Abstract
Heart failure is one of the major causes of death in the Western world because cardiac muscle loss is largely irreversible and can lead to a relentless decline in cardiac function. Novel therapies are needed since the only therapy to effectively replace lost myocytes today is transplantation of the entire heart. The advent of embryonic and induced pluripotent stem cell (ESC/iPSC) technologies offers the unprecedented possibility of devising cell replacement therapies for numerous degenerative disorders. Not only are ESCs and iPSCs a plausible source of cardiomyocytes in vitro for transplantation, they are also useful tools to elucidate the biology of stem cells that reside in the adult heart and define signaling molecules that might enhance the limited regenerative capability of the adult human heart. Here, we review the extracellular factors that control stem cell cardiomyogenesis and describe new approaches that combine embryology with stem cell biology to discover drug-like small molecules that stimulate cardiogenesis and potentially contribute to the development of pharmaceutical strategies for heart muscle regeneration.
Keywords: Cardiogenesis, Small molecules, Drug discovery, Regeneration
Introduction
There are approximately 1.2 million new incidences of heart failure every year in the USA, making it the major cause of mortality [1]. The underlying problem, in essence, is that up to a billion cardiomyocytes can be lost after myocardial infarction and that endogenous regeneration cannot restore the heart’s function, resulting in increased risk for death by heart disease. Consequently, over the past two decades, there has been an intense research effort into the development of heart cell replacement therapies, with the major emphasis on generation of cardiomyocytes. In principal, cardiomyocytes could be replaced using exogenous sources such as human embryonic stem cells (hESCs) or human induced pluripotent stem cells (hiPSCs) [2]. hESC-derived cardiomyocytes resemble immature human fetal cardiomyocytes by multiple criteria, including electro-physiology [3–5], calcium handling [4, 6, 7], force generation [5, 7], contractile protein expression and myofibrillar structure [8], and cardiomyocytes from hiPSCs appear similar [9]. Because hESC-derived cardiomyocytes have the potential, albeit limited, to engraft into surgical models of heart disease [10, 11], they and hiPSC-derived cardiomyocytes have been considered for replacement therapies. However, despite encouraging advances, the use of hESC/hiPSC-derived cardiomyocytes for basic developmental research and large-scale applications, such as high throughput screening, toxicology testing, and large animal pre-clinical studies, has been hampered by their poor yield from typically heterogeneous (and expensive) stem cell cultures. A number of naturally occurring, diffusible signaling molecules and intracellular mediators are known to drive cells in the developing heart field to form a functional heart tube during embryogenesis. Judicious testing of such factors by several groups has led to optimized defined conditions for production of cardiomyocytes [10, 12, 13]. Although such advances quantitatively improved the proportion of the cells that differentiate into cardiomyocytes, efficient, large-scale differentiation towards heart cells remains challenging. A potentially greater challenge for hESC- or hiPSC-derived cardiomyocytes comes from the fact that these cardiomyocytes have to be transplanted into the injured heart where they would need to survive and become functionally and safely integrated into the patient’s myocardium. Aside from the need to develop effective and safe delivery modalities, hESC/hiPSC-derived cardiomyocytes are functionally immature and the signals that drive maturation are largely unknown.
One of the major advances in cardiology research has been the discovery that the adult mammalian heart is endowed with regenerative capacity, albeit insufficient to offset the cell death in heart disease. Originally controversial, recent pulse-chase studies have confirmed that new myocytes arise in the mammalian heart [14, 15]. Four potential cell sources have been identified to date: (1) a subpopulation of mononuclear cardiomyocytes may be able to de-differentiate and proliferate in certain conditions. Indeed it has been reported that cardiomyocytes can be induced to re-enter cell cycle by either removing cell cycle inhibitors, or by over-expressing cell cycle activators, or by providing growth factors, such as neuregulin1 and periostin or in particular culture conditions, but it is not clear to which extent this phenomenon occurs in vivo [16–18]. (2) Circulating progenitor cells derived from the bone marrow may be recruited to the heart in response to an injury and acquire, at least in part, some regenerative capacity [19]. (3) It was recently shown that epicardial cells respond to injury by re-activating the embryonic program, generating multipotent mesenchymal cells that could contribute to regeneration, via vasculogenesis and to the regulation of myocardial tissue remodeling [20, 21]. (4) Several populations of progenitor cells have been identified in the heart, based on the expression of specific markers (c-kit, Sca1, Isl1) [22–24], or on the basis of functional properties, such as the ability to efflux the vital dye Hoechst (side population) [25] or to spontaneously migrate from explants and grow in three-dimensional structures called cardiospheres [26, 27].
