Abstract
Thioredoxins (Trxs) are a multigenic family of proteins in plants that play a critical role in redox balance regulation through thiol-disulfide exchange reactions. There are 10 members of the h-type Trxs in rice (Oryza sativa), and none of them has been clearly characterized. Here, we demonstrate that OsTRXh1, a subgroup I h-type Trx in rice, possesses reduction activity in vitro and complements the hydrogen peroxide sensitivity of Trx-deficient yeast mutants. OsTRXh1 is ubiquitously expressed in rice, and its expression is induced by salt and abscisic acid treatments. Intriguingly, OsTRXh1 is secreted into the extracellular space, and salt stress in the apoplast of rice induces its expression at the protein level. The knockdown of OsTRXh1 results in dwarf plants with fewer tillers, whereas the overexpression of OsTRXh1 leads to a salt-sensitive phenotype in rice. In addition, both the knockdown and overexpression of OsTRXh1 decrease abscisic acid sensitivity during seed germination and seedling growth. We also analyzed the levels of hydrogen peroxide produced in transgenic plants, and the results show that more hydrogen peroxide is produced in the extracellular space of OsTRXh1 knockdown plants than in wild-type plants, whereas the OsTRXh1 overexpression plants produce less hydrogen peroxide under salt stress. These results show that OsTRXh1 regulates the redox state of the apoplast and influences plant development and stress responses.
Thioredoxins (Trxs) are a class of small proteins ubiquitously distributed in different organisms. These proteins contain two redox-active Cys residues in a highly conserved redox-active site (WCGPC; Holmgren, 1989). In plants, Trxs are involved in a variety of cellular processes in two manners. First, Trxs can regulate the redox status of target proteins through thiol-disulfide exchange reactions. For example, Trxs catalyze the conversion of the salicylic acid-induced NONEXPRESSER OF PR GENES1 to a monomer upon pathogen challenge (Tada et al., 2008) and regulate cytosolic, chloroplastic, and mitochondrial enzyme activities in plant cells (Balmer et al., 2003, 2004; Yamazaki et al., 2004). Second, Trxs can enhance heat shock resistance in plants through redox-independent modes (Lee et al., 2009; Park et al., 2009).
A large family of Trxs has been identified in plants. In Arabidopsis (Arabidopsis thaliana), Trxs can be divided into six major groups (Trx f, Trx m, Trx x, Trx y, Trx h, and Trx o) according to amino acid sequence homology. Trx f, Trx m, Trx x, and Trx y are localized in chloroplasts, where they participate in the pentose phosphate and C4 pathways by regulating the activity of the carbon metabolism enzymes (Gelhaye et al., 2005; Schürmann and Buchanan, 2008). Trx f and Trx m have also been localized in nonphotosynthetic organs, which suggests that they might have novel functions in heterotrophic tissues related to cell division, germination, and plant reproduction (de Dios Barajas-López et al., 2007). The Trx o members are localized in the mitochondria and regulate fundamental processes of plant mitochondria (Laloi et al., 2001; Balmer et al., 2004). Trxs h are primarily distributed in the cytoplasm, but they have also been found in the mitochondria (Gelhaye et al., 2004a), nucleus (Serrato et al., 2001), extracellular matrix of Nicotiana alata (Juárez-Díaz et al., 2006), and phloem sap of rice (Oryza sativa; Ishiwatari et al., 1995).
In Arabidopsis, there are 10 h-type putative Trxs, and little is known about them as compared with the well-described chloroplastic Trxs (Gelhaye et al., 2004b). Nevertheless, some studies of h-type Trxs have recently been conducted. The host-selective toxin victorin can induce programmed cell death in Arabidopsis. The active-site mutant AtTRXh5 displays a loss of victorin sensitivity. Further examination of this mutation revealed that the second Cys in the active site is not required for AtTRXh5-mediated victorin sensitivity, which suggests that AtTRXh5 participates in the victorin pathway through an atypical mechanism (Sweat and Wolpert, 2007). Interestingly, AtTDX (a plant-specific Trx-like protein) and AtTRXh3 have dual functions. In the low-Mr forms, they possess a disulfide reductase function, whereas in the high-Mr forms, the chaperone function predominates. Furthermore, overexpression of one of the two Trxs in Arabidopsis confers heat resistance to the plant that occurs primarily through its chaperone function (Lee et al., 2009; Park et al., 2009). Recently, AtTRXh9 was implicated in plant growth and development and found to move from cell to cell, which extends the known boundaries of Trx and suggests that it may play a role in intercellular communication (Meng et al., 2010). GmTrx, a soybean (Glycine max) h-type Trx, contributes to nodule development and maintains the symbiotic state in soybean roots (Lee et al., 2005). In N. alata, NaTrxh is localized in the extracellular matrix of the stylar transmitting tract and reduces S-RNase in vitro (Juárez-Díaz et al., 2006).
The apoplast, which is the portion of the plant cell outside of the cell membrane, plays crucial roles in cell division (Tian et al., 2009), cell differentiation (Takeda et al., 2003), pollen germination (Ge et al., 2011), plant-pathogen interaction (Misas-Villamil and van der Hoorn, 2008), and abiotic stress responses (Dietz, 1997; Dani et al., 2005; Moschou et al., 2008; Zhang et al., 2009a; Song et al., 2011). In the last decade, extensive literature has accumulated that demonstrates a role for the apoplastic redox state in the regulation of plant growth, hormone action, gene expression, systemic acquired resistance, and signal transduction (Pei et al., 2000; Murata et al., 2001; Rodríguez et al., 2002; Schopfer et al., 2002; Mittler et al., 2011). The apoplast of plant cells contains many antioxidants, such as ascorbate, which have been implicated in redox state regulation of the apoplast (Vanacker et al., 1998; Pignocchi and Foyer, 2003). Previously, we identified an h-type Trx, OsTRXh1, in the rice root apoplast during salt stress by two-dimensional electrophoresis (Zhang et al., 2009a). OsTRXh1 is also known as rice phloem protein13-1 (Ishiwatari et al., 1995), Os1 (Gelhaye et al., 2005), and OsTrx23 (Nuruzzaman et al., 2008; Xie et al., 2009a). OsTRXh1 is reportedly expressed in companion cells within the mature phloem (Ishiwatari et al., 1995, 1998). Recently, another report showed that OsTRXh1 was induced by cold stress and negatively regulated the kinase activities of Oryza sativa Mitogen-activated Protein Kinase3 (OsMPK3) and OsMPK6 in vitro through a redox-dependent mechanism (Xie et al., 2009).
In this work, we show that OsTRXh1 was secreted into the apoplast and that its expression at the protein level was induced by salt stress in the apoplast of rice. We also demonstrate that OsTRXh1 possessed disulfide reductase activity in vitro and in vivo. In addition, we found that it influenced plant architecture and was involved in the stress response through regulating the balance of reactive oxygen species (ROS) in the rice apoplast. These results indicate that OsTRXh1 plays a critical role in Trx-associated redox state regulation and plant growth and stress responses.
RESULTS
OsTRXh1 Is an h-Type Trx
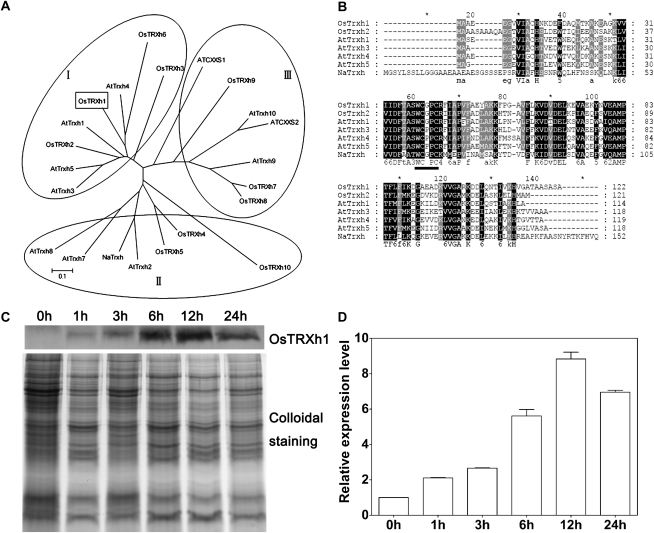
Previously, we identified an h-type Trx, OsTRXh1, which was significantly induced in the rice root apoplast under salt stress (Zhang et al., 2009a). Trx type h is classified into three subgroups based on primary amino acid sequences, subcellular localization, and intracellular activities (Gelhaye et al., 2005; Renard et al., 2011). To better understand the features of OsTRXh1, a phylogenetic tree was constructed with rice and Arabidopsis h-type Trxs and an N. alata-secreted Trx. The results show that OsTRXh1, together with OsTRXh2, OsTRXh3, OsTRXh6, AtTRXh1, AtTRXh3, AtTRXh4, and AtTRXh5, belongs to the h-type Trx subgroup I (Fig. 1A). The amino acid sequence alignment of OsTRXh1, OsTRXh2, AtTRXh1, AtTRXh3, AtTRXh4, AtTRXh5, and NaTrxh shows that OsTRXh1 has the common structure of Trxs and a conserved redox-active site (WCGPC; Fig. 1B). To verify the expression pattern of OsTRXh1 in the apoplast, apoplast proteins were extracted from rice seedlings by the vacuum infiltration method and analyzed by western blot with an antibody against OsTRXh1. We observed that the protein expression level of OsTRXh1 was significantly increased after treatment with 200 mm NaCl for 6 h, and a maximum level of expression was obtained after 12 h (Fig. 1, C and D).
