Abstract
Reactive oxygen species (ROS) are ubiquitous signaling molecules in plant stress and development. To gain further insight into the plant transcriptional response to apoplastic ROS, the phytotoxic atmospheric pollutant ozone was used as a model ROS inducer in Arabidopsis (Arabidopsis thaliana) and gene expression was analyzed with microarrays. In contrast to the increase in signaling via the stress hormones salicylic acid, abscisic acid, jasmonic acid (JA), and ethylene, ROS treatment caused auxin signaling to be transiently suppressed, which was confirmed with a DR5-uidA auxin reporter construct. Transcriptomic data revealed that various aspects of auxin homeostasis and signaling were modified by apoplastic ROS. Furthermore, a detailed analysis of auxin signaling showed that transcripts of several auxin receptors and Auxin/Indole-3-Acetic Acid (Aux/IAA) transcriptional repressors were reduced in response to apoplastic ROS. The ROS-derived changes in the expression of auxin signaling genes partially overlapped with abiotic stress, pathogen responses, and salicylic acid signaling. Several mechanisms known to suppress auxin signaling during biotic stress were excluded, indicating that ROS regulated auxin responses via a novel mechanism. Using mutants defective in various auxin (axr1, nit1, aux1, tir1 afb2, iaa28-1, iaa28-2) and JA (axr1, coi1-16) responses, ROS-induced cell death was found to be regulated by JA but not by auxin. Chronic ROS treatment resulted in altered leaf morphology, a stress response known as “stress-induced morphogenic response.” Altered leaf shape of tir1 afb2 suggests that auxin was a negative regulator of stress-induced morphogenic response in the rosette.
Environmental challenges activate a complex signaling network initiating processes that ultimately determine plant stress tolerance. Both abiotic and biotic stresses cause increased production of reactive oxygen species (ROS), such as superoxide, hydrogen peroxide (H2O2), singlet oxygen, and hydroxyl radical. In addition to being hazardous by-products of metabolism, ROS are important ubiquitous signaling molecules with diverse roles depending on the specific ROS, their subcellular localization, and the stress in question. Plants have evolved sophisticated antioxidant systems to cope with increased ROS concentrations, but interestingly, they also possess enzymatic tools to themselves produce ROS for both intracellular and intercellular signaling purposes (Mittler et al., 2011). Apoplastic ROS can be produced by plasma membrane-localized NADPH oxidases (RESPIRATORY BURST OXIDASE HOMOLOGS [RBOHs]) and by cell wall peroxidases in response to several pathogens (Torres, 2010). ROS production by RBOHD is also induced by heat, wounding, salt stress, high light, and cold (Miller et al., 2009). The mechanisms by which cells sense extracellular ROS, leading to intracellular signaling, are not yet identified. The gaseous ROS ozone (O3) enters leaves through stomata and degrades in the apoplast into superoxide and H2O2, which also cause the activation of RBOHD and RBOHF (Joo et al., 2005; Vahisalu et al., 2010). Therefore, O3 can be used to deliver a precise and controlled apoplastic ROS burst for the study of signaling events shared by a multitude of stresses.
ROS-induced signaling is entwined with plant hormonal responses. Ethylene (ET) biosynthesis is an early O3 response, and later, salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) are produced (Overmyer et al., 2005). ET and SA signaling promote enhanced ROS production and programmed cell death (PCD), which all together form a self-amplifying loop. JA attenuates this cycle by reducing ROS production downstream of ET and cell death. This form of PCD has relevance to both abiotic stress symptom formation and resistance to biotic stress (Overmyer et al., 2000). ABA is important especially as the regulator of stomatal closure and O3 entry (Vahisalu et al., 2008, 2010; Brosché et al., 2010). Recently, also the connections between oxidative stress and the classical plant hormone auxin have gained attention. Defects in the antioxidative capacity of a thioredoxin and glutathione mutant resulted in altered auxin homeostasis and development (Bashandy et al., 2010). Iglesias et al. (2010) have shown that auxin receptor mutants were more tolerant to H2O2, methyl viologen (paraquat [PQ]), and salinity stress. Suppression of auxin signaling mediates pathogen tolerance via SA-auxin antagonism (Wang et al., 2007) or pathogen-inducible microRNA393 (miR393), which targets several auxin receptors for degradation (Navarro et al., 2006). Expression of auxin-responsive genes is decreased by H2O2 treatment via mitogen-activated protein kinase activation (Kovtun et al., 2000). Ultimately, prolonged stress exposure leads to altered growth patterns, including more compact growth, reduced cell division, and increased lateral growth (Potters et al., 2007, 2009). This response, termed “stress-induced morphogenic response” (SIMR), is proposed to be regulated through interaction between ROS and auxin; however, the molecular mechanisms governing SIMR are not well defined (Potters et al., 2009). Some Arabidopsis (Arabidopsis thaliana) mutants (e.g. rcd1) have been proposed to exhibit constitutive SIMR and may offer a tool to study the mechanisms regulating SIMR (Teotia et al., 2010).
Full genome array analysis of abiotic stress-treated plants allows the possibility to uncover new aspects of plant adaptation to stress (Urano et al., 2010). Transcriptomics studies have been previously performed on O3-exposed Arabidopsis (Ludwikow and Sadowski, 2008); however, they lacked temporal resolution and/or full genome arrays. In this study, an array representing 21,071 genes was used to get a complete view of O3-induced changes in gene expression across five time points. The transcriptomic data revealed that various aspects of auxin homeostasis and signaling were modified by apoplastic ROS. This provides a comprehensive view on how a ROS-auxin interaction could regulate plant responses, including SIMR. A rapid and transient decrease in the activity of the auxin-sensitive synthetic reporter promoter DR5 suggests that ROS regulate auxin signaling time dependently. The O3-derived changes in the expression of auxin signaling genes partially overlapped with pathogen responses and SA signaling, but detailed analysis revealed these to be mechanistically distinct. The role of auxin signaling, biosynthesis, inactivation, and transport in ROS responses was further studied in acute and chronic oxidative stress causing PCD and SIMR, respectively.
RESULTS
Gene Expression of Ecotype Columbia in Response to O3
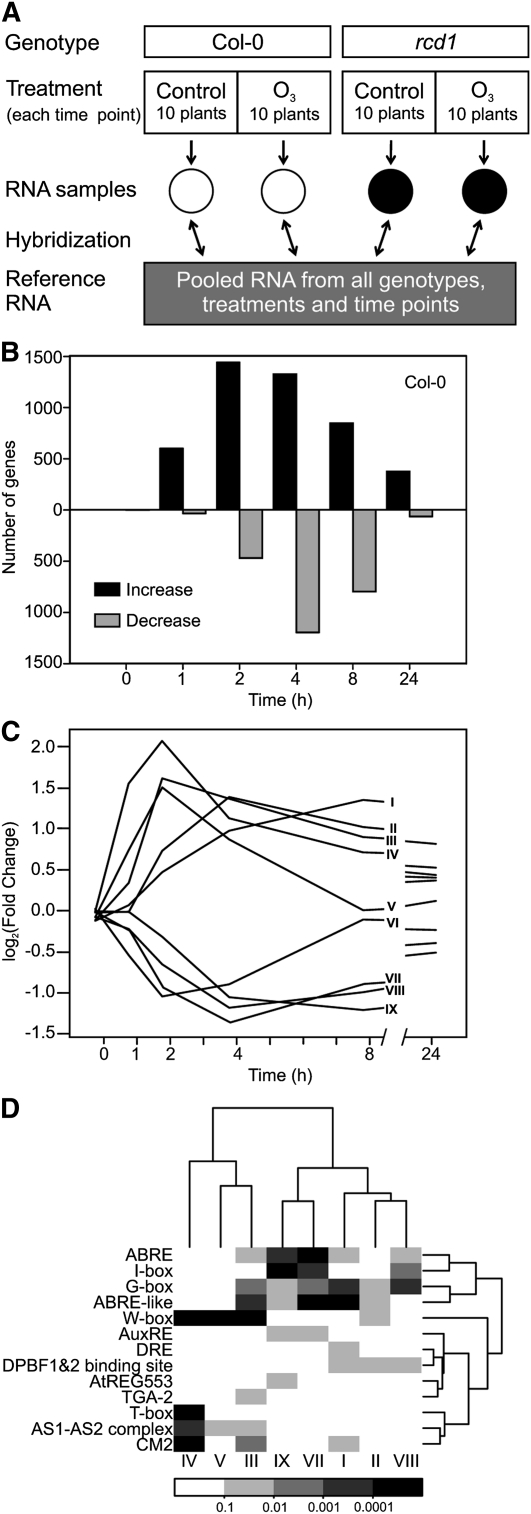
Plant transcriptional responses to apoplastic ROS formation were elucidated in a time series array experiment using O3 as a tool to produce an apoplastic ROS burst. ROS-induced changes in transcript levels were analyzed before (0 h), during (1, 2, and 4 h), and after (8 and 24 h) the 6-h 350 nL L−1 O3 treatment, which allowed the separation of both early and late responses. The experimental design utilized a common reference sample of pooled RNA from all time points of O3-tolerant ecotype Columbia (Col-0) and O3-sensitive rcd1 (Overmyer et al., 2000). Three biological repeats and a technical dye-swap replicate for every sample were hybridized against the common reference (Fig. 1A). Identification of O3-regulated genes was done by fitting a linear mixed model to each gene using the genotype, time, and treatment as explanatory variables, followed by computation of contrasts: fold changes with respect to the same time point at control conditions, and their statistical significance. One benefit of the repeated measures design (i.e. the time course) is that differential gene expression can be measured more accurately. In Col-0, 3,635 genes had at least 2-fold higher or lower mRNA abundance at single or multiple time points when compared with their respective clean-air controls (Supplemental Table S1). Using this criterion, transcript levels of only two genes differed at 0 h between the control and O3 treatments (Supplemental Table S1), validating the experimental setup and analysis work flow. O3-regulated genes identified in this study were compared with other publicly available array data, including two experiments with the Affymetrix platform (ATH1) representing altogether three time points (Supplemental Fig. S1). Strikingly, 2,211 genes were O3 responsive only in our data, of which a subset (295 genes) could be attributed to different array platforms (i.e. they are not present on the ATH1 chip; Supplemental Fig. S1). Late time points (8 and 24 h) present only in this study included 315 and 75 transcripts, respectively, regulated exclusively at these time points (Supplemental Table S1), which suggested that the majority of novel genes responsive to apoplastic ROS identified in this study were due to multiple (early) sampling time points together with biological and technical repeats and robust statistical analysis. Expression of only 98 genes was increased and that of three genes was decreased by apoplastic ROS at all time points from 1 to 24 h (Supplemental Table S1), which indicated a dynamic temporal regulation of genes throughout the time points studied. ROS-induced gene expression of the rcd1 mutant will be discussed elsewhere.
Figure 1.
Apoplastic ROS-responsive genes in a time series microarray experiment. A, Diagram showing the experimental setup. The experiment described was repeated three times. B, Number of genes with at least 2-fold expression changes (log2 ratio ± 1, q < 0.05) in O3-treated Col-0 compared with control plants per each time point (0, 1, 2, 4, 8, and 24 h). Altogether, 3,635 transcripts were responsive to apoplastic ROS. C, Clustering of genes responsive to apoplastic ROS across all time points identified nine expression profiles. Profiles I to V included genes with mainly increased expression, and profiles VI to IX included genes with mainly decreased expression. D, Promoter motif analysis for the nine expression profiles identified within 500-bp promoters of O3-regulated genes. The color scale represents statistical significance. Motif sequences are provided in Supplemental Table S3.
