Abstract
The SPINDLY (SPY) gene was first identified as a negative regulator of plant gibberellic acid (GA) signaling because mutation of this gene phenocopies plants treated with an overdose of bioactive GA and results in insensitivity to a GA inhibitor during seed germination. The SPY gene encodes an O-linked N-acetylglucosamine transferase that can modify the target protein and modulate the protein activity in cells. In this study, we describe the strong salt and drought tolerance phenotypes of Arabidopsis (Arabidopsis thaliana) spy-1 and spy-3 mutants in addition to their GA-related phenotypes. SPY gene expression was found to be drought stress inducible and slightly responsive to salt stress. Transcriptome analysis of spy-3 revealed that many GA-responsive genes were up-regulated, which could explain the GA-overdosed phenotype of spy-3. Some stress-inducible genes were found to be up-regulated in spy-3, such as genes encoding late embryogenesis abundant proteins, Responsive to Dehydration20, and AREB1-like transcription factor, which may confer stress tolerance on spy-3. CKX3, a cytokinin (CK) catabolism gene, was up-regulated in spy-3; this up-regulation indicates that the mutant possesses reduced CK signaling, which is consistent with a positive role for SPY in CK signaling. Moreover, overexpression of SPY in transgenics (SPY overexpressing [SPY-OX]) impaired plant drought stress tolerance, opposite to the phenotype of spy. The expression levels of several genes, such as DREB1E/DDF1 and SNH1/WIN1, were decreased in SPY-OX but increased in spy-3. Taken together, these data indicate that SPY plays a negative role in plant abiotic stress tolerance, probably by integrating environmental stress signals via GA and CK cross talk.
The SPINDLY (SPY) gene was first identified by ethyl methanesulfonate (EMS) mutagenesis screening for mutants resistant to paclobutrazol (PAC), an inhibitor of GA synthesis in plants, and isolated by map-based cloning (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996). Unlike GA-insensitive mutants, most of the identified spy mutants phenocopy wild-type plants treated with GA. Relative to wild-type plants, spy mutants exhibit lighter green-colored leaves, early flowering, increased stem elongation, and partial male sterility, suggesting that SPY functions as a negative regulator to repress GA signaling in plants, and mutation of the SPY gene gives rise to an elevated GA response.
GA is one of the most important plant hormones and has been known to play major roles in various aspects of plant biology, including seed germination, leaf expansion, stem and root elongation, flowering time, and fruit development. A major breakthrough in our understanding of GA perception and signal transduction was achieved in the last 10 years, when several components of the GA signaling pathway were identified. Initially, a GA signal is perceived by a soluble GA receptor, GA-Insensitive Dwarf1 (GID1; Ueguchi-Tanaka et al., 2005). A null mutation in the single GID1 gene in rice (Oryza sativa) leads to an extremely short and GA-insensitive plant. In Arabidopsis (Arabidopsis thaliana), when three homologs of GID1 (GID1a, GID1b, and GID1c) were disrupted, plants also displayed a similar dwarf phenotype (Griffiths et al., 2006; Iuchi et al., 2007), indicating that GID1 has an essential role in GA signaling. Following GID1 binding to bioactive GA, the signal is transduced to the DELLA proteins, leading to their degradation (Hirano et al., 2008). The DELLA proteins are conserved repressors of GA signaling that modulate all aspects of GA-induced growth and development in plants. DELLA proteins were named for an N-terminal conserved DELLA domain, which is essential for GA-dependent proteasomal degradation (Willige et al., 2007). In Arabidopsis, there are five DELLA proteins: GA Insensitive (GAI), Repressor of ga1-3 (RGA), RGA-like1 (RGL1), RGL2, and RGL3, and they form a subfamily of the GRAS family of putative transcription regulators (Pysh et al., 1999; Richards et al., 2001). The N-terminal DELLA and VHYNP domains are necessary for protein interactions with GID1 (Griffiths et al., 2006). Loss-of-function mutations of DELLA proteins can rescue the dwarf phenotype of the GA biosynthesis mutant ga1-3 (Dill and Sun, 2001; King et al., 2001), indicating that DELLA proteins function as negative regulators in GA signaling. Another important component of GA signal transduction is an F-box protein, SLY1, that functions as a subunit of the E3 ubiquitin ligase complex SCFSLY1. SLY1 interacts with GAI and RGA in a yeast two-hybrid system and mediates their ubiquitination and degradation (Dill et al., 2004; Fu et al., 2004; Tyler et al., 2004). Mutations in the GA1 locus block GA biosynthesis prior to the formation of ent-kaurene. The phenotypes of ga1 mutants, including dwarfism, germination failure, male sterility, and incomplete petal development, can be reversed by exogenous GA application (for review, see Richards et al., 2001).
The SPY gene is expressed throughout the plant and can be detected not only in all organs where the phenotypes of spy mutants have been observed but also in the roots, indicating a role for the gene in root development (Swain et al., 2002). The SPY protein is predominantly localized to the nucleus, where it modifies components of the GA signaling pathway (Swain et al., 2002). Cytosolic SPY activity was reported to promote cytokinin (CK) responses and to repress GA signaling (Maymon et al., 2009). The SPY protein consists of 915 amino acids, including multiple tetratricopeptide repeats (TPRs) at the N terminus and a Ser and Thr O-linked GlcNAc (O-GlcNAc) transferase (OGT) domain at the C terminus (Jacobsen et al., 1996). Each TPR motif consists of a highly degenerate sequence of 34 amino acids with eight loosely conserved residues. The TPR motifs and OGT domain are important for mediating protein-protein interactions and correct assembly for the enzyme activity, respectively (Kreppel and Hart, 1999; Lubas and Hanover, 2000; Tseng et al., 2001, 2004). Similar to animal OGTs, in vitro-expressed SPY protein possesses OGT activity (Thornton et al., 1999). OGTs transfer a GlcNAc monosaccharide to the O-linkage of Ser or Thr of cytosolic and nuclear proteins. In animals, more than 1,000 O-GlcNAc-modified proteins have been identified, and this kind of protein modification is believed to regulate many basic cellular and disease processes. In some cases, O-GlcNAc modification and phosphorylation occur at the same site on the substrate protein, leading to the hypothesis that these two modification processes compete with each other to fine-tune substrate protein activity under different circumstances (Comer and Hart, 2001; Wells et al., 2001; Slawson and Hart, 2003; Love and Hanover, 2005). In Arabidopsis, there are only two genes encoding O-GlcNAc transferases, SPY and SECRET AGENT (SEC; Hartweck et al., 2002). Mutations in or knockout of the SPY gene leads to elevated GA responses, indicating that SPY functions as a negative regulator of GA signaling, whereas loss-of-function mutations of the SEC gene do not result in any obvious phenotypic changes in plants. It has been suggested that SPY plays a primary role in GA signaling. Double mutants containing loss-of-function mutations in both the SPY and SEC genes are embryonically lethal (Hartweck et al., 2002, 2006). Although the SPY gene has been identified as a negative regulator of GA signaling, the molecular action of SPY in GA signaling and the functions of plant OGTs remain largely unknown. Although the role of SPY in GA-related processes is indisputable, it is also known that SPY is involved in other cellular processes. The spy mutants exhibit altered phyllotaxy, CK responses, light responses, and reduced hypocotyl and rosette growth, none of which are directly related to GA responses (Swain et al., 2001; Tseng et al., 2004; Greenboim-Wainberg et al., 2005).
