Abstract
The phytohormone abscisic acid (ABA) regulates stress responses and controls numerous aspects of plant growth and development. Biosynthetic precursors and catabolites of ABA have been shown to trigger ABA responses in physiological assays, but it is not clear whether these are intrinsically active or whether they are converted into ABA in planta. In this study, we analyzed the effect of ABA precursors, conjugates, and catabolites on hormone signaling in Arabidopsis (Arabidopsis thaliana). The compounds were also tested in vitro for their ability to regulate the phosphatase moiety of ABA receptor complexes consisting of the protein phosphatase 2C ABI2 and the coreceptors RCAR1/PYL9, RCAR3/PYL8, and RCAR11/PYR1. Using mutants defective in ABA biosynthesis, we show that the physiological activity associated with ABA precursors derives predominantly from their bioconversion to ABA. The ABA glucose ester conjugate, which is the most widespread storage form of ABA, showed weak ABA-like activity in germination assays and in triggering ABA signaling in protoplasts. The ABA conjugate and precursors showed negligible activity as a regulatory ligand of the ABI2/RCAR receptor complexes. The majority of ABA catabolites were inactive in our assays. To analyze the chemically unstable 8′- and 9′-hydroxylated ABA catabolites, we used stable tetralone derivatives of these compounds, which did trigger selective ABA responses. ABA synthetic analogs exhibited differential activity as regulatory ligands of different ABA receptor complexes in vitro. The data show that ABA precursors, catabolites, and conjugates have limited intrinsic bioactivity and that both natural and synthetic ABA-related compounds can be used to probe the structural requirements of ABA ligand-receptor interactions.
Abscisic acid (ABA) is a plant hormone that controls a broad range of processes in plants and is most widely recognized for its involvement in plant adaptation to abiotic stress (Wasilewska et al., 2008). Drought or salinity lead to an elevation of the ABA stress signal via enhanced ABA biosynthesis, possible ABA release from storage forms, and reduced ABA catabolism in the plant. ABA induces the closure of leaf stomata to minimize water loss through transpiration, and the phytohormone ultimately mediates stress tolerance adaptation. In addition to its recognized role in stress responses, ABA regulates plant growth and development, including seed maturation, maintenance of dormancy, secondary root formation, and leaf size.
The molecular mechanism of ABA signal transduction has been a subject of great interest over the years (Hirayama and Shinozaki, 2007; Novikova et al., 2009; Cutler et al., 2010; Raghavendra et al., 2010). Recently, members of the Bet V 1 superfamily of proteins (known as RCAR/PYR1/PYL) have been identified as bona fide ABA receptors in Arabidopsis (Arabidopsis thaliana; Ma et al., 2009; Park et al., 2009; Santiago et al., 2009a). Binding of ABA to RCAR/PYR1/PYL directly inhibits the activity of type 2C protein phosphatases (PP2C), which act as negative regulators of ABA signaling. RCAR/PYR1/PYL can form tight complexes with PP2Cs (Nishimura et al., 2010), which are stabilized by ABA, and the heteromeric protein complex allows for high-affinity interaction with ABA (Ma et al., 2009; Santiago et al., 2009b). The class of ABA-binding proteins is evolutionarily conserved and present in both lower and higher plants (Saavedra et al., 2010). Structural studies of RCAR/PYR1/PYL and PP2C protein complexes have revealed an obstruction of the active site of PP2C by RCAR/PYR1/PYL in the presence of ABA and provide the mechanistic rationale for the enzymatic inactivation of PP2C (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009a; Yin et al., 2009; Shibata et al., 2010).
There are 14 homologous RCARs in Arabidopsis, and six clade A PP2Cs have been implicated in ABA signaling events (Fujii et al., 2009; Cutler et al., 2010; Raghavendra et al., 2010). Thus, numerous combinations of the receptor complexes may exist. Some of these have been shown to differ in their sensitivity to ABA and are postulated to target different downstream components (Szostkiewicz et al., 2010). Multiple receptor complexes could thereby fine-tune a multitude of ABA responses. The nature of the ligand could also be critical to target specific receptor complexes, as exemplified by the identification of the ABA agonist pyrabactin [4-bromo-N-(2-pyridinylmethyl)-1-napthalenesulfonamide] that selectively binds to PYR1/RCAR11 (Park et al., 2009; Melcher et al., 2010). It is well established that natural structural derivatives of ABA have biological activity (Zaharia et al., 2005b).
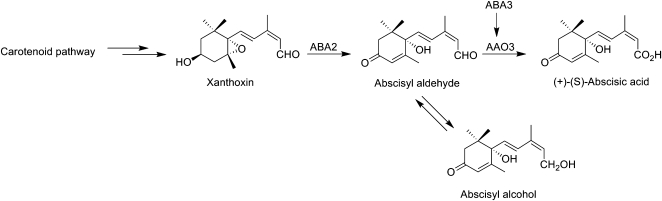
ABA biosynthesis occurs through the carotenoid pathway via a series of oxidation and isomerization reactions followed by the cleavage of C40 carotenoid to yield the sesquiterpene xanthoxin (Fig. 1). Xanthoxin is converted to ABA aldehyde via an alcohol dehydrogenase enzyme (ABA2), which is subsequently oxidized to ABA via abscisic aldehyde oxidase (AAO3; Nambara and Marion-Poll, 2005). ABA3 activity is required to generate the molybdenum cofactor for the functional AAO3 (Bittner et al., 2001; Xiong et al., 2001). In a minor pathway, ABA aldehyde is also converted to ABA alcohol, which can purportedly be oxidized by a P450 monooxygenase to ABA (Rock et al., 1991). For ABA-deficient mutants impaired in the oxidation of ABA aldehyde to ABA, this shunt pathway is considered to be an important source of ABA.
Figure 1.
ABA biosynthetic pathway. See text for a detailed description.
Physiological analyses of ABA precursors have yielded variable results depending on the assay and the compound tested. In stomatal closure assays, xanthoxin was found to be active, although at a much higher concentration than ABA to obtain the same level of response (Raschke et al., 1975; Yamomoto and Oritani, 1996). Both ABA alcohol and aldehyde were highly effective at inducing stomatal closure, whereby the aldehyde was more potent than the alcohol (Uehara et al., 1975). ABA aldehyde and alcohol were reported to have moderate ABA-like activity at inducing freezing tolerance in bromegrass (Bromus inermis) cell cultures (Robertson et al., 1994). In a wheat (Triticum aestivum) seed germination assay, ABA aldehyde was found to substitute for natural ABA, but ABA alcohol was much less effective (Hays et al., 1996). On the other hand, ABA alcohol was more effective than both ABA aldehyde and ABA at inducing oleosin gene expression (Yamomoto and Oritani, 1996). A rationale for the high bioactivity of these ABA precursors is either their conversion to the active hormone within the plant and/or an intrinsic ABA-like activity (Raschke et al., 1975; Parry et al., 1991; Yamomoto and Oritani, 1996).
