Abstract
While the term `protein structure' is commonplace, it is increasingly appreciated that proteins may not possess a single, well-defined structure: some regions of proteins are intrinsically disordered. The role these intrinsically disordered regions (IDRs) play in protein function is an area of significant interest. In particular, because proteins containing IDRs are largely involved in processes related to molecular recognition, the question arises whether IDRs are important in these recognition events. It has been observed that IDRs are enriched in transcription factors (TFs) in comparison with other proteins, and we sought to explore this enrichment more precisely, with an eye toward functional dissection of the prevalence and locations of IDRs in different classes of TFs. Specifically, we considered the occurrences of 76 classes of DNA-binding domains (DBDs) within a comprehensive set of 1,747 human, sequence-specific TFs. For each DBD class, we analyzed whether a significant level of disorder was present within the DBD itself, the N-terminal or C-terminal sequence flanking the DBD, or both flanking sequences. We found that although the DBDs themselves exhibit significant order, the regions flanking the DBDs exhibit significant disorder, which suggests a functional role for such IDRs in TF DNA binding. These results may have important implications for studies of TFs not just in human but across all eukaryotes, and suggest future studies focused on testing the roles of N- and C-terminal flanking regions in determining or modulating the DNA binding a affinity and/or specificity of the associated TFs.
Keywords: intrinsically disordered region, DNA-binding domain, transcription factor
1. Introduction
The function of a protein is encoded in its amino acid sequence (i.e., primary structure). However, protein activity typically depends on the protein being folded properly into its component secondary structure elements (e.g., alpha helices, beta sheets) and the overall, global conformation of the protein (i.e., tertiary structure). Protein structure can be determined experimentally at high resolution either by X-ray crystallography or by nuclear magnetic resonance (NMR). X-ray crystallography is widely used, but cannot provide information on the conformation of regions that are either highly dynamic or unstructured in the crystal. In contrast, while NMR can provide information about flexibility and dynamics in proteins, it is currently limited to smaller proteins.
Through a combination of structural and biochemical studies, it has become increasingly appreciated that a protein may not adopt a single, well-defined `structure', a term connoting a measure of rigidity. Rather, a protein may sample an ensemble of global conformations; parts of the protein may be largely constantly structured across this ensemble, while other parts may be quite variable or flexible across the ensemble. These latter regions are sometimes termed `intrinsically disordered regions' (IDRs), though they may adopt a more structured conformation upon interaction with another molecule, whether a protein, DNA, or other ligand [1].
Proteins are largely involved in processes related to molecular recognition (e.g., binding, signaling, complex formation, enzymatic catalysis), and IDRs may enable these recognition events either directly (e.g., serving as the recognition domain of a protein) or indirectly (e.g., serving as a hinge that allows two ordered regions of a protein to come together to effect recognition). For this reason, IDRs have been studied rather extensively over the past decade, and a large number of computational methods have been developed for the prediction of IDRs on the basis of amino acid sequence, though this remains an imperfect art (see [2] for a review).
In this study, we were interested in exploring the role(s) that IDRs might play in the recognition tasks of transcription factors (TFs) in particular. Computational explorations have found that IDRs are generally more prevalent in TFs than would be expected by chance, especially in eukaryotes [3–5]. As a specific example, careful molecular studies have shown that a region of fifteen amino acids within the DNA-binding domain (DBD) of the estrogen receptor (ER) is disordered in solution, and makes contacts with DNA (and with another ER DBD monomer), as shown in a co-crystal structure of the ER DBD bound to DNA [6]. Moreover, IDRs outside the homeodomain DBD have also been found to impact the DNA-binding affinity of the Drosophila TF Ubx [7]. In addition, the region N-terminal to the proximal accessory region of the Saccharomyces cerevisiae C2H2 zinc finger TF Adr1 is disordered in solution (even after binding DNA) and increases the a affinity for non-specific DNA, mainly by increasing the DNA association rate; increased affinity for non-specific DNA might allow a protein to find its specific sites more quickly after translocation from non-specific sites that are bound initially [8]. Finally, DBDs often have N- or C-terminal extensions, referred to as `arms' or `tails', that bind DNA but are disordered when free in solution [9]. Intrigued by this ensemble of findings pointing to the importance of IDRs in TFs and their interactions with DNA, we sought to explore the connection between IDRs and TF function more precisely and systematically. We were particularly interested in determining whether IDRs were more prevalent in the regions flanking the DBDs that are responsible for the binding of sequence-specific TFs to DNA.