Figure 1.
Multiple sequence alignment of h-type Trxs and OsTRXh1 expression in rice apoplast under salt stress. A, Phylogenetic tree of rice and Arabidopsis h-type Trxs and an N. alata-secreted Trx. The radiation phylogenetic tree was constructed using the MEGA program and the neighbor-joining method. B, Amino acid sequence alignment of OsTRXh1 with OsTRXh2, AtTRXh1, AtTRXh3, AtTRXh4, AtTRXh5, and NaTrxh was performed using the ClustalX program. The conserved redox-active site is indicated with a black line. C, Western-blot analysis of OsTRXh1 expression in the apoplast of 10-d-old rice plants treated with 200 mm NaCl for 0, 1, 3, 6, 12, and 24 h. Apoplastic proteins were extracted using a vacuum infiltration method, and 5 μg of the extracts was loaded for SDS-PAGE. Sensitive colloidal Coomassie blue G-250 staining was used as a loading control. D, Densitometric analysis of the OsTRXh1 western-blot shown in C. The data represent means ± se of three independent experiments.
OsTRXh1 Possesses Trx Activity in Vitro
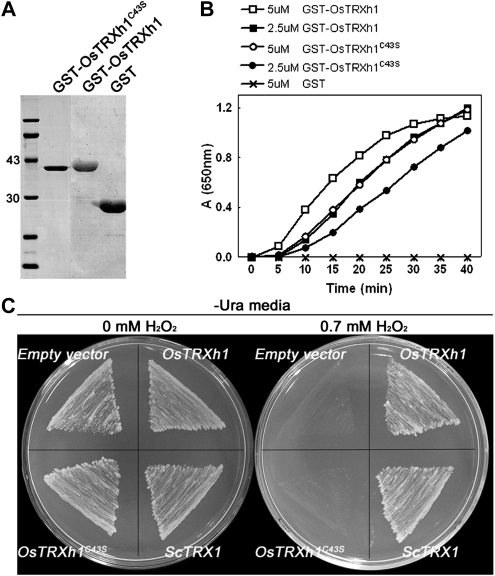
All Trx proteins have a conserved active site (WCxPC, where x = G or P) that contains two redox-active Cys residues, which play important roles in the reduction of targets (Verdoucq et al., 1999; Motohashi et al., 2001). To determine the redox activity of OsTRXh1 in vitro, we expressed the wild-type OsTRXh1 and the OsTRXh1C43S mutant (Cys-43 was mutated to Ser) as N-terminal glutathione S-transferase (GST)-tagged fusion proteins in Escherichia coli and purified them from bacterial lysates by affinity chromatography using immobilized glutathione (Fig. 2A). The Trx activity of the recombinant protein was determined using the insulin reduction assay in vitro (Holmgren, 1979) with GST as a negative control. OsTRXh1 demonstrated high insulin reduction activity, whereas GST displayed no activity. When the second active-site Cys was mutated to a Ser (OsTRXh1C43S), the insulin reduction activity was maintained, but it was reduced as compared with the wild-type protein (Fig. 2B).
Figure 2.
Insulin disulfide reductase activity of OsTRXh1 and H2O2 tolerance assay in the yeast mutant EMY63 expressing OsTRXh1. A, GST-OsTRXhC43S, GST-OsTRXh1, and GST were expressed in E. coli, and the purified proteins were subjected to SDS-PAGE. B, The insulin reduction rate was measured using a 650-nm optical absorption in reaction assays containing the following purified proteins: 5 or 2.5 μm GST-OsTRXh1, 5 or 2.5 μm GST-OsTRXhC43S, and 5 μm GST. C, The growth performance of the transformed yeast mutant EMY63 under synthetic dextrose/Gal/-Ura agar medium containing 0 mm H2O2 and 0.7 mm H2O2. The yeast ScTrx1 and the empty vector were used as controls.
OsTRXh1 Can Complement the Hydrogen Peroxide Sensitivity of the trx1Δ trx2Δ Yeast Mutant
The deletion of the two cytosolic Trxs in the trx1Δ trx2Δ yeast mutant leads to defects in organic sulfur auxotrophy and increased sensitivity to hydrogen peroxide (H2O2; Muller, 1991; Mouaheb et al., 1998). To test whether OsTRXh1 has the ability to complement the yeast mutant, the full-length cDNA of OsTRXh1 and the OsTRXh1C43S mutant were introduced into the yeast mutant. The empty vector YCpIF5 and the yeast native Trx ScTrx1 were used as negative and positive controls, respectively. The H2O2 tolerance of the transformed yeast cells was assayed according to previous reports (Lee et al., 2005). As shown in Figure 2C, in the presence of 0.7 mm H2O2, the negative control yeast cells transformed with the empty YCpIF5 plasmid did not grow; however, the positive control, which expressed ScTrx1, grew normally in the oxidant medium. The yeast mutant cells expressing the wild-type OsTRXh1 grew as well as the positive control, whereas the transformants expressing the OsTRXh1C43S mutant did not grow in the culture medium containing H2O2 (Fig. 2C). All transformed yeast mutant cells grew well in medium that did not contain H2O2 (Fig. 2C). These results indicate that OsTRXh1 had the ability to complement the H2O2 hypersensitivity of the trx1Δ trx2Δ yeast mutant but the OsTRXh1C43S mutant did not.
OsTRXh1 Is Secreted into Extracellular Regions
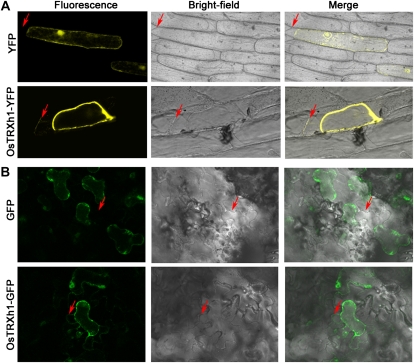
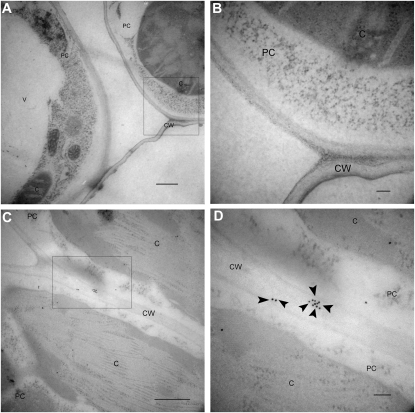
To determine the subcellular localization of OsTRXh1, fluorescently tagged OsTRXh1-YFP (for yellow fluorescent protein) and YFP-OsTRXh1 fusion proteins were transiently expressed in onion (Allium cepa) epidermal cells by particle bombardment. The protein fluorescence was observed using a confocal laser-scanning microscope. We used the empty vector pAVA321 as a negative control. The results showed that the fluorescence of OsTRXh1-YFP was detected in the cell wall after plasmolysis with mannitol, whereas YFP-OsTRXh1 and YFP fluorescence was distributed in the intracellular region and could not be detected in the extracellular region (Fig. 3A; Supplemental Fig. S1). We also constructed OsTRXh1-GFP, which was transformed into Nicotiana benthamiana leaf epidermal cells using Agrobacterium tumefaciens-mediated transformation. The transformed leaf epidermal cells were treated with sodium chloride for plasmolysis. We found that the fluorescence of OsTRXh1-GFP could be detected in the cell wall region, whereas the GFP control was distributed in the intracellular region and could not be detected in the extracellular region (Fig. 3B). To detect the localization of OsTRXh1 in wild-type rice plants, we performed immune colloidal gold experiments with antibodies against OsTRXh1. The specificity of the antibodies was examined by western blot (Supplemental Fig. S2). The results showed that OsTRXh1 was primarily localized in the cell wall (Fig. 4). Localization was also investigated by immunofluorescence experiments with antibodies against OsTRXh1. The results showed that OsTRXh1 was primarily localized in the margin of the cell in vascular bundles of leaves, leaf sheaths, and roots (Supplemental Fig. S3). Altogether, these results show that OsTRXh1 is secreted into extracellular regions.
Figure 3.
Subcellular localization of OsTRXh1. A, Onion epidermal cells were transformed by particle bombardment. YFP (top panels) and OsTRXh1-YFP (bottom panels) fluorescence was observed with a confocal microscope. The onion epidermal cells were treated with 0.9 m mannitol for plasmolysis. B, The OsTRXh1-GFP fusion protein was secreted in the cell wall in N. benthamiana leaves. The top panels show the empty vector control, and the bottom panels show OsTRXh1-GFP fluorescence. Leaf epidermal cells were treated with 1 m sodium chloride for plasmolysis. Fluorescence, bright field, and merged images are shown for each transformation condition. Arrows indicate the cell wall.
Figure 4.