The number of genes with altered transcript levels varied between time points. Especially at 1 h, but also at 2 and 24 h, increased expression was more common than decreased expression, whereas at 4 and 8 h, both were equally present (Fig. 1B). Clustering revealed that the apoplastic ROS-responsive transcriptome could be clustered into nine main expression profiles (Fig. 1C), of which profiles I to V comprise genes with increased expression and profiles VI to IX comprise genes with decreased expression (Fig. 1C). These profiles were analyzed for Gene Ontology (GO) enrichment (Supplemental Table S1). Profile I expression peaked late (8 h) and remained slightly elevated at 24 h. Characteristic of this profile were biological processes related to ubiquitin-dependent protein degradation and various abiotic stress responses, such as responses to salt and osmotic stress traditionally associated with the stress hormone ABA. Profile IV included the most rapidly regulated genes and had 74 enriched biological processes, which included several types of protein modifications (lipidation, myristoylation, and phosphorylation), signaling and response to stress, response to bacterium, and regulation of immune responses. One of the first responses detected after bacterial infection or treatment of plants with pathogen-associated molecular patterns, such as flg22, is an apoplastic ROS burst (Torres, 2010). The biological processes represented in profile IV suggest that the early ROS burst in bacterial infection and O3-derived ROS formation have similar signaling roles. Profiles II and III had somewhat lower expression and later peaks compared with profile IV. They were enriched in biological processes for secondary metabolism. Profile V included biological processes related to pollen and cell recognition. Recently, ROS have emerged as regulators of pollen tube growth (Potocký et al., 2007). Profile VI contained genes with rapidly decreased expression that at 8 h had returned to the basal level. The biological processes in this group of genes related to ion homeostasis, developmental growth, and auxin stimulus. Profiles VII, VIII, and IX had decreased expression at 4 to 24 h and included photosynthesis and other chloroplast-related processes. Additionally, profile VIII was enriched for processes related to glucosinolate metabolism. Genes belonging to each expression profile and the GO enrichment results are listed in Supplemental Table S1.
Time-Point Analysis of Biological Processes Affected by O3 Treatment
To further investigate O3-induced changes in gene expression, we analyzed biological processes enriched at each time point (Supplemental Table S2). Altogether, 502 biological processes were enriched among the genes showing increased expression in O3-exposed plants, whereas 301 biological processes were enriched among genes with decreased expression (Supplemental Table S2). Within the set of genes exhibiting increased transcript levels, enrichment was seen consistently across all time points for 81 biological processes assigned to biotic and abiotic stresses, such as defense response, response to osmotic stress, response to oxidative stress, response to temperature stimulus, and response to chitin (Supplemental Table S2). In contrast, for genes with decreased transcript levels, there was no biological process enrichment spanning all time points and only photosynthesis-related processes were enriched in four out of the five time points (Supplemental Table S2). Response to auxin stimulus and the partially overlapping process of cell morphogenesis were identified as the only enriched biological processes among genes with decreased transcript levels at 1 h (Table I), consistent with profile VI (Fig. 1C). Consequently, GO categories associated with plant hormones were studied in more detail. The variation in the number of altered transcripts for hormone-responsive genes throughout the time course was hormone specific (Table I). ABA-responsive genes were the largest group of hormone-related genes with elevated expression levels. The timing and direction of ET, SA, and JA response activation were consistent with previous work (Overmyer et al., 2003). Novel hormone responses were also detected; the regulation of brassinolide (BR) and GA responses by O3 have not been reported previously. Auxin-responsive transcripts were the largest group of hormone-regulated genes with decreased expression (Table I).
Table I. Plant hormone responses are elicited by apoplastic ROS.
The number of apoplastic ROS-regulated genes belonging to plant hormone response categories are shown. GO enrichment analysis was performed for genes with increased (log2 ratio of 1, q < 0.05) or decreased (log2 ratio of −1, q < 0.05) expression in O3-treated Col-0. The enrichment was calculated separately for each time point (0, 1, 2, 4, 8, and 24 h), and no enriched processes were present in 0-h samples (data not shown). Statistically significant (q < 0.05) numbers of O3-responsive genes annotated to a given hormone response category are depicted with asterisks and boldface. The number of individual apoplastic ROS-regulated genes belonging to each hormone response category across all time points is shown for elevated (↑) and decreased (↓) expression in separate rows. CK, Cytokinin; IAA, auxin.
| Hormone | GO Identifier | Genes on Array | O3 Response |
||||||
| Genes (%) | 1 h | 2 h | 4 h | 8 h | 24 h | ||||
| ABA | 0009737 | 314 | ↑ | 78 (25%) | 26* | 53* | 52* | 33* | 18* |
| ↓ | 22 (7%) | 0 | 10 | 16 | 10 | 1 | |||
| BR | 0009741 | 48 | ↑ | 9 (19%) | 6* | 6 | 5 | 4 | 2 |
| ↓ | 7 (15%) | 0 | 5* | 7 | 0 | 0 | |||
| CK | 0009735 | 67 | ↑ | 4 (6%) | 0 | 2 | 3 | 3 | 1 |
| ↓ | 7 (10%) | 0 | 4 | 6 | 4 | 0 | |||
| ET | 0009723 | 123 | ↑ | 29 (24%) | 12* | 22* | 14 | 7 | 7* |
| ↓ | 6 (5%) | 1 | 3 | 5 | 3 | 0 | |||
| GA | 0009739 | 105 | ↑ | 8 (8%) | 2 | 2 | 5 | 4 | 4 |
| ↓ | 14 (13%) | 1 | 8* | 10 | 7 | 1 | |||
| IAA | 0009733 | 234 | ↑ | 36 (15%) | 8 | 22 | 19 | 13 | 4 |
| ↓ | 34 (15%) | 7* | 15* | 25* | 16 | 4 | |||
| JA | 0009753 | 139 | ↑ | 39 (28%) | 12* | 30* | 32* | 19* | 19* |
| ↓ | 13 (9%) | 1 | 8 | 11 | 5 | 0 | |||
| SA | 0009751 | 127 | ↑ | 36 (29%) | 13* | 24* | 29* | 24* | 12* |
| ↓ | 7 (6%) | 1 | 4 | 4 | 3 | 1 | |||
Promoter Elements Mediating the Transcriptional Response to Apoplastic ROS
Promoter analysis was used to explore the role of cis-elements in the regulation of apoplastic ROS-mediated gene expression. A list of confirmed Arabidopsis promoter elements from the databases AGRIS (Yilmaz et al., 2011), PlantCARE (Lescot et al., 2002), and PLACE (Higo et al., 1999), together with a few added elements from the literature (for details, see “Materials and Methods”), was used for the analysis of 500-bp promoter fragments of genes regulated at each individual time point (Supplemental Table S3). The most abundant enriched element was the W-box element TTGAC (Supplemental Table S3), a target for WRKY transcription factors, which are key regulators of abiotic and biotic stress responses (Rushton et al., 2010). Due to the large number of enriched elements (48) and the redundancy in their regulatory sequence, a subset of promoter elements was chosen for a more detailed analysis within 500-bp promoters of genes in profiles I to IX (Fig. 1C). The W box was enriched in the fast and highly induced expression profiles III to V, which are enriched for responses to various biotic stresses (Fig. 1, C and D). O3 exposure leads to elevated ABA concentration, particularly at the late time point 8 h (Overmyer et al., 2008). The role of this increase in ABA has remained obscure. The promoter analysis revealed that profiles with expression changes at late time points (both increased and decreased expression) were enriched for the ABA response element (ABRE) and ABRE-like element (Fig. 1, C and D). Consistent with this, profile I had biological processes enriched for salt and osmotic stress, which are regulated through ABA signaling. Overall, this suggests that late responses in gene expression to apoplastic ROS could be regulated through increased ABA concentration and ABA signaling.
Transcriptional Regulation of the Auxin Signaling Pathway by Apoplastic ROS
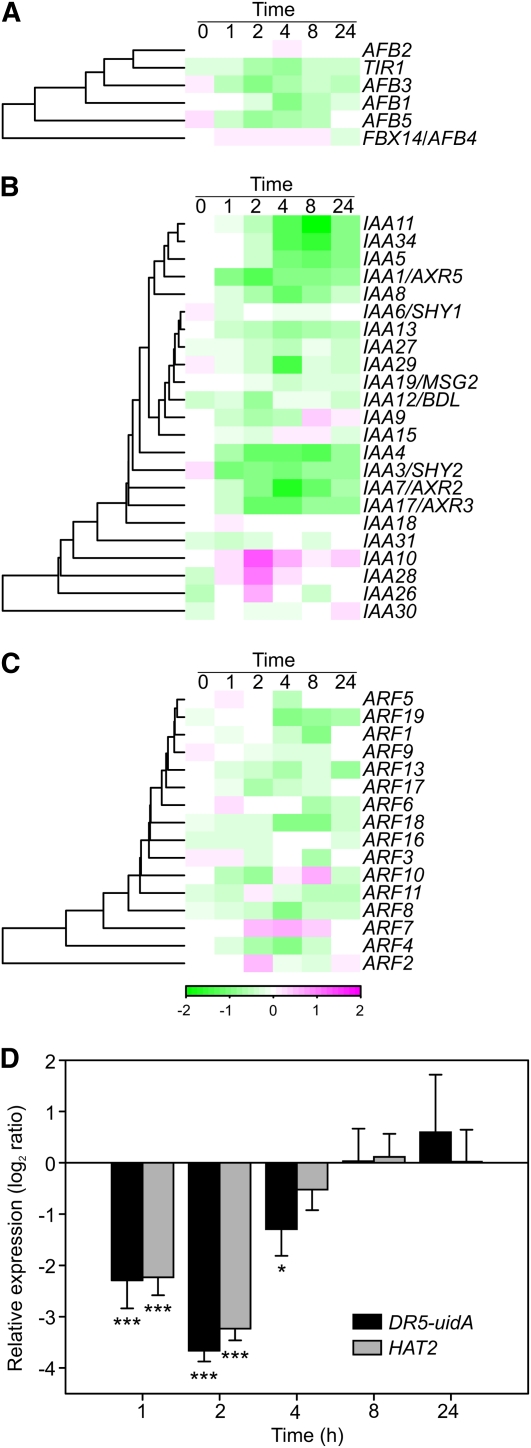
The transient decrease in the expression of auxin-related genes in O3-treated plants (Fig. 1C; Table I) prompted a more detailed study of the auxin signaling pathway. Of the TIR1-AFB auxin receptor gene family (Dharmasiri et al., 2005), TIR1, AFB1, AFB3, and AFB5 had decreased transcript levels in response to O3 (Fig. 2A). Transcript levels for Auxin/Indole-3-Acetic Acid (Aux/IAA; Liscum and Reed, 2002) genes were mainly decreased, with two exceptions being IAA10 and IAA28, whose transcript levels transiently increased at 2 h (Fig. 2B). The expression of genes encoding ARF transcription factor proteins (Okushima et al., 2005; Guilfoyle and Hagen, 2007), which mediate auxin-responsive gene expression via the Auxin-Responsive Element (AuxRE), was only marginally affected by apoplastic ROS, in both directions with no consistent trends (Fig. 2C). Signaling downstream of ARFs was studied using the auxin-responsive synthetic promoter DR5, which contains seven repeats of AuxRE, fused to the uidA reporter gene (Ulmasov et al., 1997). The uidA transcript levels were monitored with real-time quantitative reverse transcription (qRT)-PCR. An early decrease in DR5-driven uidA transcript abundance was consistently detected in O3-treated plants, similar to the expression pattern of the auxin-responsive marker gene HAT2 (Fig. 2D). The DR5-uidA and HAT2 expression levels were partially recovered already at 4 h, during the O3 treatment, and were indistinguishable from the controls by 8 h (Fig. 2D). This indicates that these AuxRE-dependent transcripts are transiently reduced in response to O3 treatment. In 3-week-old rosettes, the DR5-driven accumulation of the GUS activity encoded by DR5-uidA was distinctly localized in hydathodes and young leaves, consistent with the localization of auxin biosynthesis in leaves (Supplemental Fig. S2; Teale et al., 2006). O3 treatment caused no change in the staining pattern, as detected by histochemical GUS staining (Supplemental Fig. S2).