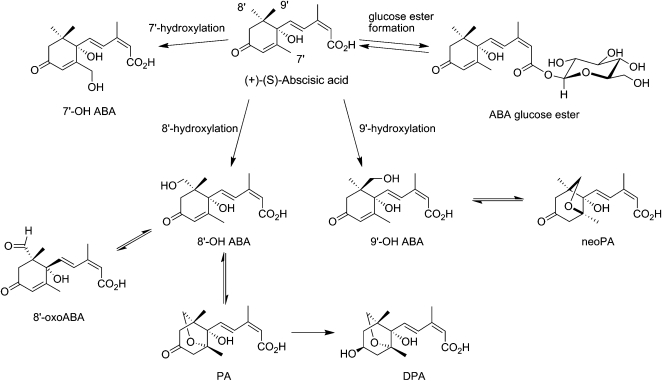
The catabolism of ABA occurs mainly through oxidative and conjugative processes (Fig. 2). Hydroxylation at each of the 7′, 8′, or 9′ methyl groups of ABA leads to three different oxidation pathways, among which 8′-oxidized metabolites are the most abundant (Nambara and Marion-Poll, 2005). Hydroxylation at the 8′ or 9′ position generates an unstable intermediate that is poised to undergo an energetically favorable rearrangement leading to the formation of an oxygen-containing ring. This reaction results in the formation of the more stable catabolites phaseic acid (PA) and neo-phaseic acid (neoPA) from the 8′- and 9′-hydroxylation products (8′- and 9′-OH ABA), respectively. PA is transformed by an unknown reductase enzyme to dihydrophaseic acid (DPA), a biologically inactive ABA catabolite. The metabolic fate of neoPA has yet to be determined. ABA and some of its oxidized catabolites (8′-OH ABA, PA, DPA, and epi-DPA) are also conjugated to Glc (Nambara and Marion-Poll, 2005; Zaharia et al., 2005b). Direct conjugation of ABA yields a storage and transport form of the phytohormone, ABA glucosyl ester (ABA GE; Jiang and Hartung, 2008). ABA can be released from this ABA conjugate by a stress-activated β-glucosidase (Lee et al., 2006). The release of ABA from the sites of biosynthesis and its uptake into cells are regulated by the ATP-dependent transporters ABCG24 and ABCG40, respectively (Kang et al., 2010; Kuromori et al., 2010).
Figure 2.
ABA catabolites and conjugates. See text for a detailed description.
Biological testing of the open forms of 8′- and 9′-hydroxylated metabolites has been problematic, owing to their propensity to undergo cyclization to PA and neoPA in vitro (Milborrow et al., 1988; Cutler et al., 1997), although the 9′-OH derivative tends to be more long lived (Zhou et al., 2004). Conditions have been identified to generate and stabilize 8′-OH ABA (8′-OH ABA) from PA so that its activity can be investigated in rapid assays (Zou et al., 1995). In such assays, freshly prepared 8′-OH ABA exhibited ABA-like activity in inducing lipid or storage protein-related gene expression (Zou et al., 1995; Jadhav et al., 2008). PA generally displays very little ABA-like activity (Balsevich et al., 1994; Hill et al., 1995; Zou et al., 1995), but in barley (Hordeum vulgare), PA shows substantial activity in the induction of ABA-responsive genes encoding barley germ agglutinin, inhibition of α-amylase activity in aleurone layers, and embryo germination inhibition (Dashek et al., 1979; Hill et al., 1992, 1995; Todoroki et al., 1995). It is postulated that the biological activity associated with PA is actually derived from 8′-OH ABA, with which it is in equilibrium (Milborrow et al., 1988). Similarly, 9′-OH ABA exhibits some ABA-like activity in seed germination assays of Arabidopsis, while neoPA is inactive (Zhou et al., 2004). In developing seeds of Arabidopsis, neoPA has been found in significant quantities (Kanno et al., 2010). Finally, 7′-OH ABA shows similar activity to ABA in inducing genes related to lipid and storage protein accumulation (Hill et al., 1995; Jadhav et al., 2008). In conifers, 7′-OH ABA is a significant ABA catabolite (Feurtado et al., 2007; Kong et al., 2009).
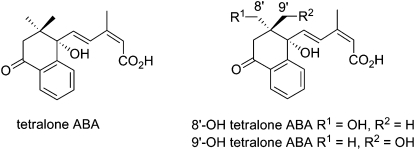
Stable synthetic mimics of 8′- and 9′-OH ABA have been designed in order to more easily ascertain the effects of these transient catabolites in biological assays (Nyangulu et al., 2006). These derivatives were based on a tetralone framework, in which the inherent stability of the aromatic ring precludes dearomatization by conjugate addition of the 8′ or 9′ hydroxyl (Fig. 3). The parent compound, tetralone ABA (Fig. 3), retains the structural elements and functional groups of ABA that are required for activity and was shown to have equal or better activity than ABA in a number of assays (Nyangulu et al., 2006). The 8′- and 9′-OH tetralone derivatives maintain good ABA-like activity; for example, both are comparable to ABA in the inhibition of maize cell growth. On the other hand, 9′-OH tetralone ABA was effective similar to ABA for inhibiting the germination of Arabidopsis seeds, while the 8′-OH tetralone was completely ineffective at lower concentrations (Nyangulu et al., 2006).
Figure 3.
Structures of synthetic tetralone ABA analogs. The 8′- and 9′-OH tetralone ABAs are stable mimics of the short-lived 8′- and 9′-OH ABA catabolites.
ABA precursors and catabolites have not been tested for their ability to act directly on the ABA receptor or other ABA signaling components. Analysis of bioactivity, to date, has been restricted to physiological assays in whole cells or intact plants. The interpretation of such assays is confounded by the variability of parameters such as compound uptake, transport, and metabolism. The identification of the ABA receptor complexes now allows for the determination of a direct signaling function for ABA precursors and catabolites. Our study reveals a hormonal activity of the stable tetralone mimic of 9′-OH ABA in ABA responses. The ABA precursors xanthoxin and ABA aldehyde have no and negligible intrinsic ABA activity, respectively. While the ABA precursors are known to exhibit considerable biological activity, evidence here corroborates their action by their in vivo conversion to ABA. Finally, our experiments provide insight into the structural requirements of ABA ligand-receptor interactions using a set of natural ABA-related compounds as probes.
RESULTS
Biological Activity of ABA Biosynthetic Precursors and Catabolites
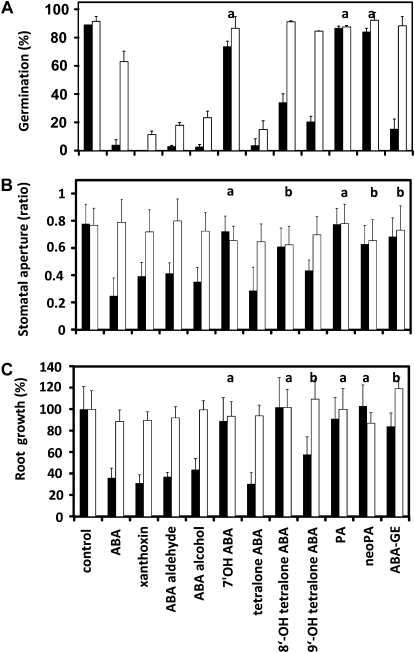
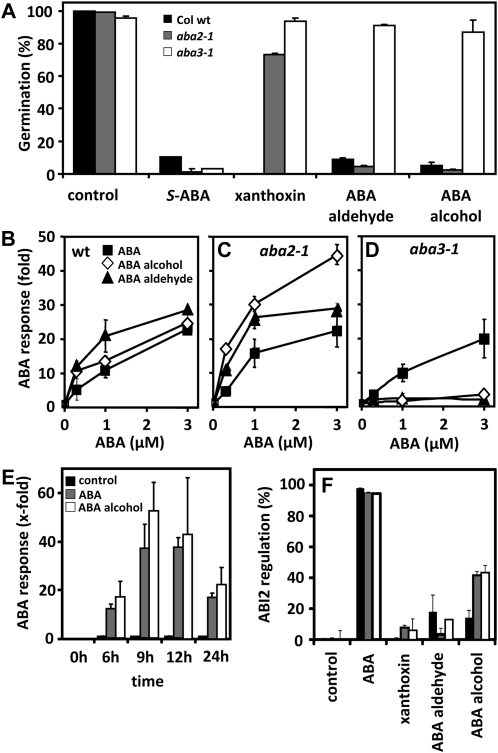
We assayed the effect of the ABA precursors xanthoxin, ABA aldehyde, and ABA alcohol as well as the ABA catabolites 7′-OH ABA, PA, neoPA, and ABA GE for their ability to inhibit germination and root growth as well as to induce stomatal closure in Arabidopsis. The specific metabolites chosen include the direct precursors of ABA after carotenoid cleavage and the first products of ABA catabolism. In addition, we tested 8′- and 9′-OH tetralone ABA as mimics of the transient ABA catabolites hydroxylated at the 8′ and 9′ positions and the parent ABA tetralone. For the germination assay, we found that the biosynthetic precursors had comparable activity to ABA and inhibited germination almost completely at a concentration of 3 μm (Fig. 4A). Conversely, the catabolites 7′-OH ABA, PA, and neoPA were ineffective for germination inhibition. ABA GE and the hydroxy tetralone ABAs demonstrated moderate ABA-like activity, with 9′-OH and ABA GE being slightly more effective than the 8′-OH tetralone, but neither compound was as active as ABA. Tetralone ABA was found to provide similar activity to ABA, which showed that the tetralone framework has little impact on ABA activity. Seeds of the mutant abi1-1 were insensitive to ABA and to the physiologically active ABA precursors and catabolites (Fig. 4A). Interestingly, seed germination of abi1-1 was more efficiently inhibited by ABA precursors than by S-ABA itself.