Subcellular localization of OsTRXh1 detected by immunogold electron microscopy. Cross-sections of wild-type leaf are shown. A and C, Xylem parenchymal cells of the wild-type rice leaf. Bars = 500 nm. B and D, High magnification of the areas indicated with boxes in A and C, respectively. Bars = 100 nm. The samples in A and B were probed with preimmune serum. The samples in C and D were detected with anti-OsTRXh1 antibody. The immune reaction was labeled with 10-nm gold-conjugated goat anti-rabbit IgG antibody. Gold particles are indicated by arrowheads. C, Chloroplast; CW, cell wall; PC, xylem parenchymal cell; V, vacuole.
OsTRXh1 Is a Salt and Abscisic Acid Response Gene and Is Extensively Expressed
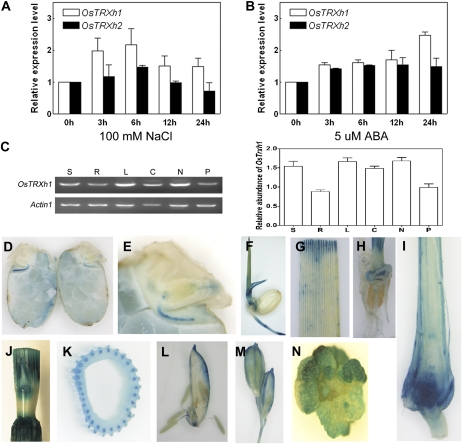
We performed real-time quantitative reverse transcription (RT)-PCR to examine the mRNA expression levels of OsTRXh1 in response to salt in 10-d-old rice seedlings. The OsTRXh1 mRNA expression levels were slightly increased after treatment with 100 mm NaCl for 3 h, and a nearly 2.2-fold increase was observed after treatment for 6 h (Fig. 5A). We also found that OsTRXh1 expression was slightly induced by abscisic acid (ABA; Fig. 5B). The expression level of its homologous gene, OsTRXh2, was lower as compared with OsTRXh1 in whole plants under the same conditions (Fig. 5, A and B).
Figure 5.
Expression pattern of OsTRXh1. A and B, Real-time Q-PCR analysis of OsTRXh1 and OsTRXh2 expression under salt stress and ABA treatment. Total RNA was extracted from 10-d-old seedlings treated with 100 mm NaCl (A) or 6-d-old seedlings treated with 5 μm ABA (B) for 0, 3, 6, 12, and 24 h. The data represent means ± se of three independent experiments. C, OsTRXh1 expression pattern and densitometric analysis in various organs: seedlings (S), roots (R), leaves (L), columns (C), nodes (N), and panicles (P). The data are means ± se of three independent experiments. D to N, GUS staining of tissue from the ProOsTRXh1:GUS transgenic lines. D and E, Expression of OsTRXh1 in the germinated seedling. F, Five-day-old seedlings. G and H, Leaf and root of 10-d-old seedlings. I, Longitudinal section of the shoots of 2-week-old seedlings. J, Stem. K, Cross-sections of the stem. L, Palea, stamen, and pistil of a floret. M, Spikelet. N, Callus.
The expression analysis of OsTRXh1 under normal conditions using RT-PCR showed that OsTRXh1 was expressed in all of the tissues examined, and the level of transcription was abundant in leaves and nodes (Fig. 5C). In addition, the OsTRXh1 promoter-driven GUS was used to examine the tissue-specific expression of OsTRXh1. During seed germination, GUS staining was detected at the radicle, germ (Fig. 5, D and E), bud, and root of 5-d-old seedlings (Fig. 5F). In seedlings, OsTRXh1 was expressed in the leaf, root, and shoot apical meristem (Fig. 5, G–I). In mature rice plants, staining was detected in the stem and node (Fig. 5, J and K). We could also detect OsTRXh1 expression in the palea, lodicule, stamen, pistil, and panicle during the reproductive phase (Fig. 5, L and M). OsTRXh1 expression was also observed in the callus (Fig. 5N). These results indicate that OsTRXh1 is a stress-responsive gene that is widely expressed in rice.
Knockdown of OsTRXh1 in Rice Causes Dwarf and Low-Tillering Phenotypes
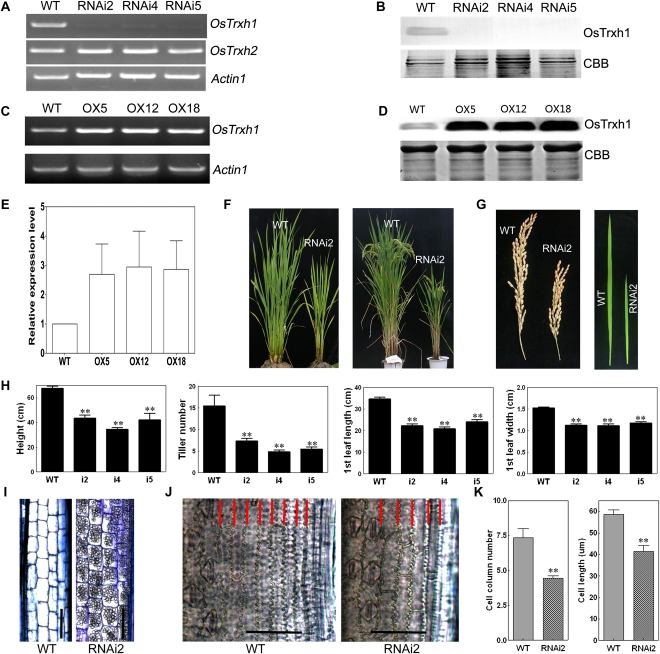
To explore the function of OsTRXh1 in rice, we constructed an OsTRXh1 RNA interference (RNAi) plasmid and introduced it into rice. We examined the T-DNA insertions of select transgenic lines by Southern-blot analysis (Supplemental Fig. S4B). To examine the specificity and efficiency of the RNAi, OsTRXh1 gene-specific RT-PCR was performed. The results showed that the transcription of OsTRXh1 was strongly decreased in the RNAi-2, RNAi-4, and RNAi-5 transgenic lines as compared with the wild type, whereas the OsTRXh1 RNAi did not affect the transcription of OsTRXh2 (Fig. 6A). We also investigated the mRNA expression levels of OsTRXh2, -3, and -4 in RNAi lines by quantitative (Q)-PCR. The results show that the expression of OsTRXh2 was slightly reduced in the RNAi line. The expression of OsTRXh3 and OsTRXh4 did not change in the RNAi line (Supplemental Fig. S5). To determine the protein expression level of OsTRXh1 in the RNAi lines, we performed a western-blot assay with an anti-OsTRXh1 polyclonal antibody. The results showed that the protein expression level of OsTRXh1 was dramatically reduced in the RNAi-2, RNAi-4, and RNAi-5 lines (Fig. 6B). This result was consistent with the results from the RT-PCR assays. We used the OsTRXh1 RNAi lines as a control to study OsTRXh1 and OsTRXh2 expression (Supplemental Fig. S6). The results show that the expression pattern of OsTRXh2 did not change in wild-type and RNAi plants following NaCl and ABA treatment.
Figure 6.
Molecular identification and phenotypic analysis of overexpression and RNAi plants. A and C, RT-PCR analysis of the expression of OsTRXh1 in wild-type (WT), overexpression (OX), and RNAi plants. ACTIN1 was used as a control. B and D, Western-blot analysis of the expression of OsTRXh1 in overexpression and RNAi plants. The blots of crude protein extracts (20 μg) were probed with antibodies directed against OsTRXh1. CBB, Coomassie Brilliant Blue R-250 staining. E, Densitometric analysis of the OsTRXh1 western blot shown in D. The data represent means ± se of three independent experiments. F, Morphology of wild-type and RNAi plants at vegetative and productive stages. G, Main panicle and leaf morphology of control and RNAi plants. H, Statistics of plant height, tiller number, leaf length, and leaf width of wild-type and RNAi plants (n = 20). I, Microscopic observation of longitudinal sections of the elongated zone of the internode of wild-type and RNAi plants. Bars = 100 μm. J, Micrographs of cleared flag leaves. The cell columns are indicated with red lines. Bars = 50 μm. K, Statistics of cell column number (n = 10) and cell length (n = 20) shown in I. Bars with two asterisks indicate P < 0.01. [See online article for color version of this figure.]
We overexpressed OsTRXh1 in rice to further examine its function. The coding sequence of OsTRXh1 under the control of the maize (Zea mays) ubiquitin promoter was introduced into wild-type plants. We obtained eight transgenic lines that were verified by Southern-blot analysis (Supplemental Fig. S4C). RT-PCR and western-blot analyses were performed to confirm the success of OsTRXh1 overexpression at the mRNA and protein levels, respectively (Fig. 6, C–E).
The wild-type, OsTRXh1 RNAi, and overexpression plants were cultivated in the field under identical conditions and appeared normal at the seedling stage (approximately 40 d after planting). However, after the formation of the fourth complete leaf, the OsTRXh1 RNAi lines showed a dwarf phenotype as compared with wild-type plants. At the vegetative and productive phases, the height and tiller number of the RNAi plants were reduced as compared with wild-type plants (Fig. 6F). The average height of the wild-type plants was nearly 68 cm, but the RNAi-2, RNAi-4, and RNAi-5 plants were only 44, 35, and 42 cm, respectively (Fig. 6H). In the wild-type plants, there were approximately 15 primary tillers at the filling stage, but the RNAi-2, RNAi-4, and RNAi-5 plants averaged 7.4, 4.9, and 5.5 tillers, respectively (Fig. 6H). The RNAi plants also had reduced inflorescence length, leaf length, and leaf width as compared with the wild-type plants (Fig. 6G). In addition, the histological analysis suggested that the cell length and columns were reduced (Fig. 6, I–K). During growth and development, the overexpression lines showed no obvious differences from the wild-type plants.