Figure 2.
Auxin signaling is altered by apoplastic ROS. A to C, O3 responses (log2 ratio) of TIR1-AFB receptors (A), Aux/IAA genes (B), and ARFs (C). D, Relative expression of DR5-uidA and HAT2 in O3-treated DR5-uidA plants was determined with qPCR. Averages from three biological replicates are shown, and error bars represent sd. Asterisks depict statistically significant differences from 0 (* P < 0.05, *** P < 0.001). DR5-uidA and HAT2 expression levels remained unchanged across time points in clean air (data not shown).
To further characterize the connection between O3-altered gene expression and auxin signaling, we identified auxin-regulated genes from several publicly available microarray data sets, which utilized different auxin concentrations and time points (for the data sets used, see “Materials and Methods”). One hundred seventy-nine genes were identified as regulated by auxin, and of these, 60 genes were at least 2-fold regulated by both auxin and apoplastic ROS. Thirty-six genes were regulated by auxin and apoplastic ROS in the same direction (increased expression by both treatments or decreased expression by both treatments), whereas 24 genes showed an inverse regulatory pattern (increased expression by one treatment type and decreased expression by the other). Several Aux/IAA and SAUR genes, the HAT2 transcription factor, and the auxin efflux carrier PIN3 were among the transcripts belonging to the latter category.
Publicly available array data were then clustered to gain further information on how these 60 auxin- and O3-responsive genes were regulated by different ROS, hormones, and abiotic and biotic stress treatments (Fig. 3A). Also included in the clustering were additional Aux/IAA genes not regulated by auxin treatment and five O3 marker genes selected from Wrzaczek et al. (2010). Three major clusters (I–III) were identified. Cluster Ia contained mainly Aux/IAA genes not regulated by auxin treatment and mixed responses to O3 treatment. Cluster Ib included genes with expression reduced by auxin, the auxin transport inhibitor TIBA, the SA analog benzothiadiazole S-methylester (BTH), O3, and other ROS-related treatments, such as methyl viologen (PQ), and H2O2. Cluster II included genes with expression increased by auxin but decreased by O3, PQ, H2O2, BTH, and TIBA. Cluster III included genes induced by all treatments (Fig. 3A). Among abiotic stress treatments, UV-B fell into the same clusters with O3, whereas heat, cadmium, and to some extent salt stress exhibited distinct expression patterns. Several biotic stress treatments had expression profiles similar to O3, although with weaker expression levels. Treatment with the translation inhibitor cycloheximide showed a similar expression profile as auxin treatment (Fig. 3A), suggesting that the changes observed in gene expression for this group of auxin-responsive genes were mediated by the altered stability or activity of some component in the auxin signaling pathway (i.e. Aux/IAA proteins or ARFs) instead of de novo synthesis of a new regulator.
Figure 3.
Regulation of auxin-related genes by plant hormones and biotic and abiotic stress. A, Clustering of transcripts regulated by auxin and apoplastic ROS. Sixty O3 and auxin-regulated transcripts (log2 ratio ± 1, q < 0.05) were identified from publicly available microarray experiments. In addition, apoplastic ROS-responsive Aux/IAA transcripts and five stress-responsive marker genes not regulated by auxin were included in the analysis (marked with asterisks). The number of AuxRE (TGTCnC) and TGACG elements was calculated within 1,000-bp promoter regions. Putative transcriptional regulation by ARFs is indicated with the corresponding ARF number. +, Increased expression; −, decreased expression; NR, not regulated. B, Expression of selected auxin or apoplastic ROS-regulated genes in Col-0, ein2, NahG, npr1, and sid2 in clean air (black bars) and after 2 h of 350 nL L−1 O3 (gray bars) determined with qPCR. Expression levels are normalized to ACTIN2. Means of two to three biological replicates are shown ± sd. Asterisks depict statistically significant differences from Col-0 in clean air (black bars) and from O3-treated Col-0 (gray bars) at P < 0.05.
Looking closer at individual genes, the expression of several Aux/IAA genes (IAA1, IAA3, IAA4, IAA5, IAA11, and IAA29) was enhanced by auxin and decreased by ROS treatments. In contrast, the expression of IAA10 and IAA28 was enhanced by ROS-inducing treatments, especially at early time points, but was poorly responsive to auxin. Cluster Ia included also IAA7, IAA8, IAA17, and IAA34, which also lacked regulation by auxin but were decreased at late time points by O3 treatment. To study the role of AuxRE-mediated regulation of transcription, the number of AuxREs in the promoters of transcripts regulated by auxin and O3 was determined (Fig. 3A) and compared with known target genes of the activator ARFs: ARF5, ARF6, ARF7, ARF8, and ARF19 (Nagpal et al., 2005; Okushima et al., 2005; Schlereth et al., 2010). Surprisingly, 33% of auxin-responsive genes did not contain an AuxRE, not even the more general AuxRE sequence (TGTCnC), in their 1-kb promoter. Because this group of genes also included seven putative targets of transcriptional activator ARFs (Fig. 3A), the promoters of these genes were checked for the presence of AuxREs even farther upstream. Indeed, 3 kb of all these genes contain at least one copy of the AuxRE (data not shown). Genes regulated by ARFs were predominantly found among the genes induced by auxin and repressed by apoplastic ROS (cluster II), possibly due to enhanced stability of a negative regulator (i.e. Aux/IAA in O3-treated plants). However, targets of these ARFs were to a lesser extent also found in the other clusters (Fig. 3A).
Apoplastic ROS Regulation of Auxin Signaling Does Not Involve SA, ET, miR393, IAA28, or Mitogen-Activated Protein Kinases
Treatment with the SA analog BTH reduces the expression of certain auxin-related genes (Wang et al., 2007; Fig. 3A, cluster II). Mutants with altered SA biosynthesis, accumulation, or signaling were used to assess the role of SA in modulating the expression of auxin-related genes in O3-treated plants. ISOCHORISMATE SYNTHASE1 (ICS1), which is disrupted in the sid2 mutant, is required for SA biosynthesis in response to O3 treatment (Ogawa et al., 2007). NahG carries the SA-degrading bacterial salicylate hydroxylase gene. The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) transcriptional coregulator is a central component of SA signaling (Dong, 2004). In addition, the role of ET was evaluated using the ethylene insensitive2 (ein2) mutant. Expression of the auxin receptors TIR1, AFB1, AFB3, and AFB5 and the auxin-inducible genes SAUR16, SAUR68, and HAT2 was studied by qPCR in Col-0, npr1, sid2, NahG, and ein2. All the genes studied exhibited decreased expression levels 2 h after the start of the O3 treatment in Col-0, similar to the microarray results obtained (Fig. 3B). This response was not changed in ein2, npr1, or sid2 mutants, which suggests that neither ET or SA signaling nor O3-induced SA biosynthesis was involved in this decline in expression (Fig. 3B). The decreased expression of auxin receptors, especially TIR1 and AFB5, was slightly compromised but not absent in NahG plants (Fig. 3B). Treatment of plants with the elicitor flg22 leads to increased levels of miR393 and subsequent cleavage of TIR1, AFB2, and AFB3 transcripts (Navarro et al., 2006). In contrast to flg22 treatment, the expression of the miR393 precursor transcript premiR393b and the mature miR393 decreased after the start of the O3 treatment in Col-0 (Supplemental Figs. S3 and S4). Another precursor, premiR393a, did not respond to O3 (Supplemental Fig. S3). Since premir393a is induced in flg22-treated plants prior to the increase of miR393 levels (Navarro et al., 2006), this result suggested that the decrease of TIR1 expression in response to flg22 and apoplastic ROS in Col-0 occurs via different mechanisms. The mitogen-activated protein kinase kinase kinase ANP1 is a proposed link between ROS signaling and auxin signaling, acting through MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3) and MPK6 (Kovtun et al., 2000). Neither of these MPKs was involved in the decreased expression of TIR1, SAUR68, or HAT2 (Supplemental Fig. S5). Based on mutant analysis, MPK4 is the proposed regulator of ROS and auxin signaling (Nakagami et al., 2006). Unfortunately, the dwarf phenotype of the mpk4 mutant makes it difficult to use in accurate measurements of O3 responses.
The induction pattern of IAA28 (Fig. 2B) suggested that it might act as a negative regulator of auxin-related genes under ROS treatment. The expression of the marker gene HAT2 was tested in two mutant alleles, gain-of-function iaa28-1 (Rogg et al., 2001) and loss-of-function iaa28-2 (SALK_129988C), of IAA28 exposed to O3 (Supplemental Fig. S6). No differences were found between IAA28 mutants and their respective controls. In conclusion, SA, ET, miR393, IAA28, MPK3, and MPK6 signaling do not appear to be involved in O3-mediated reduction in the expression of auxin-related genes.
Apoplastic ROS Regulation of Genes with Increased Expression
The expression of DR5-uidA and HAT2 in response to apoplastic ROS indicated a role for AuxRE and auxin signaling in the regulation of genes with transient decreased expression (Fig. 2D). HAT2 belongs to cluster II (Fig. 3A), characterized by expression increased by auxin and decreased by ROS. In contrast, cluster III genes were increased by both treatments as well as the SA analog BTH. These treatments all increase the expression of the marker gene GST6 through ocs and TGACG elements (Chen et al., 1996; Chen and Singh, 1999). The TGACG element is a target for TGA-type transcription factors and NPR1 (Zhou et al., 2000). To test if genes in cluster III were regulated via NPR1, SA, or ET, four genes from this cluster (PBP1, WRKY40, ZAT10, and ACS6) and GST6 were tested for apoplastic ROS regulation in ein2, sid2, npr1, and NahG (Fig. 3B). In clean-air control plants, SA, ICS1, and NPR1 were positive regulators and EIN2 was a negative regulator of PBP1 and WRKY40. In response to O3, the expression of PBP1 and GST6 was increased in npr1 compared with Col-0, indicating a negative role for SA signaling during O3, consistent with our previous model for the regulation of O3 gene expression (Wrzaczek et al., 2010). Transgenic NahG plants, which cannot accumulate SA, did not show the same O3 induction, indicating that a fine-tuned balance between signaling pathways will influence the outcome of apoplastic ROS-regulated gene expression.
