Abstract
To improve our understanding of the organization and regulation of the wheat (Triticum aestivum) gene space, we established a transcription map of a wheat chromosome (3B) by hybridizing a newly developed wheat expression microarray with bacterial artificial chromosome pools from a new version of the 3B physical map as well as with cDNA probes derived from 15 RNA samples. Mapping data for almost 3,000 genes showed that the gene space spans the whole chromosome 3B with a 2-fold increase of gene density toward the telomeres due to an increase in the number of genes in islands. Comparative analyses with rice (Oryza sativa) and Brachypodium distachyon revealed that these gene islands are composed mainly of genes likely originating from interchromosomal gene duplications. Gene Ontology and expression profile analyses for the 3,000 genes located along the chromosome revealed that the gene islands are enriched significantly in genes sharing the same function or expression profile, thereby suggesting that genes in islands acquired shared regulation during evolution. Only a small fraction of these clusters of cofunctional and coexpressed genes was conserved with rice and B. distachyon, indicating a recent origin. Finally, genes with the same expression profiles in remote islands (coregulation islands) were identified suggesting long-distance regulation of gene expression along the chromosomes in wheat.
The organization of the gene space in a genome refers to the layout of the protein-coding genes along the chromosomes (Jackson et al., 2004). With the growing number of sequenced genomes (Feuillet et al., 2011), many studies have led to the conclusion that this organization is far from random and is correlated to the genome size. For example, plants with small genome sizes, such as Arabidopsis (Arabidopsis thaliana; 125 Mb), Brachypodium distachyon (272 Mb), and rice (Oryza sativa; 389 Mb), exhibit an even distribution of their genes along their chromosomes (Arabidopsis Genome Initiative, 2000; International Rice Genome Sequencing Project, 2005; International Brachypodium Initiative, 2010), whereas for intermediate size genomes, such as those of Populus trichocarpa (485 Mb) and grape (Vitis vinifera; 487 Mb), alternation between high gene density regions and low gene density regions is observed (Tuskan et al., 2006; Jaillon et al., 2007). This tendency is even stronger in plants with large genomes, such as soybean (Glycine max; 1,115 Mb) and maize (Zea mays; 2,300 Mb), in which a positive gradient of gene density from the centromere to the telomeres has been observed (Schnable et al., 2009; Schmutz et al., 2010).
Irrespective of genome size or gene space organization, clusters of genes sharing expression profiles were identified in several plants, including Arabidopsis (Ren et al., 2005; Zhan et al., 2006) and rice (Ren et al., 2007). In addition to coexpressed genes, clusters were shown to be significantly enriched in genes sharing the same function or assigned to the same pathway in Arabidopsis, but also in cotton (Gossypium hirsutum), grape, poplar (Populus spp.), papaya, rice, and sorghum (Sorghum bicolor; Lee and Sonnhammer, 2003; Williams and Bowles, 2004; Schmid et al., 2005; Xu et al., 2008; Liu and Han, 2009).
The genome of bread wheat, Triticum aestivum, is one of the most complex plant genomes as it is allohexaploid (A, B, and D genomes) and comprises 17,000 Mb of sequence (approximately 5,700 Mb per subgenome) with >80% of repeated sequences. Consequently, molecular analyses of the wheat genome have always been very challenging, and in the absence of a reference genome sequence, little is known about the organization of the gene space. Previous studies performed on a very limited number of randomly chosen BACs suggested an uneven distribution of the genes along the wheat chromosomes (Devos et al., 2005; Charles et al., 2008). In addition, a positive gradient of gene density from the centromere to the telomeres was suggested by Akhunov et al. (2003) and Munkvold et al. (2004) based on EST mapping in wheat deletion bins and more recently by Choulet et al. (2010) determined by the annotation of megabase-sized sequences along chromosome 3B.
Previous studies in the Triticum lineage suggested that the gene space is organized in isolated genes and gene islands (i.e. gene-rich regions) whose number and distribution varied significantly between studies. In some cases, it was proposed that most of the genes are found in a few gene-rich regions (Sandhu and Gill, 2002; Erayman et al., 2004), whereas in other cases, numerous but small gene islands have been proposed (Brooks et al., 2002; Chantret et al., 2004; Wicker et al., 2005; Choulet et al., 2010; Rustenholz et al., 2010). Recently, Choulet et al. (2010) estimated that 50% of wheat intergenic distances are shorter than 43 kb and used this distance to define a gene island in the wheat genome as blocks of two to 10 genes (three genes on average) separated from other genes by about 100 to 200 kb. Moreover, hybridization of wheat BAC pools from chromosome 3B onto barley (Hordeum vulgare) expression microarrays led to the mapping of 738 genes along this chromosome and showed that the distribution of gene islands is strongly correlated with a positive gradient of gene density along wheat chromosome 3B (Rustenholz et al., 2010). Such gene islands have also been described in maize by Wei et al. (2009), who found that 56% of the intergenic distances were shorter than 20 kb, a value comparable to what has been observed in wheat (Choulet et al., 2010) after a correction of genome size. Similar gene densities in gene islands were also observed in soybean and cotton (Clough et al., 2004; Guo et al., 2008). So far, however, little is known about the formation of gene islands and the forces that maintain some genes close to each other during the evolution of genomes subjected to massive expansion in size.
In this study, we established an improved version of the chromosome 3B physical map and used it to map almost 3,000 genes whose expression patterns were tested in 15 different conditions using a newly developed wheat NimbleGen 40K unigene microarray. This transcription map of a wheat chromosome enabled us to confirm that 70% of the genes are organized in islands that are responsible for the positive gradient of gene density observed from the centromere to the telomeres. By studying their evolution, expression, and putative function, we identified islands with coexpressed and cofunctional genes, including some that are conserved with rice and/or B. distachyon, whereas others were of more recent origin. Finally, we suggest structural and functional hypotheses for the origin and the conservation of genes organized in islands in the wheat genome.
RESULTS
A Sequence-Ready Physical Map of the Chromosome 3B of Hexaploid Wheat
The first physical map of wheat chromosome 3B (995 Mb) covered 82% of the chromosome in 1,036 contigs with an average size of 783 kb (Paux et al., 2008). To perform whole-chromosome sequencing and expression profiling, the map was recently improved through the fingerprinting of 82,176 additional BAC clones and 7,440 BAC clones from the first version of the minimal tiling path (MTP). The resulting high-information content fingerprints (HICFs) were added to the original 1,036 contigs, resulting in a final assembly of 131,792 HICFs into 1,669 contigs representing 961 Mb (97% of the whole chromosome) with 19.2× coverage (http://urgi.versailles.inra.fr/cgi-bin/gbrowse/wheat_FPC_pub/). Using mapping information from the 1,443 markers already assigned to contigs (Paux et al., 2008), 919 out of the 1,669 contigs covering 740 Mb were assigned to one of the eight intervals defined by genetic deletions, so-called deletion bins (3BS8-0.78-1.00, 3BS9-0.57-0.78, 3BS1-0.33-0.57, C-3BS1-0.33, C-3BL2-0.22, 3BL2-0.22-0.50, 3BL10-0.50-0.63, and 3BL7-0.63-1.00; Table I). A subset of 9,216 BACs representing the MTP of the new version of the 3B physical map was selected and rearrayed. Sixty-four three-dimensional (plate, row, and column) pools from the MTP were produced and subsequently used in this study. These clones also are being used to sequence the wheat chromosome 3B with a BAC-by-BAC approach based on second generation sequencing (http://urgi.versailles.inra.fr/index.php/urgi/Projects/3BSeq).
Table I.