Recent research has begun to elucidate the molecular events involved in plant hormone responses and environmental stress adaptation. As an important plant phytohormone, GA has been reported to affect the plant abiotic stress response. The growth variability promoted by treatment with GA is able to reverse the inhibitory effects of salt, oxidative, and heat stresses on germination and seedling establishment. Additionally, GA promotes plant growth by inducing the degradation of the growth repressor DELLA in the nucleus, and a quadruple loss-of-function DELLA mutant is more sensitive to extremely high-salinity stress than the wild-type plants (Achard et al., 2006). It has been proposed that SPY can alter DELLA protein activity or stability via O-GlcNAc modification. Moreover, SPY can suppress GA signaling and promote CK responses in Arabidopsis (Greenboim-Wainberg et al., 2005). Interestingly, reducing CK signaling by knocking out two CK receptors, AHK2 and AHK3, can enhance plant survival rates under severe salt stress conditions (Tran et al., 2007).
In this study, we describe the salt and drought stress tolerance phenotypes of spy-1 and spy-3 mutants in addition to their GA-related phenotypes. SPY gene expression was found to be drought stress inducible and slightly responsive to salt stress, indicating a role for this gene in stress response. Transcriptome analysis of the spy-3 mutant revealed that many GA-responsive genes are up-regulated, explaining the GA-overdosed phenotype observed in the spy-3 mutant. Some stress-inducible genes were found to be up-regulated in spy-3, such as genes encoding late embryogenesis abundant (LEA) proteins, Responsive to Dehydration20 (RD20), ATP-binding cassette (ABC) transporters, and AREB1-like transcription factor under normal growing conditions. Under dehydration stress, the expression of many LEA protein genes was higher in the mutant than in the wild type, which may confer stress tolerance on the spy-3 mutants. Increased expression of the CKX3 gene was detected in the spy-3 mutant under both normal and stressed conditions, which may also contribute to plant stress tolerance. Transcriptomic comparison of spy-3 versus wild-type plants and SPY-overexpressing (SPY-OX) versus wild-type plants showed that the expression levels of DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN1E/DWARF AND DELAYED FLOWERING1 (DREB1E/DDF1) and SHINE1/WAX INDUCER1 (SHN1/WIN1) were increased by the spy-3 mutation but decreased by SPY overexpression, which may result in the reversed drought stress phenotypes. The function of the SPY gene in plant abiotic stress response and plant hormone cross talk is also discussed.
RESULTS
spy Mutants Are More Tolerant to High-Salinity Stress
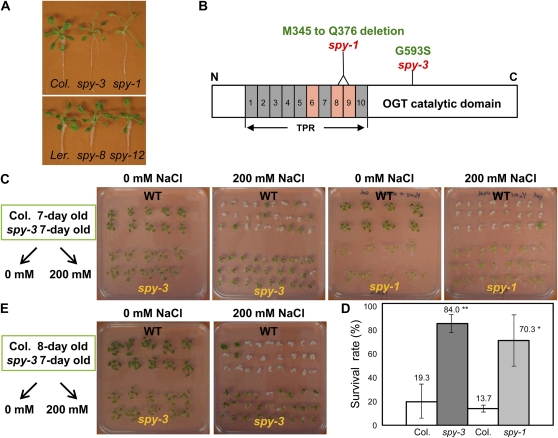
To examine whether the SPY gene also plays a role in the plant abiotic stress response, we obtained point mutation mutants of spy-1 and spy-3 (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996). Both the spy-1 and spy-3 mutants were generated by EMS mutagenesis screening of Arabidopsis ecotype Columbia (Col) for plants that were more tolerant to PAC during seed germination compared with wild-type plants. The seedlings of both mutants exhibited a GA-overdosed phenotype. The spy-1 mutant exhibited a stronger phenotype than the spy-3 mutant in terms of characteristics affected by GA responses, such as elongated stem, pale leaves, slender seedlings, and male sterility, whereas the spy-3 mutant exhibited only a mild phenotype, and this was probably due to a more severe mutation in the spy-1 mutant (Fig. 1A). The spy-3 mutation caused an amino acid substitution in the OGT domain, while the spy-1 mutation caused an in-frame deletion of 23 amino acids of the protein, located in the eighth and ninth TPRs (Jacobsen et al., 1996; Fig. 1B). We compared the ability of the spy mutants and wild-type Col plants to survive severe high-salinity stress conditions. If 7-d-old plants were transferred from normal growth conditions to 200 mm NaCl-containing plates after 7 to 10 d, less than 20% of the wild-type plants survived. In contrast, more than 80% of spy-3 and 70% of spy-1 plants continued to grow and retained a greenish color, which suggested that these two mutants were strongly salt tolerant (Fig. 1C). Statistical analysis of the data obtained in these experiments is presented in Figure 1D. Because the spy mutants grew a bit more quickly after germination than the wild-type plants, we compared the salt tolerance of 8-d-old Col and 7-d-old spy-3 to eliminate any differences due to plant age and size. Similarly, we found that the survival rate of spy-3 plants was much higher than that of the wild-type plants (Fig. 1E). In order to verify that spy mutations really altered plant stress tolerance, we obtained two additional spy alleles, spy-8 and spy-12, which were generated from an EMS mutagenesis screen for suppressors of the ga1-3 mutant (Silverstone et al., 2007). An increased resistance to the inhibitory effects of salt on germination was clearly observed in these mutants (Supplemental Fig. S1). Taken together, these data strongly suggested that mutations in the SPY locus could enhance plant resistance to salinity stress.
Figure 1.
spy mutants were more tolerant of high-salinity stress compared with wild-type plants. A, Morphological phenotypes of spy mutants. Three-week-old Col, spy-1, and spy-3 plants growing on GM agar plates were photographed. Ler, Ecotype Landsberg erecta. B, Schematic structure of the SPY protein and the locations of the mutations in the mutants. C, spy-1, spy-3, and wild-type (WT) plants grown on GM plates for 7 d and subsequently transferred onto 0.5× MS medium plates with or without 200 mm NaCl. D, The survival rate was calculated from independent experiments (n ≥ 3; ** P < 0.01, * P < 0.05, Student’s t test). E, Salt stress tolerance was compared between 8-d-old Col and 7-d-old spy-3 plants, as described in A.