Auxin Biosynthesis, Conjugation, and Transport in O3 Response
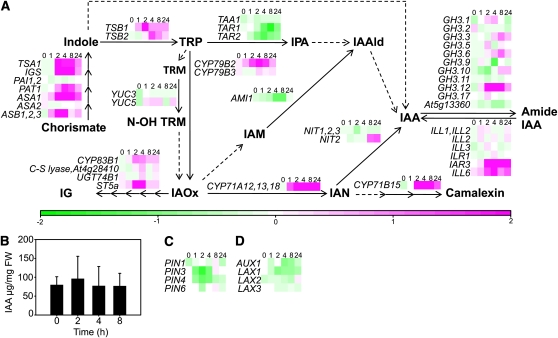
The concentration of biologically active free IAA is regulated by a multilevel network of biosynthesis, conjugation, and transport. Auxin biosynthesis can occur via several partially connected pathways, most of which are Trp dependent (Normanly, 2010). Several genes involved in Trp biosynthesis had increased expression in response to apoplastic ROS, whereas the downstream pathways varied in their response (Fig. 4A). Several Trp aminotransferases of the indole-3 pyruvic acid pathway had decreased expression, CYP79B2 in the indole-3-acetaldoxime pathway had increased expression, and YUCCA genes YUC3 and YUC5 of the tryptamine pathway were unresponsive to apoplastic ROS (Fig. 4A). IAA amido hydrolases (IAR3 and ILL6) had mainly increased expression, whereas GH3 genes (IAA amido conjugases) were regulated in both directions (Fig. 4A). Altogether, gene expression does not predict a clear outcome for the net effect of O3 exposure on auxin biosynthesis. To unambiguously determine the output of these pathways and to study possible direct auxin oxidation taking place, the concentration of free IAA was quantified from O3-treated Col-0. No difference in the concentration of biologically active free IAA was evident between clean-air and O3 treatments (2, 4, and 8 h; Fig. 4B). However, since the auxin measurements were performed on whole leaves, this does not exclude cell-specific alterations in auxin concentration. Decreased transcript levels for auxin efflux and influx carriers suggest that apolastic ROS may affect plants via modulating auxin polar transport (Fig. 4, C and D; Tognetti et al., 2011). The auxin transport inhibitor TIBA gave a similar expression profile as O3 treatment for auxin-responsive genes (Fig. 3A), supporting a role for auxin transport in the regulation of apoplastic ROS responses.
Figure 4.
Apoplastic ROS affects the expression of genes regulating auxin homeostasis without concomitant changes in IAA concentration. A, Expression of IAA biosynthesis genes, IAA amido conjugases, and IAA amido hydrolases. Biosynthesis genes and pathways are according to Normanly (2010): solid arrows indicate reactions with verified catalytic enzymes, whereas dashed arrows represent unknown metabolic steps. Multiple consecutive reactions are depicted with the respective number of arrows. The transcriptional response to apoplastic ROS is presented from all the genes with detectable expression levels. TRP = Trp; TRM = tryptamine; N-OH-TRM = N-hydroxyltryptamine; IPA = indole-3-pyruvic acid; IAAld = indole-3-acetaldehyde; IG = indole-3-methylglucosinolate; IAOx = indole-3-acetaldoxime; IAM = indole-3-acetamide; IAA = indole-3-acetic acid; IAN = indole-3-acetonitrile. All the verified (after Normanly, 2010) and putative genes involved in IAA conjugation (GH3 family) and hydrolysis are shown. B, The concentration of free IAA was quantified from Col-0 plants at 0, 2, 4, and 8 h after the onset of O3 exposure (350 nL L−1; 6 h). The experiment was repeated twice, and one representative experiment is shown. Error bars depict sd (n = 4–5). FW, Fresh weight. C, Expression of auxin efflux carriers is reduced by apoplastic ROS. D, Auxin influx carriers show a slight decrease in response to apolastic ROS.
Auxin and Cell Death
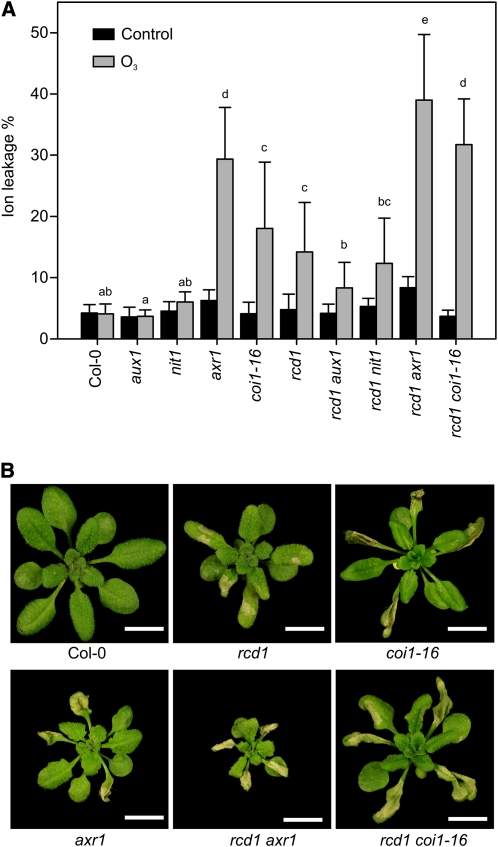
Short high-concentration O3 exposure activates PCD, resulting in tissue collapse and visible lesions in sensitive genotypes such as rcd1 (Overmyer et al., 2005). In tolerant genotypes, such as Col-0, this is restricted to a few individual cells (microlesions) with no visible damage. Several GO categories related to cell death and the regulation of cell death were significantly enriched in early O3-regulated genes (Supplemental Table S2). To investigate the role of auxin in ROS-triggered PCD, O3 sensitivity was assayed in several auxin-related mutants that do not have gross leaf abnormalities, which could obscure the analysis of stress responses. Furthermore, double mutants were constructed between rcd1 and mutants representing different aspects of auxin biology (synthesis, transport, and signaling). AUXIN RESISTANT1 (AXR1) encodes a RUB-conjugating enzyme regulating the SCF (for Skp1-Cullin-F box) activity necessary for auxin signaling (del Pozo et al., 2002). The F-box proteins TIR1 and COI1 are receptors for auxin and the bioactive JA conjugate JA-Ile, respectively, and both of their SCF complexes are targets for AXR1 regulation (Xu et al., 2002; Lorenzo and Solano, 2005). Since axr1 is defective in both auxin and JA responses (Tiryaki and Staswick, 2002), the coi1-16 mutant, which is specifically defective in only JA responses, was used to dissect auxin and JA signaling events. Both axr1 and coi1-16 were O3 sensitive, but axr1 had slightly higher levels of cell death, quantified as ion leakage (Fig. 5A). The rcd1 axr1 and rcd1 coi1-16 double mutants had additive effects on O3 sensitivity compared with the parental lines, suggesting that axr1 and coi1-16 affect different regulatory processes in PCD than rcd1 (Fig. 5A). Loss of auxin biosynthesis and influx in the nitrilase1-3 (nit1-3) and aux1-7 mutants, respectively, did not alter the ROS-induced cell death as single mutants in the tolerant Col-0 background or as double mutants in the sensitive rcd1 background (Fig. 5A). Furthermore, the auxin receptor double mutant tir1-1 afb2-3 and IAA28 mutants were not sensitive to an acute O3 exposure (data not shown). In addition to their ROS phenotypes, both axr1 and rcd1 have altered leaf morphology; interestingly, rcd1 axr1 plants were smaller than either of the parental lines, whereas rcd1 coi1-16 had a habitus similar to rcd1 (Fig. 5B). Thus, cell death and leaf morphology experiments differentiated between axr1 and coi1-16, suggesting that JA signaling is predominantly involved in the observed cell death responses and that auxin signaling is involved in the developmental responses.
Figure 5.
The axr1 mutant is sensitive to apoplastic ROS. A, O3-induced cell death was quantified as ion leakage. Three-week-old Col-0, aux-1, nit1, axr1, coi1-16, and rcd1 together with respective rcd1 double mutants were treated with O3 (6 h; 350 nL L−1) and harvested 8 h after the start of the experiment. Ion leakage was measured from control and O3-treated plants and calculated as percentage of total ions of each plant (n = 5). The experiment was repeated five times, and the results were analyzed with linear mixed models. Values shown represent mean cell death per genotype, and error bars indicate sd. Letters show statistical significance (P < 0.05) from posthoc analysis by computing contrasts from linear mixed models and subjecting the P values to single-step error correction. B, In addition to increased cell death, rcd1 axr1 plants have smaller size, whereas rcd1 coi1-16 plants do not show alterations in plant size compared with the parental lines. Plants were treated with O3 (6 h; 350 nL L−1), and photographs were taken 24 h after the start of the exposure. Bars = 1 cm.
ROS-Induced Morphological Responses
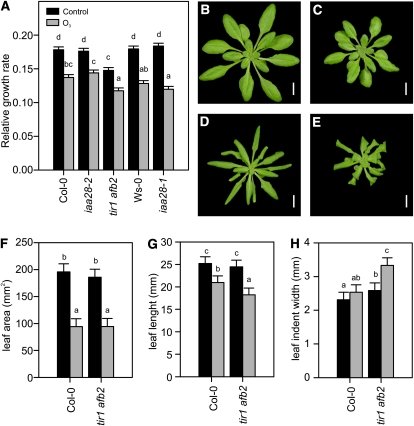
Chronic exposure to elevated O3 causes biochemical changes, reduced growth, and morphological changes in several plant species (Skärby et al., 2004; Li et al., 2006; Karnosky et al., 2007; Kontunen-Soppela et al., 2010; Street et al., 2011). Similarly, chronic exposure to several other stresses causes similar growth changes. These responses are adaptive, genetically programmed, and have recently been referred to as SIMR (Potters et al., 2007, 2009). To evaluate the long-term effects of apoplastic ROS-auxin interactions, the growth and morphology of plants were monitored in chronic O3 exposure to determine the role of auxin responses in ROS-induced SIMR. Changes in leaf morphology and the relative growth rate were used as indexes of SIMR. The auxin-insensitive receptor double mutant tir1-1 afb2-3 was used for these studies together with two mutant alleles of IAA28, an O3-inducible Aux/IAA gene (Figs. 2B and 3A; Supplemental Fig. S6). All genotypes responded to prolonged O3 treatment with a significantly decreased growth rate compared with their respective clean-air controls (Fig. 6A). The tir1 afb2 mutant exhibited a smaller rosette size and slower growth rate compared with Col-0 under both clean-air control and O3-exposed conditions (Fig. 6A). In addition to decreased growth, all genotypes exhibited changes in leaf morphology, which were documented photographically (Fig. 6, B–E) and quantified using the LAMINA leaf shape analysis software (Bylesjö et al., 2008; Fig. 6, F–H). The typical O3 response was epinastic curling in the leaf margin, as exemplified by Col-0 (Fig. 6, B and D) and quantified in the leaf area parameter (Fig. 6F). This phenotype was a specific O3 response, as it was not present in the clean-air controls. Similar to the other genotypes, the tir1 afb2 mutant exhibited curling in the leaf margins (Fig. 6, E and F); additionally, epinastic curling of the leaf tip and curvature of the mid vein were seen (Fig. 6E). Tip curling was quantified as reduced leaf length (Fig. 6G). Furthermore, tir1 afb2 was also unique in that its leaf morphology phenotypes were subtly present already in the clean-air controls (Fig. 6C), but to a much lesser extent than in O3-treated plants. This phenotype was quantified in the leaf indent width parameter (Fig. 6H). The assay used here to quantify rosette size may not report a true rosette diameter for tir1 afb2 due to its leaf phenotype. The reduced rosette size and growth rate reported in Figure 6A for tir1 afb2 are likely artificially underestimated due to leaf tip curling (Fig. 6, E and G). In support of this view, end point rosette size determinations of tir1 afb2 were significantly smaller than in Col-0, while fresh weight was not (data not shown). Therefore, the tir1 afb2 growth rate was likely not different from Col-0.