Number and density of gene loci, number of nonsyntenic genes, and number of coexpressed genes per deletion bin in wheat chromosome 3B
| 3B Deletion Bin | Contig Size Assigned per Bin | No. of Loci | Locus Density (Locus/Mb) | No. of Nonsyntenic Genes | No. of Coexpressed Genes |
| Mb | |||||
| 3BS8-0.78-1.00 | 55.7 | 205 | 3.68 | 145 | 16 |
| 3BS9-0.57-0.78 | 72.9 | 193 | 2.65 | 123 | 12 |
| 3BS1-0.33-0.57 | 124.4 | 354 | 2.85 | 216 | 22 |
| C-3BS1-0.33 | 74.2 | 184 | 2.48 | 106 | 10 |
| C-3BL2-0.22 | 56.5 | 159 | 2.81 | 89 | 9 |
| 3BL2-0.22-0.50 | 103.9 | 300 | 2.89 | 170 | 14 |
| 3BL10-0.50-0.63 | 46.5 | 140 | 3.01 | 78 | 4 |
| 3BL7-0.63-1.00 | 206.4 | 661 | 3.20 | 395 | 60 |
| Total assigned | 740.7 | 2,196 | 2.96 | 1,322 | 147 |
| Not assigned | 220.3 | 728 | 480 | 39 | |
| Total | 961.0 | 2,924 | 1,802 | 186 |
A Chromosome-Wide Survey of the Wheat Gene Space Organization
We recently demonstrated the efficiency of hybridization experiments between EST microarrays and three-dimensional pools of the MTP to assign individual genes to wheat physical maps (Rustenholz et al., 2010). However, in this first experiment, the limited number of genes present on the barley microarray (approximately 15,000) and the sequence divergence between the two species enabled only a small number of genes to be mapped to chromosome 3B. The sequence divergence was also a major limitation for the analysis of gene expression. Thus, to perform a whole-chromosome expression profiling of wheat genes on chromosome 3B, we developed an in-house wheat NimbleGen 40K unigene microarray using the Triticum aestivum National Center for Biotechnology Information (NCBI) unigene build number 55 (February 2009; http://www.ncbi.nlm.nih.gov/UniGene/UGOrg.cgi?TAXID=4565). This so-called INRA_GDEC_T.aestivum_NimbleGen_12x40K_unigenes_chip_v1 chip contains on average three 60-mer probes per gene for 39,179 unigenes out of the complete 40,349 gene set.
The array was hybridized with the 64 MTP pools of the new chromosome 3B physical map. After signal quantification, normalization, and data deconvolution (see “Materials and Methods”), 2,913 unigenes were unambiguously assigned to 3,003 loci, including 78 unigenes that were found at multiple positions on chromosome 3B (two to four positions). Out of these 2,913 genes, 2,142 (71%) were found in the vicinity (same BAC or overlapping BACs) of at least one other gene. To assess whether such neighboring genes corresponded to single or independent loci, we rebuilt the relevant EST contigs using the NCBI EST data set. We then performed pairwise sequence comparisons of the resulting unigenes as well as homology search against the rice and B. distachyon genomes. In total, 77 genes showing at least 98% sequence identity with one of their neighbors were removed from the data set as they potentially matched the same gene. Eventually, 2,836 unigenes (2,924 gene loci) were unambiguously located on wheat chromosome 3B physical map and mapped to 2,071 BACs belonging to 1,016 individual contigs of 69 kb to 3.4 Mb.
To assess the reliability of these results, hybridization data corresponding to recently sequenced and manually annotated contigs were retrieved and compared to the reference annotations (Choulet et al., 2010). Out of 62 unigenes mapping to the contigs, seven (11%) were absent from the reference sequence and thus can be considered as false positives. The remaining 55 unigenes (89%) were confirmed by BLASTN analysis, demonstrating that unigene microarray hybridization is a powerful and reliable approach to map genes to BAC contigs and investigate the gene space organization. Interestingly, 17 out the 55 matched a previously nonannotated region. Among them, 70% displayed significant expression in at least one of the 15 cDNA samples, confirming that they do correspond to transcriptional units that were missed in the first annotation. The identification by tiling microarrays of new transcriptional units that had not been identified through computational annotation has been reported already in other species, such as Escherichia coli, Arabidopsis, fruit fly (Drosophila melanogaster), human (Homo sapiens), and rice (Bertone et al., 2005; Jiao et al., 2005; Stolc et al., 2005; Gregory et al., 2008).
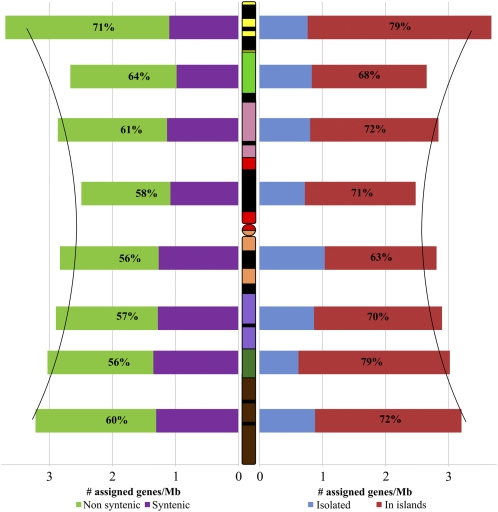
To further determine the location of genes along the chromosome, we used the deletion bin mapping information from the 1,669 BAC contigs of the 3B physical map. A total of 2,196 genes (75%) were assigned to one of the eight deletion bins, permitting us to calculate the gene density for each bin using the cumulated length of anchored contigs from the physical map in each deletion bin (Table I). The results showed that the gene density distribution is significantly correlated positively with the distance from the centromere (Pearson’s correlation coefficient r = 0.717; P value = 0.045), confirming the gradient of gene density from centromere to telomeres that was already reported by Choulet et al. (2010) and Rustenholz et al. (2010) (Fig. 1). The distal 3BS8-0.78-1.00 deletion bin revealed the highest gene density, with 3.7 genes per megabase, whereas the lowest gene density (2.5 genes/Mb) was observed in the proximal C-3BS1-0.33 deletion bin. One limitation of the hybridization-based gene mapping method is that it cannot discern whether there are one or more copies of the same gene on a BAC. Based on the assumption that tandemly duplicated genes represent approximately 30% of the genic content in the distal parts of the chromosome (Choulet et al., 2010) and on a conservative estimate of 6,000 genes for chromosome 3B (Paux et al., 2006), we extrapolated the gene density of the distal 3BS8-0.78-1.00 to 9.8 genes/Mb (one gene every 102 kb) and that of the proximal C-3BS1-0.33 deletion bin to 5.1 genes/Mb (one gene every 197 kb; see “Materials and Methods”), thereby suggesting a 2-fold increase of gene density from the centromere to the telomeres of wheat chromosome 3B as previously suggested by Choulet et al. (2010).
Figure 1.
Gene distribution along the wheat chromosome 3B. Each color on wheat chromosome 3B corresponds to a deletion bin. Yellow, 3BS8-0.78-1.00; light green, 3BS9-0.57-0.78; pink, 3BS1-0.33-0.57; red, C-3BS1-0.33; orange, C-3BL2-0.22; purple, 3BL2-0.22-0.50; dark green, 3BL10-0.50-0.63; and brown: 3BL7-0.63-1.00. The black segments correspond to heterochromatic regions identified by C-banding, the colored segments to the euchromatic regions, and the circle to centromere. On the left, the density of syntenic genes is represented by purple bars. The density of nonsyntenic genes is represented by green bars. The proportions of nonsyntenic genes per deletion bins are shown as percentages within the green bars. On the right, the density of isolated genes is represented by blue bars. The density of genes organized in island is represented by red bars. The proportions of genes organized in island per deletion bin are shown as percentages within the red bars. The two black curves represent the regression curve of gene density.