spy Mutants Consume Less Water and Survive Longer under Water Deficit Stress
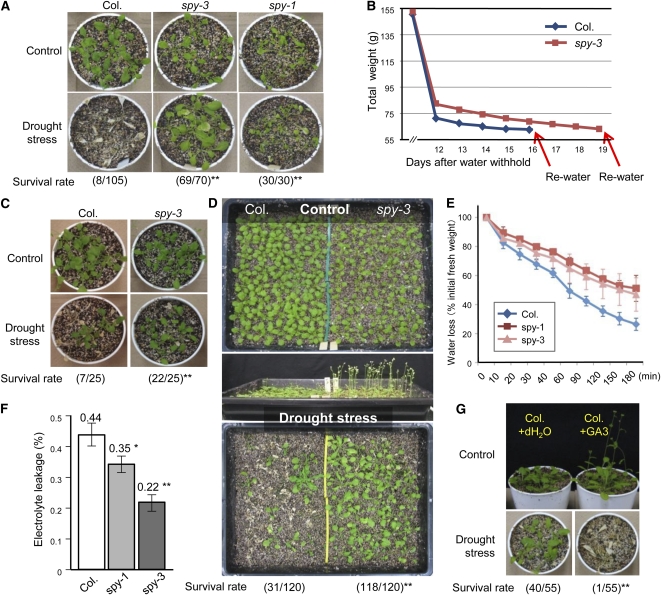
Because salt and drought stresses usually cause similar water stress in plant cells, we extended our plant survival tests to water deficit conditions. First, we compared the plant survival rates in individual soil pots. When the water supply for 4-week-old plants was terminated for 2 weeks, most of the Col plants died, whereas almost all of the spy-3 and spy-1 mutants survived (Fig. 2A). Since the rosette leaves of the 4-week-old mutants were smaller than those of wild-type plants, which could bring about the differences in transpiration and water content in the soil in different pots after withholding water, we exerted out efforts to dehydrate the plants similarly by ensuring that the soil water content dropped to the same level in the soil pots. The water supply to 4-week-old plants growing in different pots was terminated. The soil water content was recorded daily from day 12 after watering was cut off. Lower water content was observed in the soil in which wild-type plants were growing compared with soil containing spy-3 plants, indicating a higher water usage or transpiration of the wild-type plants. On day 16, when the total water amount had fallen to approximately 10% of the original in the pots containing wild-type plants, watering was resumed. Approximately 28% of the plants recovered from severe wilt. Watering was reinitiated for the spy-3 plants after 19 d of dehydration, when the soil water content in these pots had dropped to a similar level to that in the pots of the wild type (Fig. 2B). Although the plants were dehydrated for 3 d longer than the wild-type plants, 88.0% of the spy-3 plants survived (Fig. 2C).
Figure 2.
spy-1 and spy-3 mutants were more tolerant of drought stress. A, The plant survival rate under drought stress was compared in individual soil pots. B, Four-week-old Col and spy-3 plants were subjected to dehydration in pots containing the same amounts of soil. After 12 d of water withholding, the weight of each pot was recorded daily. Watering of the plants was reinitiated once they were equally dehydrated, which occurred on days 16 and 19 for wild-type and spy-3 plants, respectively. C, Representative photographs for the data in B are shown. D, Plant drought tolerance was compared in a large tray containing soil in which the plants were cultivated side by side. Water was withheld from the plants for approximately 14 d, after which the significant difference was observed between spy-3 and wild-type plants. E, Water loss rates of detached spy-1, spy-3, and wild-type shoots. Wild-type Col is shown as the control. The mean and sd were obtained from four plants in each assay. F, Leaf electrolyte leakage was compared among spy-1, spy-3, and Col plants after 2.5 h of dehydration stress. G, Three-week-old Col plants treated with 50 μm GA3 or water after germination were subjected to drought stress, and their survival rates were compared. For all drought tolerance experiments, photographs were taken after 1 week of reinitiated watering. Survival rates were calculated from at least three independent experiments. In all panels, * indicates P < 0.05 and ** indicates P < 0.01 (by Student's t test).
We also compared plant survival in same tray. In this experiment, the wild-type plants and spy-3 mutants were planted side by side in one soil plot in which the soil water content was more comparable between plants than that in different pots. The earlier bolting and flowering phenotypes of the spy-3 mutant as compared with the wild-type plants were typical GA-elevated phenotypes (Fig. 2D). In our drought experiments, water was usually withheld when the spy-3 mutant was flowering. Water consumption at this stage is considered to be most crucial for the plant. However, we again observed a much higher survival rate of the spy-3 mutants compared with the wild-type plants (Fig. 2D). Additionally, the water loss from detached leaves of spy-3 and wild-type plants was evaluated using an assay that is considered to be less affected by the size of the plants. The aerial parts of 15-d-old plants were placed in empty petri dishes and periodically weighed. As expected, both the spy-1 and spy-3 plants were better able to retain water than the wild-type plants (Fig. 2E). The leaf electrolyte leakage of the mutants and the wild type was compared after dehydrating samples for 2.5 h in petri dishes. Significantly lower ion leakage was observed in the spy mutants as compared with the wild-type plants (Fig. 2F). From all these observations, we concluded that loss-of-function mutation of the SPY gene led to increased water stress tolerance in comparison with the wild type, which was probably due to less leaf water loss and better membrane integrity under drought stress. Because spy mutants exhibit elevated GA signaling and a GA-overdosed phenotype, we wondered whether the increased GA response caused the drought tolerance. We treated wild-type plants with either water or a bioactive GA3 solution. This treatment led, as expected, to the typical GA phenotype, characterized by earlier flowering and light green coloring, which is similar to spy-3 mutants. The plants were then subjected to drought stress. Interestingly, we found that, unlike the spy-3 mutants, the GA-treated plants were much more sensitive to water stress, with a much lower survival rate compared with the water-treated plants (Fig. 2G). This result indicated that although SPY is involved in the GA response, the salt and drought stress tolerance observed in the SPY mutation was probably not due to the increased GA signaling.
SPY Gene Expression Is Stress Inducible
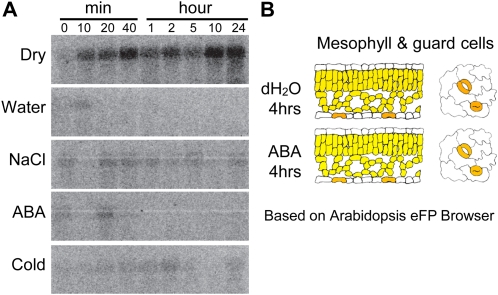
Because the analysis of salt and drought tolerance indicated a role for the SPY gene in the plant stress response, we extended our study to determine whether SPY expression is stress inducible. We found that SPY expression was responsive to dehydration stress. After a 10-min dehydration treatment, there was a clear increase in SPY gene expression, and this remained high for up to 24 h. Gene expression of SPY was slightly responsive to NaCl treatment but not to treatment with abscisic acid (ABA). The responsiveness of the gene to cold was not clear (Fig. 3A). From the eFP Browser database, which is a digital resource that indicates gene expression information based on collective microarray analyses, we found that SPY is expressed at higher levels in guard cells relative to epidermal cells, but this expression does not respond to ABA treatment (Fig. 3B).
Figure 3.
A, Expression of the SPY gene under various stress treatments. Twenty micrograms of total RNA from 3-week-old plants that had been treated as indicated was used for RNA-blot hybridization with a gene-specific probe. B, SPY was preferentially expressed in guard cells in leaf tissue based on data from the Arabidopsis eFP Browser.