Remodeling of the myocardium after injury might not create a conducive environment for cardiac regeneration. In response to a myocardial infarction, necrotic tissue is rapidly replaced by a fibrotic scar, and the hostile inflammatory milieu and the lack of oxygen in the ischemic area may limit the survival and proliferation of endogenous progenitor cells, possibly enhancing scar formation and even diverting endogenous stem cells towards a fibrogenic lineage [28, 29]. In this context, endogenous mechanisms of regeneration could be enhanced by the identification of small molecules acting on specific pathways, leading to improved cellular resistance to oxidative stress and to increased proliferation and commitment of progenitor populations to cardiomyocyte and other cardiopoietic lineages, including vascular endothelial cells to increase blood supply to the injured tissue.
Development of small molecule drugs that could act on adult cardiac progenitors is hampered by currently fragmentary knowledge of the stem and progenitor cells in the heart and the processes that direct them to differentiate into functional myocytes. Studies of endogenous adult cardiac stem or progenitor cells show that they not only express gene and protein markers of stem cells but that they also express markers that reveal their commitment to the cardiomyogenic lineage, raising the possibility that these cells might respond to some of the same signals that promote later stages of hESC/hiPSC cardiomyogenic differentiation (e.g., when committed progenitors are induced to form cardiomyocytes or other myocardial cell types) [15, 30, 31]. Thus, it seems likely that signaling pathways that control later stages of ESC/iPSC differentiation might also drive endogenous cardiomyocyte regeneration. Therefore, small molecules that promote cardiomyocyte production from hESC/hiPSC-derived progenitors might also enhance endogenous regeneration.
There are four main research strategies to discover natural and synthetic molecules that stimulate endogenous myocardial regeneration. The first is to define the mechanisms for cardiac self-repair in lower vertebrates such as zebrafish [28, 32]. Since cardiac regeneration in zebrafish occurs by reactivation of the cell cycle in order to produce new heart muscle (by replication of pre-existing adult cardiomyocytes) [33, 34], aspects of cell cycle reactivation might be exploited for human regeneration. The goal of this approach is similar to that of the second strategy, which is to study the cell cycle constraints of adult human ventricular myocytes in order to discover drugs that would override the post-mitotic phenotype [35, 36]. A third approach is to characterize stem or progenitor cells in adult hearts, including the cellular signals that control their proliferation and differentiation in the damaged heart [15, 30, 31]. Because purifying large numbers of endogenous cardiac cells for high throughput applications remains challenging, and the nature of the cells still engender controversy, a fourth strategy uses hESCs and hiPSCs as an entry point into the discovery of drug-like small molecules to stimulate endogenous repair (illustrated in Fig. 1). Specifically, cardiogenic progenitor cells derived from hESCs and hiPSCs are used in screens to identify drug-like small molecules that could then be developed into reagents to identify druggable targets, and eventually drugs themselves, to enhance myocardial regeneration. Below, we focus on the practical considerations surrounding the implementation of this latter approach and its prospects for yielding drugs for therapeutic regeneration.
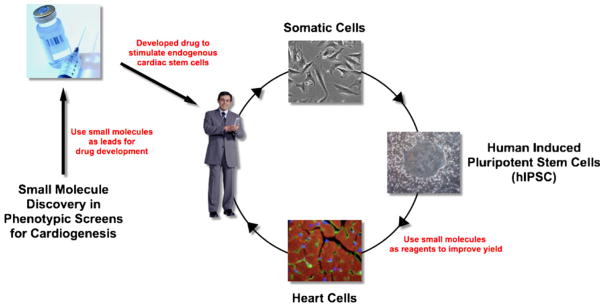
Fig. 1.
Schematic overview of proposed therapies for cardiac regeneration. Small molecule discovery experiments are emerging to generate novel tool compounds that can be used to identify and evaluate drug targets for regeneration of heart tissue from endogenous stem or progenitor cell sources. Reagent compounds can also be used in a second paradigm to produce cells for potential tissue engineering and transplantation research and clinical applications
Cardiac Differentiation Signals: What is Known?