2. Materials and Methods
2.1. Constructing the TF and non-TF control sets of proteins
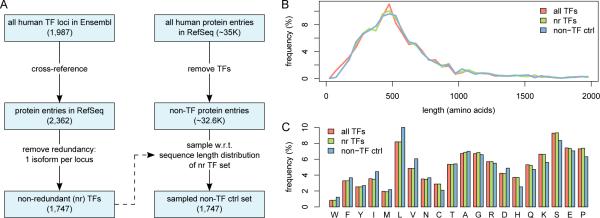
We created two non-redundant datasets of human proteins: a TF set and a non-TF set for use as a control. The procedure for constructing these sets and ensuring their non-redundancy is described below and summarized in Figure 1A.
Fig. 1.
(A) A schematic of the pipeline for generating the TF set and the non-TF control set. (B) Sequence length distributions of the TF set (nr TFs), the non-TF control set, and the set of all human TFs (with redundancy). (C) The amino acid compositions of the TF set, the non-TF control set, and the set of all human TFs. Amino acids are listed from most order-promoting to most disorder-promoting, according to [13]. It is apparent from the histogram that compared to proteins in general, TFs have fewer order-promoting residues (e.g., W, F, Y, I, M, L, V) and more disorder-promoting residues (e.g., P, E, S, K, Q, H).
We assembled the TF set from a published repertoire of human TFs [10]. In their study, Vaquerizas and colleagues manually curated and identified 1,987 TF-coding human genomic loci in the Ensembl database [11]; the list includes 1,960 high-confidence entries and 27 entries curated as probable. We cross-referenced these Ensembl loci against the RefSeq database (release 47) [12] to obtain 2,362 protein isoforms associated with 1,747 genes. To reduce sequence redundancy and thus potential bias, if multiple isoforms were associated with the same gene, we selected only the longest. This resulted in a final total of 1,747 unique TF protein sequences, and in subsequent analysis, we call this our TF set.
We assembled our non-TF control set by downloading all human proteins from RefSeq, and excluding the 2,362 TF-associated isoforms from above, which yielded a total of 32,567 non-TF proteins. To match the size and sequence length distribution of our TF set, we randomly sampled 1,747 proteins from the 32,567 according to the empirical sequence length distribution of the TF set; to ensure non-redundancy during this process, at each iteration we required that the sampled protein come from a locus not previously sampled. Therefore, the resulting control set contains 1,747 unique non-TF protein sequences.
2.2. Comparing the TF and non-TF control sets of proteins
To ensure that the non-TF set represents a well-constructed control for the TF set, we compared various properties of the two sets. First, we compared the sequence length distributions of the TF set and the non-TF control set, in addition to the set of all human TFs (i.e., with redundancy). As shown in Figure 1B, no apparent di erences exist between the sequence length distributions in the TF set, the non-TF control set, and the set of all human TFs.
Next, we compared the amino acid compositions of the TF set, the non-TF control set, and the set of all human TFs (Figure 1C). The amino acid composition of sequences in IDRs has been shown to be significantly di erent from that in ordered regions [14], and IDRs have been shown to have high prevalence in TFs [4], so we might expect compositional di erences between the TF sets and the non-TF control set. Indeed, compared to the non-TF control set, both TF sets are enriched in disorder-promoting amino acids (e.g., P, E, S, K, Q, H), and depleted in order-promoting amino acids (e.g., W, F, Y, I, M, L, V) [13, 14], as expected. However, the amino acid compositions of our non-redundant TF set and the set of all human TFs are nearly identical, suggesting that our procedure for removing redundancy introduces no significant compositional bias.
2.3. Identifying DNA-binding domains (DBDs) and their locations within proteins
Our goal is to investigate the prevalence and locations of IDRs within human TFs, and in particular, the spatial relationships between IDRs and DBDs in TFs. To identify all sequence-specific DBDs that occur within human TFs, we started with the entire set of human proteins from RefSeq and identified every Pfam domain [15] that was contained in a human protein with a p-value below 0.05. We manually filtered for those domains whose text descriptions in the Pfam or InterPro [16] databases indicated that the domain mediates sequence-specific DNA binding, resulting in 76 domains which we henceforth call Pfam DBDs.
Using HMMER [17] with default parameters, we searched for the locations of matches to Pfam DBDs within our TF set. We found 71 of the 76 Pfam DBDs matched to proteins in our TF set, with 32 DBDs appearing more than five times. Of the 1,747 proteins in our TF set, 669 contained only a single DBD, while another 642 contained multiple DBDs; proteins with multiple DBDs are typically those containing multiple zinc fingers, which are annotated as separate domains even if they occur in tandem within a protein. Indeed, the TF with the highest number of DBDs is zinc finger protein 91 (RefSeq: NP 003421), which contains 31 zf-C2H2 (zinc finger, C2H2-type) domains. The zf-C2H2 domain is interesting in its own right as it is by far the most prevalent domain in our TF set, appearing a total of 4,154 times, almost 20 times as often as the next most prevalent domain.
