Abstract
The introduction of the Reduced height (Rht)-B1b and Rht-D1b semidwarfing genes led to impressive increases in wheat (Triticum aestivum) yields during the Green Revolution. The reduction in stem elongation in varieties containing these alleles is caused by a limited response to the phytohormone gibberellin (GA), resulting in improved resistance to stem lodging and yield benefits through an increase in grain number. Rht-B1 and Rht-D1 encode DELLA proteins, which act to repress GA-responsive growth, and their mutant alleles Rht-B1b and Rht-D1b are thought to confer dwarfism by producing more active forms of these growth repressors. While no semidwarfing alleles of Rht-A1 have been identified, we show that this gene is expressed at comparable levels to the other homeologs and represents a potential target for producing novel dwarfing alleles. In this study, we have characterized additional dwarfing mutations in Rht-B1 and Rht-D1. We show that the severe dwarfism conferred by Rht-B1c is caused by an intragenic insertion, which results in an in-frame 90-bp insertion in the transcript and a predicted 30-amino acid insertion within the highly conserved amino-terminal DELLA domain. In contrast, the extreme dwarfism of Rht-D1c is due to overexpression of the semidwarfing Rht-D1b allele, caused by an increase in gene copy number. We show also that the semidwarfing alleles Rht-B1d and Rht-B1e introduce premature stop codons within the amino-terminal coding region. Yeast two-hybrid assays indicate that these newly characterized mutations in Rht-B1 and Rht-D1 confer “GA-insensitive” dwarfism by producing DELLA proteins that do not bind the GA receptor GA INSENSITIVE DWARF1, potentially compromising their targeted degradation.
The introduction of semidwarfing genes into rice (Oryza sativa) and wheat (Triticum aestivum) was a major factor in breeding higher yielding varieties during the Green Revolution (for review, see Hedden, 2003). The higher yields were associated with improved lodging resistance and the resulting ability to tolerate higher rates of inorganic nitrogen-based fertilizer (Gale and Youssefian, 1985). The decrease in stem stature resulted in an increase in assimilate partitioning to developing ears, enabling greater floret survival at anthesis and increased grain numbers per ear (Youssefian et al., 1992). Different Reduced height-1 (Rht-1) genes, and combinations, gave optimal yields depending on the height of the background variety (Flintham et al., 1997).
In the wheat and rice semidwarf mutants, reduced stature is conferred through alterations in growth mediated by the GA phytohormones. This class of plant hormones has a well-characterized role in controlling stem elongation, as illustrated by the reduced stature of GA biosynthetic and signaling mutants in many plant species (Ross, 1994). In hexaploid wheat, dwarfing has been achieved mainly through the introduction of the Rht alleles Rht-B1b and Rht-D1b, now found in the majority of varieties grown worldwide (Evans, 1998), which confer decreased responsiveness to GA (Gale and Marshall, 1973; Pinthus et al., 1989). Six additional alleles at Rht-B1 and Rht-D1 have been identified by genetic mapping, four on chromosome 4B and two on chromosome 4D, which produce dwarfs with a broad range of plant height (Fig. 1; Gale and Youssefian, 1985; Borner et al., 1996; Flintham et al., 1997). For example, the Rht-B1b and Rht-D1b alleles produce only about a 20% reduction in height, while Rht-B1c and Rht-D1c are about half the size of the appropriate tall controls (Hoogendoorn et al., 1990; Flintham et al., 1997). The Rht-B1 and Rht-D1 genes were shown by Peng et al. (1999) to encode DELLA proteins, transcriptional regulators that act to repress GA signaling (for review, see Sun, 2010). Peng et al. (1999) found that Rht-B1b and Rht-D1b contain single nucleotide substitutions that introduce premature stop codons in the N-terminal coding region. They suggested that translational reinitiation could lead to the production of N-terminally truncated proteins that confer dwarfism through increased repression of GA signaling.
Figure 1.
Phenotypes of Rht-B1 and Rht-D1 dwarfing alleles in NILs. Wheat NILs (var Mercia) were grown to maturity. The photograph is from the John Innes Centre archives (produced by Tony Worland). [See online article for color version of this figure.]
In many cases, environmental and developmental signals promote plant growth by increasing the levels of bioactive GAs (for review, see Yamaguchi, 2008), which promote the degradation of the DELLA proteins, thus causing the transcriptional changes that mediate the growth responses (for review, see Sun, 2010). Studies in Arabidopsis (Arabidopsis thaliana) and rice have demonstrated that targeted degradation of DELLAs is initiated by GA binding within a pocket of the GA receptor, GA INSENSITIVE DWARF1 (GID1; Ueguchi-Tanaka et al., 2005; Murase et al., 2008; Shimada et al., 2008). This leads to closing of the N-terminal lid of GID1, allowing DELLAs to bind through their conserved N-terminal DELLA/TVHYNP motifs. GID1/DELLA binding enables recognition by the GID2/SLEEPY1 F-box component of an SCF Ub E3 ligase, leading to ubiquitination of DELLAs and their subsequent 26S proteasome-mediated degradation (Fu et al., 2002; Sasaki et al., 2003; Dill et al., 2004; Griffiths et al., 2006; Feng et al., 2008; Hirano et al., 2010). DELLA gain-of-function mutations, which result in reduced GA sensitivity and hence dwarfism, have been identified in many plant species, including rice and Arabidopsis. The majority of these mutations produce amino acid substitutions or deletions in the DELLA/TVHYNP motifs that affect binding to GID1 (Peng et al., 1997; Dill et al., 2001; Chandler et al., 2002; Asano et al., 2009), but not their transcriptional regulatory activity, which resides in the C-terminal GRAS domain (Silverstone et al., 1998; Chandler et al., 2002; Itoh et al., 2002; Hirano et al., 2010). The resulting reduction in GA-induced degradation of the mutant DELLAs confers dwarfism through enhanced repression of growth (Dill et al., 2001; Gubler et al., 2002; Itoh et al., 2002). The N-terminally truncated DELLA proteins that are proposed to be translated from Rht-B1b and Rht-D1b transcripts would similarly lack part of the conserved motifs that are necessary for GA-induced degradation, allowing them to accumulate and repress growth (Peng et al., 1999). However, currently, there is no biochemical evidence to support this hypothesis.
A more detailed understanding of how the Rht-1 genes confer dwarfism could potentially allow the development of novel alleles with improved specificity for agronomic traits. For example, in semiarid environments where deep sowing is necessary to access moisture for grain germination, the shorter coleoptiles of Rht-B1b and Rht-D1b cause poor emergence (Rebetzke et al., 2007). In order to obtain further information on Rht-1 gene structure and dwarfism, we have analyzed at the molecular level five of the six previously uncharacterized alleles at this locus. As well as discovering novel mutations, our results indicate that the severity of dwarfism in Rht-D1c is due to increased expression of the Rht-D1b allele. We also provide further evidence that the mutant RHT-1 proteins are unable to interact with the GA receptor, TaGID1, which would result in growth repression that is less responsive to GA. To determine the importance for the control of stem elongation of Rht-A1, for which no dwarfing alleles have been identified, we have cloned Rht-A1a and demonstrated that it displays a very similar expression profile to Rht-B1a and Rht-D1a.