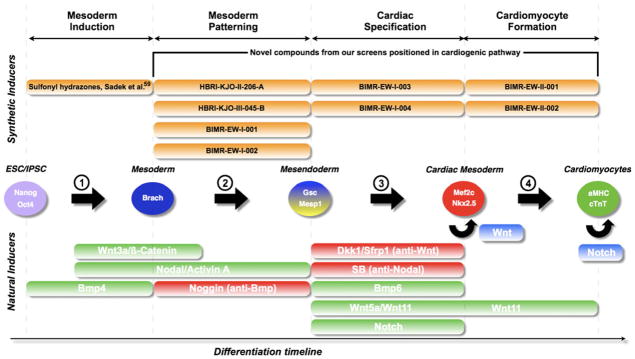
ESCs proceed through four sequential steps to form cardiomyocytes, namely: (1) formation of mesoderm, (2) patterning towards anterior mesoderm or cardiogenic mesoderm, (3) formation of cardiac mesoderm, and (4) (seemingly) spontaneous development and maturation to cardiomyocytes (Fig. 2). Each of these steps can be characterized by temporally expressed marker genes, facilitating the analysis of signaling networks in each discrete step: mesoderm can be marked by Brachyury/T, cardiogenic mesoderm can be marked by Goosecoid (Gsc) or Mesp1, early cardiac mesoderm can be characterized by Nkx2.5 and Mef2c, and, lastly, maturing cardiomyocytes are visualized by spontaneous beating and the presence of contractile proteins such as α-myosin heavy chain (αMHC) and cardiac troponin T (cTnT).
Fig. 2.
Natural and chemical biology of cardiogenesis in ESCs/iPSCs. A schematic representation of the steps that are required to generate cardiomyocytes from ESCs or iPSCs involving: (1) mesoderm induction, (2) mesoderm patterning, (3) formation of cardiogenic mesoderm, and (4) formation of cardiomyocytes. Biological inducers are mapped to the temporal steps in green (activation of the pathway), red (inhibition of the pathway), and blue (maintenance of a progenitor). Chemical inducers are mapped in orange (unpublished work and published compounds)
The signals that control mesoderm formation have been well characterized in the mammalian and amphibian embryo, and as a consequence, numerous studies have been able to demonstrate that the addition of Wnts, Bmps, and the TGFβ family member Nodal efficiently induces mesoderm in ESCs (Fig. 2, step 1) [37, 38]. Additionally, some of the signals that pattern embryonic mesoderm toward cardiogenic mesoderm have been found to act on both mouse and human ESCs (Fig. 2, step 2) [39–41].
Even though some of the signaling events required for steps 3 and 4 in cardiac differentiation have been unraveled in the embryo, little is known about how these might be applied to ESC cardiogenesis or whether they can be applied to heart regeneration. Nodal and Wnt inhibition regulate formation of cardiomyocytes in Xenopus and chick embryos [42–45] and appear to be crucial for mouse ESC (mESC) differentiation into cardiomyocytes [46–48]. Also, Notch was identified as a factor driving the induction of cardiogenesis from an ESC-derived mesoderm subpopulation through an indirect mechanism regulated by a combination of the growth factors Wnt5a, Bmp6, and Sfrp1 (Fig. 2, step 3) [49].
Differentiation of committed cardiac progenitors to beating cardiomyocytes is the final step in a series of differentiation cues to cardiomyocytes from ESCs, and is a poorly understood mechanism that often occurs spontaneously in vitro but might be controlled by factors such as Wnt11 (Fig. 2, step 4) [47].
In addition to recapitulating embryonic signals that control early events of cardiac differentiation, strategies that could improve cardiomyocyte yields through enhancing replication of committed progenitors might also be useful. For example, canonical Wnt signals are able to expand the pool of Nkx2.5+, Isl1+ early cardiac progenitors, providing a promising outlook for increasing yields of cardiomyocytes from cardiogenic mesoderm [50, 51]. Also, BMPs and FGFs control a balance between differentiation and proliferation, respectively, mediated by Msx1 [52]. Furthermore, activation of the Notch pathway in immature cardiomyocytes can prolong their period of replicative competence [53, 54], representing a different way to increase cardiomyocyte yields from ESCs.