2.4. Predicting intrinsically disordered regions (IDRs) and their locations within proteins using multiple existing methods
To perform our analysis, we first needed to predict the ordered and disordered regions within proteins using existing computational tools. Since this remains a bit of an imperfect art, we took care to ensure that our conclusions would not be overly dependent on the predictions of any single choice of method. Consequently, we chose to use three distinct disorder prediction tools, each demonstrated to perform with high accuracy [2]: PONDR VSL2 [18], DISOPRED2 [19], and PreDisorder 1.1 [20].
PONDR VSL2 (also called DisProt VSL2) was evaluated as the top-ranked disorder predictor in CASP7 in 2006 [21], and PreDisorder was ranked among the top methods in disorder prediction during CASP8 in 2008 and CASP9 in 2010. These methods employ a variety of techniques to analyze sequence and structural information for IDR prediction: PONDR VSL2 uses support vector machines (SVMs) to separately address prediction problems in short versus long sequence regions, and then merges the results using a logistic regression model; DISOPRED2 is also based on SVMs, and compared to other prediction methods, the main difference is that it is directly trained on the whole sequence using various combinations of binary-encoded amino acid sequence, secondary structure predictions, and sequence profiles; and PreDisorder 1.1 is based on an ab initio prediction method along with a meta-prediction method.
2.5. Defining disorder features: spatial relationships of IDRs relative to DBDs within TFs
Given the annotated DBDs and the predicted disorder regions in the TF set and the non-TF control set, we sought to systematically analyze the association between TF DBDs and predicted IDRs by testing for enrichment of IDRs at different locations relative to DBDs. Specifically, we were interested in IDRs within the DBD itself, as well as the regions flanking the DBD, and we developed five distinct `disorder features': we say that a DBD is disordered if at least a fraction f of its residues are predicted to be disordered; we say that the N-terminal flank of a DBD is disordered if at least a fraction f of the 30 residues flanking the DBD in the N-terminal direction are predicted to be disordered; analogously, we say that the C-terminal flank of a DBD is disordered if at least a fraction f of the 30 residues flanking the DBD in the C-terminal direction are predicted to be disordered; we say that both flanks of a DBD are disordered if both the N-terminal and C-terminal flanks are disordered; and finally, we say that an entire TF is disordered if at least a fraction f of all of its residues are disordered. We wanted to be fairly stringent in identifying these disorder features, so that we could focus on those with the highest confidence; therefore, we chose the value of 0.8 for f.
2.6. Calculating statistical significance of disorder features
To assess whether the prevalence of disorder features within and flanking DBDs was unusually high or low, we needed to determine a suitable measure of significance. Moreover, since different computational tools predict IDRs at different rates (see Section 3.2 below), our significance measure needed to enable the comparison of results across methods, and not be biased by methods that are systematically more or less likely to predict disorder within proteins.
We thus developed two different null models to test for the significance of our disorder features (e.g., disordered DBD, N-terminal flank, or C-terminal flank). The first null model pretended that the location of a DBD occurred uniformly at random within each sequence, and was based on the TF set. The second null model also pretended that the location of a DBD occurred uniformly at random in each sequence, but was based on the non-TF control set. In summary, these two null models—in which the location of a DBD was chosen uniformly at random—were designed to test whether the spatial relationships between IDRs and DBDs were statistically significant or simply occurred by chance.
With each null model providing a baseline expectation for how often a disorder feature might be found by chance, we could then compute a significance measure based on the p-value from a hypergeometric distribution (i.e., Fisher's exact test). For each disorder feature we considered, we computed two separate p-values, one for each null model. Consistency of significance across the two different null models thus gave us some confidence that our results were robust to the specific choice of null model.