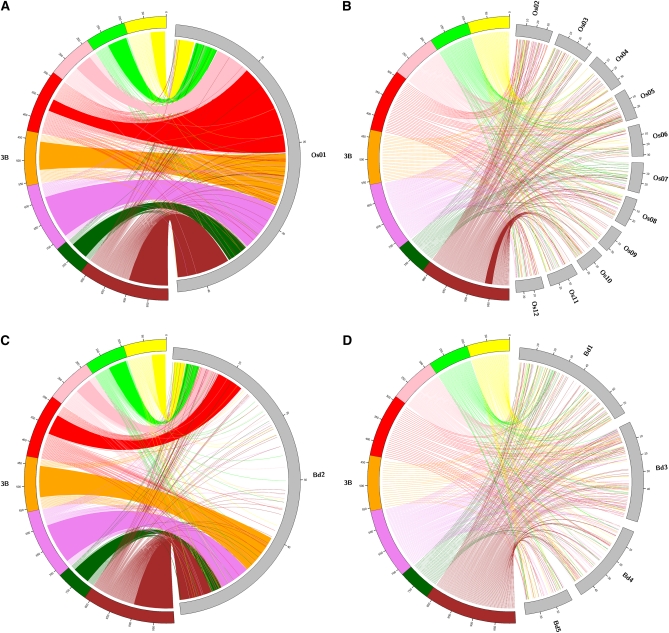
The information on gene positions in BACs and bins was used further to assess the level of synteny between wheat and rice or B. distachyon. The term synteny refers to genes in different organisms located on a chromosomal region originating from a common ancestor (Tang et al., 2008). Out of the 2,836 wheat chromosome 3B unigenes, 1,519 (54%) and 1,564 (55%) orthologous genes were found in rice and B. distachyon, respectively. Out of them, 877 (58%) and 1,016 (65%) were syntenic (i.e. orthologous to genes located on the orthologous rice chromosome 1 and B. distachyon chromosome 2, respectively; Fig. 2). By comparing the distribution of the syntenic genes with that of the nonsyntenic genes, we found that the gradient of gene density along chromosome 3B is mainly due to the presence of nonsyntenic genes (Pearson’s correlation coefficient r = 0.934; P value = 7E-4); whereas syntenic genes have no impact on the overall gradient (Pearson’s correlation coefficient r = 0.246; P value = 0.557; Table I; Fig. 1; Supplemental Table S1). In addition, the density of nonsyntenic genes correlated to the crossing-over rate estimated by Saintenac et al. (2009) (Pearson’s correlation coefficient r = 0.733; P value = 0.039).
Figure 2.
Syntenic relationships between wheat chromosome 3B and rice and B. distachyon chromosomes. A, Syntenic genes between wheat chromosome 3B (3B) and rice orthologous chromosome 1 (Os01). The axes along the chromosomes indicate the physical position in Mb. The eight deletion bins of wheat chromosome 3B are represented in colors as in Figure 1. Each line starting in a wheat deletion bin and ending on rice chromosome 1 represents a gene assigned to the deletion bin for which an orthologous gene on rice chromosome 1 was identified. The color of the line corresponds to the color of the deletion bin to which the gene was assigned. The colored blocks represent blocks of microsynteny, i.e. blocks of genes which location on wheat chromosome 3B and rice chromosome 1 is conserved. The genes within each deletion bin were ordered according to the order of their orthologs on rice chromosome 1. B, Nonsyntenic genes between wheat chromosome 3B (3B) and the other rice chromosomes. A block of genes between 3BL7 deletion bin and rice chromosome 10 was drawn as these genes are conserved with the same order than their orthologs on rice chromosome 10. C, Syntenic genes between wheat chromosome 3B (3B) and B. distachyon orthologous chromosome 2 (Bd2). D, Nonsyntenic genes between wheat chromosome 3B (3B) and the other B. distachyon chromosomes.
Recently, Choulet et al. (2010) defined gene islands as genes separated by <43 kb. Based on this definition, they estimated that 75% of the wheat genes belong to gene islands. In this study, a similar proportion (70%; 2040/2924) was observed when considering genes on the same or overlapping BACs. Thus, to further investigate the gene space organization, we considered gene islands as regions in which genes are located on the same or on overlapping BACs. These 2040 genes were found to be part of 709 gene islands composed of two to 30 genes (average = 2.9 ± 1.6, median = 2). Such a proportion of genes in islands is higher than the one expected from a random distribution as revealed by 10,000 random samplings without replacement of 2,924 gene locations on the chromosome 3B BACs (average = 57.9 ± 1.0%, P value < 0.0001). The remaining 884 genes (30%) were considered as isolated genes whose density was shown to be uniform along chromosome 3B and not correlated with the distribution of total gene density (χ2 test, P value = 0.347; Pearson’s correlation coefficient r = -0.077, P value = 0.857). The density of genes in islands varies among deletion bins (ranging from 63% in C-3BL2-0.22 to 79% in 3BL10-0.50-0.63) and is positively correlated with the distribution of the total gene density (Pearson’s correlation coefficient r = 0.951, P value = 3E-4; Fig. 1; Supplemental Table S1). Moreover, this density is also correlated with the density of nonsyntenic genes (Pearson’s correlation coefficient r = 0.898; P value = 0.002); whereas no correlation is found with the density of syntenic genes (Pearson’s correlation coefficient r = 0.207; P value = 0.622; Fig. 3; Supplemental Table S1). Thus, these results confirm that the gradient of gene density observed along the chromosome 3B results from an increase of genes in islands along the chromosome axis from centromere to telomeres. These results also indicate that the gene islands originated from genome rearrangements and were not formed by genes that were already close to each other in the ancestral grass genome.
Figure 3.
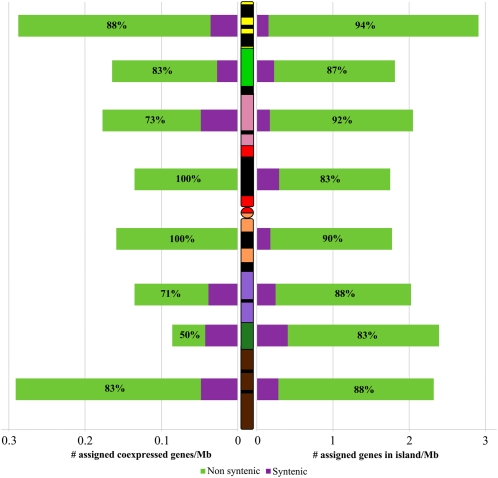
Proportion of syntenic and nonsyntenic gene islands and coexpression clusters along the wheat chromosome 3B. The purple bars represent the density of genes part of an island or a coexpression cluster composed by syntenic genes only. The green bars represent the density of genes part of an island or a coexpression cluster in which at least one nonsyntenic gene is present. On the left, the density of genes part of a coexpression cluster is shown. The proportions of genes part of coexpression cluster in which at least one nonsyntenic gene is present are shown as percentages within the green bars. On the right, the density of genes in island is shown. The proportions of genes in island in which at least one nonsyntenic gene is present are shown as percentages within the green bars.
Transcription Mapping of Chromosome 3B
To study the relationships between gene space organization and gene expression, we established a transcription map of chromosome 3B by hybridizing 15 cDNA samples that originated from five wheat organs (root, leaf, stem, spike, and grain) at three developmental stages onto the INRA_GDEC_T.aestivum_NimbleGen_12x40K_unigenes_chip_v1 microarray. After signal quantification and data normalization, transcript profiles were drawn for 32,284 unigenes (82%), including 2,515 of the 2,836 (89%) that were physically located on chromosome 3B. The remaining 6,895 unigenes (321 from 3B) did not show any significant hybridization signals in any of the 15 samples.
The expression profiles were validated for eight genes by quantitative reverse transcription-PCR (QRT-PCR; see “Materials and Methods”). The QRT-PCR expression profiles of seven genes were highly correlated with the profiles established by hybridization (Pearson’s correlation coefficient r > 0.7), whereas for one gene, no amplification was obtained, probably due to technical problems in primer design.
After hierarchical clustering of the unigenes located on chromosome 3B with a Pearson’s correlation coefficient threshold of 0.641 (P value = 0.01), 153 groups comprising 2,515 unigenes with similar expression profiles in the 15 samples were established. A total of 1,727 of them corresponded to genes that were located previously in islands on chromosome 3B. Among those, 186 (11%) show similar expression profiles with their neighbor(s), defining 80 so-called coexpression clusters of two to 21 genes (average = 2.3 ± 2.1, median = 2; Fig. 4). This proportion (11%) is significantly higher than the value obtained after 10,000 random samplings without replacement of the gene locations on the BACs (average = 3.9 ± 0.6%, P value < 0.0001), thereby suggesting selection for such coexpression clusters. In addition, the density of coexpressed genes in islands was significantly nonuniform along chromosome 3B (χ2 test, P value = 0.009) with more coexpressed genes in islands than expected in the two most distal deletion bins (13% in 3BS8-0.78-1.00 and 15% 3BL7-0.63-1.00; Table I). Of the 80 coexpression clusters identified on chromosome 3B, 63 (79%) contained at least one nonsyntenic gene, whereas only 17 (21%) were comprised of syntenic genes only (Fig. 3).
Figure 4.