Figure 4.
Arabidopsis responses to (S)-ABA, ABA precursors, and ABA catabolites. A, Germination of Ler (black bars) and abi1-1 (white bars) seeds (n > 100) in the presence of 3 μm (S)-ABA, ABA precursors, ABA catabolites, and ABA GE (six experiments). B, Aperture (ratio) of Ler (black bars) and abi1-1 (white bars) stomatal opening in the presence of 3 μm (S)-ABA and ABA precursors and catabolites. C, Root growth of Ler (black bars) and abi1-1 (white bars) seedlings (n > 100) in the absence or presence of ABA or ABA-related compounds (3 μm). Five-day-old seedlings were transferred to medium containing ABA-related compounds, and the difference in root length was measured 3 d after incubation at 23°C. The 9′-OH tetralone ABA and 7′-OH ABA samples were racemic. Data shown are means ± sd. Data for Ler are highly significant to the control (P < 0.001), unless otherwise indicated: a no significant difference (P ≥ 0.05); b significant difference (P ≤ 0.01).
Stomata of epidermal peels of Arabidopsis closed in response to ABA and in the presence of the ABA precursors xanthoxin, ABA alcohol, and ABA aldehyde (Fig. 4B). These compounds showed no or negligible activity in guard cells of abi1-1. Among the ABA catabolite group, only the 9′-OH tetralone derivative revealed significant activity. Similar results were obtained with respect to the effect of the compounds on root growth (Fig. 4C). The ABA biosynthetic precursors were comparable in activity to ABA, and among the catabolites, only the 9′-OH tetralone ABA and ABA GE were weakly active, with root growth at about 58% and 73% that of the control, respectively. Again, abi1-1 revealed a clearly insensitive phenotype toward all these chemicals, indicating a specific targeting of the ABA response pathway by the ABA-related molecules. Taken together, the ABA catabolites showed a surprisingly consistent action in different ABA responses. The effect of the 8′-OH tetralone ABA and ABA GE varied, having a moderate activity on seed germination and no significant effect on root growth and stomatal regulation, respectively.
The bioactivity observed for the ABA precursors in all three ABA responses prompted us to ascertain whether the ABA-like activity comes from an inherent signal function of these compounds or from bioconversion into ABA. Arabidopsis mutants impaired in the ABA biosynthetic pathway provide a tool to disrupt conversion to ABA and thus allow examining a signal function of ABA precursors. We chose to analyze the inhibitory action of these compounds on seed germination because the ABA precursors proved to be very effective in inhibiting Arabidopsis seeds (Fig. 4A). The aba2-1 mutant is impaired in the bioconversion of xanthoxin to ABA aldehyde, while the aba3-1 mutant is deficient in the synthesis of the molybdenum cofactor necessary for AAO3 activity (Fig. 1). AAO3 catalyzes the last step of ABA biosynthesis, the conversion of ABA aldehyde to ABA. In the aba2-1 mutant, the effect of xanthoxin on germination was markedly reduced, with a germination rate of 73% of the mutant compared with less than 1% of the wild type (Fig. 5A). Seeds of aba3-1 were even more insensitive toward xanthoxin, and the germination rate was 93%, similar to aba3-1 seeds not exposed to xanthoxin (95%). ABA alcohol and aldehyde retained their activity in aba2-1 as compared with that in wild-type seeds. Both ABA precursors showed no significant effect in the aba3-1 background.
Figure 5.
Influence of ABA and ABA precursors on seed germination and ABA-dependent reporter gene expression. A, Germination of wild-type (wt), aba2-1, and aba3-1 seeds (n > 100) in the presence of 3 μm (S)-ABA or ABA precursors. Germination was scored after 2 d at 4°C followed by 3 d at 23°C. B to D, Activation of ABA-dependent gene expression by ABA, ABA alcohol, and ABA aldehyde was monitored using the reporter construct pRD29B::LUC in Arabidopsis ecotype Columbia wild-type (B), aba2-1 (C), and aba3-1 (D) protoplasts. Each data point represents the mean of three independent transfections. E, Time course of pRD29B::LUC up-regulation in wild-type protoplasts in the presence of either 3 μm (S)-ABA or ABA alcohol. Each data point represents the mean of three independent transfections. The control represents the protoplast experiment in the absence of ABA or its precursors. F, Regulation of protein phosphatase activity by RCAR1 (black bars), RCAR3 (gray bars), or RCAR11 (white bars) in the presence of (S)-ABA, xanthoxin, ABA aldehyde, and ABA alcohol at a ligand concentration of 10 μm.
These findings indicate that most, if not all, of the activity associated with the ABA precursors in wild-type Arabidopsis is due to their conversion to ABA within the plant. To substantiate this conclusion, we tested ABA, ABA aldehyde, and ABA alcohol for their ability to induce ABA-responsive reporter gene expression. The reporter gene consists of the ABA-responsive promoter RD29B driving the expression of firefly luciferase (Moes et al., 2008). Arabidopsis mesophyll protoplasts are able to perform the final steps of ABA biosynthesis (Bianco-Trinchant and Le Page-Degivry, 1998). Hence, Arabidopsis protoplasts from wild-type, aba2-1, and aba3-1 leaves were transfected with the reporter gene and exposed to ABA-related compounds. We found that the biosynthetic precursors were very active in inducing an ABA response and elicited expression of the reporter to even higher levels than ABA in wild-type cells (Fig. 5B). In aba2-1 mutant protoplasts, both ABA aldehyde and alcohol induced luciferase expression (Fig. 5C), similar to their bioactivity in the germination assay of aba2-1 seeds (Fig. 5A). Neither compound was able to induce ABA-responsive reporter gene expression in aba3-1 protoplasts, while ABA yielded induction levels in aba3-1 similar to the wild type (Fig. 5D). This confirms that ABA precursors must be converted to ABA in order to be physiologically active.