Drought-Inducible Expression of Some Genes Is Enhanced in the spy-3 Mutant
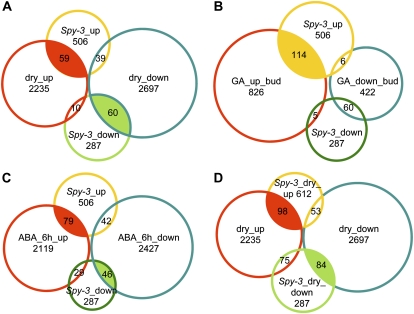
Because the spy mutants exhibited increased tolerance to both salinity and water deficit stresses, we compared genome-wide gene expression changes between the spy mutants and wild-type plants (Col ecotype). For this analysis, gene expression was compared between spy-3 and wild-type plants, because the spy-3 mutant displayed a strongly tolerant phenotype with the fewest morphological changes. The expression of 506 genes was found to be enhanced in the spy-3 mutant compared with the wild type, and 287 genes were down-regulated in the mutant. The criteria for identifying up- or down-regulated genes were a fold change absolute (FCA) ≥ 2 and P < 0.05 (Supplemental Table S1). To obtain an overview of the genes with altered expression, the top 200 up- or down-regulated genes were submitted to Genevestigator to determine their expression in response to various treatments, including chemical, biotic, abiotic, and hormone treatments. The 200 most strongly up-regulated genes did not show any predominant response to the various treatments, except that many genes were found to be drought stress inducible (Supplemental Fig. S2A). However, among the 200 most strongly down-regulated genes, many were found to be ABA or drought repressible (Supplemental Fig. S2B). Furthermore, microarray analysis of Col plants growing in soil that were dehydrated for 3 d, which identified drought-inducible genes genome wide (Maruyama et al., 2009), showed that of the 506 genes up-regulated in the spy-3 mutant, 59 were drought stress inducible, suggesting that these genes may contribute to the enhanced drought tolerance of the spy mutant (Fig. 4A; Supplemental Table S2).
Figure 4.
Venn diagrams of gene expression alterations in spy-3 plants under normal or dehydration stress conditions based on microarray analysis. A, Overlap between Col plants dehydrated for 3 d and nondehydrated spy-3 plants. B, Overlap between genes up- or down-regulated by GA with those of untreated spy-3 plants. C, Overlap between Col plants treated for 6 h with ABA and untreated spy-3 plants. D, Overlap between Col plants dehydrated for 3 d and spy-3 plants dehydrated for 2 h.
Expression of GA-Responsive Genes Is Increased in the spy-3 Mutant
Because the spy-3 mutant displays many of the same phenotypes as GA-treated plants, we were interested to determine whether the expression of GA-responsive genes was altered in the spy-3 mutant. Microarray analysis of ga1-3 mutants versus wild-type plants using flower buds identified 826 GA-responsive genes (Cao et al., 2006). When we compared our data with these results, we found that of the 506 genes up-regulated in the spy-3 mutant, 114 were GA responsive, including RGL1 (Fig. 4B; Supplemental Table S3). Such a large overlap in gene expression may provide a good explanation for the GA-overdosed phenotype of the spy-3 mutant. In addition to these 114 genes, there were at least another three GA-responsive genes found to be up-regulated in the spy-3 mutant: RGL2, GA 2-oxidase, and GA-responsive gene (At5g59845). Although DELLA proteins are degraded by treatment with GA, the expression of these genes was found to be slightly up-regulated in the spy-3 mutant.
We also examined the correlation between gene expression in the spy-3 mutant and ABA-treated Col plants. According to the microarray analysis of the Col plants treated with ABA for 6 h, there were 2,119 and 2,427 genes that were up- and down-regulated, respectively (Fujita et al., 2009). Of the 2,119 ABA-up-regulated genes, the expression of 79 was found to be increased, and of the 2,427 ABA-down-regulated genes, the expression of 46 was decreased in the spy-3 mutant (Fig. 4C). Compared with the overlap observed between the genes up-regulated in both GA-treated and spy-3 mutant plants, those up-regulated in ABA-treated plants overlapped with those up-regulated in the spy mutants much less, indicating that the spy-3 mutation has strong effects on GA-dependent gene expression, whereas it does not considerably affect ABA-responsive gene expression.
Transcriptome Alterations in Dehydrated spy-3 Mutants Compared with Wild-Type Plants
We also compared the gene expression changes in spy-3 and wild-type plants that were dehydrated for 2 h. Overall, the expression of 612 genes was found to be enhanced in the spy-3 mutant compared with Col, and 720 genes were down-regulated in the mutant, as determined by selecting genes with FCA ≥ 2 and P < 0.05 (Supplemental Table S4). The top 200 up- or down-regulated genes were analyzed using Genevestigator. Similarly, among the top 200 up-regulated genes, many were found to be drought stress inducible (Supplemental Fig. S3A). However, among the top 200 down-regulated genes, there were many genes that were ABA or drought stress repressible, which was similar to the results of the microarray analysis of the untreated spy-3 mutants. Unexpectedly, many biotic stress-inducible genes were revealed to be down-regulated in the spy-3 mutant (Supplemental Fig. S3B). Compared with the results of the microarray analysis of Col plants growing in soil that were dehydrated for 3 d (Maruyama et al., 2009), we found that of the 612 up-regulated genes, 98 were drought stress inducible (Fig. 4D; Supplemental Table S5). Importantly, 16 LEA protein genes were found to be up-regulated in the spy-3 plants that were dehydrated for 2 h, and all of them were drought stress inducible, which might contribute to the drought stress tolerance of plants. Similarly, the expression of five lipid transfer proteins and seven transporter genes was found to be enhanced in the spy-3 mutants dehydrated for 2 h. Some transcription factor- and protein enzyme-encoding genes were also revealed to be transcriptionally enhanced.
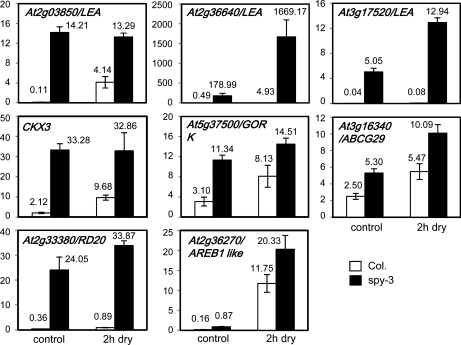
To verify the microarray results, the expression levels of several genes that are potentially related to water stress response in the spy-3 mutant were monitored by quantitative reverse transcription (qRT)-PCR analyses. Three LEA protein-encoding genes, At2g03850, At2g36640, and At3g17520, were determined to be significantly up-regulated under both normal growing conditions and stress conditions in the spy-3 mutant as compared with wild-type plants (Fig. 5). These LEA proteins may confer the stress-tolerant phenotype of the spy-3 mutant. Two genes encoding transporters, an ABC transporter gene (ABCG29) and an outward K+ channel gene (GORK) that may be involved in leaf water transpiration or ABA response, were revealed to be up-regulated in the spy-3 mutant (Fig. 5). Moreover, the CKX3 gene encoding a CK oxidase/dehydrogenase was found to be 15.6- and 3.4-fold up-regulated in the spy-3 mutant under normal and dehydration conditions, respectively, according to the qRT-PCR analyses (Fig. 5). Recently, it was reported that overexpression of the CKX3 gene, which leads to a reduction in CK content, enhanced the drought and salt tolerance of Arabidopsis and tobacco (Nicotiana tabacum) transgenic plants (Werner et al., 2010; Nishiyama et al., 2011). Increased expression levels of two other stress-related genes, RD20 and an AREB1-like gene (At2g36270), were also found in the mutant under both normal and stress conditions (Fig. 5). Taken together, we concluded that the microarray data are reliable and reproducible. The enhanced expression of LEA genes, genes encoding transporters, stress-related genes, and CKX3 might contribute to the salt- and drought-tolerant phenotypes of the spy-3 mutant. It is worth mentioning that among the 720 genes down-regulated in the dehydrated spy-3 mutants, the expression of 84 was found to be drought repressible. Many genes encoding disease resistance proteins, Leu-rich repeat protein kinases, and receptor-like protein kinases were transcriptionally down-regulated, which was consistent with the results of Genevestigator analysis of the gene expression responsiveness of the top 200 down-regulated genes (Supplemental Table S4).