Overexpression of OsTRXh1 Leads to a Salt-Sensitive Phenotype
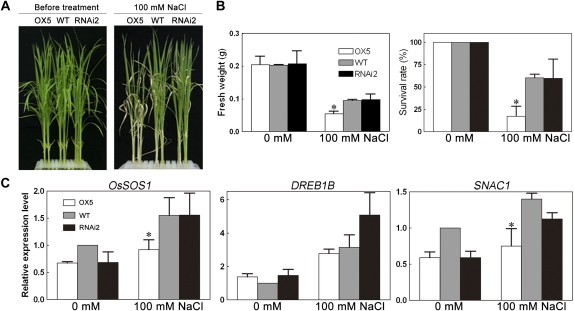
Because OsTRXh1 expression was induced by salt, we wanted to examine whether OsTRXh1 is involved in the regulation of salt tolerance. Homozygous plants of the RNAi and overexpression lines were selected. The seedlings were cultured in Hoagland liquid medium for 10 d and subsequently transferred to Hoagland medium containing 100 mm NaCl for salt stress treatment. The OsTRXh1 overexpression plants showed a salt-sensitive phenotype as compared with the wild-type and RNAi plants (Fig. 7A). After salt stress treatment for 14 d, the survival rate of the OsTRXh1 overexpression plants was only 17%, whereas the wild-type and RNAi plants had 60% and 59% survival rates, respectively, under the same conditions (Fig. 7B). The fresh weight of the OsTRXh1 overexpression plants was also reduced by almost 50% as compared with the wild-type and RNAi plants (Fig. 7B). The results of the salt tolerance test for other transgenic lines were consistent with those obtained above (Supplemental Fig. S7B). All transgenic plants showed no difference from the wild type in normal Hoagland medium (Fig. 7A; Supplemental Fig. S7A). These results suggested that the OsTRXh1 overexpression seedlings were more sensitive to salt, whereas the OsTRXh1 RNAi seedlings were similar to the wild type in salt stress sensitivity.
Figure 7.
Examination of salt tolerance in OsTRXh1 overexpression and RNAi plants. A, Seedling growth performance of wild-type (WT), OsTRXh1 overexpression (OX), and RNAi plants after transfer to Hoagland liquid medium containing 0 or 100 mm NaCl for 14 d. B, Fresh weight and survival rates of wild-type, OsTRXh1 overexpression, and RNAi seedlings after transfer to Hoagland liquid medium containing 0 or 100 mm NaCl for 14 d. The data represent means ± se of three independent experiments, with 32 seedlings per experiment. Bars with asterisks indicate P < 0.05 versus wild-type seedlings. C, Real-time Q-PCR analysis of the expression levels of stress-responsive genes in wild-type, OsTRXh1 overexpression, and RNAi plants. Total RNA was extracted from leaves of 10-d-old plants grown under normal conditions or treated with 100 mm NaCl for 30 min. Each value represents the mean ± se of three independent experiments. Bars with asterisks indicate P < 0.05 versus wild-type seedlings. [See online article for color version of this figure.]
To test whether OsTRXh1 plays a role in the regulation of salt-induced gene expression, we examined the expression levels of the stress-responsive genes OsSOS1, SNAC1, and DREB1B during the initial phase of salt stress in wild-type, OsTRXh1 overexpression, and RNAi seedlings by real-time Q-PCR analysis. As shown in Figure 7C, after treatment with 100 mm NaCl, the expression of OsSOS1 and SNAC1 in the OsTRXh1 overexpression lines was significantly lower than in the wild-type plants, whereas DREB1B expression was only slightly lower than in the wild-type plants. The expression of OsSOS1, SNAC1, and DREB1B in the RNAi line was not significantly changed as compared with wild-type plants.
Knockdown or Overexpression of OsTRXh1 Leads to an ABA-Insensitive Phenotype
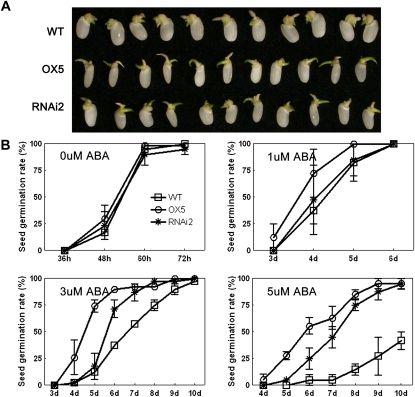
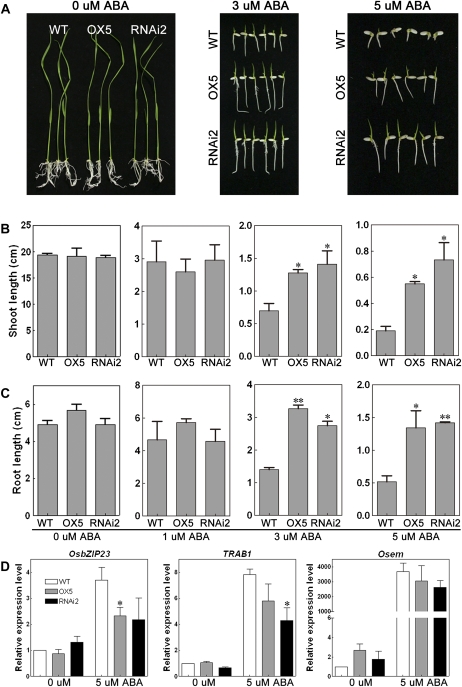
Because OsTRXh1 was also induced by ABA, we tested the ABA sensitivity of the transgenic plants. Homozygous seeds were cultured on one-half-strength Murashige and Skoog (1/2 MS) medium containing 5 μm ABA. The knockdown or overexpression of OsTRXh1 resulted in an ABA-insensitive phenotype (Fig. 8A). When treated with 1 μm ABA, the OsTRXh1 RNAi and wild-type seeds showed similar germination rates that were slightly lower than the germination rate of the OsTRXh1 overexpression seeds (Fig. 8B). After treatment with 3 μm ABA, the germination rates of OsTRXh1 overexpression and RNAi seeds were nearly 92% and 87%, respectively, at 7 d after seed imbibition, whereas the wild-type seeds had a germination rate of only 57% under the same conditions (Fig. 8B). Using 5 μm ABA in the medium, the OsTRXh1 overexpression and RNAi seeds also showed significantly higher germination rates (63% and 45%, respectively) than the wild type (5%; Fig. 8B). There were no differences in the germination rates between the wild-type, OsTRXh1 overexpression, and RNAi seeds in normal medium (Fig. 8B). In addition, the shoot and root lengths were measured 10 d after seed imbibition (Fig. 9A). The OsTRXh1 overexpression and RNAi plants showed no significant difference in growth rates under 1 μm ABA treatment as compared with the wild-type plants (Fig. 9, B and C). The statistical analysis showed that the OsTRXh1 overexpression and RNAi plants had significantly longer shoots and roots than the wild-type plants in the presence of 3 or 5 μm ABA in the medium (Fig. 9, B and C). The other transgenic lines also showed an ABA-insensitive phenotype (Supplemental Fig. S8). All transgenic plants showed no difference from the wild type in normal medium (Fig. 9; Supplemental Fig. S8).
Figure 8.
Overexpression or knockdown of OsTRXh1 leads to an ABA-insensitive phenotype and an ABA-induced inhibition of seed germination. A, Germination performance of wild-type (WT), OsTRXh1 overexpression (OX), and RNAi seeds on 1/2 MS medium containing 5 μm ABA at 6 d after initiation. B, Germination rate of wild-type, OsTRXh1 overexpression, and RNAi seeds on 1/2 MS medium containing 0, 1, 3, and 5 μm ABA. The data represent means ± se of three independent experiments, with 30 seeds per experiment. [See online article for color version of this figure.]
Figure 9.
OsTRXh1 overexpression and RNAi plants showed an increased growth rate under ABA treatment. A, Growth performance of wild-type (WT), OsTRXh1 overexpression (OX), and RNAi seedlings on 1/2 MS medium containing 0, 3, and 5 μm ABA. B and C, Shoot and root length of wild-type, OsTRXh1 overexpression, and RNAi seedlings on 1/2 MS medium containing 0, 1, 3, and 5 μm ABA. The data represent means ± se of three independent experiments, with 30 seedlings per experiment. Bars with asterisks indicate P < 0.05 versus wild-type seedlings; two asterisks indicate P < 0.01. D, Real-time Q-PCR analysis of the expression levels of ABA-responsive genes in wild-type, OsTRXh1 overexpression, and RNAi seeds. Total RNA was extracted from seeds germinated for 96 h after imbibition under normal medium or medium containing 5 μm ABA. Each value represents the mean ± se of three independent experiments. Bars with asterisks indicate P < 0.05 versus wild-type seedlings. [See online article for color version of this figure.]