Correlation maps of gene expression profiles. The correlations of gene expression profiles are represented using the green-magenta scale shown. Light green corresponds to positive correlation and light magenta to negative correlation. A, Correlation map of chromosome 3B. The chromosome 3B is schematically depicted as in Figure 1. The contigs were ordered as described in “Materials and Methods.” The green clusters on the diagonal line correspond to coexpression clusters. B, Zoom of the orange squared region in A. Some coexpression clusters are circled in yellow. A coregulation island composed of two coexpression clusters with correlated expression profiles distantly is circled in white.
In an attempt to assign putative functions to the wheat chromosome 3B unigenes, we used the Gene Ontology (GO) of rice and B. distachyon orthologs. GO function terms were associated with 1,175 unigenes out of the 2,836 unigenes located on wheat chromosome 3B (41%) among which 585 genes were located in gene islands. The average level in the GO classification was 2.81 for these unigenes (median = 3). To search for differential distribution of gene functions along the chromosome, we calculated the proportion of each GO term in each deletion bin. Two GO terms displayed a significant nonuniform distribution along the chromosome: binding (GO:0005488; χ2 test, P value = 0.025) and transporter activity (GO:0005215; χ2 test, P value = 0.034). The distribution of the transporter activity GO term along chromosome 3B was negatively correlated with the distance from centromere and the total gene density (Pearson’s correlation coefficient r = −0.832 and −0.941, P value = 0.010 and 5E-4, respectively). Of the 585 genes in islands associated with a GO term, 292 (50%) located in 125 gene islands shared at least one GO term with their neighbor(s). Compared with the average of 39.4% ± 2.4% obtained after 10,000 random samplings without replacement of the gene locations on BACs, this significantly higher value (P value < 0.0001) strongly suggests that genes sharing the same function tend to occur at adjacent or nearby positions in the genome. Furthermore, 91% (53/58) of the 58 coexpressed genes in islands with a GO term also shared one or more GO term(s) with their neighbor(s), a percentage that is significantly higher than the average observed by randomization of 10,000 samplings without replacement of the gene locations on the BACs (average = 30.7% ± 19.8%, P value = 0.008). Thus, we conclude that the coexpression clusters identified on wheat chromosome 3B are significantly enriched in genes sharing the same GO term(s), hereafter referred to as cofunctional genes.
A striking example is a gene-rich island located in the distal bin of the long arm of chromosome 3B (3BL7-0.63-1.00), which contains 30 genes distributed over 474 kb. Out of these 30 genes, 16 are found in the same order on the nonsyntenic rice chromosome 10. This region contains mainly housekeeping genes, such as genes coding for chlorophyll-associated proteins, ribosomal proteins, or phytochrome. Twelve of these 16 genes are coexpressed and cofunctional, three are coexpressed only, and one did not share any functional feature with any other member of the group. Transcriptomic data of rice chromosome 10 (see “Materials and Methods”) revealed that the orthologous genes in rice are also highly coexpressed (data not shown). Therefore, despite a large genome rearrangement that led to a syntenic break, microcollinearity as well as coregulation was maintained after wheat and rice diverged 50 to 70 million years ago. Surprisingly, this cluster is not conserved in B. distachyon as 20 of the 30 genes were not found in the genome sequence and the 10 remaining ones were located on three different and nonsyntenic chromosomes (namely, 1, 3, and 5).
Finally, the transcription correlation map of chromosome 3B (Fig. 4B) revealed correlations between the expression patterns of 18 physically distant coexpression clusters that were named “coregulation islands.” These were composed of two to seven distant coexpression clusters with the same expression profiles (as defined by the 153 profiles mentioned previously). These coregulation islands involved 54 of the 80 coexpression clusters (68%) and 132 of the 186 coexpressed genes (71%). In addition to their expression profiles, most of the genes involved in these coregulation islands also shared the same GO terms. The density of genes involved in coregulation islands was positively correlated significantly with the distance from the centromere (Pearson’s correlation coefficient r = 0.730, P value = 0.040). For example, the telomeric 3BS8-0.78-1.00 deletion bin showed 16 genes as part of coexpression clusters with all of them also involved in a coregulation island, whereas only 40% of the 10 coexpressed genes from the centromeric C-3BS1-0.33 deletion bin were involved in a coregulation island. In terms of synteny, eight coregulation islands were comprised exclusively of nonsyntenic genes, nine contained both syntenic and nonsyntenic genes, and only one was composed entirely of syntenic genes.
DISCUSSION
Genome Rearrangements Shaped the Wheat Genome through the Formation of Gene Islands
By hybridizing a newly developed wheat NimbleGen 40K unigene microarray with MTP BAC pools from the sequence-ready version of the chromosome 3B physical map as well as with 15 cDNA samples originating from five different tissues at three developmental stages, we established the first transcription map of a wheat chromosome. The 3B transcription map contains 2,924 gene loci corresponding to 2,836 unigenes, 2,515 of which are associated with transcript profiles and 1,175 with GO terms.
This unique resource allowed us to perform the most comprehensive structural and functional analysis of the wheat gene space to date and provided new evidence to support a model in which genes are distributed along the entire length of chromosomes and not specifically clustered in a few gene rich regions. Indeed, our 3,000 loci transcription map confirmed that the gene space spans the entire length of chromosome 3B, with a 2-fold gradient of gene density between the centromere and the telomeres. In addition, it suggests that 70% of the genes are organized in small gene islands composed of two to three genes on average and that the nonuniform distribution of genes in islands might explain the gene density gradient. These results are in complete agreement with previous studies based on EST mapping (Munkvold et al., 2004), individual BAC sequencing (Devos et al., 2005; Charles et al., 2008), megabase-sized contig sequencing (Choulet et al., 2010), and hybridization-based gene mapping onto physical map (Rustenholz et al., 2010).
Comparative analyses between the genes identified on wheat chromosome 3B and their orthologs on rice chromosome 1 suggested an overall synteny level of 58%, which is consistent with previous studies (La Rota and Sorrells, 2004; Munkvold et al., 2004; Varshney et al., 2005; Bilgic et al., 2007; Stein et al., 2007). The level of synteny was slightly higher with B. distachyon chromosome 2 (65%), as expected based on the more recent divergence time (30 million years) between these two species compared to rice (>50 million years; Bossolini et al., 2007; International Brachypodium Initiative, 2010). Detailed analysis in each deletion bin revealed a negative gradient of synteny conservation toward the telomeres. In addition, it also revealed a clear negative correlation between the level of conserved synteny and the proportion of genes in islands. All in all, our data strongly suggest that the gradient of gene density along the chromosome results from an increase in the proportion of genes in islands toward the telomeres and that most of these islands originate from genome-specific rearrangements that occurred after the divergence between the different grass lineages.
Gene islands, also referred to as gene-rich regions, are common features of large and highly repetitive plant genomes, especially at the distal parts of chromosomes (Paterson et al., 2009; Schnable et al., 2009; Schmutz et al., 2010). Different evolutionary scenarios have been proposed to explain the formation of these islands, including passive mechanisms that do not imply gene movement but rather differential insertion or deletion of transposable elements (TEs) into the genome to create gene islands from an ancestral backbone. A first example of these passive mechanisms was suggested for sorghum where homogeneous expansion of the genome combined with preferential deletions of TEs in gene-rich regions led to a gene space where most of the genes are found toward the end of the chromosomes (Paterson et al., 2009). This scenario is different for wheat according to Choulet et al. (2010) who sequenced and annotated 18 Mb of megabase-sized contigs on chromosome 3B and found that solo-long terminal repeats (LTRs) that are marks of LTR retrotransposon deletion through recombination were found mainly in large gene-poor regions. A second passive mechanism was proposed for different species, including maize, in which TEs insert preferentially in particular parts of the genome depending on their integrase affinity with specific chromatin protein (Baucom et al., 2009). This preferential insertion pattern was also observed in wheat, where LTR retrotransposons such as gypsy and copia display a clear insertion bias into each other (Paux et al., 2006) and therefore might be responsible for the formation of gene islands involving only syntenic genes.