3. Results
3.1. Comparing the three methods for predicting IDRs within proteins
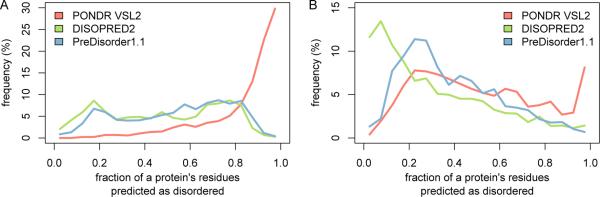
We used three different disorder prediction tools to predict IDRs in both the TF set and the non-TF control set. Though the purpose of this paper is to make use of existing prediction methods and not to evaluate them (which has already been done by others, for example [2, 21]), it is important to have at least an overall sense of how each method is performing on our protein sets. A summary of the results of the three methods is shown in Figure 2 and listed in Table 1. In Figure 2, we compare the fraction of each protein's residues predicted as disordered by each method. In Table 1, we calculate the total percentage of protein residues predicted as disordered by each method, along with the average length of each predicted IDR. The figure and table reveal that all three methods consistently predict proteins in the TF set to have a greater fraction of disordered residues, more disordered residues, and longer IDRs than proteins in the non-TF control set, confirming earlier findings that IDRs are enriched in TFs.
Fig. 2.
Distributions of the fraction of each protein's residues predicted as disordered by each method for the proteins in (A) the TF set and (B) the non-TF control set.
Table 1.
Statistics summarizing disorder predictions on all the residues of all the proteins in both the TF set and the non-TF control set using three different disorder prediction tools.
| TF set | non-TF ctrl set | |||
|---|---|---|---|---|
|
| ||||
| % of residues predicted in IDRs | average length of IDRs | % of residues predicted in IDRs | average length of IDRs | |
|
| ||||
| PONDR VSL2 | 83.2% | 106 | 53.3% | 39 |
| DISOPRED2 | 47.4% | 44 | 34.1% | 36 |
| PreDisorder 1.1 | 50.1% | 19 | 38.3% | 18 |
As an aside, it is apparent that PONDR VSL2 is far more likely than the other two methods to call a residue as disordered, in both the TF set and the non-TF control set, suggesting that the method is probably operating at a different point on its receiver operating characteristic (ROC) curve, with high sensitivity but also perhaps a relatively high false positive rate [21]. In addition, the average length of IDRs predicted by PONDR VSL2 is higher than the other two methods, which may be related to the previous point, but may also be because the method uses different SVMs to predict IDRs in short and long sequences separately.
3.2. Assessing significance of order or disorder within and flanking human TF DBDs
To systematically study the associations between IDRs and DBDs, for each occurrence of a DBD class within a human TF, we calculated 30 different p-values: the significance under two different null models (based on the TF set and the non-TF control set) of five different kinds of disorder features (DBD, N-terminal flank, C-terminal flank, both flanks, and entire TF) as computed by three different prediction methods (PONDR VSL2, DISOPRED2, and PreDisorder 1.1). For each combination of null model and feature, we say that the feature exhibits significant disorder under that null model if at least two of the three prediction methods predict disorder at p-value ≤ 0.005; on the other hand, we say that the feature exhibits significant order under that null model if at least two of the three prediction methods predict disorder at p-value ≥ 0.995. Note that it is certainly possible for a feature to be neither significantly ordered nor significantly disordered under a particular null model.
Although we computed whether features exhibited significant order or disorder across all Pfam DBDs occurring in our TF set, to avoid artifacts due to small sample size, we restricted our subsequent analysis to the 32 DBD classes with at least five occurrences in the TF set. Many of the most frequent DBD classes, including the 10 most prevalent ones, are structurally similar and can be roughly classified into two groups: (1) those containing zinc fingers, and (2) those containing a basic helix-turn-helix type of domain, domains in which helices are separated by loops (e.g., Homeobox, HLH, Fork_head, Ets). The enrichment analysis results for these 32 DBD classes are listed in Table 2; at the bottom of the table, we also included the Pfam domains Basic, AT_hook, and P53 (Basic and AT_hook are included because we mention them below in comparison to another study; P53 is a well-studied DBD included for general interest).
Table 2.
Enrichment analysis of significantly occurring ordered and disordered regions within and flanking human TF DBDs.