Figure 6.
Chronic O3 reduces plant growth. A, Two-week-old Col-0, iaa28-2, tir1 afb2, Ws-0, and iaa28-1 plants were exposed to O3 daily (6 h; 350 nL L−1). For growth rate analysis, rosette diameter was determined by finding the minimal circle that contained all leaves using ImageJ image-analysis software (http://rsbweb.nih.gov/ij/) before and after 7 d of O3 exposure. Relative growth rate was calculated by fitting a linear model to plant size (for details, see “Materials and Methods”). B to E, Four-week-old control clean-air and ozone-exposed plants. Under control clean-air conditions, Col-0 plants (B) are larger than tir1 afb2 plants (C), which have constitutively smaller size and slightly curled leaves. After 2 weeks of O3 exposure, Col-0 exhibits stunted growth and curled leaves (D). Leaf curling in response to O3 is more prominent in tir1 afb2 plants (E). Bars = 1 cm. F to H, Leaf shape parameters area (F), length (G), and mean indent width (H) were quantified using the LAMINA software (Bylesjö et al., 2008). All experiments were repeated twice with similar results, and the data were analyzed with linear models. Values shown represent mean relative growth rate, and error bars indicate sd of the linear model (n ≥ 6).
We conclude that chronic O3 decreased growth rate independent of auxin signaling. ROS-dependent growth control in the root has been previously shown to be independent of auxin (Tsukagoshi et al., 2010). In contrast, SIMR, measured as altered leaf morphology, was exaggerated in O3-treated tir1 afb2 mutant plants, suggesting that O3-induced SIMR is auxin regulated.
DISCUSSION
ROS are essential signaling molecules in plant development and in response to biotic and abiotic stresses (Jaspers and Kangasjärvi, 2010; Torres, 2010; Mittler et al., 2011). Unfortunately, the easy-to-use abbreviation “ROS” is in many cases also misleading, since individual ROS (i.e. O3, H2O2, superoxide, and singlet oxygen) have different outcomes on signaling pathways and the subcellular localization of ROS production (i.e. apoplast, cytosol, chloroplast, peroxisome, or mitochondria) can have a profound effect on the signaling pathways activated (Gadjev et al., 2006; Wrzaczek et al., 2010; Giraud et al., 2011). Here, we have used O3 to study the role of apoplastic ROS in signaling in an extended time series microarray experiment. Compared with previously published O3 array experiments, the use of multiple time points allowed the identification of far more apoplastic ROS-responsive transcripts (Supplemental Fig. S1), and expression profiles with GO enrichment analysis allowed the identification of novel apoplastic ROS-regulated biological processes (Fig. 1; Supplemental Tables S1 and S2).
The early response to apoplastic ROS consisted mainly of genes with increased expression and belonged to GO categories that represent signaling and defense (Supplemental Table S2). Interestingly, the GO category “regulation of transcription” was significant only at late time points, suggesting that the early changes in O3-induced gene expression were executed by preexisting components regulating the transcriptional response. Genes with a peak in expression at 8 h (profile I; Fig. 1C) were enriched for various processes related to protein degradation and abiotic stresses such as salt and osmotic stress. This indicates that two different processes are ongoing: a reorganization of the proteome by targeted degradation, and an activation of ABA signaling. The latter is supported by a large increase in ABA concentration at 8 h of O3 treatment (Overmyer et al., 2008) and enrichment of the ABRE in promoters of the genes in profiles I, III, and VII to IX (Fig. 1D). The role for increased protein degradation at 8 h could be to reset the system. Prolonged activation of defense responses is detrimental to plants (Jirage et al., 2001), and the massive increase of defense-related genes seen early (profiles III–V; Fig. 1C), could be offset by degradation of the translated proteins a few hours later.
Apoplastic ROS Rapidly Decrease Auxin Signaling
Several hormones are involved in the regulation of apoplastic ROS responses (Overmyer et al., 2003). Analysis of GO categories for each of the major plant hormones showed significant enrichment of genes related to SA, JA, ET, BR, and ABA (Table I) in ROS-induced genes. In contrast, the GO categories for GA and IAA (also BR) were enriched among genes with decreased expression (Table I). This suppression of genes belonging to the GO category “response to auxin stimulus” together with recent findings showing that auxin and ROS are regulators of plant development during stress (Potters et al., 2007, 2009; Tognetti et al., 2011) prompted us to study auxin signaling in more detail. The highly sensitive auxin reporter construct DR5-uidA indicated a 4-fold decrease in auxin response already at 1 h of O3 treatment (Fig. 2D). It is possible that this results from a decrease in auxin concentration due to decreased biosynthesis or increased inactivation via conjugation or even direct auxin oxidation during oxidative stress (Normanly, 2010). This was addressed by auxin measurements, and the method used is mass specific (i.e. detects the free active form but not conjugated or oxidized auxins). The concentration of free active auxin did not change, suggesting that auxin homeostasis is not involved. However, since only one form of auxin was measured from whole rosettes, the possibility of cell type-specific changes and increased auxin flux (i.e. simultaneous increases in both biosynthesis and inactivation) cannot be excluded. Alternatively, the rapid decrease in auxin response mentioned above may be conditioned by regulation at the protein level in the preformed auxin signaling cascade (i.e. the TIR1/AFB, Aux/IAA, and ARF proteins). Recent studies have shown that a decrease in auxin signaling is required for pathogen and abiotic stress tolerance. F-box auxin receptors play an important role in this desensitization process via several different mechanisms. The flagellin-triggered decrease in the expression of TIR1, AFB1, AFB2, and AFB3 takes place via both miR393-dependent and independent mechanisms (Navarro et al., 2006). SA signaling decreased the expression of auxin-responsive genes and TIR1 likely through the stabilization of Aux/IAA proteins independent of miR393 (Wang et al., 2007). Our results suggest a novel miR393- and SA-independent mechanism that modulates auxin signaling in response to apolastic ROS. Different tir1 afb double mutant combinations offered tolerance to salinity and decreased accumulation of ROS (Iglesias et al., 2010), and similarly, tir1 and tir1 afb2 mutants in the Col-0 background were as O3 tolerant as Col-0 (data not shown). The axr1 mutant, which is defective in both SCFTIR/AFB and SCFCOI1 complex function (Tiryaki and Staswick, 2002), was sensitive to apolastic ROS, apparently due to its JA insensitivity phenotype. However, the lack of JA signaling in axr1 is only partial, and Llorente et al. (2008) reported enhanced expression levels of JA marker genes TAT1 and LOX3 in axr1 mutants in response to pathogen infection. The rcd1 mutant has been proposed to exhibit constitutive SIMR (Teotia et al., 2010). Importantly, rcd1 axr1 had a morphological phenotype not present in rcd1 coi1-16, suggesting that auxin, but not JA, enhances the SIMR-like morphological phenotypes of this mutant. Together, these results suggest that JA negatively regulates ROS-induced PCD and auxin ROS-induced SIMR.
Transient Decrease in DR5-uidA May Be Attributable to Increased Aux/IAA Protein Stability
The transient decrease in DR5-uidA and HAT2 expression indicated that auxin signaling via AuxRE was transiently decreased in response to apoplastic ROS. According to the current model of auxin signaling, short-lived Aux/IAA proteins are degraded upon auxin stimulus, which allows auxin-dependent gene expression to occur via ARF transcription factors. At which step in the signaling pathway are ROS acting? Given the fast response in DR5-uidA expression, it is unlikely that de novo synthesis of a new signaling protein is involved; instead, altered stability of Aux/IAA proteins, which was also observed in response to BTH treatment (Wang et al., 2007) and flg22 (Navarro et al., 2006), would lead to decreased output from the auxin signaling pathway (Fig. 7). Furthermore, this mechanism would inhibit the de novo synthesis of Aux/IAAs and consequently allow auxin signaling to return to prestress levels, as was observed with DR5-uidA and HAT2 expression (Figs. 2D and 7). Apoplastic ROS could also directly regulate the activity or localization of ARFs via yet unknown mechanisms.
Figure 7.
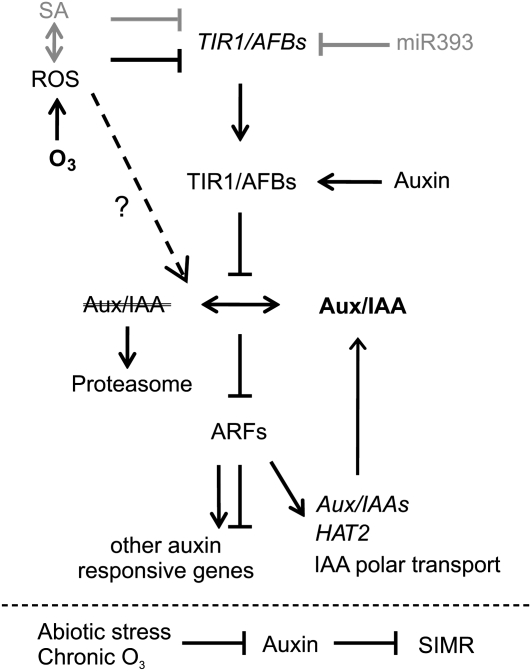
The auxin signaling pathway is modulated by apoplastic ROS. The binding of auxin to TIR1/AFB receptors leads to the degradation of AUX/IAA repressors via the 26S proteasome, allowing the activation of ARF transcription factors and changes in gene expression. O3 treatment leads to the production of apoplastic ROS, which could suppress the auxin pathway by decreasing the expression of TIR1/AFBs independently of miR393 and SA. O3 may affect the stability of Aux/IAAs (e.g. IAA10 and IAA28), or ARF activity could be directly modulated independently of auxin F-box proteins. ARFs regulate auxin-dependent gene expression, which includes Aux/IAA transcripts. Decreased levels of Aux/IAA transcripts provide a feedback mechanism counteracting the increased Aux/IAA stability. In addition, apoplastic ROS modulated the expression of several genes involved in auxin signaling, polar transport, and biosynthesis. Chronic O3 exposure leads to SIMR, which is under negative regulation of the auxin signaling pathway.