Figure 5.
qRT-PCR analysis of the expression of genes that were identified as being up-regulated in the spy-3 mutant under normal growing and 2-h-dehydrated conditions. Primer sequences are listed in Supplemental Table S11.
Overexpression of the SPY Gene Reduces Plant Drought Stress Tolerance
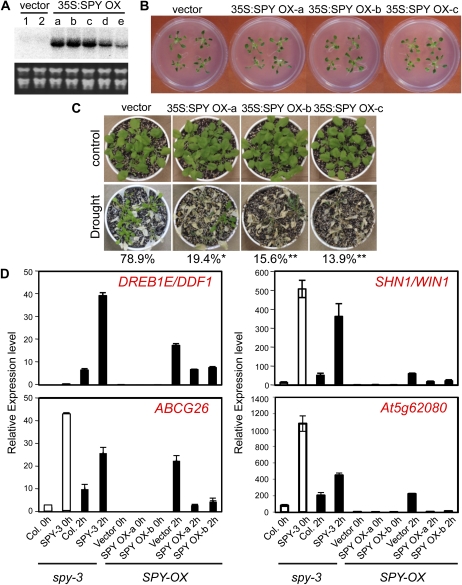
To further elucidate the function of the SPY gene in the plant water deficit stress response, we generated transgenic Arabidopsis plants overexpressing the SPY gene using the 35S constitutive promoter (35S:SPY-OX). Several lines of transgenic plants were obtained, and transgene expression was checked by RNA gel-blot analysis (Fig. 6A). In contrast with spy mutants, the SPY overexpressors did not exhibit dramatic morphological changes (Fig. 6B). Three lines of 35S:SPY-OX plants, a, b, and c, with comparably high levels of transgene expression, were tested for plant drought stress tolerance. Water was withheld from 4-week-old plants for approximately 2 weeks, and the plants were then watered again. The survival rate was recorded and compared between the vector-transformed control and 35S:SPY-OX lines. While 78.9% of the control plants survived, 19.4%, 15.6%, and 13.9% of the 35S:SPY-OXa, -b, and -c lines, respectively, survived after the drought stress treatment. Statistical analysis of the plant survival rates from three replicate experiments is shown in Figure 6C and Supplemental Table S6. It suggested that drought stress tolerance was decreased in these three types of 35S:SPY-OX transgenic plants.
Figure 6.
Overexpression of SPY reduced plant drought stress tolerance. A, RNA gel-blot analysis of transgene expression in vector and 35S:SPY-transformed plants; 10 μg of total RNA was loaded. B, Morphological phenotype of 3-week-old SPY-OX plants as compared with the wild type. C, Survival rate of vector-transformed and SPY-OX lines. The averaged data were obtained from three independent experiments (** P < 0.01, * P < 0.05, Student’s t test). D, qRT-PCR analysis of the expression of genes that were up-regulated in spy-3 mutants but down-regulated in SPY-OX plants. White bars indicate the relative expression level in the untreated plants, and black bars indicate the relative expression level in the 2-h-dehydrated plants. Primer sequences are listed in Supplemental Table S11.
We were further interested in exploring alterations of gene expression in SPY-OX plants because the expression levels of hundreds of genes were changed in the spy-3 mutants. Mutations in spy-3 affected GA and CK signaling and led to a variety of morphological changes, whereas SPY overexpression only negatively altered plant stress tolerance without any obvious morphological effects. Analyzing SPY-OX plants may facilitate the identification of a direct relationship between SPY function and plant stress tolerance. Two additional microarray experiments were constructed comparing two independent 35S:SPY-OX-a and -c lines with vector-transformed transgenic plants. Transcriptomic changes were compared under both normal and 2-h-dehydrated conditions. In total, 48 genes were up-regulated and 156 genes were down-regulated (FCA ≥ 2, P < 0.05) in SPY-OX plants during normal growth. Among them, the expression of the SPY gene was 12.1-fold higher in the transgenic plants as compared with the wild-type plants (Supplemental Table S7). According to the same criteria, 66 and 184 genes were found to be up- and down-regulated in 2-h-dehydrated SPY-OX plants (Supplemental Table S8). From both microarray experiments, more genes were found to be down-regulated than up-regulated in the SPY-OX plants, indicating a negative, genome-wide effect of SPY on gene expression regulation. Based on the drought-tolerant phenotype of the spy-3 mutants and sensitive of SPY-OX plants, we were particularly interested in the genes that were up-regulated in spy-3 and down-regulated in the SPY-OX plants, which might be the genes most affected by the SPY protein. Out of 156 down-regulated genes in the SPY-OX plants, 19 genes were found to be up-regulated in the spy-3 mutant under normal growing conditions (FCA ≥ 2, P < 0.05). Six of these genes are GA-up-regulated genes in Arabidopsis buds, but none of these genes is GA repressive; this result is consistent with a negative role for SPY in GA signaling (Supplemental Table S9). Indeed, there is no overlap of genes up-regulated in the SPY-OX plants that are also down-regulated in the spy-3 plants.