| DBD | N-terminal flank | C-terminal flank | both flanks | whole TF sequence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| No. | DBD (Pfam) | TF family | average DBD length (res.) | number of DBDs in TFs | TF set | non-TF Ctrl set | TF set | non-TF Ctrl set | TF set | non-TF Ctrl set | TF set | non-TF Ctrl set | TF set | non-TF Ctrl set |
| 1 | PF00096 | zf-C2H2 | 23.1 | 4154 | OR | OR | OR | OR | OR | OR | OR | OR | OR | OR |
| 2 | PF00046 | Homeobox | 56.3 | 216 | OR | OR | ID | ID | ID | ID | ID | ID | ID | ID |
| 3 | PF00010 | HLH | 53.3 | 100 | OR | OR | ID | ID | ID | ID | ID | ID | – | ID |
| 4 | PF00505 | HMG_box | 68.0 | 56 | OR | OR | ID | ID | ID | ID | ID | ID | – | ID |
| 5 | PF00250 | Fork_head | 98.2 | 47 | OR | OR | ID | ID | ID | ID | ID | ID | ID | ID |
| 6 | PF00105 | zf_C4 | 70.2 | 45 | OR | OR | ID | ID | ID | ID | ID | ID | OR | – |
| 7 | PF00249 | Myb_DNA-binding | 47.2 | 43 | OR | OR | – | – | – | ID | – | – | – | – |
| 8 | PF00170 | bZIP_1 | 64.2 | 34 | OR | OR | ID | ID | – | – | ID | ID | ID | ID |
| 9 | PF00178 | Ets | 85.0 | 27 | OR | OR | ID | ID | ID | ID | ID | ID | – | – |
| 10 | PF00320 | GATA | 35.1 | 20 | – | OR | ID | ID | ID | ID | ID | ID | – | ID |
| 11 | PF00907 | T-box | 187.6 | 18 | – | – | ID | ID | ID | ID | ID | ID | – | – |
| 12 | PF01530 | zf-C2HC | 31.0 | 14 | ID | ID | ID | ID | ID | ID | ID | ID | ID | ID |
| 13 | PF02319 | E2F_TDP | 73.8 | 13 | – | – | ID | ID | – | – | – | ID | – | – |
| 14 | PF00313 | CSD | 68.6 | 12 | – | – | – | – | – | – | – | – | – | – |
| 15 | PF05485 | THAP | 89.2 | 12 | – | – | ID | ID | ID | ID | ID | ID | – | – |
| 16 | PF01422 | zf-NF-X1 | 21.5 | 11 | – | – | – | – | – | – | – | – | – | – |
| 17 | PF03165 | MH1 | 109.9 | 11 | – | – | ID | ID | ID | ID | ID | ID | – | – |
| 18 | PF07716 | bZIP_2 | 54.0 | 10 | – | – | ID | ID | – | – | ID | ID | – | – |
| 19 | PF00292 | PAX | 125.6 | 9 | – | – | ID | ID | ID | ID | ID | ID | – | – |
| 20 | PF00098 | zf-CCHC | 17.9 | 8 | – | – | – | ID | – | ID | – | ID | – | – |
| 21 | PF00808 | CBFD_NFYB_HMF | 63.1 | 8 | – | – | – | ID | – | – | – | – | – | – |
| 22 | PF04218 | CENP-B_N | 52.5 | 8 | – | – | – | – | – | – | – | – | – | – |
| 23 | PF00751 | DM | 47.0 | 7 | – | – | ID | ID | ID | ID | ID | ID | – | ID |
| 24 | PF01342 | SAND | 79.0 | 7 | – | – | ID | ID | ID | ID | ID | ID | – | – |
| 25 | PF02257 | RFX_DNA_binding | 72.7 | 7 | – | – | ID | ID | – | – | – | – | – | – |
| 26 | PF02864 | STAT_bind | 251.9 | 7 | – | – | – | – | – | – | – | – | – | – |
| 27 | PF02892 | zf-BED | 50.1 | 7 | – | – | ID | ID | ID | ID | ID | ID | – | – |
| 28 | PF10401 | IRF-3 | 174.0 | 7 | – | – | – | – | – | – | – | ID | – | – |
| 29 | PF00447 | HSF_DNA-bind | 104.2 | 6 | – | – | – | – | ID | ID | ID | ID | – | – |
| 30 | PF04516 | CP2 | 227.2 | 6 | – | – | – | – | – | – | – | – | – | – |
| 31 | PF03299 | TF_AP-2 | 208.2 | 5 | – | – | – | ID | – | – | ID | ID | – | – |
| 32 | PF05044 | Prox1 | 224.0 | 5 | – | – | ID | ID | – | – | ID | ID | – | – |
|
| ||||||||||||||
| PF01586 | Basic | 91.0 | 4 | ID | ID | ID | ID | ID | ID | ID | ID | ID | ID | |
| PF00870 | P53 | 196.3 | 3 | – | – | ID | ID | ID | ID | ID | ID | – | – | |
| PF02178 | AT_hook | 109.0 | 1 | ID | ID | ID | ID | ID | ID | ID | ID | ID | ID | |
Notes:
The DBDs with at least 5 occurrences in the TF set are listed in the table, together with Basic, P53, and AT_hook.
ID indicates significant disordered DBDs, DBD flanks, or TFs (in at least two of three methods, p-value ≤ 0.005).