RESULTS
Identification and Sequence Analysis of the Rht-A1 Gene
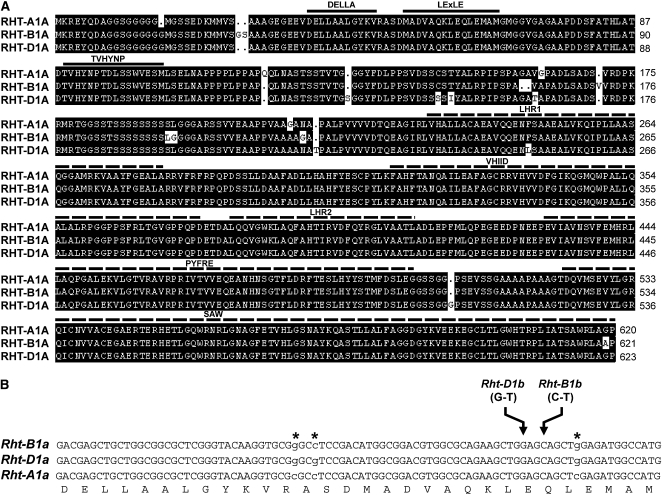
Although the existence of the homeologous DELLA gene, Rht-A1, is indicated (Peng et al., 1999; Febrer et al., 2009), its contribution to GA-dependent growth is currently unclear. In order to address this, a full-length putative Rht-A1a sequence was amplified by PCR using cDNA derived from seedling leaf tissues (var Cadenza). Across the predicted coding region, the amino acid sequence is approximately 96% identical to corresponding RHT-B1A and RHT-D1A sequences (Fig. 2A). Furthermore, nucleotide positions 289 to 769 of the coding region are identical to an Rht-A1 sequence amplified from a Chinese Spring bacterial artificial chromosome library (Febrer et al., 2009), making it likely that this represents the Rht-A1 gene. To confirm the chromosomal location of this gene, specific PCR primers were designed to each Rht-1 homeolog and used to amplify genomic DNA (gDNA) from chromosome 4 aneuploid wheat lines (Brewer et al., 1969). Whereas all three homeologs were amplified to a comparable level using the wild-type Chinese Spring gDNA template, the putative Rht-A1 sequence was not amplified from the template lacking chromosome 4A (N4AT4D; Supplemental Fig. S1), consistent with it representing the Rht-A1 gene on chromosome 4A.
Figure 2.
Predicted RHT-1 sequences. A, Amino acid sequence alignment of RHT-A1A, RHT-B1A, and RHT-D1A proteins. The amino acid sequences are predicted from nucleotide sequences amplified from var Cadenza. Gaps introduced to improve the sequence alignment are indicated by dots. Conserved N-terminal regulatory motifs (DELLA, LExLE, and TVHYNP) are indicated by thick solid lines above the sequences. The C-terminal (LHR1, VHIID, LHR2, PFYRE, and SAW) functional domains are indicated by thick dashed lines above the sequences. B, Comparison of the nucleotide sequences of the Rht-1 homeologs across the conserved N-terminal coding region. SNPs between the Rht-1 homeologs are indicated by lowercase letters and asterisks above the sequences. The Rht-B1b and Rht-D1b point mutations are shown, and their positions are indicated by arrows above the sequences. The predicted amino acid sequences of RHT-A1A, RHT-B1A, and RHT-D1A are identical in this region.
The predicted RHT-A1 amino acid sequence contains all known motifs that are conserved in functional DELLA proteins. These include the DELLA, LExLE, and TVHYNP motifs within the N-terminal regulatory domain (Fig. 2A), which are responsible for binding to GID1 to initiate GA-mediated degradation (Murase et al., 2008; Shimada et al., 2008), and a highly conserved C-terminal GRAS domain that is important for functionality (Peng et al., 1997; Silverstone et al., 1998; Hirano et al., 2010). The conservation in these regions, and the expression profile of Rht-A1 (see below), strongly suggest that Rht-A1, in addition to Rht-B1 and Rht-D1, encodes a functional DELLA protein.
Expression Analysis of the Rht-1 and GA Biosynthetic Genes during Wheat Stem Elongation
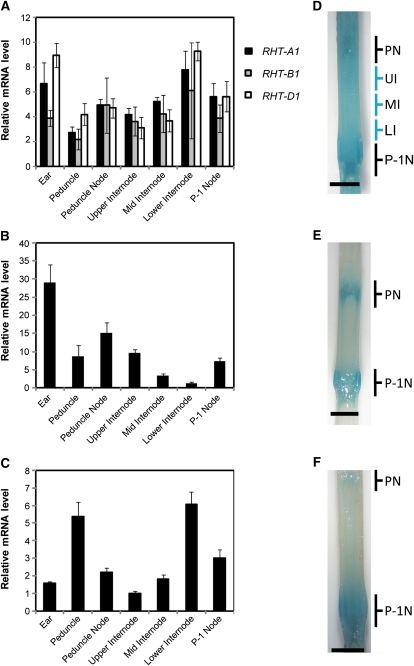
Differences in the expression profiles of the three Rht-1 homeologs during stem elongation could, in part, explain the phenotypic diversity observed in the Rht-1 mutants and the absence of identified semidwarfing alleles of Rht-A1 that are analogous to Rht-B1b or Rht-D1b. Transcript levels for the three homeologs were monitored in different regions of an elongating stem (var Cadenza) at a stage prior to the main period of peduncle expansion (Supplemental Fig. S2) using quantitative real-time reverse transcription (qRT)-PCR with homeolog-specific primers (Fig. 3A). The results indicated that Rht-A1 is expressed with a similar profile to Rht-B1 and Rht-D1 in all regions of the stem that were analyzed. All three homeologs are expressed at a slightly higher level in the peduncle and lower section of the P-1 internode, the two tissues that are undergoing cell expansion, but otherwise the expression is similar in other stem tissues. A slight increase in the levels of expression of the Rht-1 homeologs is observed in the ear compared with most regions of the stem. In contrast, on the basis of RNA-blot hybridization, Appleford et al. (2006) reported considerable tissue specificity in the expression of the GA-biosynthesis genes, TaGA20ox1 and TaGA3ox2. We confirmed this by qRT-PCR, which showed highest levels of expression of TaGA20ox1 in the ear and peduncle node (Fig. 3B), while TaGA3ox2 is most highly expressed in the peduncle and lower part of the P-1 internode (Fig. 3C).
Figure 3.
Expression analysis of the three Rht-1 homeologs and two GA biosynthesis genes in the upper expanding tissues of the developing wheat stem. A to C, Relative expression levels of Rht-A1, Rht-B1, and Rht-D1 (A), TaGA20ox1 (B), and TaGA3ox2 (C) in different regions of the extending wheat stem at 7 weeks post germination. The P-1 internode was divided into three equal sections for tissue collection. The mRNA levels of these genes were determined by qRT-PCR from three biological replicates for each sample type, normalized against UBIQUITIN. Results are plotted as the ratio to the lowest detectable level ± se. D to F, Localization of GUS activity under the control of the Rht-A1 (D), TaGA20ox1 (E), and TaGA3ox2 (F) promoters in an elongating wheat stem at the same stage of development as analyzed by qRT-PCR. The positions of the peduncle node (PN) and the P-1 node (P-1N) are indicated. The positions of the upper (UI), middle (MI), and lower (LI) internodes are indicated in D. Bars = 5 mm.