Here, we observed that a significant percentage of gene islands are composed of nonsyntenic genes, thereby suggesting more active mechanisms where genes are duplicated, then translocated into the genome, and added to the ancestral gene backbone to create gene islands. This mechanism has been proposed for wheat by Choulet et al. (2010) based on evidence suggesting TE-mediated interchromosomal gene duplications. First, there was a significant correlation between the proportion of nonsyntenic genes and the age of TEs, as an estimate of their activity: the younger the TEs, the lower the synteny level. Second, four of the 158 annotated genes were included fully into CACTA transposons, and the authors estimated that in total approximately 200 gene fragments could result from TE-mediated gene capture on chromosome 3B, similar to what has been reported for the sorghum genome (Paterson et al., 2009). Even if TEs can be involved in gene amplification and mobilization in wheat, it is highly unlikely that this mechanism led to the movement of almost 4,000 genes, as estimated by the overall synteny level of 52% to 58% between wheat and rice. Wicker et al. (2010) reached the same conclusion in a recent study where they performed a three-way comparison of B. distachyon, rice, and sorghum genomes. In addition to the TE-driven gene capture, the authors proposed a mechanism where foreign fragments containing genes were introduced as filler DNA to repair double-strand breaks (DSBs) that occurred upon TE insertion or during recombination. Interestingly, our study clearly shows a strong correlation between the proportion of nonsyntenic genes and the crossing-over rate, suggesting a putative relationship between gene movement and recombination.
Thus, we conclude that gene islands in wheat originate from a combination of different factors, including the preferential insertion of LTR retrotransposons into each other that expand intergenic regions, TE-mediated gene movements, and repair of DSBs produced by TEs or recombination. However, the relative contribution of these different factors remains to be determined, and future access to reference sequences of wheat chromosomes that are currently underway under the umbrella of the International Wheat Genome Sequencing Consortium (http://www.wheatgenome.org/Projects/IWGSC-Bread-Wheat-Projects) will help to address these questions.
Selection for Clusters of Coexpressed and Cofunctional Genes Have Shaped the Gene Order in Wheat
In addition to the mechanisms underlying the creation of gene island, functional selection pressures, such as for gene regulation and coexpression, have been proposed as driving forces for maintaining genes physically close to each other in islands (Hurst et al., 2004; Batada et al., 2007; Babu et al., 2008; Chen et al., 2010; Choulet et al., 2010). Numerous studies have detailed the two main mechanisms involved in the cis-regulation of adjacent coexpressed genes. First, promoters, whether bidirectional or not, enhancers, and regulatory sequences binding transcription factors can induce coexpression of multiple genes within short distances (Hurst et al., 2004; Babu et al., 2008; Chen et al., 2010). These regulatory elements could lead to polycistronic transcripts, i.e. transcripts containing two or more open reading frames, implying perfect coexpression as observed in Arabidopsis (Thimmapuram et al., 2005; Chen et al., 2010). However, Chen et al. (2010) suggested that such direct interactions are enhanced at gene distances below 400 bp in Arabidopsis. For large intergenic distances, chromatin modifications are likely favored to induce coexpression of multiple genes as they allow regions to switch between repression (heterochromatin) and activation of transcription (euchromatin; Hurst et al., 2004; Batada et al., 2007; Babu et al., 2008; Chen et al., 2010).
In the human, mouse (Mus musculus), Arabidopsis, and rice genomes, the percentage of adjacent coexpressed genes ranges from 2% to 10% with two to four genes involved (Ren et al., 2005, 2007; Sémon and Duret, 2006; Zhan et al., 2006). In fruit fly, Spellman and Rubin (2002) found 20% of adjacent genes that were part of large coexpression domains involving 10 to 30 genes. Here, we found that gene islands were significantly enriched in genes having the same transcription profiles. This proportion may be even higher if we consider that we only analyzed one-third of the putative 8,000 genes located on chromosome 3B. In particular, the hybridization-based approach cannot detect tandemly duplicated genes that have been shown to be more prone to coexpression than adjacent nontandemly duplicated genes (Williams and Bowles, 2004; Ren et al., 2005; Zhan et al., 2006). We also found a few coexpressed gene clusters that were conserved between wheat and rice or B. distachyon, suggesting that genes maintained their proximity during the evolution and that common regulatory mechanisms are also conserved between plant genomes. Other examples have been reported of coexpressed and cofunctional gene pairs conserved between Arabidopsis, rice, and poplar (Krom and Ramakrishna, 2008; Liu and Han, 2009). However, in addition to the syntenic genes, we found that more than two-thirds of the coexpression clusters also contained nonsyntenic genes when compared to rice and/or B. distachyon genes. As discussed previously, these clusters likely originate from gene movement mediated by TE or from DSB repair and could have acquired shared expression patterns. If this is the case, then we can hypothesize that when a gene is translocated from one genome region to another, it may be under specific selection pressure to acquire an expression pattern that correlates strongly with the expression patterns of the domain of integration. A similar phenomenon has been described previously by Gierman et al. (2007) who analyzed GFP reporter constructs inserted at different chromosomal loci in the human genome. They found that the GFP expression levels corresponded to the transcriptional activity of the integration domain, demonstrating that a gene inserted in the vicinity of another gene has a significant probability to share the expression profile with its neighbor. Based on our current data, we cannot estimate the relative contribution of each type of regulation (direct interactions and chromatin effects) on gene coexpression in wheat. Nevertheless, this might be investigated in a near future when the whole-chromosome 3B sequence will be released.
In addition to coexpressed genes, we found a significant number of cofunctional genes, i.e. sharing the same GO term(s). Some also shared the same expression profiles, but the majority was not coexpressed. For the latter, we could hypothesize that we failed to detect their coexpression as we established the expression profiles on a limited number of cDNA samples. Furthermore, the expression profiles that we established may not reflect the exact expression profiles of genes located on chromosome 3B since microarray hybridization does not allow discrimination between the homoeologous A, B, and D copies that are potentially present and transcribed for each individual gene in hexaploid wheat. However, studies in various model organisms demonstrated that organization in clusters was not only found for coexpressed genes but also for genes involved in the same metabolic pathway and/or associated in protein-protein complexes (Cohen et al., 2000; Lee and Sonnhammer, 2003; Krom and Ramakrishna, 2008; Xu et al., 2008). The mechanisms involved in neighbor gene regulation described above cannot explain the tendency of noncoexpressed cofunctional genes to occur in adjacent or nearby locations in the genome. To some extent, clusters of coexpressed and/or cofunctional genes might result from neutral coevolution (Sémon and Duret, 2006). However, assuming that such neutral coevolution would result in a random distribution of coexpression and/or cofunction clusters, our results suggest that clusters in wheat, at least partially, result from natural selection as they are found more often than would be expected if by chance. Similar conclusions have been reached already in other organisms (for review, see Hurst et al., 2004); therefore, it is very likely that rearrangements in the wheat genome have led to the formation of coexpression and/or cofunctional clusters that were maintained due to some selective advantage, as proposed by Batada et al. (2007), Sémon and Duret (2006), and Babu et al. (2008). The fact that these clusters are more abundant in the distal parts of chromosomes corresponding to the most dynamic regions of the genome (Eichler and Sankoff, 2003; See et al., 2006) suggests a role of this phenomenon in wheat evolution and adaptation.
The correlation map of the wheat chromosome 3B also enabled us to identify 18 putative intrachromosomal coregulation islands, i.e. clusters of genes that share the same expression profiles distantly on the same or different chromosomes. The density of genes involved in coregulation islands was positively correlated with the distance from centromere. These findings suggest possible mechanisms underlying long-distance regulation of gene expression in wheat. Hurst et al. (2004) and Babu et al. (2008) described various mechanisms for long-range regulation in three dimensions in contrast to the cis-regulation of adjacent genes. In wheat, the interphase chromosomes adopt a regular Rabl configuration, a highly polarized pattern with the two chromosome arms lying next to each other and the centromeres and telomeres located at opposite poles of the nuclei (Dong and Jiang, 1998; Cowan et al., 2001; Santos et al., 2002). Strikingly, this configuration departs from the classical cloud-shaped nuclei observed in many organisms during interphase. Based on these findings, we hypothesize that the Rabl organization might be involved in the positioning of genes within the three-dimensional chromatin structure of the wheat genome, therefore promoting long-distance coregulation of gene clusters. Chromosome conformation mapping experiments, such as in situ hybridization or chromosome conformation capture (3C; Shaw, 2010), will be necessary to test this hypothesis.