Results from the 2-h-dehydrated samples showed that among the 184 genes down-regulated in the SPY-OX plants, three were up-regulated in the spy-3 mutant (FCA ≥ 2, P < 0.05; Supplemental Table S10). These genes are SHN1/WIN1, ABCG26, and APETALA3 (AP3). Two additional genes, DREB1E/DDF1 and At5g62080, were verified to be inversely regulated by SPY overexpression and spy-3 mutation based on further qRT-PCR analyses. The expression of all these genes was found to be drought stress inducible (Fig. 6D). SHN1/WIN1 encodes an AP2/ERF transcription factor belonging to the A-6 subgroup. Overexpression of SHN1/WIN1 increased the leaf cuticular wax content, leading to reduced water loss and enhanced tolerance to drought stress (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007). DREB1E/DDF1 is also an AP2/ERF transcription factor, classified as a member of the A-1 subgroup, that can bind the DRE sequence in the promoter of dehydration-responsive genes (Sakuma et al., 2002). Transgenic plants overexpressing the DREB1E/DDF1 gene exhibited improved tolerance to drought, cold, and heat stresses (Kang et al., 2011). The up-regulated expression of SHN1/WIN1 and DREB1E/DDF1 may contribute to the maintenance of higher water content, resulting in stronger drought stress tolerance of the spy-3 mutant (Fig. 2). Although expression of the ABCG26 and ABCG29 genes was increased in the spy-3 mutant, only ABCG26 transcription was repressed by overexpression of SPY under both normal and stress conditions (Figs. 5 and 6D). The function of ABCG26 was reported to be associated with male fertility and pollen maturation (Quilichini et al., 2010). Together with the AP3 gene, ABCG26 might cause the flowering-related phenotypes of the spy mutant.
DISCUSSION
To properly respond to environmental signals, plants need to integrate external and internal signals and a complex network of signal transduction pathways. In recent years, different components involved in these signal cascades have been identified, including genes involved in stress-induced transcriptional regulation and complex cross talk among different hormones at either the biosynthesis or action level (Fujita et al., 2006; Shinozaki and Yamaguchi-Shinozaki, 2007). GA is an important plant phytohormone involved in several processes, including seed germination, vegetative growth, flowering induction, and fruit development (Sun and Gubler, 2004). The SPY gene was first identified as a negative regulator in GA signaling, because the mutant displayed a set of GA-treated phenotypes, such as insensitivity to PAC during germination, early flowering, partial male sterility, and pale green color (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996). Although the involvement of SPY in GA signaling is indisputable, it has also been recognized that SPY is involved in other cellular processes. spy mutants also display phenotypes, such as altered leaf phyllotaxy, CK responses, light responses, and reduced hypocotyls and rosette growth, that are not directly related to GA responses (Swain et al., 2001; Tseng et al., 2004; Greenboim-Wainberg et al., 2005). Additionally, double mutation of the SPY and SEC genes, the only two OGTs in plants, leads to lethality, which may not be caused by GA treatment. These phenomena support the hypothesis that SPY has an unidentified function in one or more processes unrelated to GA signaling. Here, we demonstrated that mutation of spy led to a strong salt and drought tolerance in plants whose mechanism is independent of GA-DELLA signaling.
SPY gene expression was found to be drought stress inducible and slightly responsive to salinity stress. Furthermore, based on microarray analysis, we revealed that under normal growing conditions, in addition to the up-regulation of many GA-responsive genes, the expression of a number of drought-inducible genes was increased; these genes encode LEA proteins, RD20, ABC transporters, and AREB1-like transcription factor (Fig. 5). Under drought conditions, the expression of 16 LEA protein genes was higher in spy-3 mutants compared with wild-type plants (Supplemental Table S5). LEA proteins function in an unfolded state as ion sinks or water replacement molecules, and that water deficit stress can induce folding and conformational shifts in LEA proteins, so that they act as molecular chaperones to protect important proteins (Goyal et al., 2003; Wise and Tunnacliffe, 2004). Overexpression of DREB1A, constitutively active DREB2A, or an activated form of AREB1 (Maruyama et al., 2004; Fujita et al., 2005; Sakuma et al., 2006) in Arabidopsis resulted in up-regulation of the expression of some LEA genes, and all of these transgenic plants displayed improved drought stress tolerance, further supporting the hypothesis that LEA proteins play a role in plant water deficit tolerance. Direct evidence has shown that overexpression of a rice OsLea3 gene was able to enhance plant water deficit stress tolerance (Xiao et al., 2007). The elevated stress-responsive gene expression may confer the stress tolerance on the spy mutant.
In addition to the negative regulation of GA responses, the SPY gene has been shown to play a positive regulatory role in CK signaling. spy mutants were found to be more resistant to exogenously applied CK and exhibited inhibition of the induction of Arabidopsis TYPE A RESPONSE REGULATOR5 (ARR5) after CK treatment (Greenboim-Wainberg et al., 2005). Interestingly, according to the results of our microarray analysis, we found that the expression of CKX3 is increased 15.6- and 3.4-fold relative to the wild type in both normal and dehydrated conditions, respectively, which may cause a decrease in CK level in the spy-3 mutant (Fig. 5; Werner et al., 2003, 2010). Overexpression of the CKX3 gene resulted in reduced ARR5 and ARR7 promoter activity, a decrease in the total surface area of rosette leaves, and plant growth retardation (Werner et al., 2003; Nishiyama et al., 2011). Similarly, spy-3 mutant rosette leaves were smaller than those of wild-type plants, despite the fact that GA treatment usually increases the size of these leaves. This morphological phenotype is consistent with the increased expression of CKX3 in the spy-3 mutant. Interestingly, recent research has shown that reduction of the CK level via overexpression of the CKX3 gene in roots can improve plant drought tolerance and leaf mineral enrichment (Werner et al., 2010). Functional analyses of CK-deficient AtCKX overexpressors and atipt mutants provided direct evidence that CKs negatively regulate salt and drought stress responses in plants (Nishiyama et al., 2011). Moreover, when the CK receptors AHK2 and/or AHK3 were disrupted, both the single and double mutants were more tolerant of high-salt and drought stresses (Tran et al., 2007). Taken together, the up-regulation of CKX3 gene expression might reduce CK signaling in the spy-3 mutant, contributing to plant salt and drought stress tolerance. Although CKX3 gene expression was significantly up-regulated in the spy-3 mutant, it was not down-regulated in the SPY-OX plants. This lack of down-regulation probably explains why the stress-tolerant phenotype of SPY-OX was not as evident as that of the mutant. No remarkable tolerance to high-salinity stress was observed in the SPY-OX plants. The molecular mechanism by which SPY mediates CK signaling must be investigated further.