In order to localize the expression domains of Rht-A1, TaGA20ox1, and TaGA3ox2 more precisely, promoter-GUS transcriptional fusion constructs were generated using approximately 1.5 kb of sequence upstream of the predicted initiating ATG. Multiple transgenic wheat lines were produced for each construct showing consistent expression patterns in at least five independent T1 lines (data not shown). GUS activity was determined in the elongating wheat stems of the pRht-A1::GUS, pGA20ox1::GUS, and pGA3ox2::GUS lines at an equivalent developmental stage to that used for the qRT-PCR study described above. GUS activity in the pRht-A1::GUS lines was observed at similar levels in the nodes and P-1 internode region (Fig. 3D), whereas the pGA20ox1::GUS and pGA3ox2::GUS lines displayed GUS activity predominantly in the P-1 and peduncle nodal tissues (Fig. 3, E and F), with lower levels detectable in the P-1 internode. In contrast to the pGA20ox1::GUS line, GUS activity in the pGA3ox2::GUS line extended from the P-1 node up into the lower internode (Fig. 3F). These differences in expression profiles of GA3ox2 and GA20ox1 are supported by our qRT-PCR analysis (Fig. 3C) and the earlier study by Appleford et al. (2006). However, the level of GUS activity observed in the lower internode of the pGA3ox2::GUS lines is not entirely consistent with the high transcript levels of GA3ox2 observed by qRT-PCR. This observation potentially indicates the requirement for additional regulatory elements within the GA3ox2 gene that are necessary to direct the appropriate expression within this tissue. The expression profiles for TaGA20ox1, TaGA3ox2, and Rht-A1 inferred from the promoter-GUS reporter lines, which in most cases were consistent with the qRT-PCR data, clearly illustrate differences in the regulation of GA biosynthetic and DELLA genes during wheat stem elongation.
Characterization of Rht-B1 and Rht-D1 Mutations
In addition to Rht-B1b and Rht-D1b, six further dwarfing alleles of Rht-B1 or Rht-D1, conferring different degrees of dwarfism, have been identified by genetic mapping (Fig. 1) and reviewed by Borner et al. (1996). The phenotypes range from a mild decrease in plant stature in Rht-B1d to moderate in Rht-D1d and Rht-B1e to more severe in Rht-B1c and Rht-D1c (Fig. 1). While the molecular basis of the dwarfing mutations in Rht-B1b and Rht-D1b has been determined (Peng et al., 1999), there is currently no sequence information regarding the six further Rht-1 dwarfing alleles. Characterization of these different alleles at the molecular level should aid understanding of how different mutations at homeologous loci induce such variation in plant height. Such information would also allow the design and use of allele-specific PCR markers, permitting simple genotyping at these loci.
The high GC content within the Rht-1 genes (approximately 71% over their coding regions) made their amplification from gDNA or cDNA templates difficult. However, homeolog-specific amplification of the entire coding sequences of Rht-B1 and Rht-D1was achieved by amplifying overlapping 5′ and 3′ fragments.
Rht-B1c
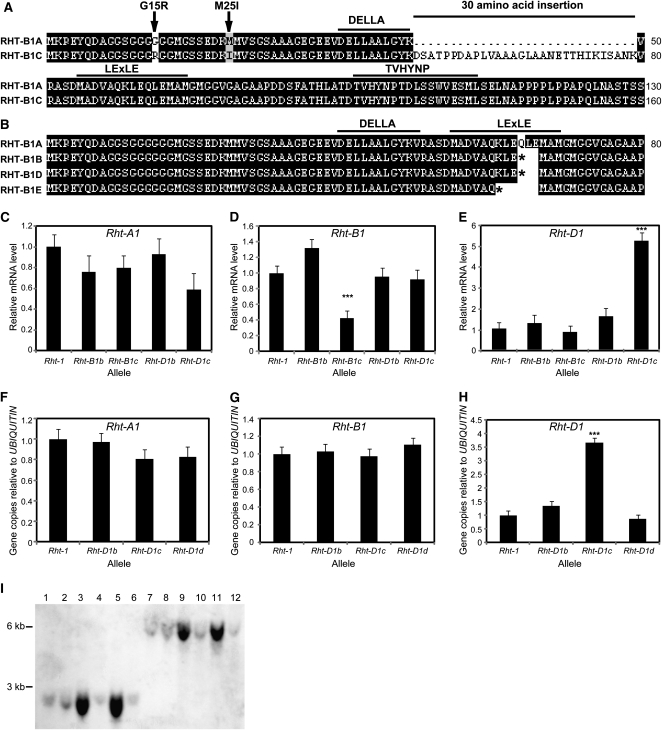
The Rht-B1 coding region was amplified using cDNA prepared from independent lines carrying the Rht-B1c allele, including the variety Tom Thumb (the original source of the dwarfing gene) and Rht-B1c near-isogenic lines (NILs) in Maris Huntsman, Maringá, and Nainari 60 (Gale and Marshall, 1973; Gale and Youssefian, 1985). Strikingly, we found that the Rht-B1c transcript carries a 90-bp in-frame insertion within the region encoding the conserved N-terminal DELLA domain (Supplemental Fig. S3). In addition, the Rht-B1c allele also contains two single nucleotide polymorphisms (SNPs) upstream of the insertion that result in amino acid substitutions (Fig. 4A; G15R and M25I substitutions). In the C-terminal GRAS domain, there were two silent mutations (codons Ala-271 [GCA to GCG] and Ala-617 [GCA to GCC]). Interestingly, a much larger insertion was present in gDNA from all lines containing Rht-B1c that were examined, although attempts to amplify across the full insert to determine its size were unsuccessful. However, PCR and sequencing indicated the presence of at least a 508-bp fragment that included the 90-bp insertion found in the transcript (Supplemental Fig. S3). Furthermore, the larger insertion contains potential splice sites at its 5′ end and at the start of the 90-bp insertion (predicted using http://genes.mit.edu/GENSCAN.html). This suggested that most of the insertion in Rht-B1c is subsequently excised during transcript processing to give an mRNA containing a 90-bp in-frame insertion and a protein with a 30-amino acid insertion within the DELLA domain (Fig. 4A). Allele-specific PCR-based markers were designed to detect the presence of the insertion in Rht-B1c, allowing it to be distinguished from other Rht-B1 alleles (Supplemental Fig. S4A).
Figure 4.