Auxin-Responsive Genes Are Targeted by Several Stresses
The mechanism by which apoplastic ROS affect auxin responses was further studied by the identification of auxin-responsive genes from several publicly available microarray experiments (Paponov et al., 2008) and comparing them with our set of O3-regulated genes by hierarchical clustering. Although some discrepancy might be brought into the comparison by the differences in plant age (i.e. auxin array experiments were performed on seedlings versus the 3-week-old plants used in the O3 array experiment) and tissue types (whole plant versus rosette only), it still gave an indication of which processes are regulated both by auxin and ROS. Approximately 30% of auxin-regulated genes were also regulated by O3, and several Aux/IAA genes were coordinately regulated by several stresses together with apoplastic ROS (Fig. 3A). The commonly occurring AuxRE is found in approximately 25% of Arabidopsis 500-bp promoters (Keilwagen et al., 2011). Auxin-responsive genes have a variable number of AuxREs in their 2-kb promoters (Lee et al., 2009), and in our analysis, they were absent in the 1-kb promoters of some auxin-responsive genes (Fig. 3A). This may have complicated the promoter element analysis and therefore explain the lack of AuxRE enrichment in the expression profile VI containing genes classified as auxin responsive (Fig. 1, C and D). However, due to the decrease in DR5-uidA expression (Fig. 2D), ARF function must at some point be altered by apoplastic ROS. ARFs can be classified into transcriptional activators and repressors according to their amino acid sequence, but the molecular functions of individual ARFs are still largely unexplored. No major changes in the expression of 16 ARFs (including all the activator ARFs) were found (Fig. 2C), indicating that ROS regulates ARFs posttranscriptionally. The large number of ARFs expressed in leaves offers interesting possibilities for the regulation of auxin signaling by specific ARF-Aux/IAA interactions (Weijers et al., 2005), by other transcription factors (Shin et al., 2007), or by brassinosteroid-dependent phosphorylation (Vert et al., 2008). It has been suggested that the ARF activator/repressor activity might be tissue dependent (Lee et al., 2009) and that competition of AuxRE binding may occur within the ARF gene family. Indeed, it remains to be elucidated by which mechanism auxin treatment decreases gene expression. Altogether, the ARF function remains incompletely defined, and future studies are needed especially to address the role of repressor ARFs and to define the Aux/IAA-ARF interactome.
The genes regulated by both auxin and apoplastic ROS were divided into three major clusters (Fig. 3A). Cluster II was characterized by two properties: genes with decreased expression by ROS and increased expression by auxin; it contained many genes with roles in auxin signaling, Aux/IAAs, and SAURs. In contrast, cluster III included genes with increased expression by both auxin and apoplastic ROS; the annotation of these genes revealed no obvious link to auxin signaling and no enrichment of the AuxRE. The stress-responsive marker gene GST6 has increased expression by auxin, SA, and H2O2 and is regulated through ocs and TGA elements (Chen et al., 1996; Chen and Singh, 1999; Zhou et al., 2000). The similarity between GST6 expression and cluster III prompted us to study the expression of several genes in response to O3 in SA and ET mutants (Fig. 3B). Transcripts analyzed included transcription factors (ZAT10 and WRKY40), ET biosynthesis (ACS6), pinoid-binding protein (PBP1), and GST6. Already in clean air, some differences were observed between the mutants: decreased expression of WRKY40 and PBP1 in sid2, npr1 and NahG and, in contrast, higher expression of these genes in ein2. This indicates that SA signaling is a positive regulator of these genes and that ET is a negative regulator in nonstressed conditions. Interestingly, the role of SA appears to be reversed during O3 treatment, and PBP1 had increased expression in npr1 relative to Col-0. This regulation is consistent with the previously proposed model, where SA and npr1 act as negative regulators of genes with increased expression during O3 signaling (Wrzaczek et al., 2010). Positive or negative interactions between ROS and SA on gene expression or cell death are context dependent (i.e. the sources and localization of ROS production). During cell death, ROS and SA are acting in a self-amplifying loop (Overmyer et al., 2003). Recently, identification of a mutant defective in mitochondrial complex II (dsr1) revealed that SA and dicamba (a synthetic auxin herbicide) activation of GST6 expression required mitochondrial ROS production (Gleason et al., 2011). Thus, despite several sources of ROS production in the cell, the plant is able to distinguish the source and integrate the ROS signal with other hormone signals. The negative interaction between SA and apoplastic ROS is reminiscent of the negative interaction between flg22 and SA signaling (Sato et al., 2010). Both positive and negative interactions between hormones have long been observed in development and during stress (Pieterse et al., 2009; Jaillais and Chory, 2010), and especially negative interactions between signaling pathways are prevalent during pathogen infection (Sato et al., 2010).
Hormone Interactions
So far, the discussion of auxin signaling has focused on mechanisms within the classic auxin signaling pathway (Fig. 7). These processes, of course, do not define a simple linear pathway; rather, they are connected in a complex web of interacting hormone signaling pathways. In response to apoplastic ROS, there is clear evidence for the involvement of phytohormones other than auxin (Table I), which suggests potential hormone-hormone interactions during the ROS response. Specifically, over the time frame of decreased transient auxin responses (1–4 h), ABA, BR, GA, ET, JA, and SA response genes were enriched. This is consistent with previously published profiles of O3-induced hormone accumulation for SA and ET (Overmyer et al., 2000, 2008), while ABA and JA signaling appear to be active prior to the actual accumulation of the corresponding hormone. To our knowledge, the accumulation of BR and GA in response to ROS has not yet been examined. In the comparison of different hormones and stress treatments on gene expression (Fig. 3A), BR treatment was the hormone with the most similar effect to auxin treatment. The roles and interactions of the stress hormones (SA, JA, ET) have been discussed at length previously (Overmyer et al., 2003). The role of classical plant hormones in stress responses has gained increasingly more attention in recent years (Peleg and Blumwald, 2011; Tognetti et al., 2011). Many hormone interactions that are relevant to this study have been observed. In biotic stress, auxin and cytokinin act as positive regulators of JA/ET-based resistance and antagonists of SA-based resistance (Robert-Seilaniantz et al., 2007). In contrast, GA has the opposite effect, antagonizing JA/ET and working cooperatively with SA (Robert-Seilaniantz et al., 2007). Auxin has long been known to enhance ET production via the transcriptional induction of some of the Arabidopsis ACS genes (Tsuchisaka and Theologis, 2004), including ACS6, which has three AuxREs in its promoter (Fig. 3A) and is the primary O3-responsive ACS gene in Arabidopsis (Vahala et al., 1998). Interestingly, the timing of the O3-induced 1-aminocyclopropane-1-carboxylic acid production peak (Overmyer et al., 2000) directly coincides with the decrease in DR5-uidA and HAT2 expression (Fig. 2D).
SIMR Caused by Apoplastic ROS
Long-term adaptation to stress involves adjusted plant growth and morphology. Altered auxin biosynthesis, conjugation, transport, and signaling are all implicated in the establishment of adaptations to environmental perturbations (Potters et al., 2009; Tognetti et al., 2011). Short-term apoplastic ROS affected gene expression at all levels of auxin homeostasis or signaling (Figs. 2–4), which might lead to long-term developmental alterations; consequently, we studied the SIMR of plants chronically exposed to O3. The chronic exposure to apoplastic ROS altered plant morphology, which was enhanced in the auxin receptor tir1 afb2 double mutant (Fig. 6). This is in contrast to previous studies that report an attenuated SIMR response in auxin receptor mutants (Iglesias et al., 2010; Zolla et al., 2010). However, previous SIMR studies have been performed in the plant root, a tissue in which many auxin effects are the opposite of those seen in the shoot. Thus, auxin appears to act as a negative regulator of SIMR in the shoot (Fig. 7). While auxin is involved in the regulation of stress morphology, it had no effect (Fig. 6A) on growth retardation, a parameter that commonly falls under the label of SIMR. This study may be unique in addressing SIMR responses in adult leaf tissues, which facilitated the dissection of growth and morphological responses. The most commonly used parameter in SIMR studies is root length, an index that reflects both changes in growth and morphology (cell expansion). Our results suggest that stress-induced growth repression and morphological responses are regulated independently. Leaf curling is caused by differential cell expansion on the top and bottom layers of the leaf. These results suggest that there is a specific cell identity determined by positional information that responds to ROS and auxin to alter the leaf developmental program. This developmental role for ROS is consistent with previous results in root development, where ROS signals are required for root hair development (Foreman et al., 2003) and for determining the boundary between the growth and differentiation zones (Tsukagoshi et al., 2010).
Collectively, ROS-auxin interactions are observed at two biological processes, gene expression and SIMR, which may be connected to each other (Fig. 7). Apoplastic ROS led to a rapid transient decrease in AuxRE-driven expression, as exemplified by DR5-uidA and HAT2. We propose that the entry point for ROS in the auxin signaling pathway is through the stabilization and/or degradation of Aux/IAAs (Fig. 7). In particular, two Aux/IAA genes, IAA10 and IAA28, were transcriptionally induced by O3, and they could mediate decreased expression of AuxRE-containing genes by specific interactions with ARFs. Mutant plants with altered IAA28 protein function did not exhibit any change in their SIMR or HAT2 gene expression response, suggesting that IAA28 is not involved in regulating these processes (Fig. 6A; Supplemental Fig. S6). However, IAA10 may be functionally redundant with IAA28, and studies with iaa28 iaa10 double mutants will be required to fully address the involvement of these proteins. What other support exists for this model of ROS-auxin interaction? The auxin transport inhibitor TIBA gave a strikingly similar result to O3 in the regulation of auxin-responsive genes (Fig. 3A), indicating that redistribution of auxin is a regulator for this set of genes. This similarity between ROS and TIBA is not restricted to gene expression, as it is also observed in SIMR (Pasternak et al., 2005).
Plant survival in a changing environment requires adaptation to the prevailing conditions. The use of ROS in combination with auxin may provide plants with an elegant mechanism to optimize plant performance during acute and chronic stresses.