OR indicates significant ordered DBDs, DBD flanks, or TFs (in at least two of three methods, p-value ≤ 0.005).
A dash (–) indicates entries that are neither significantly ordered nor significantly disordered.
The DBDs with fewer than 5 occurrences in the TF set include: Runt, TEA, Basic, HALZ, z-alpha, FYRN, FYRC, P53, ARID, DMA, AKAP95, GATA-N, P53_tetramer, Homez, XPA_N, zf-DHHC, GCM, CG-1, Vert_HS_TF, SIM_C, Rad51, HAND, Beta-trefoil, LAG1-DNAbind, PWI, zf-MYND, SAP, GCR, Oest_recep, Prog_receptor, zf-TRAF, zf-CHY, Vert_IL3-reg_TF, HSA, Rio2_N, BrkDBD, zf-RAGl, AT_hook, and TMF_DNA_bd.
The top 10 most frequently occurring DBD classes in human TFs all exhibit significant order within the DBD itself, suggesting that structural flexibility within these domains is rather limited. Strikingly, our results indicate that although the DBDs themselves exhibit significant order, the regions flanking the DBDs are likely to exhibit significant disorder. Only in the case of zf-C2H2 do the flanking regions exhibit significant order (this will be discussed further in the next section). In contrast, 26 of the other 31 DBDs exhibit significant disorder in either the N-terminal flank, the C-terminal flank, or both; and none of the other 31 DBDs exhibit significant order in either flank under either null model. This is consistent with prior studies in which it was found that DBDs are often separated by flexible linker regions, allowing TFs to bind DNA with fine control over DNA binding a nity [22, 23].
3.3. Investigating detailed spatial relationships of IDRs relative to DBDs within TFs
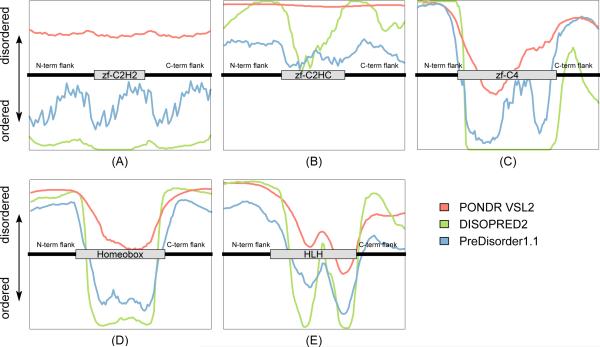
To further investigate the detailed spatial relationships of the IDR predictions of the three different methods to protein DBDs, we generated a meta-plot of the average predicted order/disorder in the vicinity of each Pfam DBD according to each prediction method. To do this, we first identified all occurrences of a Pfam DBD in the TF set, and then across all those occurrences, calculated the average (mean) order/disorder score predicted by each method at each residue within the DBD match and both of its flanks (up to 30 amino acids). In cases where a TF contained only a partial DBD match and not a full domain according to the HMMER alignment, we considered only the aligned region in our calculations. We normalized the resulting scores for the purpose of comparison across methods, and for uniformity in scale across plots for different DBD classes (Figure 3).
Fig. 3.
Shown are meta-plots for five prevalent DBDs in human TFs. (A) zinc-finger C2H2-type (length: ~23 amino acids), (B) zinc-finger C2HC-type (length: ~31 amino acids), (C) zinc-finger C4-type (length: ~70 amino acids), (D) homeodomain fold (length: ~58 amino acids), and (E) helix-loop-helix (length: ~53 amino acids).
Figure 3 displays meta-plots for five of the ten Pfam DBDs most prevalent in human TFs. Results from DISOPRED2 and PreDisorder 1.1 are fairly consistent across all five domain classes. Moreover, all three methods are in good agreement in zf-C2HC and demonstrate similar prediction trends in zf-C4, Homeobox, and HLH. Extended to all the DBDs listed in Table 2, over 67.2% of the DBD classes that are found to exhibit either significant disorder or significant order are identified as such by all three methods.
Nevertheless, some discrepancies in the results from the different methods are evident, such as zf-C2H2. The C2H2-type zinc finger domain is the most prevalent DBD class found in metazoan TFs, including in human [24]. It is also one of the most highly ordered DBDs; however, the linker regions between these C2H2 zinc finger domains are often disordered [25]. As shown in Figure 3A, PONDR VSL2 reports that the C2H2 domain occurrences in human TFs exhibit significant disorder in both the C2H2 domain itself and the adjacent N- and C-terminal flanks; however, DISOPRED2 and PreDisorder both report the opposite, namely that zf-C2H2 and its flanks exhibit significant order. Liu and colleagues carefully analyzed the diffculties of predicting intrinsic disorder in the zf-C2H2 domains and their linker regions [4]. They concluded that because many linker regions between C2H2 zinc fingers are quite short, the windowing procedures employed by some IDR prediction algorithms prevent them from being detected as disordered; the result is an artifact in which linker regions between C2H2 zinc fingers are over-predicted as being ordered.