Rht-1 allele characterization. A, Amino acid alignment of RHT-B1A and RHT-B1C. The translated RHT-B1C protein contains a 30-amino acid insertion close to the DELLA motif and two upstream amino acid substitutions (indicated with arrows above the sequences; G15R and M25I). Conserved N-terminal regulatory (DELLA, LExLE, and TVHYNP) domains are indicated by lines above the sequences. B, Amino acid alignment of the N-terminal domains of Rht-B1 alleles. Rht-B1d carries the same point mutation as Rht-B1b, while Rht-B1e expresses a point mutation that introduces a premature stop codon three amino acids farther upstream. C to E, mRNA levels for Rht-1 in 2-week-old seedlings of wheat NILs in the Mercia variety, determined by qRT-PCR using primers designed to amplify all three homeologs. Values shown are means of three biological replicates, normalized against UBIQUITIN. Results are plotted as the ratio to the level in the Rht-1 control ± se. *** Significantly different from the Rht-1 control (P < 0.01). F to H, Gene copy number of the Rht-1 homeologues in the Rht-D1 allelic series (NILs in the Mercia variety) determined by qPCR of gDNA template. The relative gDNA copy number was normalized against UBIQUITIN. Results are plotted as the ratio to the level in the Rht-1 control. Values shown are means of three biological replicates ± se. *** Significantly different from the Rht-1 control (P < 0.01). I, Hybridization of D1-2855 to a Southern blot containing gDNA isolated from wheat lines containing different Rht-D1 alleles: Rht-D1a Mercia (Rht-1) control, lanes 1 and 7; Rht-D1b Mercia NIL, lanes 2 and 8; Rht-D1c Mercia NIL, lanes 3 and 9; Rht-D1d Mercia NIL, lanes 4 and 10; Rht-D1c Ai-bian 1, lanes 5 and 11; Rht-D1d Ai-bian 1a, lanes 6 and 12, digested with DraI (lanes 1–6) or EcoRI (lanes 7–12).
In order to determine whether mutations in Rht-B1 affected Rht-1 gene expression, the relative levels of the homeologous Rht-1 transcripts were compared in Rht-B1a (Rht-1), Rht-B1b, and Rht-B1c seedlings by qRT-PCR. There were no significant differences in the levels of expression of Rht-A1 and Rht-D1 in the two Rht-B1 mutants that were analyzed (Fig. 4, C and E). Similarly, expression of Rht-B1 was largely unaltered in the Rht-B1b mutant compared with the Rht-1 control (Fig. 4D), but its expression was significantly reduced in both the Mercia and Maris Huntsman Rht-B1c NILs (Fig. 4D; Supplemental Fig. S5B). The reduction in Rht-B1 transcript levels in Rht-B1c is potentially due to a lower splicing efficiency during mRNA processing.
Rht-B1d and Rht-B1e
Rht-B1d is derived from the Japanese variety Saitama 27 (Worland and Petrovic, 1988). A recent study by Pestsova et al. (2008) using a PCR-based assay indicated the presence of the Rht-B1b point mutation in the Rht-B1d allele. We confirmed this result by amplifying and sequencing the Rht-B1 gene from genomic DNA extracted from Rht-B1d seedlings (Fig. 4B). Surprisingly, no further mutations were found in the coding region of this gene. Plants carrying the Rht-B1d mutation are less severely dwarfed than those carrying Rht-B1b alone (Fig. 1; Worland and Petrovic, 1988), which suggests that another, undetected, mutation outside of the Rht-B1 coding region is affecting plant height.
Another source of GA-insensitive dwarfism that is used commercially has its origins in the Russian variety Bezostaya, which carries the Rht-B1e allele (Worland, 1986; Borner et al., 1996). PCR-based assays indicated that Rht-B1e is distinct from Rht-B1b (Pestsova et al., 2008). To characterize this allele, the Rht-B1 gene from two Rht-B1e semidwarf lines, Krasnodari dwarf and Bezostaya dwarf, was amplified and sequenced, revealing a novel mutation introducing a stop codon three codons upstream of the Rht-B1b point mutation (Fig. 4B). An A-to-T nucleotide substitution changes the Lys-61 codon (AAG) to a translational stop codon (TAG). This is in an almost identical position to both the Rht-B1b and Rht-D1b mutations, where similar point mutations result in the conversion of Rht-B1 codon 64 and Rht-D1 codon 61 to stop codons (Fig. 4B; Peng et al., 1999). No further mutations in the Rht-B1e coding sequence were detected. We have designed PCR-based markers that detect the presence of this mutation and distinguish it from both the Rht-B1a and Rht-B1b sequences (Supplemental Fig. S4B). Based on the similarity in nature and its position within the coding region, it is likely that the Rht-B1e mutation confers its GA-insensitive semidwarf phenotype in a manner analogous to Rht-B1b, potentially through the production of an N-terminally truncated protein product that represses GA signaling, as hypothesized by Peng et al. (1999).
Rht-D1c and Rht-D1d
The extreme dwarfing allele Rht-D1c is derived from the Chinese variety Ai-bian 1, which is also the source of another Rht-D1 allele, Rht-D1d (Borner et al., 1996). This latter allele arose spontaneously from an Rht-D1c population and was distinguished by an increase in height compared with the Rht-D1c plants (Fig. 1). We used genomic DNA extracted from Ai-bian 1, Rht-D1c, and Rht-D1d as templates to amplify the Rht-D1 coding region using homeolog-specific primers. Sequence analysis revealed that all of these lines carry the Rht-D1b mutation, as suggested by screening using PCR markers (Pestsova et al., 2008). Since no further mutations were detected, the sequences of Rht-D1b, Rht-D1c, and Rht-D1d are identical across the coding region. However, the far more severe dwarf phenotype of Rht-D1c lines compared with Rht-D1b suggests that it may carry an additional mutation outside the coding region that alters its expression level. This was investigated by comparing the transcript abundance for the three Rht-1 homeologs in seedlings of Rht-D1a, Rht-D1b, Rht-D1c, and Rht-D1d by qRT-PCR using specific primers. There were no significant differences in the levels of expression of Rht-A1 and Rht-B1 in the Rht-D1 mutants compared with Rht-D1a (Fig. 4, C and D). However, while Rht-D1 was expressed at similar levels to Rht-1 in Rht-D1b (Fig. 4E), its expression level in Rht-D1c was about 4-fold higher (Fig. 4E). We then investigated whether the increased Rht-D1 expression in Rht-D1c is due to the presence of extra copies of Rht-D1 by performing real-time qPCR on gDNA isolated from Rht-D1a (tall), Rht-D1b, Rht-D1c, and Rht-D1d lines. The copy number of Rht-A1 and Rht-B1 was found to be very similar in all these lines (Fig. 4, F and G). However, while the Rht-D1 copy number in Rht-D1b and Rht-D1d was nearly identical to that in the Rht-D1a control (Fig. 4H), it was about 4-fold higher in Rht-D1c (Fig. 4H), providing a good correlation between gene copy number and expression level. To confirm this increase in Rht-D1 copy number in Rht-D1c, we hybridized a Rht-D1-specific DNA probe to a Southern blot containing EcoRI- or DraI-digested gDNA isolated from an Rht-D1 allelic series. The probe (D1-2855) was generated by amplifying a DNA fragment between 2,855 and 3,097 bp downstream of the Rht-D1-initiating ATG. Hybridization of D1-2855 to gDNA fragments from Rht-1, Rht-D1b, and Rht-D1d (Mercia NIL and Aibian 1A progenitor) produced similar band intensities (Fig. 4I), suggesting equal gene copy numbers in these lines. In contrast, the intensity of signal for the equivalent gDNA fragments from Rht-D1c (observed in both Mercia NIL and Aibian 1 progenitor) was highly elevated (Fig. 4I), supporting an increase in Rht-D1 copy number. Taken together, these findings strongly suggest that the severe dwarfing caused by the Rht-D1c allele results from the presence of multiple gene copies of Rht-D1b, producing elevated transcript levels and potentially higher amounts of the GA-insensitive DELLA repressor. While Rht-D1d is potentially derived from Rht-D1c (Borner et al., 1991), it does not appear to have an increase in Rht-D1 copy number compared with the Rht-D1a control (or Rht-D1b; Fig. 4H). It is likely, therefore, that the substantial increase in height of Rht-D1d compared with Rht-D1c (Borner et al., 1991, 1996) is due to this reduction in the number of gene copies of Rht-D1b.