MATERIALS AND METHODS
Plant Material and O3 Treatment
For microarray experiments, Arabidopsis (Arabidopsis thaliana) wild-type Col-0 and rcd1-1 seeds were sown on a 1:1 peat:vermiculite mixture, stratified for 2 d, and grown in controlled environment chambers (Weiss Bio1300; Weiss Gallenkamp [http://www.weiss-gallenkamp.com/]) with a 12-h/12-h (day/night) cycle, temperature of 22°C/19°C, and relative humidity of 70%/90%. One-week-old plants were transplanted into individual pots (5 × 5cm) and subirrigated twice per week. O3 experiments (6 h of 350 nL L−1) were performed with 3-week-old plants. Control and O3-treated individual plants were collected at 0, 1, 2, 4, 8, and 24 h after the start of the O3 treatment and flash frozen in liquid nitrogen. The experiment was repeated three times, in addition to which a fourth identical repeat was used as the common reference RNA. For SIMR experiments, 2-week-old plants were exposed for 4 to 6 h per day at 350 nL L−1 O3 for 14 d. O3-exposed and parallel clean-air control plants were photographed on days 1, 7, 14, and 15 for size determinations and were harvested on day 15 for fresh weight measurements. Rosette size was measured as the minimum circle that contains all rosette leaves using the ImageJ image-analysis program (http://rsbweb.nih.gov/ij/) and used for growth rate analysis. Size and shape parameters of individual middle-aged leaves were quantified with the LAMINA leaf shape determination program (Bylesjö et al., 2008). Six plants per genotype were used for experiments repeated twice with similar results. For electrolyte leakage, plants were O3 treated with 6 h of 350 nL L−1, allowed to recover for 2 h in clean air, and rosettes were collected into 15 mL of MilliQ water (n = 5). Cell death from control and O3-treated plants was quantified as described by Ahlfors et al. (2009). The experiment was repeated five times, and the data were analyzed with linear models. All plant photographs were taken against a dark background, which was digitally made uniformly black. The axr1-3, aux1-7, nit1-3, and iaa28-2 (SALK_129988C) mutants were obtained from the European Arabidopsis Stock Centre (http://arabidopsis.info/), and the coi1-16, tir1-1 afb2-3, and iaa28-1 mutants were kind gifts from John Turner, Mark Estelle, and Bonnie Bartel, respectively. Double mutants were constructed with rcd1 or coi1-16 as the pollen acceptor. Double mutants were initially screened for the visible rcd1 phenotype (curly leaves and compact rosette) or the methyl jasmonate-insensitive root growth of coi1-16. Subsequently, all mutations were identified through cleaved-amplified polymorphic sequence or derived cleaved-amplified polymorphic sequence markers: rcd1-1 (CCGTTTTCGTCAACATCACA and CTGCAGACTGCCCTTATTTCAA; SspI); axr1-3 (TTAGGCTTCTTTTTCCTGTGTT and AAAACCAACTTAACGTTTGCATGTCGA; SalI); aux1-7 (GAAGCCACCGTTCTTTATGC and CAAAACCCCAAAAGAGAAAAA; Eco31I); nit1-3 (ATCGTCGATGCTTCACATTG and ATCATGTTCTTTGTCGTGGTAC; KpnI); and coi1-16 (TGTGAAGGTCGGTGACTTTG and AGTTTTCGGGGAAAAACCAG; Hpy188III). All double mutants were verified in the F3 generation. DR5-uidA plants (Ulmasov et al., 1997) were grown and O3 treated together with ein2, sid2, npr1, and NahG plants. The experiment was repeated three times. (Repeat 2 was omitted for npr1 due to outliers, and the qPCR results were confirmed with two additional repeats [data not shown].) GUS staining was performed according to Weigel and Glazebrook (2002). IAA measurements of O3-treated Col-0 samples harvested in liquid nitrogen 0, 2, 4, and 8 h after the start of O3 exposure were repeated twice with similar results. Hormones were extracted and quantified with the vapor-phase extraction method described by Schmelz et al. (2004). Gas chromatography-mass spectrometry analysis was performed on an Agilent 6890N/5973N with G1088B electronics upgrade as a splitless injection in single ion monitoring mode as described by Montesano et al. (2005). Ions 130, 135, 189, and 194 were monitored with 100-ms dwell time. The inlet and transfer line temperatures were 230°C and 300°C, respectively. A Restek Rxi-5Sil MS capillary column (30 m × 0.25 mm × 0.25 μm) with 10-m integrated precolumn was used. The column was held at 40°C for 1 min after injection, then heated by 15°C min−1 to 250°C, held for 4 min, and heated by 20°C min−1 to a 300°C final temperature (held for 3 min) with helium as the carrier gas (flow of 1 mL min−1).
RNA Extraction and Microarray Hybridizations
Individual plants in Eppendorf tubes were ground with cooled metal balls in a tissue lyser (Retsch MM300; www.retsch.com), after which frozen plant powder for metabolite analysis was weighed into a separate tube. The remaining leaf powder (three to 10 plants from each genotype, time point, and treatment) was pooled for RNA extraction. RNA was extracted with the Spectrum Total RNA Extraction Kit (Sigma-Genosys). The RNA integrity was analyzed on a formaldehyde gel, and no RNA degradation was observed. Sixty micrograms of RNA was used for the cDNA synthesis reaction and subsequently divided into two tubes for labeling with different dyes (Cy3 and Cy5). RNA samples were hybridized against a common reference RNA sample obtained from an identical experiment. The experiment was repeated three times. A full description of the experimental and hybridization conditions as well as all raw array data are available in the Array Express database (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-662. The MWG (http://www.mwg-biotech.com/) 50-mer oligomicroarrays were identical to Ahlfors et al. (2009) and Jaspers et al. (2009). The oligonucleotide sequences were reannotated to The Arabidopsis Information Resource (TAIR) 9 database with the BLASTn algorithm (Swarbreck et al., 2008). A matching gene was defined to be either an ungapped perfect match of more than 37 nucleotides or a perfect match of more than 44 nucleotides with one gap. If several genes fulfilled the criterion, the oligonucleotide was termed “ambiguous”; if no matches were found, the oligonucleotide was labeled “nomatch.” Altogether, the remapping resulted in 21,071 genes, 2,634 ambiguous oligonucleotides, and 738 nomatch oligonucleotides. Only oligonucleotides binding specifically to a single gene were used in further analysis.
Microarray Data Analysis
Microarray data preprocessing and analysis was carried out using scripts in R, version 2.12.1. Preprocessing of the microarrays was carried out as described by Jaspers et al. (2009). A linear mixed model was constructed to model the expression of each gene using the nlme package (Pinheiro and Bates, 2000). The model consisted of factor-level effects from time (0, 1, 2, 4, 8, 24 h), genotype (Col-0, rcd1), treatment (O3, control), and dye swap. The repeated-measured design of the experiment was taken into account by including a random effect for biological repeats. The model contrasts were computed from the linear mixed model using the multcomp package (Bretz et al., 2010), with one-step error correction of P values. The P values for each gene and contrast were subjected to a false discovery rate correction of the P values (Storey, 2003). Genes expressed with log2 fold change of ±1 and a q value of less than 0.05 were considered differentially expressed.
For clustering of expression profiles, pair-wise distances between the time series of differentially expressed genes were computed as described by Toh and Horimoto (2002). The pair-wise distance matrix was then clustered using affinity propagation (Frey and Dueck, 2007), as implemented in the R package apcluster. The algorithm was initialized with 10 random initializations with self similarity of −21.10333. The solution giving the best net similarity was selected.
GO enrichment analysis was carried out for the differentially expressed genes clustered in each of the expression profiles using the annotation of TAIR10 (ftp://ftp.arabidopsis.org/home/tair/Ontologies/) and scripts in R. The enrichment was analyzed using the Fisher exact test. A Benjamini-Hochberg false discovery rate correction of the P values was applied for each GO category. GO enrichment also was performed separately for differentially expressed genes at each time point.
Real-Time qRT-PCR
Verification of the microarray results and additional gene expression analysis was performed with qRT-PCR. RNA was isolated and treated with DNase I as described by Jaspers et al. (2009). RT was performed with 5 μg of RNA with RevertAid Premium RT and Ribolock RNase inhibitor (Fermentas), and the reaction was diluted to a final volume of 200 μL. qRT-PCR was performed in triplicate using 1 μL of cDNA template per reaction with primers, iQ SYBR GREEN supermix (Bio-Rad), and water. The cycle conditions with Bio-Rad CFX were as follows: one cycle initiating with 95°C for 10 min, 39 cycles with 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, and ending with a melting-curve analysis. Primer sequences and amplification efficiencies determined with the Bio-Rad CFX Manager program from a cDNA dilution series are given in Supplemental Table S4. The raw cycle threshold values were normalized to ACTIN2 (At3g18780). Fold change and P values were computed with scripts in R using linear mixed models. Based on a likelihood ratio test statistic, a random effect for each biological repeat was incorporated (P < 0.05 was considered significant); otherwise, a standard linear model was used. Contrasts were computed with the multcomp package (Bretz et al., 2010) with single-step P value correction.
The quantification of miR393 was done from RNA isolated with Tri Reagent (Molecular Research Center) according to the method by Chen et al. (2005). One microgram of RNA was used for cDNA synthesis (16°C for 30 min, 42°C for 30 min, and 85°C for 5 min) with a miR393-specific stem-loop primer (Feng et al., 2010). Undiluted cDNA was used for miR393 quantification with the primers in Supplemental Table S4, and miR393 expression was normalized to ACTIN2.
Promoter Analysis
For promoter analysis, the 500-, 1,000-, and 3,000-bp promoter sequences of TAIR10 were downloaded from http://www.arabidopsis.org/. A list of 196 known binding motifs was collected from AGRIS (Yilmaz et al., 2011), PlantCARE (Lescot et al., 2002), and PLACE (Higo et al., 1999) and from recent publications reporting new binding motifs (CM2; Doherty et al., 2009), auxin motif AtREG553 (Yamamoto et al., 2011), and AuxRE TGTCnC (Lee et al., 2009). Matching of the motifs was carried out with scripts in R for both plus and minus DNA strands of the promoter areas. Enrichment of motifs was determined with the Fisher exact test.
Analysis of Publicly Available Gene Expression Data
A data set of publicly available experiments using the Affymetrix ATH1-121501 platform was collected from several data sources (NASCArrays; http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl [ABA, NASCARRAYS-176; IAA, NASCARRAYS-175; 2,4,6-T and TIBA, NASCARRAYS-186; cycloheximide, NASCARRAYS-189; MG132, NASCARRAYS-190; SA, NASCARRAYS-192; brassinolide, NASCARRAYS-179; BTH, NASCARRAYS-392]; ArrayExpress; http://www.ebi.ac.uk/microarrayas/ae/ [methyl jasmonate, E-ATMX-13; PQ, E-ATMX-28; IAA, E-MEXP-1256]; and Gene Expression Omnibus; http://www.ncbi.nlm.nih.gov/geo/ [H2O2, GSE5530; IAA, GSE1491; Salt, GSE5623; Heat, GSE19603; High light, GSE7743; UV-B, GSE3533; Cd, GSE22114; Flg22, GSE5615; Pseudomonas, GSE5685]). The data were analyzed with scripts in R. The raw .cel files were first normalized with robust multiarray average normalization, and for each experiment, the log2 base fold changes of treatment versus control were computed. The preprocessed data were clustered using Bayesian hierarchical clustering as described by Wrzaczek et al. (2010).
Identification of auxin-regulated transcripts was done from robust multiarray average-normalized .cel files: E-GEOD-1491 (5 μm IAA for 1 h), E-MEXP-1256 (20 μm IAA for 2 h), and NASCARRAYS-175 (1 μm IAA for 1 h, 3 h, and 30 min). Genes with at least 2-fold change and P < 0.05 in one or several of the experiments analyzed were considered as auxin responsive. A total of 196 genes filled these criteria, of which 179 genes were present on the MWG oligonucleotide array.
For comparison of O3 array data, the following public data sets were analyzed as above (3 h at 350 nL L−1 for E-MEXP-1863, 6 h at 350 nL L−1 for E-MEXP-1863, and 1 + 3 h at 200 nL L−1 for GSE5722).
ARF target gene lists were identified from the following sources: ARF7 and ARF19 targets (227 genes) from Okushima et al. (2005), ARF6 and ARF8 targets (18 genes) after Nagpal et al. (2005), and ARF5 targets (97 genes) after Schlereth et al. (2010). SAUR64 (At1g29450), whose expression level mildly decreased in response to apoplastic ROS, was present in all of the three original lists, whereas 32 targets were shared by two lists. Altogether, these gene lists contained 308 genes, of which 200 were present on the oligonucleotide array. A total of 125 putative ARF targets were found to be regulated by apoplastic ROS. Seventy-eight ARF targets were in common with the auxin-regulated gene list.
Analysis of SIMR Data
SIMR data were analyzed with scripts in R. First, a logarithm of raw rosette diameter data was computed as described by Hoffmann and Poorter (2002). Then, a linear mixed regression model was estimated from the data with logarithmic diameter as the dependent variable and measurement day (1 or 7 d; continuous), treatment (O3 or control; categorical), and genotype (Col-0, Wassilewskija [Ws-0], iaa28-1, iaa28-2, tir1 afb2; categorical) as covariates. The repeated-measures design of the experiment was taken into account by including a plant-specific random effect. Biological repeats were further modeled with a random effect. The cross-effect of treatment and genotype was not significant (likelihood ratio P value of 0.88) and was removed from the model. Differences and false discovery rate-corrected P values between relative growth rates were computed using the multcomp package (Bretz et al., 2010). The LAMINA data were analyzed with a linear mixed model having hierarchical random effects for plant and leaf within a plant. A logarithm of the data was taken before modeling to improve the linear model fit of the data.