3.4. Analyzing spatial relationships for some DBD classes prevalent in human TFs
3.4.1. Zinc fingers
Zinc fingers are small structural motifs whose folds are stabilized by coordination of one or more zinc ions. Zinc fingers can be classified according to their zinc-coordinating residues and folds. In Figures 3A–C, we show our IDR prediction results for the three major zinc finger domain classes found in human TFs: zf-C2H2 (the most prevalent DBD class in human TFs), zf-C4 (also referred to as nuclear receptors), and zf-C2HC. Although all three classes contain zinc fingers, we find variability in their regions of order and disorder. As discussed above, the C2H2 zinc finger domain is itself believed to be highly ordered, with individual ordered zinc fingers separated by highly flexible linker regions [25]. We find that the C4 domain exhibits significant order within the DBD itself, but significant disorder in flanking regions. In contrast, we find that the C2HC domain exhibits significant disorder in both the DBD and flanking regions.
3.4.2. Homeobox
Homeobox (homeodomain fold) is the second-most abundant DBD class within human TFs. The homeodomain fold consists of an approximately 60 amino acid helix-turn-helix structure in which three alpha helices are connected by short loop regions. Our results (Figure 3D) extend the results of a prior study [7] that found multiple intrinsically disordered sequences located outside the homeodomain DBD of the Drosophila TF Ubx, that allow Hox family members (i.e., a subclass of TFs with Homeobox DBDs) to bind DNA with high affinity but relatively low specificity [26, 27].
3.4.3. HLH
HLH (basic helix-loop-helix) is the third-most abundant DBD class within human TFs, and is characterized by two α-helices connected by a loop. TFs that have this domain typically bind DNA as either homo- or hetero-dimers, with each monomer contacting DNA through a helix containing basic residues that facilitate DNA binding [28]. As shown in Figure 3E, all three methods report that HLH exhibits significant order within the domain itself, but significant disorder in both the N- and C-terminal flanking regions. Our results also indicate that a short but highly disordered region may frequently occur in the middle of the HLH domain, consistent with prior observations that the linker regions and the loop region of HLH proteins are of higher flexibility, allowing dimerization by folding and packing one smaller helix against the other one [28].
4. Discussion
In this study, we used three different computational disorder prediction methods to investigate the prevalence of IDRs within DBDs and in their flanking regions across essentially the entire repertoire of human, sequence-specific TFs and their associated Pfam DBDs. Our choice of multiple prediction methods was motivated by a desire to be able to draw robust conclusions that were not dependent on any one particular method.
Previously it was found that TFs are enriched for IDRs [3, 4]. At the same time, DBDs responsible for TF binding did not seem themselves to be particularly enriched for IDRs. For example, of the 25 DBDs studied in [4], only the Basic and AT_hook domains exhibited high amounts of disorder; however, those domains are not particularly prevalent in human TFs, occurring in our TF set just four times and one time, respectively.a We were intrigued by the possibility that the enrichment of IDRs observed in TFs might be at least partly due to disorder in the regions flanking DBDs; under such a hypothesis, DBDs can be thought of as islands of order flanked by regions of disorder.
Our results support exactly such a hypothesis: the most prevalent DBDs in human TFs exhibit significant order, but the flanking regions of these DBDs generally exhibit significant disorder. Similarly, although DBDs of intermediate prevalence (occurring between 5 and 20 times in our TF set) do not appear often enough to exhibit either significant order or disorder within the domains themselves, most of them still exhibit significant disorder in one or both flanking regions.