Dwarfing Mutations in RHT-B1 Abolish the Interaction with TaGID1 in Yeast Two-Hybrid Assays
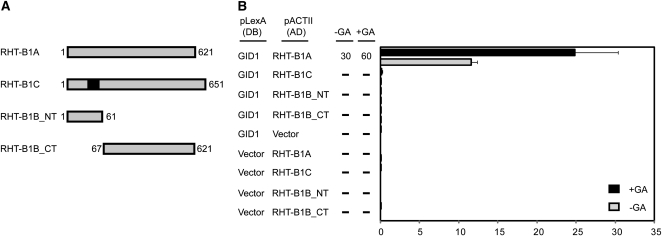
Binding of bioactive GAs within a pocket in the GID1 protein induces a conformational change that permits interaction between the DELLA and GID1 proteins (Murase et al., 2008; Shimada et al., 2008). This interaction with GID1 involves several conserved amino acids within the DELLA N-terminal region. We hypothesize that disruption of these conserved motifs in mutant RHT-1 proteins would prevent their interaction with GID1 and allow the proteins to accumulate, despite adequate endogenous GA levels. In the case of Rht-1 mutations that introduce premature stop codons, it has been suggested that translational reinitiation occurs, producing an N-terminally truncated protein product that lacks the GID1 interaction motifs (Peng et al., 1999; Willige et al., 2007). However, an alternative explanation is that the reduced GA sensitivity results from expression of the short N-terminal peptide, which could sequester GID1 and thereby reduce its effective concentration. In an attempt to distinguish between these possibilities and to examine the impact of the 30-amino acid insertion in RHT-B1C, we used yeast two-hybrid assays to assess the interaction between the predicted RHT-B1B and RHT-B1C mutant proteins and GID1. A GID1 bait construct was prepared by amplifying a TaGID1 sequence from wheat cDNA. The TaGID1 sequence is 95% identical to the barley (Hordeum vulgare) GA receptor, GSE1 (Chandler et al., 2008), strongly suggesting that it encodes a functional GA receptor. The prey constructs containing the domains indicated in Figure 5A were cotransformed into yeast with the TaGID1 bait, and the interactions were assessed in the absence or presence of bioactive GA3. The full-length RHT-B1A interacted strongly with TaGID1 in yeast even in the absence of GA, and this interaction was enhanced by the presence of GA3 (Fig. 5B). This observation provides support for TaGID1 being a functional GA receptor that binds RHT-B1A, potentially targeting it for degradation via a pathway that appears conserved in both dicots and monocots (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Willige et al., 2007). RHT-B1C, which contains the 30-amino acid insertion within the DELLA domain, failed to interact with TaGID1 in the absence or presence of GA (Fig. 5B). The predicted N- and C-terminally truncated RHT-B1B protein products also did not interact with TaGID1 in the yeast two-hybrid assays, irrespective of whether GA3 was included (Fig. 5B).
Figure 5.
Interaction between wild-type and mutant forms of RHT-B1 and TaGID1 in yeast two-hybrid assays. A, Schematic diagram showing full-length and mutant forms of RHT-B1, which were expressed as LexA activation domain (AD) fusions in yeast. Numbers indicate the amino acid positions within RHT-B1. The black box in the RHT-B1C schematic diagram indicates the position of the 30-amino acid insertion. The full-length TaGID1 was expressed as a LexA DNA-binding domain (DB) fusion protein in yeast. B, Interactions between activation domain and DNA-binding domain protein fusions were determined in cotransformed L40 yeast cells by scoring growth on His− medium containing 3-aminotriazole (3-AT; 0, 2, 5, 10, 30, and 60 mm). Data presented are maximum concentrations of 3-AT at which growth was observed; dashes signify no growth on 2 mm 3-AT. Quantitative values for each interaction were determined by measuring β-Gal activity in the presence and absence of 100 μm GA3. Values are means of three biological replicates ± se.
DISCUSSION
The major contribution to global wheat production from the introduction of the Rht-B1b and Rht-D1b semidwarfing genes in the 1960s is well documented (Evans, 1998). However, it was not until 1999 that the mutant genes were characterized at the molecular level (Peng et al., 1999), and the biochemical basis for the resulting semidwarf phenotype has still not been demonstrated. In addition to Rht-B1b and Rht-D1b, a number of other dwarfing alleles at the Rht-B1 and Rht-D1 loci have been described, several having applications in agriculture (Borner et al., 1996). We have characterized three further dwarfing Rht-B1 alleles and two at the Rht-D1 locus and provide evidence to support the proposed biochemical basis for the phenotype in these and the Rht-B1b and Rht-D1b mutants. No dwarfing allele has been identified in Rht-A1, and it was unclear whether this gene was expressed. Our results here indicate that Rht-A1 is expressed at similar levels to the other Rht-1 homeologs. Furthermore, the nucleotide sequence in the N-terminal coding region encompassing the premature stop codons in Rht-B1b, Rht-B1e, and Rht-D1b is identical in all three genes (Fig. 2B), which should allow the introduction of similar stop codons into Rht-A1. The similarity in phenotype between the Rht-B1b, Rht-D1d, and Rht-B1e mutants suggests that mutations introducing a premature stop within at least three codons (codons 61, 62, and 64; Fig. 2B) will confer GA insensitivity. The absence of additional Rht-B1 and Rht-D1 alleles with novel mutations at these positions illustrates the rarity of these mutational events. Therefore, it is possible that corresponding mutations in the Rht-A1 gene will also confer GA insensitivity but have yet to occur or be selected. However, it is also conceivable that these mutations do not confer an obvious phenotype due to changes in the functional role of Rht-A1 compared with the other Rht-1 homeologs. To distinguish between these two scenarios, it will be important to introduce the equivalent Rht-A1 mutations using targeted mutagenesis-based approaches.