It is interesting that both ABA aldehyde and ABA alcohol show enhanced luciferase up-regulation when compared with ABA in both the wild type and the aba2-1 mutant protoplasts (Fig. 5, B and C). A time-course profile comparing gene induction for ABA alcohol with ABA demonstrates that at all time points, the alcohol outperforms ABA, but the differences in gene regulation are more apparent at 9 h of exposure (Fig. 5E). This observation can be rationalized as due to a greater concentration of ABA present at the site of action for the samples treated with ABA alcohol than by ABA treatment itself.
In Vitro Activity of ABA Biosynthetic Precursors and Catabolites
The observed ability of ABA precursors and catabolites to mediate ABA responses is likely caused either by an enhancement of active ABA pools or by an inherent signal function of the compounds themselves. The analysis of ABA biosynthesis mutants supports an efficient bioconversion of the ABA precursors into ABA. However, a physiological role of these compounds as ABA-like signals cannot formally be excluded. This is because the intracellular availability of administered compounds is controlled by rates of uptake, transport, and metabolism, which confounds the interpretation of whole plant or protoplast assays.
To address this issue, we undertook to evaluate the capacity of ABA metabolites and derivatives thereof to directly regulate RCAR/PP2C complexes in vitro. We choose RCAR1 because it is highly specific for S-ABA discriminating R-ABA by a factor of more than 100 (Ma et al., 2009). RCAR3 together with ABI1 and ABI2 provide the highest ABA affinity known so far (Szostkiewicz et al., 2010). RCAR11 has been chosen because of the more sluggish binding features accommodating also pyrabactin and being far less discriminative toward ABA (Park et al., 2009; Peterson et al., 2010). First, we screened xanthoxin, ABA aldehyde, ABA alcohol, and ABA for their effects on the phosphatase activity of purified ABI2 in the presence of RCAR1 (Fig. 5F). ABA, supplied at a level of 10 μm, completely abolished the phosphatase activity of ABI2, demonstrating the formation of a functional trimeric complex consisting of ligand, RCAR1, and ABI2. ABA aldehyde and ABA alcohol exhibited a slight reduction of 10% and 18% PP2C activity in the presence of RCAR1, respectively. Xanthoxin was inactive. On the other hand, an analysis of protein complexes of ABI2 and RCAR3 or RCAR11 revealed an approximately 43% inhibition of PP2C activity by ABA alcohol. ABA aldehyde slightly inhibited the ABI2/RCAR1 and ABI2/RCAR11 complexes (16% and 13% inhibition, respectively), while no significant activity (threshold of 6%) was observed for RCAR3. Xanthoxin did not significantly regulate PP2C activity in the presence of RCAR1, RCAR3, and RCAR11. These data support the notion that there are minor differences in ligand recognition of the different RCARs analyzed. Among the ABA precursors examined, only ABA alcohol has the potential to be of physiological relevance as an ABA-like signal.
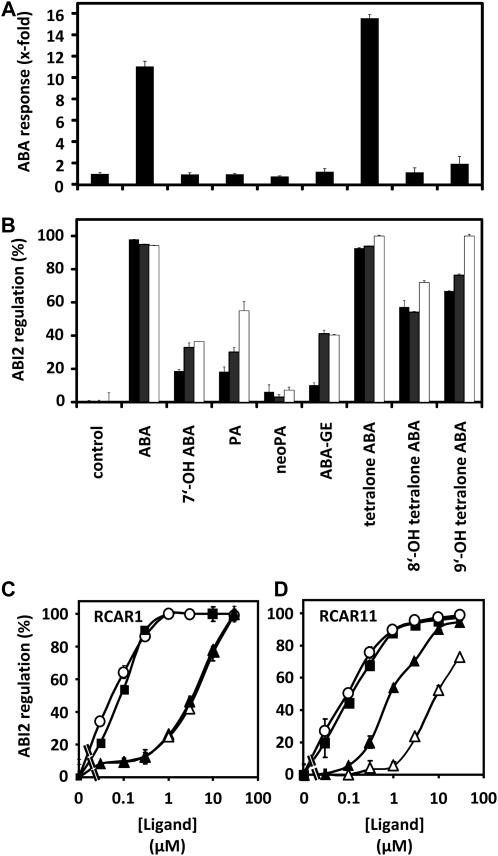
In order to assess whether ABA catabolites had an intrinsic ABA-like activity, we tested their action both on ABA signaling in protoplasts and on purified ABA receptor complexes. We examined the catabolites 7′-OH ABA, PA, neoPA, and ABA GE for their effects on ABA-responsive gene expression and found that only ABA GE weakly induced the reporter in protoplasts by a factor of 1.2, which is much lower than the 11-fold induction observed with ABA (Fig. 6A). Analysis of these catabolites with ABA receptor complexes, however, identified 7′-OH ABA, PA, and ABA GE to be moderately active in regulating ABI2 activity in the presence of RCAR1, RCAR3, or RCAR11 at a ligand concentration of 10 μm (Fig. 6B). neoPA was not active in the regulation of ABI2 (less than 6%). The activity of ABA catabolites observed, however, is rather low in comparison with ABA. At a ligand concentration of 1 μm, ABA inhibited PP2C by approximately 90%, while the ABA precursors and catabolites showed no significant effect with the exception of ABA GE (9%–16% ABI2 regulation; Supplemental Fig. S1). The in vivo analysis is in agreement with the lack of and low activity observed in the physiological assays for neoPA and ABA GE, respectively (Fig. 4). 7′-OH ABA and PA have minor ABA-like activities on ABA receptor complexes not evident from analyses of ABA responses in isolated cells and whole plants.
Figure 6.
A, Regulation of the ABA reporter construct pRD29B::LUC by natural ABA catabolites and hydroxy tetralone ABA analogs at 1 μm concentration. Each data point is the mean of three independent transfections. The control represents the protoplast experiment in the absence of ABA or its catabolites. B, Regulation of protein phosphatase activity by RCAR1 (black bars), RCAR3 (gray bars), and RCAR11 (white bars) in the presence of (S)-ABA, ABA catabolites, and ABA tetralone mimics at a ligand concentration of 10 μm. C, Ligand-dependent inhibition of ABI2 in the presence of RCAR1. Half-maximal inhibition of ABI2 occurred at 90 nm (S)-ABA (black squares), 55 nm (S)-tetralone ABA (white circles), 4 μm 8′-OH tetralone ABA (white triangles), and 3.5 μm 9′-OH tetralone ABA (black triangles). D, Ligand-dependent inhibition of ABI2 in the presence of RCAR11. Half-maximal inhibition occurred at 120 nm (S)-ABA (black squares), 95 nm (S)-tetralone ABA (white circles), 8 μm 8′-OH tetralone ABA (white triangles), and 0.9 μm 9′-OH tetralone ABA (black triangles). The 9′-OH tetralone ABA sample was racemic. All phosphatase assays were performed at a PP2C level of 0.05 μm and at a molar ratio of RCAR and ABI2 of 2:1.
