, Walker C. (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105: 1413–1421 [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. (2008) Mycorrhizal Symbioses, Ed 2. Academic Press, London [Google Scholar]
- Stekel DJ, Git Y, Falciani F. (2000) The comparison of gene expression from multiple cDNA libraries. Genome Res 10: 2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger H, Walker C, Schüssler A. (2009) ‘Glomus intraradices DAOM197198’, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol 183: 1176–1187 [DOI] [PubMed] [Google Scholar]
- Takeda N, Haage K, Sato S, Tabata S, Parniske M. (2011) Activation of a Lotus japonicus subtilase gene during arbuscular mycorrhiza is dependent on the common symbiosis genes and two cis-active promoter regions. Mol Plant Microbe Interact 24: 662–670 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Cammue BP, Thevissen K. (2002) Plant defensins. Planta 216: 193–202 [DOI] [PubMed] [Google Scholar]
- Tisserant E, Kohler A, Dozolme-Seddas P, Balestrini R, Benabdellah K, Colard A, Croll D, da Silva C, Gomez SK, Koul R, et al. (2011) The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol (in press) [DOI] [PubMed] [Google Scholar]
- Uehlein N, Fileschi K, Eckert M, Bienert GP, Bertl A, Kaldenhoff R. (2007) Arbuscular mycorrhizal symbiosis and plant aquaporin expression. Phytochemistry 68: 122–129 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierheilig H, Coughlan AP, Wyss U, Piche Y. (1998) Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64: 5004–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieweg MF, Frühling M, Quandt HJ, Heim U, Bäumlein H, Pühler A, Küster H, Andreas MP. (2004) The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol Plant Microbe Interact 17: 62–69 [DOI] [PubMed] [Google Scholar]
- Wulf A, Manthey K, Doll J, Perlick AM, Linke B, Bekel T, Meyer F, Franken P, Küster H, Krajinski F. (2003) Transcriptional changes in response to arbuscular mycorrhiza development in the model plant Medicago truncatula. Mol Plant Microbe Interact 16: 306–314 [DOI] [PubMed] [Google Scholar]
- Young ND, Udvardi M. (2009) Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol 12: 193–201 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Blaylock LA, Harrison MJ. (2010) Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 22: 1483–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]