The three genes known to be involved in the ABA signaling pathway, OsbZIP23 (Xiang et al., 2008), TRAB1 (Hobo et al., 1999; Kagaya et al., 2002), and Osem (Hattori et al., 1995), were used for real-time Q-PCR analysis. The results showed that, after treatment with 5 μm ABA for 96 h, the induction of OsbZIP23, TRAB1, and Osem was reduced in the OsTRXh1 overexpression and RNAi seeds as compared with the wild-type seeds (Fig. 9D). Taken together, these results indicate that the ABA pathway was impaired in the OsTRXh1 overexpression and RNAi lines.
OsTRXh1 Regulates Apoplastic ROS Accumulation in Rice
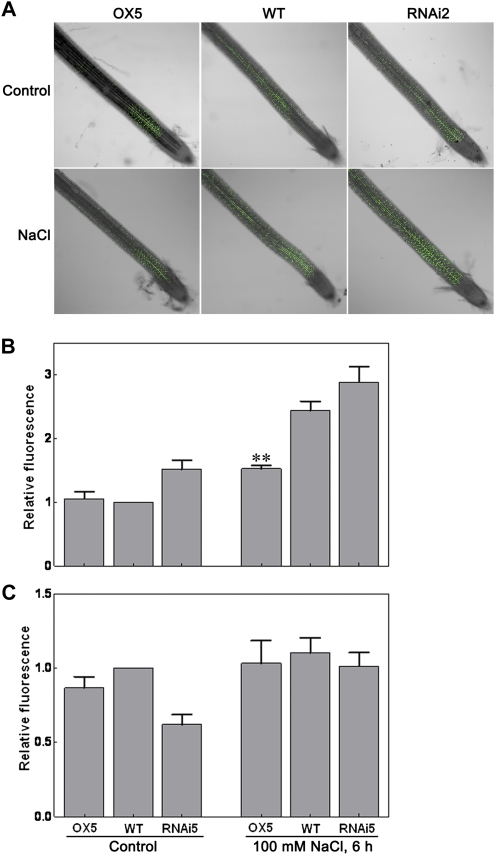
To determine the redox state of transgenic plants, we examined the H2O2 content of 60-d-old transgenic plants using Amplex Red reagent. The results showed that the H2O2 content in the overexpression plants was slightly lower than that in the wild-type plants. In contrast, the RNAi plants contained more H2O2 (Supplemental Fig. S9). The OsTRXh1 expression at the protein level was induced by salt stress in the apoplast of rice, suggesting that OsTRXh1 might regulate the apoplastic redox state. Measurements of ROS in the apoplast of 10-d-old seedlings using a cell-impermeable sensitive fluorogenic dye (OxyBURST Green H2HFF BSA; Fig. 10A) revealed that ROS was accumulated in the roots of wild-type seedlings treated with salt (Fig. 10B). Furthermore, we found that salt treatment resulted in a lower ROS accumulation in the apoplast of overexpression plants than in that of wild-type plants, whereas the RNAi plants contained more H2O2 in the apoplast (Fig. 10B). We also detected a slight increase of cytosolic ROS in wild-type, overexpression, and RNAi seedlings treated with 100 mm NaCl (Fig. 10C). These results implicate OsTRXh1 in the regulation of the redox state of rice apoplast.
Figure 10.
Apoplastic and cytosolic ROS in situ localization under normal conditions or salt stress treatment. A and B, The accumulation of ROS in apoplast of wild-type (WT), OsTRXh1 overexpression (OX), and RNAi plants. Seedlings were exposed to 100 mm NaCl and then stained with OxyBURST Green H2HFF BSA for 30 min. A, Fluorescence images. B, Fluorescence intensity. The data represent means ± se of measurements taken from at least 15 roots for each experiment. C, Cytosolic ROS detection under normal conditions or salt stress treatment. The seedlings were exposed to 100 mm NaCl and then stained with OxyBURST Green H2DCFDA for 20 min. The data represent means ± se of measurements taken from at least 15 roots for each experiment. Bar with asterisks indicates P < 0.05; two asterisks indicate P < 0.01.
DISCUSSION
OsTRXh1 Is a Redox-Active h-Type Trx
Previously, we analyzed the soluble apoplast proteins of rice roots by two-dimensional electrophoresis. OsTRXh1 is an h-type Trx that was identified using mass spectrometry (Zhang et al., 2009a). The expression of OsTRXh1 at the protein level was significantly increased in the apoplast under salt stress (Fig. 1, C and D) but was less induced by salt stress at the transcription level. These results imply that protein secretory pathways and protein synthesis and turnover rates play important roles in apoplastic protein accumulation (Song et al., 2011). There are 10 members of Trx h in rice, and none of them have been well characterized. In Arabidopsis, AtTRXh1, AtTRXh3, AtTRXh4, and AtTRXh5 reduce insulin in the presence of dithiothreitol (DTT) in vitro (Yamazaki et al., 2004). Here, we showed that the wild-type OsTRXh1 and the OsTRXh1C43S mutant protein could also reduce insulin (Fig. 2B). This result is consistent with previous reports that showed that OsTRXh1 and the phloem sap proteins catalyzed the reduction of the disulfide bonds of insulin in the presence of DTT (Ishiwatari et al., 1995). In Arabidopsis, not all of the h-type Trxs are able to complement the H2O2 sensitivity of the trx1Δ trx2Δ yeast mutant (Bréheĺin et al., 2000), which indicates that some of the h-type Trxs are not involved in H2O2 scavenging but might function in other roles. We found that the wild-type OsTRXh1 complemented the H2O2 sensitivity of the trx1Δ trx2Δ yeast mutant but the OsTRXh1C43S mutant did not (Fig. 2C). Previous studies have shown that the two active-site Cys residues of Trx are indispensable for the reduction of protein disulfide bridges: the first Cys of the active site attacks the S-S bond, thereby reducing one Cys and forming a disulfide bridge with the second Cys of the target; subsequently, the second Cys of the Trx active site attacks the newly formed disulfide intermediate, reducing the target protein and oxidizing Trx itself (Kallis and Holmgren, 1980; Wynn et al., 1995). The examination of insulin reduction in vitro demonstrated that the second reaction might be performed by DTT, which caused the OsTRXh1C43S mutant to have partial insulin reduction activity (Fig. 2B). However, the second Cys residue is required for the yeast mutant complementation, implying that it is important for OsTRXh1 Trx activity in vivo. Overall, these results suggest that OsTRXh1 is a typical and redox-active Trx.
OsTRXh1 Can Be Secreted into the Extracellular Regions
Although h-type Trxs are generally localized in the cytosol, they have also been found in the mitochondria (Gelhaye et al., 2004a), the nucleus (Serrato et al., 2001), and the extracellular matrix of N. alata (Juárez-Díaz et al., 2006). OsTRXh1 was reported to be expressed in companion cells and moved from the injected cell into surrounding cells (Ishiwatari et al., 1998). Using transient expression systems, we showed that OsTRXh1 is secreted into the apoplast. Two YFP fusion constructs were used for the subcellular localization studies. The results showed that OsTRXh1-YFP fluorescence was detected in the cell wall after plasmolysis with mannitol, whereas YFP-OsTRXh1 was distributed in the intracellular region and could not be detected in the extracellular region (Fig. 3A; Supplemental Fig. S1). This result indicated that OsTRXh1 is secreted into apoplast and that the N terminus of OsTRXh1 might play an important role in apoplast localization. The apoplast localization of OsTRXh1 was also confirmed by immunogold and immunofluorescence experiments (Fig. 4; Supplemental Fig. S3). A novel Trx in N. alata that belongs to Trx h subgroup II, NaTrxh, was localized in the extracellular matrix of the stylar transmitting tract and could reduce S-RNase in vitro (Juárez-Díaz et al., 2006). In our study, we found that OsTRXh1 not only exists in the apoplast but also regulates the apoplastic redox state in rice (Fig. 10).
OsTRXh1 Is Expressed Extensively and Is Involved in Plant Growth and Development
Using RT-PCR and GUS staining, we found that OsTRXh1 was expressed abundantly and extensively (Fig. 5, C–N). The expression characteristics of OsTRXh1 suggested that it might play an important role in plant growth and development. There is currently no genetic evidence that h-type Trxs are involved in plant oxidative stress responses, which is probably due to functional redundancy between the members (Meyer et al., 2005). In plants, the production and removal of ROS are strictly controlled (Apel and Hirt, 2004). However, a variety of environmental stimuli can perturb the equilibrium between the production and scavenging of ROS, which results in an oxidative burst (Prasad et al., 1994; Tsugane et al., 1999; Chung et al., 2008; Zhang et al., 2009b). As a result, plants reduce the transcript levels of certain carbohydrate metabolism and photosynthesis-related genes that suppress plant growth (Sakamoto et al., 2004). In 60-d-old OsTRXh1 RNAi plants, the knockdown of OsTRXh1 led to the dwarf and low-tillering phenotypes in the late developmental stages, and we also found that the knockdown of OsTRXh1 increases H2O2 levels (Supplemental Fig. S9). The phenotypes of 60-d-old rice plants might be caused by a long-term abnormal ROS level (Fig. 6E; Supplemental Fig. S9).