Because our knowledge concerning the molecular actions of the SPY protein has been limited by the inability to reliably detect modified proteins that are direct targets of the SPY protein (Olszewski et al., 2010), we generated SPY-OX plants and compared transcriptome alterations in spy-3 versus wild-type plants and SPY-OX versus wild-type plants to identify the direct effects of the SPY gene on the transcriptome, especially in relation to plant stress response. The SPY-OX transgenic plants were morphologically similar to the wild-type plants but were more sensitive to drought stress than the wild-type plants (Fig. 5). Comparative transcriptomic analyses uncovered genes that showed inverse transcriptional regulation by SPY overexpression and spy-3 mutation under both normal and drought stress conditions (Fig. 6D; Supplemental Tables S9 and S10). The expression of DREB1E/DDF1 and SHN1/WIN1 was up-regulated in the spy-3 mutant but down-regulated in SPY-OX plants. Both genes positively contribute to plant drought or salt tolerance. The DREB1E/DDF1 gene not only activated stress-responsive gene expression but also enhanced plant tolerance to drought, cold, and heat stresses (Kang et al., 2011). Moreover, the DREB1E/DDF1 gene can activate GA2ox7, a gene encoding a GA-deactivation enzyme, under salinity stress. The expression of this gene may reduce the active GA content in the plant, thus reducing plant growth under stress (Magome et al., 2008). GA2ox7 gene expression was also consistently decreased 1.68-fold as the DREB1E/DDF1 gene was reduced in SPY-OX plants. Our results provide evidence that overexpression of the SPY gene can suppress the GA response, supporting the hypothesis that SPY functions as a negative regulator of GA signaling. The SPY gene may not directly regulate the gene activity involved in GA metabolism but may instead regulate transcription factors that control metabolic gene expression. SHN1/WIN1 is also an AP2/ERF transcription factor, which can activate cuticular wax synthesis, alter cuticle properties, and increase plant drought stress tolerance. SHN1/WIN1 may act by controlling the expression of LACS2, a long-chain acyl-CoA synthetase gene (Aharoni et al., 2004; Kannangara et al., 2007). Recently, plant cuticle composition was found to be essential for osmotic stress response and tolerance. The cuticle not only functions as a physical barrier to minimize water loss but also mediates abiotic stress signaling (Wang et al., 2011). The expression of SHN1/WIN1 was significantly increased in the spy-3 mutant and down-regulated in SPY-OX plants, which may result in the different drought-related phenotypes of these two kinds of plants. The expression of CKX3 and some LEA protein genes was up-regulated by the spy-3 mutation but not down-regulated by SPY overexpression, which probably explains why the SPY-OX plants were not only sensitive to drought but also to salt stress in comparison with wild-type plants (data not shown).
In recent decades, great progress has been made regarding the characterization of GA signal reception and transduction, due largely to the identification of the upstream GID1 GA receptors and the downstream GA-targeted DELLA proteins (Ueguchi-Tanaka et al., 2005; Harberd et al., 2009). GA promotes plant growth by destroying a class of transcription factors, the DELLA proteins, that function as repressors to restrain plant growth. DELLA proteins are potential candidate targets of SPY (Silverstone et al., 1998; Olszewski et al., 2002). The O-GlcNAc modification, mediated by SPY, was proposed to compete with phosphorylation to modulate DELLA protein abundance and activity in response to GA or environmental signals (Itoh et al., 2005). It has been proposed that salt stress inhibits plant growth by reducing endogenous bioactive GA levels, leading to the accumulation of DELLAs (Achard et al., 2006). Studies have demonstrated that the growth restraint conferred by DELLA proteins is beneficial and promotes plant survival under severe salinity stress conditions. A quadruple DELLA mutant lacking GAI, RGA, RGL1, and RGL2 has been found to be less tolerant to severe salt stress and to exhibit suppression of the salt tolerance conferred by ga1-3 (Achard et al., 2006). Unexpectedly, in various spy mutants, the level of RGA protein was consistently higher than in wild-type plants, although spy mutants displayed GA-treated phenotypes (Silverstone et al., 2007). Higher levels of DELLA proteins in the spy mutants indicate that the absence of the O-GlcNAc modification might enhance the stability of the proteins but likely reduces their activity in terms of GA signaling. It is also possible that abundant DELLA proteins might confer plant stress tolerance on the spy-3 mutant. Additionally, many biotic-responsive genes were down-regulated in the 2-h-dehydrated spy-3 mutant (Supplemental Fig. S3B; Supplemental Table S4). Whether the mutant is more susceptible to biotic attack is unknown.
Research on O-GlcNAc modification is more advanced in mammalian cells than in plants. O-GlcNAc modification modulates signaling by influencing gene expression, protein degradation, and trafficking in cells and also plays roles in nutrient sensing, cell cycle progression, and stress response (Hart et al., 2007). In mammalian cells, a rapid and global increase in O-GlcNAc-ylation on many proteins upon exposure to stress stimuli was found, indicating a positive role for OGTs (Hart et al., 2007). Here, we characterized a negative role for SPY, a plant OGT that plays a negative role in plant survival under severe salinity and drought stresses because the spy mutants were more tolerant to these stresses. Unlike mammals and insects, plants have two distinct genes encoding OGTs, SPY and SEC, and both genes have unique and overlapping roles (Olszewski et al., 2010). The activity of a single OGT gene is essential for cell viability in mammals (Hart et al., 2007). In plants, the simultaneous deletion of the SPY and SEC genes is embryo lethal, whereas single mutations of each gene produce different phenotypes. A phylogenetic analysis of all eukaryotic OGT genes found that mammalian OGTs are SEC-like but not SPY-like proteins (Olszewski et al., 2010). The gene function of SEC in plant stress and/or hormone response and the relationship between these two gene functions merit further investigation.
MATERIALS AND METHODS
Plant Materials and Transgenic Plant Construction
Arabidopsis (Arabidopsis thaliana) plants were grown, transformed, and treated as described previously (Qin et al., 2008). spy-1 and spy-3 mutant seeds were obtained from the Arabidopsis Biological Resource Center (catalog nos. CS6266 and CS6268, respectively). spy-8 and spy-12 mutant seeds were kindly provided by Dr. Tai-ping Sun. The full-length coding sequence of the SPY gene was constructed downstream of the 35S promoter with an Ω enhancer sequence in the pGKX vector (Qin et al., 2008) and transformed into Col plants for the purpose of overexpressing this gene.
RNA Gel-Blot and Real-Time RT-PCR Analyses
Total RNA was isolated with the RNAiso (Takara) reagent from 3-week-old plants. RNA gel-blot analysis was performed as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). A SPY-specific probe was prepared by PCR amplification of the C-terminal end of the SPY gene from nucleotide 2,003 to the final nucleotide. For real-time RT-PCR analysis, 1 μg of total RNA was used for the first-strand cDNA synthesis using SuperScript III transcriptase (Invitrogen). The resultant cDNA solution was diluted five times, and a 1-μL solution was used as the template in subsequent real-time PCR. PCR was performed in a 10-μL volume using the ABI7500 system and SYBR Green I mixture (Takara). Each reaction was performed in triplicate to obtain the average and sd for the expression level of each gene.
Microarray Analysis and Statistical Analysis
Genome-wide expression studies with Arabidopsis 44K Oligo Microarrays (Agilent Technologies) were performed using 3-week-old wild-type and spy-3 plants. Gene expression was compared between wild-type and spy-3 plants under both unstressed and 2-h dehydration stress conditions. For each sample (pooled from six plants), 200 ng of total RNA was isolated using the RNAiso reagent (Takara) and used for the analysis. Biological replication was performed by analyzing the samples obtained from two independent treatments (for both the control and 2-h dehydration stress conditions). For each experiment, two slides were analyzed to perform a Cy3 and Cy5 dye swap, and a total of eight slides were hybridized for the four experiments. Statistical analysis of the microarray data to integrate and normalize each spot’s signal intensity was performed using the Lowess method with the Feature Extraction 9.5 program (Agilent Technologies). The statistical analysis was carried out using ArrayAssist software (Stratagene). The Welch t test was used as a parametric test, and the Benjamini and Hochberg false discovery rate for multiple testing corrections was used with a threshold set at P < 0.05 to identify reliable genes. Following this statistical analysis, we generated lists of the genes showing significant differences in expression between wild-type and spy-3 plants under normal conditions and after exposure to a 2-h dehydration stress. Of the genes that were considered to show significant expression differences based on the previous tests, those that exhibited an absolute fold change greater than 2.0 were selected. Such genes were considered to be up-regulated in spy-3 plants relative to wild-type plants. All of the microarray data are available at http://www.ebi.ac.uk/arrayexpress/ under the accession numbers E-MEXP-3171 and E-MEXP-3362.