Here, by constructing a transcription map of wheat chromosome 3B, we gained significant insights in the organization, evolution, and function of the wheat gene space. Our results strongly suggest that the wheat genome has evolved more rapidly and more dramatically than the model genomes of rice and B. distachyon. The development of other physical maps from the hexaploid wheat genome currently underway within the International Wheat Genome Sequencing Consortium framework (www.wheatgenome.org) will enable similar studies at the whole-genome scale in the near future, while the sequence of chromosome 3B, which is currently underway (http://urgi.versailles.inra.fr/index.php/urgi/Projects/3BSeq), should provide the foundation for progressing further in our understanding of gene expression regulation and evolution in wheat.
MATERIALS AND METHODS
Construction of the Second Version of the Wheat Chromosome 3B Physical Map and Production of MTP BAC Pools
A chromosome 3B-specific BAC library containing 82,176 clones was constructed as described by Simková et al. (2011). The 82,176 BAC clones together with the 7,440 MTP BAC clones originating from the first version of the physical map were fingerprinted using a slightly modified HICF SNaPshot protocol that uses a combination of five type II restriction enzymes and capillary electrophoresis on automated sequencers (Paux et al., 2008). A total of 78,840 high-quality fingerprints were obtained using the FPB software (Scalabrin et al., 2009) and analyzed with fingerprinted contigs program (Soderlund et al., 1997, 2000) to be combined with the first version of the chromosome 3B physical map containing 1,036 contigs. Briefly, the initial build of chromosome 3B was performed by incremental contig building with a cutoff of 1e-75 and a tolerance of 4. These were subsequently run through keyset-to-fingerprinted contigs program, single-to-end, and end-to-end merging (match: 1; from end: 55) at six successively higher cutoffs terminating at 1e-45. The DQer function was used after each merge to break up all contigs that contained >10% of questionable (Q) clones (step: 3). Sixty-four three-dimensional (plate, row, and column) MTP BAC pools were produced following the procedure described by Paux et al. (2008).
The Wheat NimbleGen 40K Unigene Microarray
The wheat (Triticum aestivum) NimbleGen 40K unigene microarray was designed using the Triticum aestivum NCBI unigene set build number 55 counting 40,349 unigenes, resulting from the assembly of 960,174 ESTs and mRNAs (http://www.ncbi.nlm.nih.gov/UniGene/UGOrg.cgi?TAXID=4565).
Unigene sequences were masked based on a k-mer frequency analysis. A 17-mer-based mathematically defined repeat index was built with Tallymer (Kurtz et al., 2008) using 2 Gb from Illumina reads produced from sorted 3B chromosomal DNA (accession no. ERA000182). K-mer frequency of all unigene sequences was thus computed using this mathematically defined repeat index. Seventeen-mers repeated five times or more in the index were masked to exclude repeated motifs from the probe design. The masked sequences were submitted to NimbleGen (Roche NimbleGen) for probe design using proprietary algorithms. In total, 39,179 unigenes were represented by at least one 60-mer probe: 39,019 unigenes with three probes, 78 with two probes, and 82 with one probe. These probes were synthesized onto 12×135K arrays. The chip has been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under the name INRA_GDEC_T.aestivum_NimbleGen_12x40K_unigenes_chip_ v1 (ref. A-MEXP-1928).
Microarray Hybridizations
Total RNAs of hexaploid wheat cv Chinese Spring were extracted in duplicate from five organs (root, leaf, stem, spike, and grain) at three developmental stages each (beginning, middle, and end of the development) and were reverse transcribed as detailed by Choulet et al. (2010).
The 64 MTP pools were sonicated to obtain an average fragment size between 500 and 2,000 bp. The cDNA and the MTP pool labeling were carried out using the NimbleGen Dual-Color DNA labeling kit (Roche NimbleGen) according to the manufacturer’s procedure for gene expression analysis. Dual-color hybridizations were performed on each plex of the arrays. The labeled cDNAs and MTP pools were hybridized independently. The pairs of samples were randomly chosen and allocated to a plex. A dye swap was performed for all the duplicate cDNAs making four repetitions for each of the 15 samples. Hybridizations and washing of the arrays were performed according to the manufacturer’s procedure for gene expression analysis (Roche NimbleGen). The arrays were scanned using the InnoScan 900AL scanner (Innopsys). Data were extracted from scanned images using NimbleScan 2.5 software (Roche NimbleGen), which allows for automated grid alignment, extraction, and generation of data files. All data has been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/; accession no. E-TABM-1095) under MIAME guidelines (http://www.mged.org/Workgroups/MIAME/miame.html).
Normalization and Data Deconvolution of the MTP Pool Data
The normalization and the data deconvolution were performed using automated scripts developed with the R software (www.r-project.org). First intensity values were checked for each gene and each MTP pool. If one probe was found to be an outlier among the three probes per unigene, it was deleted from further analysis. Data from each MTP pool were then made comparable with each other by subtracting the median to each intensity value and then by dividing by the sd. Three files were generated corresponding to the normalized intensities for the plate, row, and column pools. The three pool types were treated separately. Two complementary methods with three stringency thresholds each were then used to detect the positive signals.
The first method, called The Mean + X × Standard Deviation method, required the calculation of the median for the intensities of probes corresponding to the same gene and to the same pool. Then, the same method rather than the automated scoring method described by Rustenholz et al. (2010) was applied to the data with minor modifications. Indeed, the coefficients multiplying the sd were modulated and the plate and column pools shared the same coefficients (coefficients for high thresholds: plate and column = 2.8 and row = 2.5; coefficients for medium thresholds: plate and column = 2.4 and row = 2.3; coefficients for low thresholds: plate and column = 2.2 and row = 2.1).
The second method, called the t test method, used Student’s t tests for each gene to compare intensities of one pool to the intensities of the others. Variance was considered to be equal. The three stringency levels corresponded to the three P value thresholds (high, P value threshold = 0.01; medium, P value threshold = 0.025; and low, P value threshold = 0.05).
Data deconvolution was carried out independently for both methods and stringency levels as described by Rustenholz et al. (2010). All BAC addresses found with the high thresholds of both methods were retained. Then, for the medium thresholds, only the unigenes located on the overlap of two BACs that were not previously identified by the high thresholds were added. Finally, for the low thresholds, only the unigenes located on the overlap of two BACs and that were not previously identified by the high and medium thresholds were added.
Transcriptomic Data Analyses and Validation
The normalization of the transcriptomic data were performed using automated scripts developed with the R software (www.r-project.org). First, each image was divided into 30 identical zones. The background intensity was estimated using the median of the intensities of the empty spots and of the spots made of random sequences for each zone and subtracted to each spot intensity within the zone. Then, two successive lowess corrections were undertaken to correct the dye and the duplicate biases. Afterward, the corrected intensity values were checked for each gene, and the aberrant values were deleted. Student’s t tests were performed to compare the corrected intensity values for each gene with the corrected background intensity values. The P value threshold was set at 0.05. The median of the corrected intensity values was calculated for each significantly expressed gene and was considered as the expression value of the gene for the cDNA sample. To harmonize the expression values between the 15 cDNA samples, the expression values of each significantly expressed gene were subtracted by the median of the expression values of the significantly expressed genes for the cDNA sample and then divided by the sd. All the expression values were finally made positive, and the expression value of a gene that was not significant expressed in a cDNA sample was set to 0.
The validation of the expression profiles established through microarray hybridization was carried out on eight genes. The QRT-PCR experiments were performed on the 30 cDNA samples (15 samples extracted in duplicate) in duplicate on the LightCycler 480 (Roche Diagnostics) using the LightCycler 480 SYBR Green I Master mix (Roche Diagnostics) following the manufacturer’s procedure. The data were analyzed using the LightCycler480 software release 1.5.0 (Roche Diagnostics) with the absolute quantification/fit points method. The median of the four values obtained for each sample was calculated to be compared with the expression data from microarray hybridization.
Hierarchical clustering was performed using the Hierarchical Clustering Explorer 3.0 software (http://www.cs.umd.edu/hcil/hce/hce3.html) with the complete linkage method and the Pearson correlation coefficient. The minimal similarity to establish the clusters was set to 0.641, which is a Pearson correlation significant at the P value threshold of 0.01.