OsTRXh1 Is Involved in Salt and ABA Responses by Regulating the Apoplastic ROS Accumulation
In the salt stress experiments, the seedlings of the OsTRXh1 overexpression lines showed a salt-sensitive phenotype (Fig. 7A; Supplemental Fig. S7). To clarify the mechanism underlying this salt sensitivity, we detected apoplastic ROS contents and salt stress-specific response genes in wild-type, OsTRXh1 RNAi, and overexpression plants. The results show that salt treatment caused decreased ROS accumulation in the apoplast of overexpression plants as compared with that in wild-type plants, whereas the RNAi plants contained more ROS in the apoplast (Fig. 10B). The ROS signals were important for the induction of the stress response genes. In our experiments, we found that the expression of the salt-responsive genes OsSOS1 and SNAC1 was reduced in the OsTRXh1 overexpression plants as compared with wild-type and RNAi plants under salt stress. Salt induces the expression of stress response genes that are important for adaptation to salinity (Zhu, 2002). OsSOS1 is a homolog of the Arabidopsis SALT OVERLY SENSITIVE1 (SOS1) gene and can complement the salt sensitivity of the sos1-1 mutant in Arabidopsis (Martínez-Atienza et al., 2007). In rice, the overexpression of SNAC1, which encodes a NAC transcription factor, showed significantly improved drought and salt tolerance (Hu et al., 2006). These results suggest that overexpression lines did not respond to salt stress because of their inability to accumulate the ROS required for stress signaling.
ROS is also an essential second messenger in ABA signaling (Neill et al., 2002); thus, we examined the seed germination and growth rates of the OsTRXh1 RNAi and overexpression plants under ABA treatment. The results showed that the rates of germination and growth of OsTRXh1 overexpression lines were higher than those in the wild-type plants under ABA treatment (Figs. 8 and 9; Supplemental Fig. S8). The overexpression of OsTRXh1 led to a lower apoplastic ROS level, which could cause the ABA-insensitive phenotype. In Arabidopsis, AtrbohD and AtrbohF are two guard cell-expressed NADPH oxidase catalytic subunit genes that are important enzymes during ROS production. In the AtrbohD/F double mutants, ROS production was impaired under ABA treatment, which resulted in the impairment of ABA-induced stomatal closing and the inhibition of seed germination and root elongation (Kwak et al., 2003). Interestingly, the RNAi lines had the same phenotype as the overexpression lines. Furthermore, we found that the induction of the ABA-responsive genes OsbZIP23, TRAB1, and Osem was reduced in both the OsTRXh1 overexpression and RNAi plants as compared with the wild-type plants under ABA treatment (Fig. 9D). The expression of endogenous OsTRXh1 and homologous OsTRXh genes was examined using real-time Q-PCR analysis. The results showed that expression of the endogenous OsTRXh1 or homologous OsTRXh genes in whole plants was not affected in the overexpression lines (Supplemental Fig. S10). In addition, we examined the localization of OsTRXh1 in the RNAi and overexpression plants using immunofluorescence experiments with antibodies against OsTRXh1. The results showed that OsTRXh1 was drastically reduced in the RNAi plants as compared with the wild-type plants (Supplemental Fig. S11). Although the phenotypes were the same between RNAi and overexpression lines, the expression of OsTRXh1 and the ROS content were different. Abiotic stress induces the accumulation of ROS, which has dual effects on the plant cell. ROS could either function as an important signaling molecule at low concentrations or become toxic by inducing oxidative stress when it increases above a certain level (Apel and Hirt, 2004; Buchanan and Balmer, 2005; Møller et al., 2007). We speculated that RNAi plants were insensitive to ABA because of a consistently high ROS stressful condition, which would prevent proper ABA stress perception by decreasing sensitivity to stress. In the overexpression plants, we could detect increased fluorescence in the vascular bundles as compared with the wild-type plants, but the OsTRXh1 expression level was reduced in the epidermis and the sclerenchyma in the overexpression plants. This reduction might contribute to the observed phenotypes of the overexpression plants (Supplemental Fig. S11).
Based on previous reports and the results in this study, we assumed that salt and ABA treatment would cause apoplastic ROS generation in plants. In addition, the ROS elimination system in the apoplast maintains ROS at a suitable level. In plant cells, ROS causes damage, but it also acts as a signaling molecule (Mittler et al., 2011). These two effects of ROS jointly determine plant stress responses and seed germination (Supplemental Fig. S12). The plant apoplast redox state is primarily regulated by ascorbic acid, which comprises a unique plant antioxidant pool (Vanacker et al., 1998; Pignocchi and Foyer, 2003). Superoxide dismutase and other antioxidant enzymes are also involved in apoplast ROS elimination (Vanacker et al., 1998). In this study, we found that OsTRXh1 was secreted into extracellular regions (Figs. 3 and 4). OsTRXh1 was drastically reduced in the apoplast of RNAi plants and increased in the overexpression plants (Fig. 6). Consistent with the expression level of OsTRXh1, we found that salt treatment reduced ROS accumulation in the apoplast of overexpression plants, whereas the RNAi plants contained more H2O2 in the apoplast, as determined using the apoplastic-specific ROS dye H2HFF-BSA (Fig. 10). These results indicate that OsTRXh1 could be secreted into the apoplast to regulate the redox state of plants. OsTRXh1 is also localized in the cytosol, and we know that the cytosol redox state is also regulated by salt stress; therefore, whether the regulation of apoplastic ROS is apoplastic-OsTRXh1 specific or a consequence of OsTRXh1 activity in the cytosol remains a question. To examine this question, we investigated variations in ROS using the cell membrane-permeable dye H2DCFDA to determine the cytosolic redox state of transgenic plants under the same conditions. We detected a slight increase of cytosolic ROS in wild-type, overexpression, and RNAi seedlings when treated with 100 mm NaCl (Fig. 10). A comparison of treated and untreated plants revealed that the RNAi line showed the highest apoplastic ROS level and the lowest cytosolic ROS level in both the treated and untreated groups. These results indicate that the modulation of apoplastic ROS is not a direct consequence of the cytosolic ROS. These results also imply that apoplastic ROS might increase the intracellular antioxidant capacity. In tobacco (Nicotiana tabacum), decreasing the apoplastic redox state impaired the cytoplasmic capacity to eliminate H2O2, which suggests that the redox networks in the apoplast and cytoplasm are closely linked (Pignocchi et al., 2006). ROS generation and scavenging in the plant cell are complex processes in the ROS dynamic regulation network (Apel and Hirt, 2004). Based on our results, we speculate that OsTRXh1 could regulate apoplastic ROS accumulation in rice. How OsTRXh1 influences the redox state of the apoplast remains to be explored. The identification of OsTRXh1 target proteins and the characterization of the regeneration system in the apoplast that maintains OsTRXh1 in the reduced form will give us a better understanding of the molecular mechanism of OsTRXh1 function.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa japonica ‘Nipponbare’) seedlings were grown in a greenhouse at 28°C/25°C under a 16-h-light/8-h-dark cycle and 50% humidity (Kawasaki et al., 2001). Rice plants were cultivated in the experimental field in Shijiazhuang and Pingshan of Hebei Province in China under natural growing conditions.
Vector Construction and Rice Transformation
To generate the RNAi vector, a 359-bp OsTRXh1 fragment was amplified with primers RNAiF and RNAiR and was ligated to the pTCK303 vector in an inversely oriented manner (Wang et al., 2004). To generate ProOsTRXh1:GUS, an 1,147-bp DNA fragment upstream of the OsTRXh1 ATG (start codon) was amplified with primers proF and proR and was ligated to the same pCAMBIA1300 vector in which the GUSa gene had previously been cloned. To generate Ubi:OsTRXh1, the coding sequence of OsTRXh1 was amplified with the primers OXF and OXR and cloned into the pCAMBIA1300 vector, in which the Ubi-1 promoter replaced the cauliflower mosaic virus 35S promoter. All binary vector constructs were transformed by Agrobacterium tumefaciens strain EHA105. The A. tumefaciens-mediated transformation of embryonic callus was conducted to obtain transgenic rice according to previous reports (Yang et al., 2004). All primer sequences used above are listed in Supplemental Table S1.
Phylogenic and Sequence Analyses
Using MEGA software (version 4.1; http://www.megasoftware.net/mega4/mega41.html; Tamura et al., 2007), a radial phylogenetic tree was constructed using full-length protein sequences by the neighbor-joining method with the following parameters: bootstrap (1,000 replicates; random seed), pair-wise deletion, and Poisson correction. The amino acid sequences were aligned using the ClustalX 2.0 program with default settings and adjusted manually using GeneDoc (version 2.6.002) software (Pittsburgh Supercomputing Center; http://www.psc.edu/biomed/genedoc/).