The array experiment accession number is E-MTAB-662.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of this study data (apoplastic ROS-responsive genes in Col-0) obtained with a 21K oligonucleotide array (MWG) with publicly available full-genome O3 experiments with ATH1.
Supplemental Figure S2. GUS staining images of DR5-uidA plants.
Supplemental Figure S3. Expression of premir393a and premiR393b in response to apoplastic ROS determined with qPCR.
Supplemental Figure S4. Expression of miR393 in O3-treated plants.
Supplemental Figure S5. Expression of HAT2, SAUR68, and TIR1 in Col-0, mpk3, and mpk6 plants after 2 h of O3 treatment (350 nL L−1) determined with qPCR.
Supplemental Figure S6. Expression of early (SAG21, HAT2, IAA10, IAA28, and ARF7) and late (IAA7 and IAA11) O3-responsive genes in Ws-0, iaa28-1, Col-0, and iaa28-2 analyzed with qPCR.
Supplemental Table S1. Genes with statistically significant change of expression (q < 0.05) in O3-treated plants.
Supplemental Table S2. GO enrichments in O3-treated plants separately for each time point.
Supplemental Table S3. Promoter element sequences and enrichment results from this study.
Supplemental Table S4. Primers used for qPCR.
Acknowledgments
We thank Tuomas Puukko and Leena Laakso for excellent technical assistance. We acknowledge the Finnish DNA Microarray Center (Turku Centre for Biotechnology) for manufacturing the microarrays and TAIR for running the BLAST algorithm for microarray probe annotations.
References
- Ahlfors R, Brosché M, Kollist H, Kangasjärvi J. (2009) Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J 58: 1–12 [DOI] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP. (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretz F, Hothorn T, Westfall T. (2010) Multiple Comparisons Using R. CRC Press, Boca Raton, FL [Google Scholar]
- Brosché M, Merilo E, Mayer F, Pechter P, Puzõrjova I, Brader G, Kangasjärvi J, Kollist H. (2010) Natural variation in ozone sensitivity among Arabidopsis thaliana accessions and its relation to stomatal conductance. Plant Cell Environ 33: 914–925 [DOI] [PubMed] [Google Scholar]
- Bylesjö M, Segura V, Soolanayakanahally RY, Rae AM, Trygg J, Gustafsson P, Jansson S, Street NR. (2008) LAMINA: a tool for rapid quantification of leaf size and shape parameters. BMC Plant Biol 8: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Chao G, Singh KB. (1996) The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J 10: 955–966 [DOI] [PubMed] [Google Scholar]
- Chen W, Singh KB. (1999) The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J 19: 667–677 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. (2004) NPR1, all things considered. Curr Opin Plant Biol 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Feng XM, You CX, Qiao Y, Mao K, Hao YJ. (2010) Ectopic overexpression of Arabidopsis AtmiR393a gene changes auxin sensitivity and enhances salt resistance in tobacco. Acta Physiol Plant 32: 997–1003 [Google Scholar]
- Foreman J, Demidchik V, Bothwell JHF, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JDG, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Frey BJ, Dueck D. (2007) Clustering by passing messages between data points. Science 315: 972–976 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Uggalla V, Whelan J. (March 24, 2011) REDOX regulation of mitochondrial function in plants. Plant Cell Environ http://dx.doi.org/10.1111/j.1365-3040.2011.02293.x [DOI] [PubMed] [Google Scholar]
- Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB. (2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci USA 108: 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann WA, Poorter H. (2002) Avoiding bias in calculations of relative growth rate. Ann Bot (Lond) 90: 37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias MJ, Terrile MC, Bartoli CG, D’Ippólito S, Casalongué CA. (2010) Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol Biol 74: 215–222 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Chory J. (2010) Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol 17: 642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P, Blomster T, Brosché M, Salojärvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F, et al. (2009) Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 60: 268–279 [DOI] [PubMed] [Google Scholar]
- Jaspers P, Kangasjärvi J. (2010) Reactive oxygen species in abiotic stress signaling. Physiol Plant 138: 405–413 [DOI] [PubMed] [Google Scholar]
- Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J. (2001) Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J 26: 395–407 [DOI] [PubMed] [Google Scholar]
- Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV. (2005) Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnosky DF, Werner H, Holopainen T, Percy K, Oksanen T, Oksanen E, Heerdt C, Fabian P, Nagy J, Heilman W, et al. (2007) Free-air exposure systems to scale up ozone research to mature trees. Plant Biol (Stuttg) 9: 181–190 [DOI] [PubMed] [Google Scholar]
- Keilwagen J, Grau J, Paponov IA, Posch S, Strickert M, Grosse I. (2011) De-novo discovery of differentially abundant transcription factor binding sites including their positional preference. PLoS Comput Biol 7: e1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontunen-Soppela S, Riikonen J, Ruhanen H, Brosché M, Somervuo P, Peltonen P, Kangasjärvi J, Auvinen P, Paulin L, Keinänen M, et al. (2010) Differential gene expression in senescing leaves of two silver birch genotypes in response to elevated CO2 and tropospheric ozone. Plant Cell Environ 33: 1016–1028 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J. (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Park JW, Lee HW, Kim J. (2009) Genome-wide analysis of the auxin-responsive transcriptome downstream of iaa1 and its expression analysis reveal the diversity and complexity of auxin-regulated gene expression. J Exp Bot 60: 3935–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Mane SP, Sioson AA, Robinet CV, Heath LS, Bohnert HJ, Grene R. (2006) Effects of chronic ozone exposure on gene expression in Arabidopsis thaliana ecotypes and in Thellungiella halophila. Plant Cell Environ 29: 854–868 [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW. (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Llorente F, Muskett P, Sánchez-Vallet A, López G, Ramos B, Sánchez-Rodríguez C, Jordá L, Parker J, Molina A. (2008) Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol Plant 1: 496–509 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Solano R. (2005) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8: 532–540 [DOI] [PubMed] [Google Scholar]
- Ludwikow A, Sadowski J. (2008) Gene networks in plant ozone stress response and tolerance. J Integr Plant Biol 50: 1256–1267 [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2: ra45. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Montesano M, Brader G, Ponce de León I, Palva ET. (2005) Multiple defence signals induced by Erwinia carotovora ssp. carotovora elicitors in potato. Mol Plant Pathol 6: 541–549 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Soukupová H, Schikora A, Zárský V, Hirt H. (2006) A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Normanly J. (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Nakajima N, Tamaoki M, Aono M, Kubo A, Kamada H, Saji H. (2007) The isochorismate pathway is negatively regulated by salicylic acid signaling in O3-exposed Arabidopsis. Planta 226: 1277–1285 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Brosché M, Kangasjärvi J. (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8: 335–342 [DOI] [PubMed] [Google Scholar]
- Overmyer K, Brosché M, Pellinen R, Kuittinen T, Tuominen H, Ahlfors R, Keinänen M, Saarma M, Scheel D, Kangasjärvi J. (2005) Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiol 137: 1092–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Kollist H, Tuominen H, Betz C, Langebartels C, Wingsle G, Kangasjärvi S, Brader G, Mullineaux P, Kangasjärvi J. (2008) Complex phenotypic profiles leading to ozone sensitivity in Arabidopsis thaliana mutants. Plant Cell Environ 31: 1237–1249 [DOI] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Jr, Kangasjärvi J. (2000) Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12: 1849–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JAH, Palme K. (2008) Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1: 321–337 [DOI] [PubMed] [Google Scholar]
- Pasternak T, Rudas V, Potters G, Jansen MAK. (2005) Morphogenic effects of abiotic stress: reorientation of growth in Arabidopsis thaliana seedlings. Environ Exp Bot 53: 299–314 [Google Scholar]
- Peleg Z, Blumwald E. (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14: 290–295 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. (2000) Mixed Effects Models in S and S-Plus. Springer, New York [Google Scholar]
- Potocký M, Jones MA, Bezvoda R, Smirnoff N, Zárský V. (2007) Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytol 174: 742–751 [DOI] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Jansen MAK. (2009) Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ 32: 158–169 [DOI] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Palme KJ, Jansen MAK. (2007) Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci 12: 98–105 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JDG. (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379 [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Sato M, Tsuda K, Wang L, Coller J, Watanabe Y, Glazebrook J, Katagiri F. (2010) Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. (2004) The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J 39: 790–808 [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skärby L, Ottosson S, Karlsson PE, Wallin G, Selldén G, Medin EL, Pleijel H. (2004) Growth of Norway spruce (Picea abies) in relation to different ozone exposure indices: a synthesis. Atmos Environ 38: 2225–2236 [Google Scholar]
- Storey JD. (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat 31: 2013–2035 [Google Scholar]
- Street NR, Tallis MJ, Tucker J, Brosché M, Kangasjärvi J, Broadmeadow M, Taylor G. (2011) The physiological, transcriptional and genetic responses of an ozone-sensitive and an ozone tolerant poplar and selected extremes of their F2 progeny. Environ Pollut 159: 45–54 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K. (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7: 847–859 [DOI] [PubMed] [Google Scholar]
- Teotia S, Muthuswamy S, Lamb RS. (2010) Radical-induced cell death1 and similar to RCD one1 and the stress-induced morphogenetic response. Plant Signal Behav 5: 143–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiryaki I, Staswick PE. (2002) An Arabidopsis mutant defective in jasmonateresponse is allelic to the auxin-signaling mutant axr1. Plant Physiol 130: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Mühlenbock P, Van Breusegem F. (April 26, 2011) Stress homeostasis: the redox and auxin perspective. Plant Cell Environ http://dx.doi.org/10.1111/j.1365-3040.2011.02324.x [DOI] [PubMed] [Google Scholar]
- Toh H, Horimoto K. (2002) Inference of a genetic network by a combined approach of cluster analysis and graphical Gaussian modeling. Bioinformatics 18: 287–297 [DOI] [PubMed] [Google Scholar]
- Torres MA. (2010) ROS in biotic interactions. Physiol Plant 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Tsuchisaka A, Theologis A. (2004) Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol 136: 2982–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K, Kurihara Y, Seki M, Shinozaki K. (2010) ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol 13: 132–138 [DOI] [PubMed] [Google Scholar]
- Vahala J, Schlagnhaufer CD, Pell EJ. (1998) Induction of an ACC synthase cDNA by ozone in light-grown Arabidopsis thaliana leaves. Physiol Plant 103: 45–50 [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, et al. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Puzõrjova I, Brosché M, Valk E, Lepiku M, Moldau H, Pechter P, Wang YS, Lindgren O, Salojärvi J, et al. (2010) Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J 62: 442–453 [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA 105: 9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J. (2010) Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. (2002) The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Yoshioka Y, Hyakumachi M, Maruyama K, Yamaguchi-Shinozaki K, Tokizawa M, Koyama H. (2011) Prediction of transcriptional regulatory elements for plant hormone responses based on microarray data. BMC Plant Biol 11: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E. (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 13: 191–202 [DOI] [PubMed] [Google Scholar]
- Zolla G, Heimer YM, Barak S. (2010) Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J Exp Bot 61: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]