We used the rice transcriptomic data produced by Wang et al. (2010), as they sampled the same organs that we selected on the entire life cycle of the plant. Medians were calculated for the expression data corresponding to root, leaf, stem, spike, and grain parts.
Statistical Analyses
The statistical analyses were performed using automated scripts developed with the R software (www.r-project.org).
The script developed to validate the wheat gene space organization in gene islands performed 10,000 random samplings with replacements of 2,924 BACs out of all the MTP BACs of wheat chromosome 3B. It then calculated the percentage of identical and overlapping BACs retrieved. The other scripts used to validate the coexpression level, the cofunction level, and the cofunction level of the coexpressed genes performed 10,000 random samplings without replacements of the 2,924 chromosome 3B BAC addresses where a gene was located. They then calculated either the percentage of coexpressed genes retrieved at the gene island scale, the percentage of cofunctional genes retrieved at the gene island scale, or the percentage of coexpressed and cofunctional genes retrieved at the gene island scale.
χ2 tests were performed to check the uniformity of a gene density distribution along wheat chromosome 3B. First, the average density was calculated by dividing the sum of genes (isolated or coexpressed genes) by the total length of contigs assigned to the deletion bins. Then, the number of genes per deletion bin under the uniform law was calculated by multiplying the average density with the length of contigs assigned per deletion bin. The numbers of genes observed for all the deletion bins and the numbers of genes for all deletion bins under the uniform law were compared through a χ2 test. The P value threshold was set to 0.05.
The extrapolation of the gene density assuming that the chromosome 3B carries 6,000 genes (Paux et al., 2008) and that the telomeric deletion bins are composed by approximately 30% of tandemly duplicated genes was performed as follows. First, the number of genes in the fraction of the deletion bins that are not covered by contigs was estimated assuming the same gene density within the deletion bins. Then, the percentage of genes per deletion bin out of the total number of genes on chromosome 3B was recalculated and used to extrapolate the number of genes per deletion bin, assuming that the chromosome 3B carries 6,000 genes. Finally, the number of genes in the two most telomeric deletion bins (3BS8-0.78-1.00 and 3BL7-0.63-1.00) was increased by 30% to simulate the tandemly duplicated genes. These numbers of genes were used to calculate the extrapolated gene densities per deletion bin.
Classical Pearson’s correlation coefficient tests were performed to check the correlation between various variables. The P value threshold was set at 0.05.
Correlation matrixes were established by calculating Pearson’s correlation coefficient of the expression profiles between all the pairs of genes on wheat chromosome 3B or on a specific region of rice chromosome 10 (LOC_Os10g21194 to LOC_Os10g38276). The wheat genes were ordered thanks to their assignation to a deletion bin. The wheat contigs within the deletion bins were randomly ordered. The correlation maps were established based on the correlation matrixes using the corrplot package (http://cran.r-project.org/).
Sequence Analyses
Since EST assembly used for the unigene design is not available at the NCBI Web site, we rebuilt the EST contigs to check if some could exhibit highly similar regions. FASTA sequences of ESTs belonging to the same cluster have been pooled together and assembled using Phrap (http://www.phrap.org/) to rebuild the EST contigs. Two rounds of assembly were performed: first, using default Phrap parameters; second, with relaxed stringency in order to assemble EST clusters for which two or more contigs were obtained after the first round. Then, similarity search between EST contigs was conducted using BLASTN (Altschul et al., 1997). Alignments were parsed to keep only the best hits with at least 90% of sequence identity on at least 20 bp. Pairwise comparisons of EST contigs were performed (BLASTN; E value = 1), and unigenes showing at least 98% of sequence identity, mapped to the same BAC clone, and potentially matching the same gene were removed from further analyses. BLASTN (Altschul et al., 1997) analyses of the unigene sequences provided by the NCBI and of the sequences of the probes designed within the unigene sequences against the sequenced contigs of chromosome 3B (Choulet et al., 2010) were performed (BLASTN, E value = 1E-15).The Artemis viewer (Rutherford et al., 2000) was used to find the annotation (gene, nonannotated region, or repeat) at the hit position. BLASTX (Altschul et al., 1997) analyses of the unigene sequences against the databases of all the rice (Oryza sativa) peptides (http://rice.plantbiology.msu.edu) and of all the Brachypodium distachyon peptides (http://www.brachypodium.org) were performed to identify to orthologs in the wheat genome (E value = 1). The results were parsed to keep the best hits with at least 50% of sequence similarity on at least 33 amino acids. Every unigene with a hit meeting these criteria was considered as orthologous the rice or B. distachyon gene identified. Figure 2 was drawn using Circos (Krzywinski et al., 2009) to represent the syntenic relationships between wheat chromosome 3B and the rice genome and between wheat chromosome 3B and the B. distachyon genome. The genes within each deletion bin were ordered using to the chromosome and the physical position of their orthologs. The GO annotations from rice (164,737 GO terms; 29,753 genes) and from B. distachyon (71,123 GO terms; 15,439 genes; ftp://ftp.gramene.org/pub/gramene/CURRENT_RELEASE/data/ontology/go/) were used to assess the GO of the wheat genes orthologous to rice and/or B. distachyon genes. A GO-Slim annotation (goslim_plant.obo selected) was performed using Blast2GO (http://www.blast2go.org/).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FN564426 to FN564437 and FN645450.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Correlations between gene density values.
Acknowledgments
We thank Isabelle Bertin and Nelly Cubizolles for technical assistance with the QRT-PCR experiment, Pierre Sourdille for helpful comments and discussions, and Kellye Eversole for critical reading and editing of the manuscript. Hybridization experiments were performed at Institut National de la Recherche Agronomique Clermont-Ferrand on the GENTYANE genotyping platform (http://www4.clermont.inra.fr/umr1095/Equipes/Plates-formes-techniques-et-experimentales/Genotypage-a-haut-debit).
References
- Akhunov ED, Goodyear AW, Geng S, Qi LL, Echalier B, Gill BS, Miftahudin , Gustafson JP, Lazo G, Chao S, et al. (2003) The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res 13: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Babu MM, Janga SC, de Santiago I, Pombo A. (2008) Eukaryotic gene regulation in three dimensions and its impact on genome evolution. Curr Opin Genet Dev 18: 571–582 [DOI] [PubMed] [Google Scholar]
- Batada NN, Urrutia AO, Hurst LD. (2007) Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends Genet 23: 480–484 [DOI] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL. (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Gerstein M, Snyder M. (2005) Applications of DNA tiling arrays to experimental genome annotation and regulatory pathway discovery. Chromosome Res 13: 259–274 [DOI] [PubMed] [Google Scholar]
- Bilgic H, Cho S, Garvin DF, Muehlbauer GJ. (2007) Mapping barley genes to chromosome arms by transcript profiling of wheat-barley ditelosomic chromosome addition lines. Genome 50: 898–906 [DOI] [PubMed] [Google Scholar]
- Bossolini E, Wicker T, Knobel PA, Keller B. (2007) Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J 49: 704–717 [DOI] [PubMed] [Google Scholar]
- Brooks SA, Huang L, Gill BS, Fellers JP. (2002) Analysis of 106 kb of contiguous DNA sequence from the D genome of wheat reveals high gene density and a complex arrangement of genes related to disease resistance. Genome 45: 963–972 [DOI] [PubMed] [Google Scholar]
- Chantret N, Cenci A, Sabot F, Anderson O, Dubcovsky J. (2004) Sequencing of the Triticum monococcum hardness locus reveals good microcolinearity with rice. Mol Genet Genomics 271: 377–386 [DOI] [PubMed] [Google Scholar]
- Charles M, Belcram H, Just J, Huneau C, Viollet A, Couloux A, Segurens B, Carter M, Huteau V, Coriton O, et al. (2008) Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 180: 1071–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, de Meaux J, Lercher MJ. (2010) Co-expression of neighbouring genes in Arabidopsis: separating chromatin effects from direct interactions. BMC Genomics 11: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, Schlub S, Le Paslier MC, Magdelenat G, Gonthier C, et al. (2010) Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22: 1686–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Tuteja JH, Li M, Marek LF, Shoemaker RC, Vodkin LO. (2004) Features of a 103-kb gene-rich region in soybean include an inverted perfect repeat cluster of CHS genes comprising the I locus. Genome 47: 819–831 [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM. (2000) A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet 26: 183–186 [DOI] [PubMed] [Google Scholar]