Protein Purification and Antibody Preparation
To generate GST-OsTRXh1, the coding sequence of OsTRXh1 was amplified and ligated into the pGEX-2T vector. The construct was transformed into Escherichia coli strain XA90. The recombinant proteins containing an N-terminal GST tag were purified from bacterial lysates using a GSTrap FF column according to the manufacturer’s protocol (Amersham Biosciences). The purified GST-OsTRXh1 protein was used as an immunogen to generate OsTRXh1 antibodies. The Tailun Biological Technology Company prepared the polyclonal antibody against OsTRXh1, and the specificity was tested by western-blot analysis (Supplemental Fig. S2). ImageJ (version 1.44P; U.S. National Institutes of Health) was used for densitometric analysis of the western blot.
Determination of Insulin Reduction
The insulin reduction assay was performed as described previously (Holmgren, 1979). The reaction mixture contained 100 mm potassium phosphate buffer, pH 7.0, 0.1 mm EDTA, 0.13 mm bovine insulin (Sigma I5500), and 5 μm recombinant GST-Trxs or GST. The reactions were initiated by the addition of 0.33 mm DTT. The optical absorption of the solution was measured at 650 nm at room temperature using a spectrophotometer (Hitachi U-2001).
Yeast Complementation Analysis
The Trx coding sequences were amplified and cloned downstream of the GAL1 promoter in the YCpIF5 plasmid. The constructs were transformed into yeast strain EMY63 (MATa; ade2-1 ade3-100 his3-11 leu2-3 lys2-801 trp1-1 ura3-1 trx1::TRP1 trx2::LEU2; Muller, 1991) using the lithium acetate method (Moerschell et al., 1991). The induction of Trxs and the detection of H2O2 tolerance of the transformed yeast cells were performed according to a previous report (Lee et al., 2005).
Subcellular Localization of OsTRXh1
The 35S:OsTRXh1-YFP and 35S:YFP-OsTRXh1 constructs and the empty vector pAVA321 were transformed into onion (Allium cepa) epidermal cells by particle bombardment using the Bio-Rad PDS-1000/He system according to the operator’s manual. After a 22-h incubation on MS medium in a growth chamber, the transformed epidermal cells were treated with 0.9 m mannitol to initiate plasmolysis. YFP fluorescence was observed with a confocal microscope (Zeiss LSM 510).
pCAMBIA1300 35S:OsTRXh1-GFP was constructed and transformed into A. tumefaciens strain EHA105. The bacterial infiltration of Nicotiana benthamiana leaf epidermal cells was performed as described previously. GFP fluorescence was visualized with a confocal scanning microscope at 40 to 48 h after infiltration. The transformed leaf epidermal cells were treated with 1 m sodium chloride to initiate plasmolysis.
Immunogold Localization
Leaves of wild-type rice were fixed in 4% (v/v) paraformaldehyde and 0.25% glutaraldehyde in phosphate-buffered saline (PBS), dehydrated in an ethanol and acetone series, embedded in Spurr’s resin (Structure Probe, Inc.; Low Viscosity “Spurr” Kits), divided into 80-nm sections, and collected on formvar-coated nickel grids. The sections were blocked with 1% BSA (Sigma) in 0.01 m PBS (pH 7.4; 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 2 mm KH2PO4) for 30 min and then incubated with anti-OsTRXh1 antibodies (1:1,200 dilution) at 37°C for 2 h and subsequently with anti-rabbit IgG-Gold (1:150) at 37°C for an additional 1 h. The sections were observed using electron microscopy (Hitachi H-7650).
Immunohistochemistry
Rice tissues were fixed in 4% (v/v) formaldehyde in PBS, dehydrated in an ethanol series, and embedded in Paraplast Plus (Sigma). Sections of 8 to 10 μm were blocked by incubation with 3% (w/v) nonfat dry skim milk in 50 mm potassium phosphate, pH 7.1, containing 0.5 m NaCl for 3 h. The sections were subsequently incubated with the primary rabbit anti-OsTRXh1 antibody (1:100 dilution) at 4°C overnight. The sections were then incubated with secondary goat anti-rabbit DyLight 488-fluorochrome-conjugated antibodies (1:100 dilution) for 3 h. The sections were observed using confocal microscopy (Zeiss LSM 510).
Salt and ABA Sensitivity Assays
For the salt stress experiment, 10-d-old rice seedlings were transferred to Hoagland liquid medium containing 0 or 100 mm NaCl, and the solution was replaced with fresh solution every 2 d. After 14 d of treatment, the survival rate and fresh weight were calculated. For the ABA sensitivity testing, the seeds were placed in 1/2 MS medium containing 0, 1, 3, and 5 μm ABA. The germination was observed every 24 h and considered positive based on whether the shoot length exceeded half of the seed length. All of the above experiments were repeated at least three times.
RNA Extraction and Real-Time Q-PCR
Total RNA was extracted using Trizol reagent (Invitrogen). Five hundred nanograms of total RNA was used to synthesize the first-strand cDNA with the Perfect Real-Time RT reagent kit (TaKaRa DRR037A). The cDNA was diluted 30 times with deionized water, and 5 μL was used as a template for real-time Q-PCR. SYBR Green-monitored real-time Q-PCR was performed on an ABI PRISM 7000 real-time PCR system (Applied Biosystems). The rice ACTIN1 gene was used as the internal control, and each analysis was performed in three biological replicates. The primer sequences used for real-time PCR are listed in Supplemental Table S1.
GUS Staining
The GUS staining analysis was performed according to a previous report (Jefferson et al., 1987). Various tissues of ProOsTRXh1:GUS T1 transgenic plants were vacuum infiltrated for 15 min in GUS staining buffer and then incubated for 12 h at 37°C. The stained tissues were incubated in 70% ethanol to remove the chlorophyll. The images were captured using a stereomicroscope (Nikon SMZ800).
ROS Determination
Sixty-day-old rice plants were used for H2O2 quantification using an Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Molecular Probes A22188) according to the manufacturer’s protocol. The protein concentration was measured using the Bradford protein assay. For ROS detection, 10-d-old seedlings were transferred to Hoagland liquid medium containing 100 mm NaCl for 6 h The roots of the salt-treated and untreated control seedlings were gently transferred to 2-mL tubes containing OxyBURST Green H2HFF BSA (10 μg mL−1; Molecular Probes O-13291) for the detection of apoplastic ROS levels or OxyBURST Green H2DCFDA (10 μm; Molecular Probes D-2935) for the detection of intracellular ROS levels and subsequently incubated for 30 and 20 min under dark conditions, respectively. The fluorescence images were obtained using a Zeiss LSM 510 confocal microscope with 488-nm excitation and 530-nm emission.
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: Oryza sativa japonica OsTRXh1 (Os07g0186000), OsTRXh2 (Os05g0508500), OsTRXh3 (Os09g0401200), OsTRXh4 (Os03g0800700), OsTRXh5 (Os07g0190800), OsTRXh6 (Os12g0281300), OsTRXh7 (Os01g0168200), OsTRXh8 (Os05g0169000), OsTRXh9 (Os05g0480200), OsTRXh10 (Os01g0913000), OsSOS1 (Os12g44360), DREB1B (Os09g0522000), SNAC1 (DQ394702), OsbZIP23 (AK072062), TRAB1 (Os08g0472000), Osem (OSU22102), and OsACTIN1 (Os03g0718100); Arabidopsis thaliana AtTRXh1 (AT3G51030), AtTRXh2 (AT5G39950), AtTRXh3 (AT5G42980), AtTRXh4 (AT1G19730), AtTRXh5 (AT1G45145), AtTRXh7 (AT1G59730), AtTRXh8(AT1G69880), AtTRXh9 (AT3G08710), AtTRXh10 (AT3G56420), ATCXXS1 (AT1G11530), and ATCXXS2 (AT2G40790); and Nicotiana alata NaTrxh (DQ021448).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization of YFP-OsTRXh1.
Supplemental Figure S2. Western-blot analysis using specific antibodies against OsTRXh1.
Supplemental Figure S3. Immunofluorescent detection of OsTRXh1 protein localization in wild-type plants.
Supplemental Figure S4. Southern-blot analysis of independent RNAi and overexpression transgenic lines.
Supplemental Figure S5. Real-time Q-PCR analysis of the expression levels of endogenous OsTRXh1 and homolog genes of OsTRXh in OsTRXh1 RNAi plants.
Supplemental Figure S6. Real-time Q-PCR analysis of OsTRXh1 and OsTRXh2 expression using RNAi as a control under salt stress and ABA treatment.
Supplemental Figure S7. Examination of salt tolerance of other OsTRXh1 transgenic lines.
Supplemental Figure S8. Seed germination rate and seedling growth of other OsTRXh1 transgenic lines and wild type under ABA treatment.
Supplemental Figure S9. Hydrogen peroxide content of wild-type plants, OsTRXh1 overexpression lines OX5 and OX12, and RNAi lines RNAi2 and RNAi5 cultivated for 60 d in the field.
Supplemental Figure S10. Real-time Q-PCR analysis of the expression levels of endogenous OsTRXh1 and homolog genes of OsTRXh in OsTRXh1 overexpression plants.
Supplemental Figure S11. Immunofluorescent detection of OsTRXh1 expression in RNAi and overexpression plants.
Supplemental Figure S12. A model for OsTRXh1 function in apoplastic ROS balance in plants.
Supplemental Table S1. Primers sequences used in this study.
Acknowledgments
We thank Y. Meyer for providing yeast strain EMY63 and Choong-Ill Cheon for YCpIF5 plasmid. We also thank Kang Chong for providing the pTCK303 vector.