In summary, a number of important pathways have been used to generate cardiomyocytes, including Nodal/TGFβ, Wnt, and BMP. Additional characterization of three crucial steps in cardiogenesis from ESC and iPSC could enhance the yield and maturity of in vitro generated ESC/iPSC-derived cardiomyocytes, which will benefit large-scale screens for drug discovery and drug safety as well as clinical applications of cardiomyocytes: (1) differentiation of mesendoderm to form committed cardiac mesoderm, (2) differentiation of cardiac mesoderm into cardiomyocytes, and (3) physiological maturation of cardiomyocytes.
Small Molecules: Filling the Gaps in Cardiac Biology
Small molecules are excellent tools to understand and probe the biology of cardiac differentiation of ESCs/iPSCs. They are discrete, well-characterized entities that can be delivered in known quantities and can enter the cell easily where they can modulate cellular signaling pathways. Moreover, they can be chemically improved to increase their potency, selectivity, or solubility (or other pharmaceutical properties) and can be used to probe complex molecular processes (reviewed in Xu et al. [55]). Phenotypic cell-based assays, using for example tissue-specific gene promoter reporter systems, have been developed for high throughput analysis and allow simultaneous screening of libraries comprising thousands of compounds. Although attractive from the perspective that many cellular proteins can potentially be targeted to elicit differentiation, phenotypic assays using stem cells pose challenges of biological complexity that hinders assay development, and as discussed in the next section, the identification and validation of cellular targets remains a bottleneck in the development of small molecule probes of stem cell cardiogenesis. For assay development, reproducible and efficient production of late-stage progenitors, especially from hESCs or hiPSCs, is the major bottleneck, although recent advances in directed differentiation protocols can be translated into greater throughput [10, 56, 57]. An inherent complexity of ESC-based assays is the heterogeneity of the cultures as well as the requirement for precise temporal control of signaling events to obtain specific cell types, necessitating producing progenitors of a particular stage in sufficient quantity and purity for an high throughput (HT) assay that quantifies their transition to a subsequent step in the differentiation program. The existing knowledge of signals that direct differentiation can be used to prepare cultures enriched for progenitors at a particular stage, and assays can then be developed that screen for compounds that direct transition to a subsequent cardiopoietic stage visualized by gene promoter-fluorescent protein reporter, such as enhanced green fluorescent protein (eGFP), e.g., Nkx2.5-eGFP or αMHC-eGFP [58, 59]. Although enriched for a desired progenitor, the cultures are still heterogeneous and any hits identified may act indirectly, targeting other cell types that inhibit or promote the formation of cardiac progenitors, necessitating secondary assays to distinguish direct versus indirect biological mechanisms of action.
Screening assays are classified as either high throughput screens (HTS), which are based on signal detection by a plate-reader technology, versus high-content screens (HCS), which use an automated microscope to acquire images that can be analyzed to yield multiple parameters. HCS image analysis permits intensity-thresholded masking of regions of interest that increases sensitivity and signal to background of the assay considerably [60]. More sophisticated approaches include a cytometric analysis (e.g., asking how many cells differentiate) that permits gating subset populations.