The genetic lesions of all previously characterized DELLA gain-of-function mutants are either nucleotide substitutions or in-frame deletions (Peng et al., 1997, 1999; Boss and Thomas, 2002; Chandler et al., 2002; Muangprom et al., 2005; Asano et al., 2009). Here, we demonstrate that Rht-B1c contains a novel type of DELLA mutation, a genomic insertion that results in the introduction of 30 amino acids between the DELLA and LExLE motifs, which are required for binding to the GID1 N-terminal extension (Murase et al., 2008). We were unable to amplify across the whole genomic insertion, but we identified a highly conserved 508-bp fragment that has been shown to be a repeated flanking unit of a 2,031-bp terminal repeat retrotransposon in miniature at the equivalent location in Tom Thumb (Wu et al., 2011). These tandem repeats contain splice sites such that most of the insertion is removed during RNA processing, leaving an in-frame 90-bp insertion that, as demonstrated in yeast two-hybrid assays, completely abolishes the interaction of RHT-B1C with TaGID1. It is likely, therefore, that the RHT-B1C protein acts as a repressor of GA signaling that would not be polyubiquitinated and degraded by the 26S proteasome, even in the presence of GA. In addition to the DNA insertion, Rht-B1c also contains two SNPs upstream of the insertion that result in two amino acid substitutions (Fig. 4; G15R and M25I). While it is conceivable that these may also compromise the interaction with GID1, this appears unlikely, given that this region is poorly conserved in DELLA proteins. Furthermore, these substitutions are not found in the regions implicated in the DELLA-GID1 association (Murase et al., 2008). It is likely that they reflect SNPs present in the original donor.
In contrast to Rht-B1c, the severely dwarfing Rht-D1c allele harbored no DNA insertions but contained instead the same base substitution present in the mildly dwarfing Rht-D1b allele. We could detect no other mutations within its coding sequence. The severity of the dwarf phenotype in Rht-D1c may be due to its elevated level of expression (Fig. 4E) compared with other Rht-1 alleles. This may be explained by a higher copy number for Rht-D1c (Fig. 4H), although mutations in its promoter cannot be discounted, despite none being detected within 3,686 bp of sequence upstream of the predicted start codon (data not shown). The recent partial sequencing of chromosome 3B has illustrated the high degree of transposon-mediated gene amplification in wheat (Choulet et al., 2010). It is likely that this process, potentially involving CACTA transposon-mediated gene capture and amplification at the same locus, is responsible for the increased copy number for Rht-D1c. The semidwarf Rht-D1d mutant is described as being a spontaneous mutant derived from Rht-D1c (Borner et al., 1991), although it does not contain the same increase in Rht-D1 gene copy number and may have lost these additional copies through a transposon excision-mediated process. However, the similarity in phenotype to Rht-D1b and its identical Rht-D1 copy number also raises the possibility that it is not a novel allele but is derived from an Rht-D1b contaminant within the Rht-D1c population or through out-crossing with Rht-D1b.
We have found that, in common with Rht-B1b and Rht-D1b, the previously uncharacterized Rht-B1d, Rht-B1e, Rht-D1d, and, as discussed above, Rht-D1c alleles all contain nucleotide substitutions that create premature stop codons in the N-terminal coding regions. In the cases of Rht-B1d, Rht-D1c, and Rht-D1d, the mutations are identical to those found in the respective Rht-B1b and Rht-D1b mutants. However, the introduction of an alternative stop codon in the Rht-B1e mutant extends the genetic location within which these semidwarfing DELLA gain-of-function mutations can occur. Peng et al. (1999) proposed two explanations for the GA insensitivity that results from these mutations: the expression of a short N-terminal product could block GA signaling through its interaction with TaGID1 via the remaining DELLA domain, or, alternatively, translational reinitiation could result in the production of an N-terminally truncated product that constitutively represses GA signaling due to its inability to interact with TaGID1. Our findings that both truncations completely abolish the interaction with TaGID1 in yeast assays allow the first proposal to be discounted and provide support for the second. Mutant DELLA genes that are predicted to express C-terminal truncations including the complete GID1 interaction domain have been described in Arabidopsis, rice, and barley (Peng et al., 1997; Ikeda et al., 2001; Chandler et al., 2002). The findings that all of these mutations result in loss of function rather than gain of function would further suggest that the production of an N-terminal peptide is not the cause of dwarfism in the Rht-B1b, Rht-D1b, and Rht-B1e mutants. It will be necessary to confirm the effects of these mutations on the stability of endogenous RHT-1. However, our attempts at raising polyclonal antibodies that recognize the endogenous RHT-1 proteins in wild-type or mutant backgrounds have proved unsuccessful.
In contrast to other DELLA gain-of-function mutations, in which deletion or substitution of amino acids critical for GID1 binding causes severe dwarfism (Peng et al., 1997, 1999; Chandler et al., 2002; Murase et al., 2008; Asano et al., 2009), the Rht-B1b, Rht-B1e, and Rht-D1b mutations result in only mild height reductions. It is possible that a low efficiency of translational reinitiation in the Rht-1 mutants would result in reduced levels of GA-insensitive DELLA protein being produced. This is consistent with the phenotype of Rht-D1c, which indicates that increased expression of the Rht-D1b allele is capable of enhancing the severity of dwarfism. Indeed, a gene-dose effect was observed for the Arabidopsis DELLA gene GAI and its gain-of-function mutant allele gai expressed in tobacco (Nicotiana tabacum), in which the degree of dwarfism was related to the level of expression (King et al., 2001; Hynes et al., 2003).
Comparison of the spatial expression profiles of Rht-1 with those of the GA biosynthetic enzymes TaGA20ox1 and TaGA3ox2 within the expanding P-1 internode indicated substantial differences. In agreement with the observations of Appleford et al. (2006), expression of the biosynthetic genes is strongest in growing tissues, with TaGA20ox1 expression located most strongly at the nodes, presumably within the intercalary meristem, and that of TaGA3ox2 in the meristem and elongation zones. In contrast, Rht-1 was expressed throughout the internode. These observations suggest that growth of the internode is regulated by GA production rather than response. However, although our results for the biosynthetic genes are broadly in line with those of Kaneko et al. (2003), who, on the basis of reporter gene analysis in rice, showed expression of OsGA20ox2 and OsGA3ox2 in the meristem and elongation zones of the fifth internode, the same expression pattern was also found for the DELLA gene SLR1. It is possible that differences in the developmental stages of the plants analyzed in these studies are responsible for the observed disparities between them.
Yields from current wheat cultivars containing Rht-B1b or Rht-D1b alleles may be approaching their limits (Slafer et al., 2001), while a changing climate may reduce the usefulness of these genes in the future. These issues, combined with potential restrictions on the use of growth retardants and chemical fertilizer, highlight the need to develop novel dwarfing alleles. Our genetic and biochemical characterization of dwarfing Rht-B1 and Rht-D1 alleles and the finding that Rht-A1 is expressed have provided further insights and opportunities for generating such genes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of wheat (Triticum aestivum) NILs for different alleles of Rht-B1 and Rht-D1 in the varieties Maris Huntsman, Mercia, and Bezostaya were provided by Dr. J. Flintham (John Innes Centre). Wheat chromosome 4 aneuploid lines N4AT4B, N4DT4B, and CSDT4B-α were provided by Dr. M. Ambrose from the Biotechnology and Biological Sciences Research Council small grain cereals collection in the Germplasm Resources Unit, John Innes Centre. BC7 NILs for Rht-B1c in the varieties Maringá and Nainari 60 were described by Hoogendoorn et al. (1988), and grains were provided by the Australian Winter Cereal Collection. Seed for Ai-bian 1 and Ai-bian 1a were provided by Dr. Andreas Borner (Institute of Plant Genetics and Crop Plant Research, Germany). Stem expression analysis studies and transformations were carried out in the spring wheat cv Cadenza, which carries wild-type alleles of Rht-B1 and Rht-D1. Plants were grown as described previously (Appleford et al., 2006).