The functional role played by the significant prevalence of disorder in the regions flanking DBDs of human TFs is unclear. However, we can speculate that the increased flexibility a orded by these flanking IDRs might contribute to the ability of TFs to 1) recognize target sequences in the DNA appropriately, 2) bind to a wider diversity of DNA target sequences, 3) be anchored with higher affinity to the DNA after recognizing target sequences, 4) bind to other factors and complexes positioned on the DNA or involved in transcriptional regulation, or 5) present activation domains to downstream transcriptional regulatory machinery. It should be emphasized that these possibilities are speculative; however, the results of this study suggest numerous testable hypotheses regarding the roles of N- and C-terminal regions flanking DBDs for many frequently occurring DBDs in hundreds of human TFs. For example, the importance of the predicted disorder in these flanking regions in determining or modulating the DNA binding affinity and/or specificity of the associated TFs could be investigated with protein binding microarrays (PBMs) [29, 30]. PBMs could assay the affinity and/or specificity of proteins representing the DBDs with their flanking regions, as compared to either the DBDs alone or the DBDs with mutant flanking regions predicted not to be significantly disordered. If found to contribute to the DNA binding affinity and/or specificity of TFs, IDRs that flank DBDs would broaden the scope of functional domains to be considered when evaluating the potential impact of mutations or natural polymorphisms within exomes, such as in medical sequencing projects.
This study was focused on human TFs; however, since these DBD classes are the predominant DBD classes not just in human TFs but throughout eukaryotes, the results of this study may have important implications for studies of TFs across all eukaryotes.
5. Acknowledgments
We are grateful to Lizzy Rossin for creating the initial list of Pfam DBDs in human TFs. This work was funded by grants from NIH (R01 HG003985) to M.L.B., and from NIH (P50 GM081883-01) and DARPA (HR0011-09-1-0040) to A.J.H.
Footnotes
References
- [1].Eliezer D, et al. Curr. Opin. Struct. Biol. 2009 Feb;19:23. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].He B, et al. Cell Res. 2009 Aug;19:929. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- [3].Minezaki Y, et al. J. Mol. Biol. 2006 Jun;359:1137. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- [4].Liu J, et al. Biochemistry. 2006 Jun;45:6873. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fuxreiter M, et al. Trends Biochem. Sci. 2011 Aug;36:415. doi: 10.1016/j.tibs.2011.04.006. [DOI] [PubMed] [Google Scholar]
- [6].Schwabe JW, et al. Structure. 1993 Nov;1:187. doi: 10.1016/0969-2126(93)90020-h. [DOI] [PubMed] [Google Scholar]
- [7].Liu Y, et al. J. Biol. Chem. 2008 Jul;283:20874. doi: 10.1074/jbc.M800375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schaufler LE, et al. J. Mol. Biol. 2003 Jun;329:931. doi: 10.1016/s0022-2836(03)00550-3. [DOI] [PubMed] [Google Scholar]
- [9].Crane-Robinson C, et al. Trends Biochem. Sci. 2006 Oct;31:547. doi: 10.1016/j.tibs.2006.08.006. [DOI] [PubMed] [Google Scholar]
- [10].Vaquerizas JM, et al. Nat. Rev. Genet. 2009 Apr;10:252. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- [11].Flicek P, et al. Nucleic Acids Res. 2011 Jan;39:D800. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pruitt KD, et al. Nucleic Acids Res. 2009 Jan;37:D32. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Campen A, et al. Protein Pept. Lett. 2008;15:956. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dunker AK, et al. J. Mol. Graph. Model. 2001;19:26. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- [15].Finn RD, et al. Nucleic Acids Res. 2010 Jan;38:D211. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hunter S, et al. Nucleic Acids Res. 2009 Jan;37:D211. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Eddy SR, et al. Genome Inform. 2009 Oct;23:205. [PubMed] [Google Scholar]
- [18].Peng K, et al. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ward JJ, et al. J. Mol. Biol. 2004 Mar;337:635. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- [20].Deng X, et al. BMC Bioinformatics. 2009;10:436. doi: 10.1186/1471-2105-10-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bordoli L, et al. Proteins. 2007;69(Suppl 8):129. doi: 10.1002/prot.21671. [DOI] [PubMed] [Google Scholar]
- [22].Zhou HX, et al. Biochemistry. 2001 Dec;40:15069. doi: 10.1021/bi015795g. [DOI] [PubMed] [Google Scholar]
- [23].Fukuchi S, et al. J. Mol. Biol. 2006 Jan;355:845. doi: 10.1016/j.jmb.2005.10.037. [DOI] [PubMed] [Google Scholar]
- [24].Tupler R, et al. Nature. 2001 Feb;409:832. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- [25].Pabo CO, et al. Annu. Rev. Biochem. 2001;70:313. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- [26].Gehring WJ, et al. Cell. 1994 Jul;78:211. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- [27].Hoey T, et al. Nature. 1988 Apr;332:858. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- [28].Littlewood TD, et al. Protein Profile. 1995;2:621. [PubMed] [Google Scholar]
- [29].Mukherjee S, et al. Nat. Genet. 2004 Dec;36:1331. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Berger MF, et al. Cell. 2008 Jun;133:1266. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]