Despite the challenges with ESC/IPSC based screens, a few examples of chemical mediators of cardiogenesis have been reported in mESCs, including cardiogenols, ascorbic acid, isoxazolyl-serines, sulfonyl hydrazones, and DMSO [58, 59, 61–63]. Whereas some agents (e.g., DMSO and ascorbic acid [64]) have many effects on cells, other molecules are more selective and could be developed into probes of the signaling pathways that control cardiomyocyte development. Of the published compounds, only the sulfonyl hydrazones have been characterized as to their time window of functional activity and potential biological mechanism of action [59]. They appear to act early in the differentiation program and up-regulate the expression of the mesoderm marker T/Bra, reflecting an increase in the amount of mesoderm in the cultures that ultimately leads to an increased number of cardiac cells [59]. Initially, we developed and ran mESC-based HCS using a fluorescent cardiac specific reporter in search of molecules that would promote cardiogenesis in a wide time window, namely from day 2 to day 6 [65]. The assay identified new classes of compounds but most worked at an early time frame of differentiation when mesoderm was generated and specified (i.e., days 2–4, similar to the sulfonyl hydrazones), which mostly probe well-characterized differentiation steps. This limits their use to in vitro differentiation reagents and makes them unsuitable for in vivo therapies because such early progenitors do not reside in the adult. Therefore, second-generation mESC assays with serum-free differentiation conditions were developed to probe later stages of differentiation (i.e., days 4–6 and days 6–9 when mesoderm adopts a cardiac fate or when cardiomyocytes are formed) and screened against compounds from focused libraries (library selection is discussed in more detail below). These resulted in a handful of new “hits” indicated in Fig. 2 with respect to their timing of action to illustrate that HCS phenotypic screens can yield compounds that target specific stages of differentiation. The chemical nature and mode of action will be described in a forthcoming publication. Certain of these compounds passed through a series of chemical refinements and biological characterizations to be used as tool compounds to assess the effect on myocardial regeneration and possible drug candidate development.
Is the Source of Screening Libraries Important?
The success of a compound-screening program is not based solely on the raw number of compounds screened but rather on how well the library explores “chemical diversity space” and the “drug-like” nature of the library [66, 67]; hence, the judicious choice of a library is important. The concept of chemical diversity space alludes to the variety of types of functionality and chemical template types that are present in the library. In general, platforms for developing front-line drugs and biological probes largely derive from three classes of screening libraries: combinatorial synthetic chemicals, natural products (or biomimetics), and known active agents or drugs themselves. Sometimes, synthetic, combinatorial library compounds seem to cover only a limited and quite uniform chemical space, whereas existing drugs and particularly natural products exhibit much greater chemical diversity tending to distribute into a chemical space distributed more evenly throughout the library [68]. Molecules in natural product libraries (excluding peptide libraries) are often complex structures that are generally more difficult to synthesize and not readily modified. Natural product libraries contrast with medicinal chemical libraries in generally having more stereogenic centers (which increases the complexity of the molecule and its synthesis) and are generally more rigid. Increased rigidity limits the number of possible conformations (number of spatial arrangements of the atoms relative to each others), reducing the probability of fitting within a binding pocket of a target protein. On the contrary, decreased rigidity increases the number of potential conformations and, consequently, the likelihood that one conformation binds to one protein while a second binds another, reducing target selectivity of the molecule. Other chemical differences include the increased number of aromatic moieties frequently present in combinatorial library members and in the type of heteroatoms present (i.e., O and N atoms are enriched in members of natural product libraries, and S and halogen atoms are enriched in members of synthetic libraries) that might result in drug candidate development issues down the line.
Synthetic molecules with lead-like pharmaceutical properties are good starting points for the discovery of new biologically active molecules with “drug-like” properties. “Lead-like” properties and “drug-like” properties, although not mutually exclusive, are distinct [69, 70]. Lead-like molecules are typically small (MW=200–350), with modest lipophilicity (as defined by a LogP of 1.0–3.0), a single charge, and no chemically reactive functional groups. These properties leave room for structural modifications that generally occurs in the lead refinement process. For example, addition of functionality to a lead to increase potency is associated oftentimes with an increase in molecular weight and LogP. Incorporation of favorable pharmaceutical design principles in the original library can ensure significant functional group diversity and novel architectural platforms and lead to high-quality “hits”. Useful architectural platforms include: presence of aromatic and heteroaromatic rings, few stereogenic centers, low molecular weight, and lack of chemical reactivity. Desirable physicochemical properties in library members include strong conformational biases and constraints typically observed in fused-ring systems and rigidified molecules possessing strategically placed substituents that enrich a population which can bind to a biological target. Lack of floppiness is hypothesized to minimize the loss of entropy upon binding to the target and to improve the likelihood of observing a high-quality “hit”.