Tetralone ABA Derivatives Regulate ABA Receptor Complexes
To test the effects of 8′- and 9′-OH ABA on ABI2 activity, we used the synthetic hydroxy tetralone mimics, which are unable to undergo cyclization to PA-type derivatives. For comparison, tetralone ABA was also included to determine whether this modification of the hormone skeleton impairs ABA activity. The tetralone derivatives of hydroxylated ABA molecules revealed some ABA-like activity in physiological responses, particularly in germination (Fig. 4). 9′-OH tetralone also regulated ABA signaling in protoplasts, albeit much less than ABA and tetralone ABA, while 8′-OH tetralone was inactive (Fig. 6A). The in vitro assay documented an efficient inhibition (98%) of PP2C by ABA tetralone, comparable to ABA at ligand concentrations of 10 μm and in the presence of RCAR1 (Fig. 6B). Interestingly, both 8′- and 9′-OH tetralones had ABA-like activity and blocked phosphatase activity of ABI2 to approximately 58% and approximately 67%, respectively. To corroborate this finding, the half-maximal inhibitory concentration (IC50) values of these compounds were assessed by titration of RCAR1-ABI2 with the different ligands (Fig. 6C). ABA yielded an IC50 of 90 nm, and tetralone ABA was even more potent, with a calculated IC50 of 55 nm. The 8′- and 9′-OH tetralones were around 40 times less active than ABA, with IC50 values of 4 and 3.5 μm, respectively. As we have shown in Figure 4A, both hydroxy tetralone compounds are active for inhibiting germination in Arabidopsis, with 9′-OH tetralone being slightly more effective than the 8′-OH derivative. The regulation of the ABA receptor complex by these compounds mirrors their physiological activity.
Our previous receptor studies revealed differences in the ligand activities for different RCAR members (Szostkiewicz et al., 2010). Testing of the same series of compounds in the presence of RCAR11 and ABI2 yielded differing results (Fig. 6B). ABI2 activity is completely abolished by 10 μm ABA, tetralone ABA, as well as 9′-OH tetralone ABA. In this case, divergent behaviors of 9′- versus 8′-hydroxylated derivatives are apparent, as ABI2 still retains almost 30% PP2C activity in the presence of 8′-OH tetralone ABA (10 μm). The IC50 values for these compounds in this experimental system were also determined (Fig. 6D). ABA and tetralone ABA were still the most potent regulators, with values of 120 and 95 nm, respectively. Both values are somewhat higher than for the corresponding RCAR1-ABI2 complex. The IC50 value for 8′-OH tetralone was found to be 8 μm. Maximal inhibition, however, reached only 80% at 30 μm, the highest concentration tested. Compared with RCAR1-ABI2, the 8′-OH derivative is more than two times less active in regulating PP2C activity than with RCAR11-ABI2. The 9′-OH tetralone provided an IC50 of 0.88 μm, indicating that it is four times more effective with RCAR11 than with RCAR1 for ABI2 inhibition. It is also important to note that the 9′-OH tetralone used for these experiments was racemic, meaning that half of the sample consisted of the biologically less active (R)-enantiomer. Using an enantiomerically pure sample for these assays, one would expect the activity and potency of this derivative to increase considerably.
DISCUSSION
ABA precursors and catabolites have been reported to be biologically active in diverse ABA responses and plant species. The availability of purified ABA receptor complexes has now prompted us to reexamine the bioactivity of such natural structural derivatives on ABA signaling and to explore the specificity of ligand-receptor interactions.
Natural ABA compounds were assayed for their ability to inhibit germination and root growth as well as to induce ABA-dependent gene expression in Arabidopsis. The compounds were subsequently screened for their ability to regulate the phosphatase moiety of selected ABA receptor complexes in vitro. Our results suggest that a substantial amount of the activity associated with these compounds is a result of their bioconversion to ABA within the plant. However, ABA alcohol and some hydroxylated ABA derivatives are intrinsically active in regulating PP2C activity of receptor complexes, albeit much less than ABA. We also demonstrated that in protoplasts of the wild-type Arabidopsis variety, ABA aldehyde and alcohol were able to induce ABA-responsive reporter expression to an even greater extent than ABA itself.
We found that ABA alcohol and tetralone, as well as 8′- and 9′-OH ABA tetralone, are capable of regulating the RCAR1/ABI2 receptor complex. In addition to these four ligands, the physiological analyses of ABA responses also revealed bioactivity for the ABA precursors xanthoxin and ABA aldehyde and for ABA GE. The physiological activity of xanthoxin and ABA aldehyde is readily explained by conversion of the precursors into ABA, which is experimentally substantiated by the failure of these compounds to trigger ABA responses in the aba3-1 mutant, impaired in the biosynthetic conversion of ABA aldehyde to ABA. Consistently, the aba2-1 mutant, with a deficiency in the short-chain dehydrogenase/reductase that converts xanthoxin into ABA aldehyde (González-Guzmán et al., 2002), was able to respond to ABA aldehyde but was severely impaired in the response toward xanthoxin (Fig. 5A). There was, however, a residual activity of xanthoxin to inhibit aba2-1 seed germination, which could be due either to the leakiness of the mutant phenotype (Léon-Kloosterziel et al., 1996) or to the existence of a second, minor xanthoxin-metabolizing activity. The higher activity of ABA alcohol in inducing ABA reporter expression in comparison with ABA in wild-type cells (Fig. 5B) indicates an improved bioavailability caused, for instance, by enhanced uptake via a more efficient diffusion across the plasma membrane due to the less polar nature of ABA alcohol compared with ABA (Hays et al., 1996) or via ABC transporters (Kang et al., 2010; Kuromori et al., 2010). Alternatively, the ongoing enzymatic conversion of ABA alcohol into ABA within the plant cell might provide a persistent source of active ABA, which could be more effective than a single ABA application.
The catabolites 7′-OH ABA, PA, and neoPA were not found to be active in the physiological assays. In vitro, 7′-OH ABA and PA inhibited ABI2 up to 30% and 50% in the presence of RCAR1 and RCAR11, respectively (Fig. 6B). The efficiency of 7′-OH ABA and PA to function as an ABA-like ligand, however, is very low. There was no PP2C inhibition observed at 1 μm 7′-OH ABA and PA, while the same concentration of ABA almost fully inhibited the phosphatase activity. This difference in efficacy is reflected in the different IC50 value of approximately 0.1 μm for ABA, while the IC50 value of both catabolites is around or at least 100 times higher (10 μm or greater). Interestingly, PA has been identified as a potent inhibitor of barley embryo germination comparable to ABA (Hill et al., 1992), while in Arabidopsis, embryo germination activity is negligible. It is possible that PA differentially affects ABA metabolism or certain receptor complexes in different plant species. In our analyses, ABA GE is weakly active in regulating ABA receptor complexes, similar to 7′-OH ABA and PA. However, ABA GE was clearly able to inhibit seed germination. The activity of ABA GE observed in the germination assay could reflect its moderate capacity to regulate ABA receptor complexes. More likely, the release of ABA from the glucosyl conjugate by the β-glucosidase BG1 may account for the biological activity observed. BG1 regulates stomatal responses under drought stress and converts ABA GE into ABA (Lee et al., 2006). Mutants of BG1 were not studied because of other potentially nonselective glucosidase enzymes in Arabidopsis, making it difficult to limit all ABA GE hydrolysis.
Hydroxylated ABA catabolites have shown ABA-like activity in a variety of assays. The compounds have a tendency to spontaneously cyclize to PA and PA-related compounds such as neoPA, with which they are in equilibrium (Dashek et al., 1979; Hill et al., 1992, 1995; Todoroki et al., 1995). 9′-OH ABA exhibits ABA-like activity in seed germination assays of Arabidopsis, while neoPA is inactive (Zhou et al., 2004). Moreover, the 9′-OH catabolite is known to be more stable in its active form than the corresponding 8′-OH derivative (Zhou et al., 2004). We found that a stable synthetic mimic of the 9′-OH ABA catabolite that is unable to undergo cyclization to neoPA showed significant action as an ABA-like ligand in vitro. The corresponding 8′-OH mimic was also active, although much less so. Prior to its inactivation by cyclization to neoPA, the 9′-OH ABA catabolite may play an important role in the ABA response, particularly in early seed development, as levels of neoPA are known to be higher at this stage than in more mature seeds (Zhou et al., 2004). This, together with the results presented here, suggests that 9′-OH ABA may be an important ligand for mediating certain aspects of the ABA response.