References
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB. (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100: 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jacquot JP, Manieri W, Schürmann P, Droux M, et al. (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 101: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréheĺin C, Mouaheb N, Verdoucq L, Lancelin JM, Meyer Y. (2000) Characterization of determinants for the specificity of Arabidopsis thioredoxins h in yeast complementation. J Biol Chem 275: 31641–31647 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Chung JS, Zhu JK, Bressan RA, Hasegawa PM, Shi H. (2008) Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J 53: 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani V, Simon WJ, Duranti M, Croy RR. (2005) Changes in the tobacco leaf apoplast proteome in response to salt stress. Proteomics 5: 737–745 [DOI] [PubMed] [Google Scholar]
- de Dios Barajas-López J, Serrato AJ, Olmedilla A, Chueca A, Sahrawy M. (2007) Localization in roots and flowers of pea chloroplastic thioredoxin f and thioredoxin m proteins reveals new roles in nonphotosynthetic organs. Plant Physiol 145: 946–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ. (1997) Functions and responses of the leaf apoplast under stress. Prog Bot 58: 221–254 [Google Scholar]
- Ge W, Song Y, Zhang C, Zhang Y, Burlingame AL, Guo Y. (2011) Proteomic analyses of apoplastic proteins from germinating Arabidopsis thaliana pollen. Biochim Biophys Acta 1814: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Gérard J, Jolivet Y, Gualberto J, Navrot N, Ohlsson PI, Wingsle G, Hirasawa M, Knaff DB, et al. (2004a) A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci USA 101: 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP. (2004b) The thioredoxin h system of higher plants. Plant Physiol Biochem 42: 265–271 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Navrot N, Jacquot JP. (2005) The plant thioredoxin system. Cell Mol Life Sci 62: 24–35 [DOI] [PubMed] [Google Scholar]
- Hattori T, Terada T, Hamasuna S. (1995) Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J 7: 913–925 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254: 9627–9632 [PubMed] [Google Scholar]
- Holmgren A. (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264: 13963–13966 [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103: 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwatari Y, Fujiwara T, McFarland KC, Nemoto K, Hayashi H, Chino M, Lucas WJ. (1998) Rice phloem thioredoxin h has the capacity to mediate its own cell-to-cell transport through plasmodesmata. Planta 205: 12–22 [DOI] [PubMed] [Google Scholar]
- Ishiwatari Y, Honda C, Kawashima I, Nakamura S, Hirano H, Mori S, Fujiwara T, Hayashi H, Chino M. (1995) Thioredoxin h is one of the major proteins in rice phloem sap. Planta 195: 456–463 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Díaz JA, McClure B, Vázquez-Santana S, Guevara-García A, León-Mejía P, Márquez-Guzmán J, Cruz-García F. (2006) A novel thioredoxin h is secreted in Nicotiana alata and reduces S-RNase in vitro. J Biol Chem 281: 3418–3424 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T. (2002) Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14: 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallis GB, Holmgren A. (1980) Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J Biol Chem 255: 10261–10265 [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ. (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G, Meyer Y. (2001) Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci USA 98: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Lee SS, Jang HH, Lee YM, Park JH, Park SC, Moon JC, Park SK, Kim SY, Lee SY, et al. (2009) Heat-shock dependent oligomeric status alters the function of a plant-specific thioredoxin-like protein, AtTDX. Proc Natl Acad Sci USA 106: 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Shin KH, Kim YK, Suh JY, Gu YY, Kim MR, Hur YS, Son O, Kim JS, Song E, et al. (2005) Induction of thioredoxin is required for nodule development to reduce reactive oxygen species levels in soybean roots. Plant Physiol 139: 1881–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ. (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Wong JH, Feldman LJ, Lemaux PG, Buchanan BB. (2010) A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc Natl Acad Sci USA 107: 3900–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F. (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86: 419–433 [DOI] [PubMed] [Google Scholar]
- Misas-Villamil JC, van der Hoorn RA. (2008) Enzyme-inhibitor interactions at the plant-pathogen interface. Curr Opin Plant Biol 11: 380–388 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Moerschell RP, Das G, Sherman F. (1991) Transformation of yeast directly with synthetic oligonucleotides. Methods Enzymol 194: 362–369 [DOI] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A. (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459–481 [DOI] [PubMed] [Google Scholar]
- Moschou PN, Paschalidis KA, Delis ID, Andriopoulou AH, Lagiotis GD, Yakoumakis DI, Roubelakis-Angelakis KA. (2008) Spermidine exodus and oxidation in the apoplast induced by abiotic stress is responsible for H2O2 signatures that direct tolerance responses in tobacco. Plant Cell 20: 1708–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T. (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98: 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaheb N, Thomas D, Verdoucq L, Monfort P, Meyer Y. (1998) In vivo functional discrimination between plant thioredoxins by heterologous expression in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 95: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EG. (1991) Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J Biol Chem 266: 9194–9202 [PubMed] [Google Scholar]
- Murata Y, Pei ZM, Mori IC, Schroeder J. (2001) Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S, Desikan R, Hancock J. (2002) Hydrogen peroxide signalling. Curr Opin Plant Biol 5: 388–395 [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M, Gupta M, Zhang C, Wang L, Xie W, Xiong L, Zhang Q, Lian X. (2008) Sequence and expression analysis of the thioredoxin protein gene family in rice. Mol Genet Genomics 280: 139–151 [DOI] [PubMed] [Google Scholar]
- Park SK, Jung YJ, Lee JR, Lee YM, Jang HH, Lee SS, Park JH, Kim SY, Moon JC, Lee SY, et al. (2009) Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiol 150: 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI. (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Pignocchi C, Foyer CH. (2003) Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr Opin Plant Biol 6: 379–389 [DOI] [PubMed] [Google Scholar]
- Pignocchi C, Kiddle G, Hernández I, Foster SJ, Asensi A, Taybi T, Barnes J, Foyer CH. (2006) Ascorbate oxidase-dependent changes in the redox state of the apoplast modulate gene transcript accumulation leading to modified hormone signaling and orchestration of defense processes in tobacco. Plant Physiol 141: 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TK, Anderson MD, Martin BA, Stewart CR. (1994) Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell 6: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard M, Alkhalfioui F, Schmitt-Keichinger C, Ritzenthaler C, Montrichard F. (2011) Identification and characterization of thioredoxin h isoforms differentially expressed in germinating seeds of the model legume Medicago truncatula. Plant Physiol 155: 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez AA, Grunberg KA, Taleisnik EL. (2002) Reactive oxygen species in the elongation zone of maize leaves are necessary for leaf extension. Plant Physiol 129: 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P, Liszkay A, Bechtold M, Frahry G, Wagner A. (2002) Evidence that hydroxyl radicals mediate auxin-induced extension growth. Planta 214: 821–828 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Buchanan BB. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Crespo JL, Florencio FJ, Cejudo FJ. (2001) Characterization of two thioredoxins h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol Biol 46: 361–371 [DOI] [PubMed] [Google Scholar]
- Song Y, Zhang C, Ge W, Zhang Y, Burlingame AL, Guo Y. (2011) Identification of NaCl stress-responsive apoplastic proteins in rice shoot stems by 2D-DIGE. J Proteomics 74: 1045–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweat TA, Wolpert TJ. (2007) Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis. Plant Cell 19: 673–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Kotake T, Nakagawa N, Sakurai N, Nevins DJ. (2003) Expression and function of cell wall-bound cationic peroxidase in asparagus somatic embryogenesis. Plant Physiol 131: 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tian L, Zhang L, Zhang J, Song Y, Guo Y. (2009) Differential proteomic analysis of soluble extracellular proteins reveals the cysteine protease and cystatin involved in suspension-cultured cell proliferation in rice. Biochim Biophys Acta 1794: 459–467 [DOI] [PubMed] [Google Scholar]
- Tsugane K, Kobayashi K, Niwa Y, Ohba Y, Wada K, Kobayashi H. (1999) A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 11: 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H, Carver TL, Foyer CH. (1998) Pathogen-induced changes in the antioxidant status of the apoplast in barley leaves. Plant Physiol 117: 1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoucq L, Vignols F, Jacquot JP, Chartier Y, Meyer Y. (1999) In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J Biol Chem 274: 19714–19722 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K. (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22: 409–417 [Google Scholar]
- Wynn R, Cocco MJ, Richards FM. (1995) Mixed disulfide intermediates during the reduction of disulfides by Escherichia coli thioredoxin. Biochemistry 34: 11807–11813 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, Kato H, Sasaki K, Imai R. (2009) A cold-induced thioredoxin h of rice, OsTrx23, negatively regulates kinase activities of OsMPK3 and OsMPK6 in vitro. FEBS Lett 583: 2734–2738 [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T. (2004) Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol 45: 18–27 [DOI] [PubMed] [Google Scholar]
- Yang Y, Peng H, Huang H, Wu J, Jia S, Huang D, Lu T. (2004) Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci 167: 281–288 [Google Scholar]
- Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y. (2009a) Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 149: 916–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu H, Zhang Q, Li M, Yan M, Wang R, Wang L, Welti R, Zhang W, Wang X. (2009b) Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21: 2357–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]