The last category of screening libraries (i.e., bioactive agents or drug libraries) is attractive because screening candidates have presumably undergone previous rigorous pharmaceutical property and toxicology testing and constitute excellent sources for lead refinement. During the period 1981–2002, over 1,031 new chemical entities were approved by the US FDA as drugs [71], and these drugs are generally available in library formats for screening purposes. Use of a combination of known active agents/drug molecule libraries, natural product libraries, and libraries covering a large, chemically diverse space is important in stem cell library screening research because cellular targets are unknown. Such an unbiased approach gives the best chance to identify small molecule active “hits” with sufficient potency from screening libraries and discovering new signaling pathways responsible for the biological activity monitored. Screening of such libraries in the cardiac assays discussed above has yielded several active molecules, and we now have an emerging collection of ” toolbox” molecules that can be used to specify cardiogenesis in temporally distinct steps of the cardiomyocyte developmental program.
Drug Candidate Target, Mechanism of Action, and In Vivo Efficacy
Structure–activity relationship (SAR) studies on a molecule nominated as a viable “hit” are essential to explore the chemical features that can improve the molecules’ potency, physiochemical and pharmaceutical drug-like properties. While a distinct strength of the unbiased high-content cell-based screening approach described herein is that knowledge of the target is not required and hence use of this approach can embrace numerous targets simultaneously, a complete understanding of the mode of action of the “hit” and a thorough understanding of the prospects of a lead to become a drug candidate include an appreciation of the molecular target. This is because the mechanism of action and target of the molecule can shed light on safety and clinical efficacy, as well as aid medicinal chemistry development of the lead as a drug (overview in Fig. 3). Identification of the molecular target of a lead molecule even in a nebulous system such as the cardiac differentiation paradigm is becoming feasible because of recent advances in mass spectrometry and other bioanalytical procedures. Determination of the ultimate mechanism of action of the lead may be more challenging, but considerable progress in this area of research has been made as well.
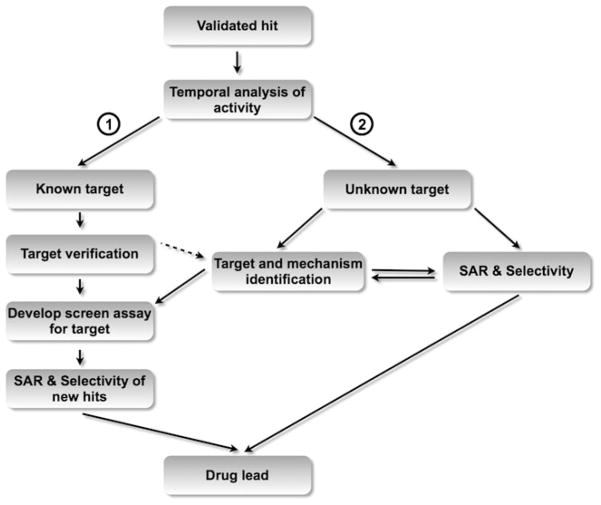
Fig. 3.
Target identification of hits. Once a “hit” has been validated, the extent of understanding of the target determines the development of a drug candidate. If a molecule with a known target is identified and the target is verified (branch 1), a new robust HTS assay for the target is developed to yield drug-like compounds that modulate the target of interest. If the target is not confirmed, these compounds are considered as unknown target molecules (dashed arrow). If the target of a hit is unknown (branch 2), the target needs to be identified first by biological and biochemical experimentation, before a drug lead can be developed either directly or through a new assay specific for that newly discovered target
The first step in characterizing the biology and identifying the target of a molecule is to understand the temporal developmental process that the lead molecules modulate. By carefully monitoring tissue-specific markers, it is possible to map a compound’s action to a particular step in the cardiogenic process. For the cardiac assays described above, gene expression, immunostaining, or fluorescent protein reporter analyses are used to visualize typical markers of cardiac fate such as Mesp1, Nkx2.5, Mef2c, or αMHC. Determining the temporal activity of a molecule in cardiac differentiation is crucial towards understanding whether a molecule would be suitable for stimulation of endogenous stem cells in vivo (Fig. 3).
A second step included in the identification of the direct target and mechanism of the lead is to examine the effect of the compound on candidate signaling pathways. “Hits” from screens of molecules with known targets (e.g., approved drugs) require minimal target analysis beyond validation and represent a relatively straightforward path to development of traditional target-based HTS or drug repurposing (Fig. 3, branch 1). Validation of the target is usually by either activation (e.g., over-expression of the target) or down-regulation (e.g., siRNA knock down). For “hits” with unknown targets (Fig. 3, branch 2), we take a candidate approach to determine if active compounds influence pathways known to modulate cardiac differentiation (discussed above). For all pathways examined, bioanalysis tools, such as the study of phosphorylation of downstream mediators and induction of direct targets as well as several candidate reporter systems in which specific gene promoter response elements drive luciferase expression, can shed light on the functional activity of the candidate molecule.