We investigated the effects of the tetralone hydroxy ABA mimics on both RCAR1/ABI2 and RCAR11/ABI2. The two receptor complexes demonstrated different interactions with the compounds tested. For example, while the synthetic tetralone analogs of 8′- and 9′-OH ABA catabolites both exhibit moderate RCAR1-mediated ABI2 inhibition, their activity profiles differ when using RCAR11 in lieu of RCAR1. The 9′ derivative was four times more effective in the presence of RCAR11 than of RCAR1, while the 8′ substituted compound was two times less effective in inhibiting RCAR11/ABI2 compared with the 9′ derivative. Similar ligand selectivity was observed with the nonphysiological (R)-ABA enantiomer, which was strongly discriminated against by RCAR1 but not by RCAR3 in receptor complexes with ABI1 or ABI2 (Szostkiewicz et al., 2010). Likewise, the ABA agonist pyrabactin regulates ABI1 in a receptor complex with RCAR11/PYR1 but not in complex with RCAR14/PYL2 (Peterson et al., 2010). Furthermore, site-directed mutagenesis of PYL2 gave rise to regulatory ligand activity of pyrabactin on ABI1/PYL2A93F (Melcher et al., 2010). Thus, minor structural changes in the protein-binding pocket can greatly affect ligand binding and/or subsequent PP2C interaction and may lead to differences in the transduction of the ABA signal and ultimately its physiological outcome.
These findings suggest that a chemical genetics strategy might be useful to gain insight into particular functions of the individual receptors, or receptor classes, whose functional redundancy is difficult to circumvent. The application of chemical probes to infer gene function will be complementary to traditional genetic approaches to provide a more in-depth understanding of the nuances of ABA signal transduction (Blackwell and Zhao, 2003; Cutler and McCourt, 2005; McCourt and Desveaux, 2010). Future studies will undertake a more comprehensive structure-activity screening of additional ABA analogs on these receptor complexes and others.
As the RCAR/PYR1/PYL-PP2C-mediated ABA response pathway is emerging as a universal stress signaling pathway, it is important to keep in mind the noteworthy roles that natural derivatives of ABA may play in species other than Arabidopsis. In a search to identify orthologs of ABA signaling molecules in liverworts (Marchantia polymorpha), the earliest diverging branch of land plants, a homolog of Arabidopsis ABI1 PP2C was identified. Its function as a regulator of ABA-dependent signaling processes was found to be conserved (Tougane et al., 2010). Furthermore, a functional PP2C in the beechnut (Fagus spp.) was found to be a negative regulator of ABA response in seeds (Lorenzo et al., 2001). This PP2C has since been shown to interact strongly with Arabidopsis PYL8/RCAR3 by yeast two-hybrid screening (Saavedra et al., 2010). The functional significance of this in the beechnut has not yet been determined, but it suggests that early events in the Arabidopsis ABA-responsive signaling pathway may be conserved in other species as well. The 14 Arabidopsis receptors can be classified into three clades based on sequence similarity, and orthologous proteins from other plant species, both higher and lower, also fit into the subgroups (Ma et al., 2009; Park et al., 2009; Saavedra et al., 2010). It is not known if there are functional differences between members of these subgroups. Future screening of ABA analogs and catabolites against members of each clade might be useful in determining these potential differences in light of the preliminary results shown here with some ABA catabolites and different RCARs.
For the receptor complexes tested here, we have generated some information on structure-activity relationships between the ligand and receptor. For example, ABA alcohol and aldehyde are structurally very similar to ABA and only differ in the oxidation state of the side chain at C-1, yet they are not able to interact with RCAR1-ABI2 to the extent that ABA does. Analysis of crystal structures of homologous receptor complexes has indicated that the C-1 carboxylate of ABA coordinates with the receptor through one direct and multiple indirect water-mediated contacts, utilizing both oxygen functionalities (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009a; Yin et al., 2009). These polar and hydrogen-bonding interactions of ABA’s carboxylate and tertiary hydroxyl with highly conserved Lys and Glu residues deep within the binding pocket appear to be important for anchoring the molecule into the active site of the receptor. Moreover, a structurally unrelated ABA agonist, pyrabactin, has also been crystallized with PYL1/RCAR12 and PYR1/RCAR11 and further reveals the importance of these interactions. In both RCAR11 and RCAR12, pyrabactin aligns itself such that its sulfonamide and pyridine functionalities are able to contact the conserved Lys and Glu residues of the protein in a similar manner as ABA (Hao et al., 2010; Melcher et al., 2010; Peterson et al., 2010; Yuan et al., 2010). Apparently, for ABA aldehyde and alcohol, the loss of one of the coordinating oxygens in the side chain drastically affects the binding of the compounds to RCAR1 and the subsequent inhibition of PP2C. We have further confirmed that changes at the 8′ and 9′ positions of ABA are well accommodated by some of the receptors and that the tetralone carbon framework is not detrimental to receptor binding. Interestingly, the tetralone ring bears structural similarity to pyrabactin’s bromonapthalene ring, which is well accommodated by RCAR11 and RCAR12 in the crystal studies. We suspect that the binding mode of the tetralone ABAs may be similar to that of pyrabactin.
In conclusion, we have shown that certain natural ABA-related compounds are capable of interacting with members of the ABA receptor family and that the extent of the interaction varies between different receptor complexes. The data provide no hint for the involvement of other postulated ABA receptors (for review, see Raghavendra et al., 2010). No single compound with ABA-like signal function failed to regulate RCAR/PP2C in our study. Moreover, the in vitro data correlate well with what is known about the physiological action of these compounds and suggest that further studies would be useful to discern the functional activities that are associated with each receptor or class of receptors. The data presented here provide a unique structure-activity screen of some ABA-related compounds and their effects on ABA signaling in the absence of confounding factors.
MATERIALS AND METHODS
Chemicals
Chemicals were obtained from Sigma-Aldrich (http://www.sigmaaldrich.com), Fluka (part of Sigma-Aldrich), Roth (http://www.carlroth.com), AppliChem (http://www.applichem.com), and J.T. Baker (http://www.mallbaker.com). ABA was purchased from Lomon Bio Technology [(S)-ABA; http://www.lomonbio.com]. Other ABA derivatives were synthesized as described in the cited literature: xanthoxin (Kuba et al., 2002), ABA alcohol and ABA aldehyde (Rose et al., 1992; ABA aldehyde was prepared immediately prior to use by oxidation of ABA alcohol), 7′-OH ABA (Nelson et al., 1991), DPA and ABA GE (Zaharia et al., 2005a), neoPA (Zhou et al., 2004), and the tetralone ABA derivatives (Nyangulu et al., 2006). PA was obtained by biotransformation of (+)-ABA in Black Mexican Sweet corn (Zea mays) as described by Balsevich et al. (1994). All materials used were enantiopure with the same stereochemistry as found in nature, with the exception of 7′-OH ABA and 9′-OH tetralone ABA, which were racemic.
Plant Material
All the Arabidopsis (Arabidopsis thaliana) lines used in this work were ecotype Columbia and Landsberg erecta (Ler). Plants used for protoplast isolation were grown for 4 weeks in a perlite-soil mixture in a controlled growth chamber at 23°C under long-day conditions with 16 h of light (250 μE m−2 s−1; Moes et al., 2008).