Gene Amplification and Sequencing
Rht-B1 or Rht-D1 mutant alleles were amplified from either gDNA or cDNA templates. Genomic DNA was extracted using a cetyl-trimethyl-ammonium bromide extraction buffer. RNA was extracted using the RNeasy Mini Kit (Qiagen) and cDNA was synthesized using the SuperScript ΙΙΙ First Strand Synthesis System (Invitrogen). Rht-B1 and Rht-D1 genes were amplified as two overlapping fragments using two sets of primers, listed in Supplemental Table S1. Amplified fragments were cloned into pGEM-T Easy vector (Promega) and sequenced. The presence of an insertion larger than the expected 90 bp in the Rht-B1 gene of Maringá Rht-B1c was first revealed by the sizes of fragments amplified using forward primers from the beginning of the gene (e.g. Rht3F5) and reverse primers from within the 90 extra nucleotides (e.g. Rht3R5). The sizes and sequences of this and other products amplified between gene and insert sequences were all consistent with a single 508-bp insertion.
Plasmid Constructs
Promoter::GUS expression constructs were created for Rht-A1, TaGA20ox1-A, and TaGA3ox2-3 by amplifying 1,529, 1,555, and 1,566 bp upstream of the predicted initiating ATG, respectively, by PCR. The amplified fragments were cloned in frame into the pPTGUS vector upstream of the uidA gene to create plasmids pRht-A1::GUS, pGA20ox1-A::GUS, and pGA3ox2-3::GUS. For the yeast two-hybrid assays, bait proteins were cloned as LexA binding domain fusion proteins in the pLexA-NLS expression vector and prey proteins as Gal4 activation domain fusions in the pACTΙΙ expression vector. In each assay, the full-length GID1 was used as the bait protein fusion. RHT-B1 prey constructs were produced in pACTII by amplifying the necessary coding regions from gDNA of Rht-1 or Rht-B1c (var Mercia).
Wheat Transformation and Histochemical Analysis of GUS Activity
Promoter::GUS reporter lines were produced by bombardment of wheat embryos with the plasmids pRHT-A1::GUS, pGA20ox1-A::GUS, and pGA3ox2-3::GUS (as described by Sparks and Jones, 2009). Expression analysis was performed on plants of the T1 generation. Plant tissues were incubated at 37°C overnight in 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid solution containing 1% Triton. Tissues were destained in 70% ethanol and examined using a light microscope.
Real-Time qRT-PCR
Gene expression analysis in the developing stem was carried out in seven separate regions from a 7-week-old plant: the ear, peduncle, peduncular node, P-1 internode split into three equal sections, and P-1 node (as described by Appleford et al., 2006). Total RNA extraction, cDNA synthesis, and qRT-PCR were performed as described previously (Rieu et al., 2008). Expression data were normalized against the UBIQUITIN control gene using the GeNORM procedure described by Vandesompele et al. (2002).
Four-day-old seedlings of wheat NILs (var Mercia and Maris Huntsman) differing in their Rht-1 alleles on chromosomes 4B and 4D were used to analyze expression of the homeologous Rht-1 genes. qRT-PCR was carried out using a DNA Engine Opticon2 Continuous Fluorescence Detector (MJ Research). RNA was extracted and cDNA was synthesized as described above. Reactions were carried out using SYBR Green Jumpstart Taq Ready Mix (Sigma-Aldrich). The average threshold cycle (CT) value for each gene was calculated from duplicate samples for each experiment. Data were analyzed using Opticon Monitor analysis software version 2.02 (MJ Research). For each target gene, expression was calculated relative to that of the reference gene, UBIQUITIN (Van Riet et al., 2006), using the ΔCT method (Pfaffl et al., 2002) and corrected for primer efficiencies.
The primers for each target gene and the UBIQUITIN endogenous control gene are listed in Supplemental Table S1. The UBIQUITIN endogenous control gene (accession no. AY297059) has been described previously (Li et al., 2008). Wheat Rht-D1 Mercia NILs were used to determine Rht-D1 gene dosage in an Rht-D1 allelic series. gDNA was extracted using cetyl-trimethyl-ammonium bromide DNA extraction buffer. qPCR was performed with the three Rht-1 homeolog-specific assays as described above but using gDNA (2 ng μL−1) as template. For each target gene, the amplification signal was normalized to UBIQUITIN as described above.
DNA Gel-Blot Hybridizations
Twenty micrograms of gDNA isolated from each line of the Rht-D1 allelic series was digested with DraI or EcoRI. Gel-blot hybridizations were performed as described previously using a digoxigenin-11-dUTP-labeled probe and chemiluminescence detection (Vaughan et al., 2006) The Rht-D1-specific probe of 243 bp (D1-2855) was amplified using primers Rht-D1_SF1 and Rht-D1_SR2 from wheat gDNA (var Mercia).
Rht-B1c and Rht-B1e PCR Markers
To screen for the presence of the Rht-B1c and Rht-B1e alleles, two PCR markers were developed. For Rht-B1c, the primer pair Rht-B1c-F1/Rht-B1c-R1 was used to amplify a 256-bp product only from lines carrying the Rht-B1c allele by PCR. For Rht-B1e, two primer sets were used: forward primer BF was used with reverse primer WR3 to amplify wild-type sequences, while PCR using BF with reverse primer MR3 amplified a 228-bp fragment only in lines carrying the Rht-B1e allele.
Yeast Two-Hybrid Assay
Saccharomyces cerevisiae strain L40 was used for all yeast two-hybrid assays (Vojtek et al., 1993). Yeast cells were cotransformed with binding domain and activation domain plasmid expression constructs, and transformants were selected on −Leu−Trp medium (Q-biogene). The ability to drive the expression of the HIS3 or LacZ reporter genes was tested as described previously (Dill et al., 2004) in the presence or absence of 100 μm GA3.
The accession numbers for the sequences described in this article are JF930277 (Rht-A1a), JF930278 (Rht-B1a), JF930279 (Rht-B1c), JF930280 (Rht-B1e), JF930281 (Rht-D1b), JF930282 (TaGA20OX1-A), and JF930283 (TaGA3OX2-3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Homeolog-specific qRT-PCR to validate the chromosomal location of Rht-A1.
Supplemental Figure S2. Wheat tissues sampled for gene expression analysis.
Supplemental Figure S3. Comparison of the nucleotide sequences of Rht-B1a and Rht-B1c alleles.
Supplemental Figure S4. PCR-based assays to detect Rht-B1c and Rht-B1e mutations.
Supplemental Figure S5. Expression analysis of the Rht-1 homeologs in wild-type wheat and the Rht-B1b and Rht-B1c mutants.
Supplemental Table S1. Oligonucleotide primers used for standard PCR and qRT-PCR.
Acknowledgments
We thank Richard Parkinson, Fiona Gilzean, and other greenhouse staff at Rothamsted Research for their support in growing plants; Prof. Huw Jones and other staff in the Cereal Transformation Unit at Rothamsted Research for support with generating wheat transgenic lines; Carol Harding (Commonwealth Scientific and Industrial Research Organization) (University of Oxford) for expert technical assistance; Prof. Nicholas Harberd (University of Oxford) for providing Rht-B1a sequence information; and Dr. John Lenton for useful discussions and critical review of the manuscript.