- Cowan CR, Carlton PM, Cande WZ. (2001) The polar arrangement of telomeres in interphase and meiosis. Rabl organization and the bouquet. Plant Physiol 125: 532–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Ma J, Pontaroli AC, Pratt LH, Bennetzen JL. (2005) Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc Natl Acad Sci USA 102: 19243–19248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Jiang J. (1998) Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res 6: 551–558 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Sankoff D. (2003) Structural dynamics of eukaryotic chromosome evolution. Science 301: 793–797 [DOI] [PubMed] [Google Scholar]
- Erayman M, Sandhu D, Sidhu D, Dilbirligi M, Baenziger PS, Gill KS. (2004) Demarcating the gene-rich regions of the wheat genome. Nucleic Acids Res 32: 3546–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C, Leach JE, Rogers J, Schnable PS, Eversole K. (2011) Crop genome sequencing: lessons and rationales. Trends Plant Sci 16: 77–88 [DOI] [PubMed] [Google Scholar]
- Gierman HJ, Indemans MH, Koster J, Goetze S, Seppen J, Geerts D, van Driel R, Versteeg R. (2007) Domain-wide regulation of gene expression in the human genome. Genome Res 17: 1286–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, Yazaki J, Ecker JR. (2008) Utilizing tiling microarrays for whole-genome analysis in plants. Plant J 53: 636–644 [DOI] [PubMed] [Google Scholar]
- Guo W, Cai C, Wang C, Zhao L, Wang L, Zhang T. (2008) A preliminary analysis of genome structure and composition in Gossypium hirsutum. BMC Genomics 9: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pál C, Lercher MJ. (2004) The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jackson S, Hass Jacobus B, Pagel J. (2004) The gene space of the soybean genome. Stalker HT, Brummer EC, Wison RF, , Legume Crop Genomics. AOCS Publishing, Champaign, IL, pp 187–193 [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. ; French-Italian Public Consortium for Grapevine Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Jia P, Wang X, Su N, Yu S, Zhang D, Ma L, Feng Q, Jin Z, Li L, et al. (2005) A tiling microarray expression analysis of rice chromosome 4 suggests a chromosome-level regulation of transcription. Plant Cell 17: 1641–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krom N, Ramakrishna W. (2008) Comparative analysis of divergent and convergent gene pairs and their expression patterns in rice, Arabidopsis, and populus. Plant Physiol 147: 1763–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Narechania A, Stein JC, Ware D. (2008) A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes. BMC Genomics 9: 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rota M, Sorrells ME. (2004) Comparative DNA sequence analysis of mapped wheat ESTs reveals the complexity of genome relationships between rice and wheat. Funct Integr Genomics 4: 34–46 [DOI] [PubMed] [Google Scholar]
- Lee JM, Sonnhammer EL. (2003) Genomic gene clustering analysis of pathways in eukaryotes. Genome Res 13: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Han B. (2009) Evolutionary conservation of neighbouring gene pairs in plants. Gene 437: 71–79 [DOI] [PubMed] [Google Scholar]
- Munkvold JD, Greene RA, Bermudez-Kandianis CE, La Rota CM, Edwards H, Sorrells SF, Dake T, Benscher D, Kantety R, Linkiewicz AM, et al. (2004) Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 168: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C. (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48: 463–474 [DOI] [PubMed] [Google Scholar]
- Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W, et al. (2008) A physical map of the 1-gigabase bread wheat chromosome 3B. Science 322: 101–104 [DOI] [PubMed] [Google Scholar]
- Ren XY, Fiers MW, Stiekema WJ, Nap JP. (2005) Local coexpression domains of two to four genes in the genome of Arabidopsis. Plant Physiol 138: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XY, Stiekema WJ, Nap JP. (2007) Local coexpression domains in the genome of rice show no microsynteny with Arabidopsis domains. Plant Mol Biol 65: 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustenholz C, Hedley PE, Morris J, Choulet F, Feuillet C, Waugh R, Paux E. (2010) Specific patterns of gene space organisation revealed in wheat by using the combination of barley and wheat genomic resources. BMC Genomics 11: 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. (2000) Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945 [DOI] [PubMed] [Google Scholar]
- Saintenac C, Falque M, Martin OC, Paux E, Feuillet C, Sourdille P. (2009) Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.). Genetics 181: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu D, Gill KS. (2002) Gene-containing regions of wheat and the other grass genomes. Plant Physiol 128: 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AP, Abranches R, Stoger E, Beven A, Viegas W, Shaw PJ. (2002) The architecture of interphase chromosomes and gene positioning are altered by changes in DNA methylation and histone acetylation. J Cell Sci 115: 4597–4605 [DOI] [PubMed] [Google Scholar]
- Scalabrin S, Morgante M, Policriti A. (2009) Automated FingerPrint Background removal: FPB. BMC Bioinformatics 10: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- See DR, Brooks S, Nelson JC, Brown-Guedira G, Friebe B, Gill BS. (2006) Gene evolution at the ends of wheat chromosomes. Proc Natl Acad Sci USA 103: 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Duret L. (2006) Evolutionary origin and maintenance of coexpressed gene clusters in mammals. Mol Biol Evol 23: 1715–1723 [DOI] [PubMed] [Google Scholar]
- Shaw PJ. (2010) Mapping chromatin conformation. F1000 Biol Rep 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simková H, Safář J, Kubaláková M, Suchánková P, Cíhalíková J, Robert-Quatre H, Azhaguvel P, Weng Y, Peng J, Lapitan NL, et al. (2011) BAC libraries from wheat chromosome 7D: efficient tool for positional cloning of aphid resistance genes. J Biomed Biotechnol 2011: 302543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Humphray S, Dunham A, French L. (2000) Contigs built with fingerprints, markers, and FPC V4.7. Genome Res 10: 1772–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Longden I, Mott R. (1997) FPC: a system for building contigs from restriction fingerprinted clones. Comput Appl Biosci 13: 523–535 [DOI] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. (2002) Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N, Prasad M, Scholz U, Thiel T, Zhang HN, Wolf M, Kota R, Varshney RK, Perovic D, Grosse I, et al. (2007) A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor Appl Genet 114: 823–839 [DOI] [PubMed] [Google Scholar]
- Stolc V, Li L, Wang X, Li X, Su N, Tongprasit W, Han B, Xue Y, Li J, Snyder M, Gerstein M, Wang J, et al. (2005) A pilot study of transcription unit analysis in rice using oligonucleotide tiling-path microarray. Plant Mol Biol 59: 137–149 [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. (2008) Synteny and collinearity in plant genomes. Science 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Thimmapuram J, Duan H, Liu L, Schuler MA. (2005) Bicistronic and fused monocistronic transcripts are derived from adjacent loci in the Arabidopsis genome. RNA 11: 128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Varshney RK, Sigmund R, Börner A, Korzun V, Stein N, Sorrells ME, Langridge P, Graner A. (2005) Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci 168: 195–202 [Google Scholar]
- Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J, et al. (2010) A dynamic gene expression atlas covering the entire life cycle of rice. Plant J 61: 752–766 [DOI] [PubMed] [Google Scholar]
- Wei F, Stein JC, Liang C, Zhang J, Fulton RS, Baucom RS, De Paoli E, Zhou S, Yang L, Han Y, et al. (2009) Detailed analysis of a contiguous 22-Mb region of the maize genome. PLoS Genet 5: e1000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Buchmann JP, Keller B. (2010) Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res 20: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Zimmermann W, Perovic D, Paterson AH, Ganal M, Graner A, Stein N. (2005) A detailed look at 7 million years of genome evolution in a 439 kb contiguous sequence at the barley Hv-eIF4E locus: recombination, rearrangements and repeats. Plant J 41: 184–194 [DOI] [PubMed] [Google Scholar]
- Williams EJ, Bowles DJ. (2004) Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res 14: 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Kohel RJ, Song G, Cho J, Alabady M, Yu J, Koo P, Chu J, Yu S, Wilkins TA, et al. (2008) Gene-rich islands for fiber development in the cotton genome. Genomics 92: 173–183 [DOI] [PubMed] [Google Scholar]
- Zhan S, Horrocks J, Lukens LN. (2006) Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J 45: 347–357 [DOI] [PubMed] [Google Scholar]