Bioassays of Stomatal Closure in Epidermal Strips
Strips of abaxial epidermis were prepared from Arabidopsis leaves by mounting the epidermal sections on glass coverslips with the help of a medical adhesive, Telesis V (Premiere Products). These were transferred to 3-cm-diameter petri dishes containing 3 mL of incubation medium (10 mm MES-KOH, pH 6.15, and 50 mm KCl) for stomatal opening. The strips were exposed to white light (150 μmol m−2 s−1) for 2 h, with the light filtered through a water jacket. Photon flux was measured with a Li-Cor quantum sensor (Li-Cor Instruments). The temperature was maintained at 25°C ± 10°C. Test compounds were added to the medium, and the strips were kept under the same conditions for another 2 h before measuring the stomatal aperture. The width of the stomatal aperture was measured with a research microscope (Nikon Eclipse TE 200) fitted with a camera and connected to an image-analysis system.
Seed Germination and Root Elongation Assays
Under sterile conditions, 100 to 150 seeds were plated on Murashige and Skoog agar medium containing tested compounds and incubated at 4°C for 2 d in the dark to break dormancy. The plates were then transferred to a culture room with continuous light (60 μE m−2 s−1) at 22°C. After 3 d, seeds were examined with a stereo microscope. Seeds were counted, and germination rate was calculated as percentage of the total number of seeds. For root elongation, 5-d-old seedlings were transferred in a row to half-strength Murashige and Skoog agar-containing plates with different combinations of treatments and kept in a vertical position at 22°C in continuous light for 3 d. Root tip position was marked every 24 h, and root lengths were measured with the millimeter scale using a microscope (Moes et al., 2008).
Plasmid Constructs
The pRD29B::LUC reporter plasmid used in this work has been described previously (Moes et al., 2008; Ma et al., 2009). RCAR1/3 and ABI1/2 constructs used in this study were generated as described by Ma et al. (2009) and Szostkiewicz et al. (2010).
For heterologous expression, the cDNA of RCAR11 (At4g17870) was amplified with the primer pair 5′-GAGTCGCATGCCTTCGGAGTTAACACCAGAAG-3′ and 5′-GACTCAGATCTCGTCACCTGAGAACCACTTCCGTC-3′. The PCR fragment was subsequently cloned into the pQE70 vector (Qiagen), yielding pQE70-RCAR11 (cloning via BglII and SphI sites).
Expression and Purification of RCARs and PP2Cs
His-tagged RCAR1/3/11 and ABI2 proteins were expressed in Escherichia coli strain M15 (Ma et al., 2009). Cells were grown overnight in 50 mL of Luria Bertani medium and used for inoculations of 1 L of culture. The cells were grown at 37°C with shaking until an optical density at 600 nm of 0.5 to 0.6 was reached. Protein expression was induced by administration of isopropyl-β-d-thiogalactopyranoside (0.5 mm final concentration). The cells were harvested at 4°C and 4,000g for 30 min at 2 h (PP2Cs) and 4 h (RCARs) after induction. The cell pellet was used directly for purification. The pellet was lysed in 10 mL of lysis buffer (50 mm NaH2PO4, 300 mm NaCl, and 5 mm imidazole, pH 8.0) and treated with lysozyme (1 mg mL−1 final concentration) for 30 min. Cells were subsequently disrupted by sonication on ice (six times for 10 s). The protein lysate was obtained after centrifugation at 4°C and 25,000g for 30 min and loaded onto a nickel-Tris(carboxymethyl)ethylene diamine column (Macherey-Nagel; http://www.macherey-nagel.com). To remove unspecifically bound proteins, 8 mL of washing buffer (50 mm NaH2PO4, 300 mm NaCl, and 20 mm imidazole, pH 8.0) was applied to the column. Proteins of interest were eluted with 4 mL of elution buffer (50 mm NaH2PO4, 300 mm NaCl, and 250 mm imidazole, pH 8.0) and dialyzed two times against dialysis buffer (100 mm Tris-HCl, 100 mm NaCl, and 2 mm dithiothreitol, pH 7.9). One-milliliter fractions of eluate were collected, and fraction 2 was used in the assays.
Phosphatase Assays
Phosphatase activity was measured using 4-methyl-umbelliferyl-phosphate as a substrate (Ma et al., 2009). Values are means ± sd of four replicates. Control experiments of ABI2 activity in the presence of ABA precursors or catabolites showed no changes (less than 3%) in activity in the absence of RCARs.
Protoplast Analysis
Preparation and analysis of Arabidopsis protoplasts was performed as described (Moes et al., 2008). Arabidopsis protoplasts were transfected with 10 μg of DNA of the reporter construct (pRD29B::LUC) and 2 μg of p35S::GUS plasmid as a control for internal normalization of the expression. Protoplast suspensions were incubated in the presence or absence of ABA, ABA analogs, and ABA precursors after transfection.
Statistical analysis was done using the Mann-Whitney U test (http://elegans.swmed.edu/˜leon/stats/utest.html).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g01360 (RCAR1), At5g53160 (RCAR3), At4g17870 (RCAR11), At5g57050 (ABI2), At4g26080 (ABI1), At1g52340 (ABA2), and At1g16540 (ABA3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Regulation of ABI2 protein phosphatase by RCAR1, RCAR3, and RCAR11 in the presence of (S)-ABA, ABA catabolites, and ABA tetralone mimics at a ligand concentration of 1 μm.
Acknowledgments
We thank Dr. L. Irina Zaharia for providing us with samples of ABA catabolites and Drs. Michele Loewen, Adrian Cutler, and Farhah Assaad for critical review of the manuscript.