References
- Appleford NEJ, Evans DJ, Lenton JR, Gaskin P, Croker SJ, Devos KM, Phillips AL, Hedden P. (2006) Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat. Planta 223: 568–582 [DOI] [PubMed] [Google Scholar]
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M. (2009) Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol Genet Genomics 281: 223–231 [DOI] [PubMed] [Google Scholar]
- Borner A, Lehmann CO, Mettin J, Plaschke J, Schlegel G, Schlegel R, Melz G, Thiele V. (1991) GA-insensitivity of ‘Aibian 1a’: pleiotropic effects of isogenic Rht-lines. Ann Wheat Newsletter 37: 59–60 [Google Scholar]
- Borner A, Plaschke J, Korzun V, Worland AJ. (1996) The relationships between the dwarfing genes of wheat and rye. Euphytica 89: 69–75 [Google Scholar]
- Boss PK, Thomas MR. (2002) Association of dwarfism and floral induction with a grape ‘Green Revolution’ mutation. Nature 416: 847–850 [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Sing CF, Sears ER. (1969) Studies of isozyme patterns in nullisomic-tetrasomic combinations of hexaploid wheat. Proc Natl Acad Sci USA 64: 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Harding CA, Ashton AR, Mulcair MD, Dixon NE, Mander LN. (2008) Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Mol Plant 1: 285–294 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Marion-Poll A, Ellis M, Gubler F. (2002) Mutants at the Slender1 locus of barley cv Himalaya: molecular and physiological characterization. Plant Physiol 129: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, Schlub S, Le Paslier MC, Magdelenat G, Gonthier C, et al. (2010) Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22: 1686–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP. (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LT. (1998) Feeding the Ten Billion: Plants and Population Growth. Cambridge University Press, Cambridge, UK [Google Scholar]
- Febrer M, Wilhelm E, Al-Kaff N, Wright J, Powell W, Bevan MW, Boulton MI. (2009) Rapid identification of the three homoeologues of the wheat dwarfing gene Rht using a novel PCR-based screen of three-dimensional BAC pools. Genome 52: 993–1000 [DOI] [PubMed] [Google Scholar]
- Feng SH, Martinez C, Gusmaroli G, Wang Y, Zhou JL, Wang F, Chen LY, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham JE, Borner A, Worland AJ, Gale MD. (1997) Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J Agric Sci 128: 11–25 [Google Scholar]
- Fu X, Richards DE, Ait-Ali T, Hynes LW, Ougham H, Peng J, Harberd NP. (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14: 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Marshall GA. (1973) Insensitivity to gibberellin in dwarf wheats. Ann Bot (Lond) 37: 729–735 [Google Scholar]
- Gale MD, Youssefian S. (1985) Dwarfing genes in wheat. Russell GE, , Progress in Plant Breeding. Butterworths, London, pp 1–35 [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV. (2002) Gibberellin signaling in barley aleurone cells: control of SLN1 and GAMYB expression. Plant Physiol 129: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. (2003) The genes of the Green Revolution. Trends Genet 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. (2010) Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22: 2680–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogendoorn J, Pfeiffer WH, Rajaram S, Gale MD. (1988) Adaptive aspects of dwarfing genes in CIMMYT germplasm. Proceedings of the Seventh International Wheat Genetics Symposium, Cambridge, UK, July 13–19, 1988 Institute of Plant Science Research, Cambridge, UK, pp 1093–1100 [Google Scholar]
- Hoogendoorn J, Rickson JM, Gale MD. (1990) Differences in leaf and stem anatomy related to plant height of tall and dwarf wheat (Triticum aestivum L). J Plant Physiol 136: 72–77 [Google Scholar]
- Hynes LW, Peng J, Richards DE, Harberd NP. (2003) Transgenic expression of the Arabidopsis DELLA proteins GAI and gai confers altered gibberellin response in tobacco. Transgenic Res 12: 707–714 [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M. (2003) Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J 35: 104–115 [DOI] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP. (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Meng FR, Yin J, Liu HY, Si ZF, Ni ZF, Sun QX, Ren J, Niu HB. (2008) Isolation and comparative expression analysis of six MBD genes in wheat. Biochim Biophys Acta 1779: 90–98 [DOI] [PubMed] [Google Scholar]
- Muangprom A, Thomas SG, Sun TP, Osborn TC. (2005) A novel dwarfing mutation in a Green Revolution gene from Brassica rapa. Plant Physiol 137: 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Pestsova EG, Korzun V, Borner A. (2008) Validation and utilisation of Rht dwarfing gene specific markers. Cereal Res Commun 36: 235–246 [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinthus MJ, Gale MD, Appleford NEJ, Lenton JR. (1989) Effect of temperature on gibberellin (GA) responsiveness and on endogenous GA1 content of tall and dwarf wheat genotypes. Plant Physiol 90: 854–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebetzke GJ, Richards RA, Fettell NA, Long M, Condon AG, Forrester RI, Botwright TL. (2007) Genotypic increases in coleoptile length improves stand establishment, vigour and grain yield of deep-sown wheat. Field Crops Res 100: 10–23 [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Ross JJ. (1994) Recent advances in the study of gibberellin mutants. Plant Growth Regul 15: 193–206 [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T. (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slafer GA, Abeledo LG, Miralles DJ, Gonzalez FG, Whitechurch EM. (2001) Photoperiod sensitivity during stem elongation as an avenue to raise potential yield in wheat. Euphytica 119: 191–197 [Google Scholar]
- Sparks CA, Jones HD. (2009) Biolistics transformation of wheat. Methods Mol Biol 478: 71–92 [DOI] [PubMed] [Google Scholar]
- Sun TP. (2010) Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol 154: 567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YIC, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Riet L, Nagaraj V, Van den Ende W, Clerens S, Wiemken A, Van Laere A. (2006) Purification, cloning and functional characterization of a fructan 6-exohydrolase from wheat (Triticum aestivum L.). J Exp Bot 57: 213–223 [DOI] [PubMed] [Google Scholar]
- Vaughan SP, James DJ, Lindsey K, Massiah AJ. (2006) Characterization of FaRB7, a near root-specific gene from strawberry (Fragaria x ananassa Duch.) and promoter activity analysis in homologous and heterologous hosts. J Exp Bot 57: 3901–3910 [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. (1993) Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74: 205–214 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland AJ. (1986) Gibberellic-acid insensitive dwarfing genes in southern European wheats. Euphytica 35: 857–866 [Google Scholar]
- Worland AJ, Petrovic S. (1988) The gibberellic-acid insensitive dwarfing gene from the wheat variety Saitama-27. Euphytica 38: 55–63 [Google Scholar]
- Wu J, Kong X, Wan J, Liu X, Zhang X, Guo X, Zhou R, Zhao G, Jing R, Fu X, Jia J. (2011) Dominant and pleiotropic effects of a GAI gene in wheat results from lack of interaction between DELLA and GID1. 157: 2120–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Youssefian S, Kirby EJM, Gale MD. (1992) Pleiotropic effects of the GA-insensitive Rht dwarfing genes in wheat: effects on leaf, stem, ear and floret growth. Field Crops Res 28: 191–210 [Google Scholar]