Salt Stress Tolerance Test
For the plant seedling tolerance test, seeds were sown on germination medium (GM) agar plates and poststratified at 4°C for 4 d. After growing on GM plates for 7 d, the plants were transferred onto 0.5× Murashige and Skoog (MS) agar plates containing 200 mm NaCl. The plates were maintained at 22°C under a 16-h-light/8-h-dark cycle until visual symptoms could be observed and photographed. For the plant salt tolerance test at the germination stage, the seeds were sown on MS agar plates with 150 mm NaCl and poststratified at 4°C for 4 d. Then, they were transferred to 22°C under a 16-h-light/8-h-dark cycle until visual symptoms could be observed and photographed.
Drought Stress Tolerance Test
Briefly, 3-week-old plants grown on GM agar plates were transferred to soil. After 1 week of conditioning, the plants were subjected to a water-withholding treatment for approximately 14 d, after which the most obvious differences were observed between the wild-type and mutant plants. The plant survival rate was recorded after 1 week of reinitiated watering and recovery. For the GA treatment, Col plants were transferred to soil after 3 d of germination on GM agar plates and sprayed with a 50 μm GA3 solution or water every other day for 3 weeks. The water supply was then cut off for approximately 2 weeks for the dehydration treatment. To ensure that the pots in which Col and spy-3 plants were growing contained equal water levels during the dehydration stress treatment, the plants were transferred into pots containing the same amount of soil saturated by water. The initial weight of each pot was controlled, and the total weight of each pot was measured daily after 12 d of dehydration stress, so as to calculate and record the water content of each pot daily. Watering was resumed when the average total weights of the pots containing Col and spy-3 were comparable. For the plant water loss assay, the aerial parts of 2-week-old spy-1, spy-3, and Col plants were placed in empty petri dishes, and the plant weights were periodically recorded. For each assay, the average and sd were calculated from four plants. The leaf electrolyte leakage was determined as described by Qin et al. (2004).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_111987 (SPINDLY).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. spy-8 and spy-12 mutants were more tolerant to salt stress during seed germination.
Supplemental Figure S2. Heat map of transcriptomic analysis of normally growing spy-3 versus Col plants.
Supplemental Figure S3. Heat map of transcriptomic analysis of 2-h-dehydrated spy-3 versus Col plants.
Supplemental Table S1. A total of 506 up-regulated and 287 down-regulated genes in 0-h spy-3 plants.
Supplemental Table S2. A total of 59 drought stress-inducible genes up-regulated in 0-h-dehydrated spy-3 plants.
Supplemental Table S3. A total of 114 GA-up-regulated genes in spy-3 plants.
Supplemental Table S4. A total of 612 up-regulated and 720 down-regulated genes in 2-h-dehydrated spy-3 plants.
Supplemental Table S5. A total of 98 drought-inducible genes up-regulated in 2-h-dehydrated spy-3 plants.
Supplemental Table S6. Drought tolerance test of SPY-OX plants.
Supplemental Table S7. Up- or down-regulated genes in SPY-OX plants under normal growing conditions.
Supplemental Table S8. Up- or down-regulated genes in SPY-OX plants after 2 h of dehydration stress.
Supplemental Table S9. Genes up-regulated in the spy-3 mutant but down-regulated in SPY-OX plants under normal growing conditions.
Supplemental Table S10. Genes up-regulated in the spy-3 mutant but down-regulated in SPY-OX plants under 2-h dehydration conditions.
Supplemental Table S11. Primer list for qRT-PCR.
Acknowledgments
We thank E. Murai, K. Amano, K. Yoshiwara, and H. Kishi for their excellent technical support and M. Toyoshima for skillful editorial assistance.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J. (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer FI, Hart GW. (2001) Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry 40: 7845–7852 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T. (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Goyal K, Tisi L, Basran A, Browne J, Burnell A, Zurdo J, Tunnacliffe A. (2003) Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J Biol Chem 278: 12977–12984 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D. (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C. (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446: 1017–1022 [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Genger RK, Grey WM, Olszewski NE. (2006) SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J Exp Bot 57: 865–875 [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE. (2002) Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Ueguchi-Tanaka M, Matsuoka M. (2008) GID1-mediated gibberellin signaling in plants. Trends Plant Sci 13: 192–199 [DOI] [PubMed] [Google Scholar]
- Itoh H, Sasaki A, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Hasegawa Y, Minami E, Ashikari M, Matsuoka M. (2005) Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol 46: 1392–1399 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim YC, Iuchi A, Kuromori T, Ueguchi-Tanaka M, Asami T, Yamaguchi I, Matsuoka M, Kobayashi M, et al. (2007) Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J 50: 958–966 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93: 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Kim J, Kim B, Jeong H, Choi SH, Kim EK, Lee HY, Lim PO. (2011) Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci 180: 634–641 [DOI] [PubMed] [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Höfte H, Pauly M, Riechmann JL, Broun P. (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP. (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreppel LK, Hart GW. (1999) Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem 274: 32015–32022 [DOI] [PubMed] [Google Scholar]
- Love DC, Hanover JA. (2005) The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE 2005: re13. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Hanover JA. (2000) Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem 275: 10983–10988 [DOI] [PubMed] [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K. (2008) The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 56: 613–626 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, et al. (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 150: 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymon I, Greenboim-Wainberg Y, Sagiv S, Kieber JJ, Moshelion M, Olszewski N, Weiss D. (2009) Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J 58: 979–988 [DOI] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski NE, West CM, Sassi SO, Hartweck LM. (2010) O-GlcNAc protein modification in plants: evolution and function. Biochim Biophys Acta 1800: 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LS, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J 50: 54–69 [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ. (2010) ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-Ali T, Harberd NP. (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52: 67–88 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T. (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP. (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C, Hart GW. (2003) Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr Opin Struct Biol 13: 631–636 [DOI] [PubMed] [Google Scholar]
- Sun TP, Gubler F. (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, Olszewski NE. (2001) Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol 126: 1174–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, Thornton TM, Gopalraj M, Olszewski NE. (2002) SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol 129: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton TM, Swain SM, Olszewski NE. (1999) Gibberellin signal transduction presents...the SPY who O-GlcNAc’d me. Trends Plant Sci 4: 424–428 [DOI] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Salomé PA, McClung CR, Olszewski NE. (2004) SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16: 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Swain SM, Olszewski NE. (2001) Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol 126: 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP. (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Xiong L, Li W, Zhu JK, Zhu J. (2011) The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 23: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW. (2001) Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291: 2376–2378 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. (2010) Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22: 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ, Tunnacliffe A. (2004) POPP the question: what do LEA proteins do? Trends Plant Sci 9: 13–17 [DOI] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L. (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115: 35–46 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]