References
- Balsevich JJ, Cutler AJ, Lamb N, Friesen LJ, Kurz EU, Perras MR, Abrams SR. (1994) Response of cultured maize cells to (+)-abscisic acid, (−)-abscisic acid, and their metabolites. Plant Physiol 106: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco-Trinchant J, Le Page-Degivry MT. (1998) ABA synthesis in protoplasts of different origin in response to osmotic stress. Plant Growth Regul 25: 135–141 [Google Scholar]
- Bittner F, Oreb M, Mendel RR. (2001) ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem 276: 40381–40384 [DOI] [PubMed] [Google Scholar]
- Blackwell HE, Zhao Y. (2003) Chemical genetic approaches to plant biology. Plant Physiol 133: 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler AJ, Squires TM, Loewen MK, Balsevich JJ. (1997) Induction of (+)-abscisic acid 8′ hydroxylase by (+)-abscisic acid in cultured maize cells. J Exp Bot 48: 1787–1795 [Google Scholar]
- Cutler S, McCourt P. (2005) Dude, where’s my phenotype? Dealing with redundancy in signaling networks. Plant Physiol 138: 558–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dashek WV, Singh BN, Walton DC. (1979) Abscisic acid localization and metabolism in barley aleurone layers. Plant Physiol 64: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtado JA, Yang J, Ambrose SJ, Cutler AJ, Abrams SR, Kermode AR. (2007) Disrupting abscisic acid homeostasis in western white pine (Pinus monticola Dougl. Ex D. Don) seeds induces dormancy termination and changes in abscisic acid catabolites. J Plant Growth Regul 26: 46–54 [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL. (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q, Yin P, Yan C, Yuan X, Li W, Zhang Z, Liu L, Wang J, Yan N. (2010) Functional mechanism of the abscisic acid agonist pyrabactin. J Biol Chem 285: 28946–28952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays DB, Rose P, Abrams SR, Moloney MM. (1996) Biological activity of optically pure C-1 altered abscisic acid analogs in Brassica napus microspore embryos. J Plant Growth Regul 15: 5–11 [Google Scholar]
- Hill RD, Durnin D, Nelson LAK, Abrams GD, Gusta LV, Abrams SR. (1992) Effects of (+/−)phaseic acid on developing embryos of barley (Hordeum vulgare, L. cv. Bonanza) cultured in vitro. Seed Sci Res 2: 207–214 [Google Scholar]
- Hill RD, Liu JH, Durnin D, Lamb N, Shaw A, Abrams SR. (1995) Abscisic acid structure-activity relationships in barley aleurone layers and protoplasts (biological activity of optically active, oxygenated abscisic acid analogs). Plant Physiol 108: 573–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12: 343–351 [DOI] [PubMed] [Google Scholar]
- Jadhav AS, Taylor DC, Giblin M, Ferrie AM, Ambrose SJ, Ross AR, Nelson KM, Zaharia LI, Sharma N, Anderson M, et al. (2008) Hormonal regulation of oil accumulation in Brassica seeds: metabolism and biological activity of ABA, 7′-, 8′- and 9′-hydroxy ABA in microspore derived embryos of B. napus. Phytochemistry 69: 2678–2688 [DOI] [PubMed] [Google Scholar]
- Jiang F, Hartung W. (2008) Long-distance signalling of abscisic acid (ABA): the factors regulating the intensity of the ABA signal. J Exp Bot 59: 37–43 [DOI] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, Seo M. (2010) Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of abscisic acid biosynthesis, abscisic acid transport and hormone interactions. Plant Cell Physiol 51: 1988–2001 [DOI] [PubMed] [Google Scholar]
- Kong L, Abrams SR, Owen SJ, Van Niejenhuis A, Von Aderkas P. (2009) Dynamic changes in concentrations of auxin, cytokinin, ABA and selected metabolites in multiple genotypes of Douglas-fir (Pseudotsuga menziesii) during a growing season. Tree Physiol 29: 183–190 [DOI] [PubMed] [Google Scholar]
- Kuba M, Furuichi N, Katsumura S. (2002) Stereocontrolled synthesis of carotenoid oxidative metabolites, (−)-loliolide, (−)-xanthoxin and their stereoisomers. Chem Lett 31: 1248–1250 [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I. (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Rodríguez D, Nicolás G, Rodríguez PL, Nicolás C. (2001) A new protein phosphatase 2C (FsPP2C1) induced by abscisic acid is specifically expressed in dormant beechnut seeds. Plant Physiol 125: 1949–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- McCourt P, Desveaux D. (2010) Plant chemical genetics. New Phytol 185: 15–26 [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Xu Y, Ng LM, Zhou XE, Soon FF, Chinnusamy V, Suino-Powell KM, Kovach A, Tham FS, Cutler SR, et al. (2010) Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol 17: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milborrow BV, Carrington NJ, Vaughan GT. (1988) The cyclization of 8′-hydroxy abscisic acid to phaseic acid in vivo. Phytochemistry 27: 757–759 [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E. (2008) Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J 54: 806–819 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nelson LAK, Shaw A, Abrams SR. (1991) Synthesis of (+)-, (−)- and (+/−)-7′-hydroxyabscisic acid. Tetrahedron 47: 3259–3270 [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova GV, Stepanchenko NS, Nosov AV, Moshkov IE. (2009) At the beginning of the route: ABA perception and signal transduction in plants. Russ J Plant Physiol 56: 806–823 [Google Scholar]
- Nyangulu JM, Nelson KM, Rose PA, Gai Y, Loewen M, Lougheed B, Quail JW, Cutler AJ, Abrams SR. (2006) Synthesis and biological activity of tetralone abscisic acid analogues. Org Biomol Chem 4: 1400–1412 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry AD, Blonstein AD, Babiano MJ, King PJ, Horgan R. (1991) Abscisic-acid metabolism in a wilty mutant of Nicotiana plumbaginifolia. Planta 183: 237–243 [DOI] [PubMed] [Google Scholar]
- Peterson FC, Burgie ES, Park SY, Jensen DR, Weiner JJ, Bingman CA, Chang CE, Cutler SR, Phillips GN, Jr, Volkman BF. (2010) Structural basis for selective activation of ABA receptors. Nat Struct Mol Biol 17: 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Raschke K, Firn RD, Pierce M. (1975) Stomatal closure in response to xanthoxin and abscisic acid. Planta 125: 149–160 [DOI] [PubMed] [Google Scholar]
- Robertson AJ, Reaney M, Wilen RW, Lamb N, Abrams SR, Gusta LV. (1994) Effects of abscisic acid metabolites and analogs on freezing tolerance and gene expression in bromegrass (Bromus inermis Leyss) cell cultures. Plant Physiol 105: 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CD, Heath TG, Gage DA, Zeevaart JA. (1991) Abscisic alcohol is an intermediate in abscisic acid biosynthesis in a shunt pathway from abscisic aldehyde. Plant Physiol 97: 670–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PA, Abrams SR, Shaw AC. (1992) Synthesis of chiral acetylenic analogs of the plant hormone abscisic acid. Tetrahedron Asymmetry 3: 443–450 [Google Scholar]
- Saavedra X, Modrego A, Rodríguez D, González-García MP, Sanz L, Nicolás G, Lorenzo O. (2010) The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol 152: 133–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. (2009a) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL. (2009b) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Shibata N, Kagiyama M, Nakagawa M, Hirano Y, Hakoshima T. (2010) Crystallization of the plant hormone receptors PYL9/RCAR1, PYL5/RCAR8 and PYR1/RCAR11 in the presence of (+)-abscisic acid. Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 456–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E. (2010) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Todoroki Y, Hirai N, Koshimizu K. (1995) 8′,8′-Difluoro- and 8′,8′,8′-trifluoroabscisic acids as highly potent, long-lasting analogues of abscisic acid. Phytochemistry 38: 561–568 [Google Scholar]
- Tougane K, Komatsu K, Bhyan SB, Sakata Y, Ishizaki K, Yamato KT, Kohchi T, Takezawa D. (2010) Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha. Plant Physiol 152: 1529–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara Y, Ogawa T, Shibata K. (1975) Effects of abscisic acid and its derivatives on stomatal closing. Plant Cell Physiol 16: 543–546 [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J. (2008) An update on abscisic acid signaling in plants and more.... Mol Plant 1: 198–217 [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK. (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamomoto H, Oritani T. (1996) Stereoselectivity in the biosynthetic conversion of xanthoxin into abscisic acid. Planta 200: 319–325 [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. (2009) Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 16: 1230–1236 [DOI] [PubMed] [Google Scholar]
- Yuan X, Yin P, Hao Q, Yan C, Wang J, Yan N. (2010) Single amino acid alteration between valine and isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2. J Biol Chem 285: 28953–28958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia LI, Galka MM, Ambrose SJ, Abrams SR. (2005a) Preparation of deuterated abscisic acid metabolites for use in mass spectrometry and feeding studies. J Labelled Comp Radiopharm 48: 435–445 [Google Scholar]
- Zaharia LI, Walker-Simmons MK, Rodriguez CN, Abrams SR. (2005b) Chemistry of abscisic acid, abscisic acid catabolites and analogs. J Plant Growth Regul 24: 274–284 [Google Scholar]
- Zhou R, Cutler AJ, Ambrose SJ, Galka MM, Nelson KM, Squires TM, Loewen MK, Jadhav AS, Ross AR, Taylor DC, et al. (2004) A new abscisic acid catabolic pathway. Plant Physiol 134: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Abrams GD, Barton DL, Taylor DC, Pomeroy MK, Abrams SR. (1995) Induction of lipid and oleosin biosynthesis by (+)-abscisic acid and its metabolites in microspore-derived embryos of Brassica napus L. cv Reston (biological responses in the presence of 8′-hydroxyabscisic acid). Plant Physiol 108: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]