If known signaling pathways are not targeted, an informatics approach can be considered to reconstruct cellular signaling affected by the hit. Briefly, profiling of gene expression and phosphoproteins of small molecule-treated samples can be assembled into signaling networks by computer modeling [72, 73]. Gene expression profiling within a couple of hours after treatment of cells may provide clues, albeit indirect, on pathways that might be activated. Scrutiny of the phosphoproteome, in contrast, has the potential of directly unveiling the entire pathway. Given the central role played by protein kinases in controlling cell differentiation, interrogation of the phosphoproteome by mass spectrophotometry has been successful in helping unravel the control of cell behavior, including stem cell pluripotency [74, 75]. While this unbiased approach overcomes the limitations of Western blotting and other antibody-based approaches that probe a small target set limited by available reagents, there remain many challenges in generating samples and in the analysis of the total phosphoproteome dataset. Practical considerations that minimize throughput include: difficulty and cost of scaling up the assay to obtain large protein samples, sample processing time of about 1 month per sample, large variance between replicates and cost per sample. Additionally, the datasets generated require extensive statistical analysis and validation before signaling network reconstruction can proceed, but this can be accomplished through the use of protein–protein interaction databases, kinase prediction algorithms, and literature mining [74].
Use of affinity reagents also has considerable utility in identifying the target of lead compounds. In this regard, extensive SAR studies of candidate compounds can provide an understanding of regions of the molecule that could be amenable to chemical manipulation and elaboration of linkers to affinity moieties. For example, a useful strategy is to attach biotin through a linker system to a region of the molecule that is non-essential from the standpoint of potency [76]. Such affinity reagents aid in mapping the signaling activity of the molecule via biochemical analyses, including target pull downs and competition assays. In theory, pull down and competition assays seem very straightforward; however, several important drawbacks should be considered. For example, because libraries for screening often contain promiscuous compounds affecting several targets, multiple targets may be identified and this requires extra validation of each target. Moreover, one should be aware that such compounds may bind to different targets with different affinities, and that the actual target may not be identified due to strong binding to an abundant protein that is irrelevant to the mechanism of action.
Once candidate targets are obtained utilizing the above described strategies, additional studies are then required to verify the targets by inhibition of factors or pathways downstream of the small molecules discovered by implementing either siRNAs or well-known chemical inhibitors where available (for example, specific kinase inhibitors).
In summary, systems biology and affinity reagent technologies for target identification are beginning to show promise but still tend to be lengthy and tedious (and require more development) (Fig. 3, branch 2). Screening smaller scale libraries of well-characterized compounds that are highly selective for a specific target and in that way facilitate the target identification process immensely is an alternate approach that should be considered in the overall target identification strategy (Fig. 3, branch 1). After target identification, conventional approaches can be employed to validate the signaling pathway or protein as a target for consideration in a drug development pipeline, including validation in animal models and relevance to human disease.
An important early stage validation includes verification that the target or pharmacologically optimized tool compound is efficacious in a relevant animal disease model. Although a clinical endpoint, e.g., heart function, should be evaluated, endpoints more proximal to the effect of the compound or target give verifiable intended mechanism of action. As an example, cardiac stem cell progenitor differentiation or proliferation can be measured in transgenic mice harboring a reporter that allows visualization of progenitor cells, such as the Nkx2.5-eGFP mouse [77]. Flow cytometry and immunocytochemistry can reveal the potential effects of candidate compounds on proliferation and differentiation of committed Nkx2.5+ progenitors in vitro as well as in vivo. In fact, a modest increase in GFP-positive cells was observed in the adult mouse heart in response to myocardial infarction (Sean Wu, Massachusetts General Hospital, Boston, personal communication), suggesting that this could be a valuable model to evaluate the activation of the endogenous repair response in the adult heart.
Beyond Research Tool—“Hit” to “Drug Lead”
The moderate throughput phenotypic screening outlined above is envisioned as yielding tool compounds to investigate targets, which in turn would be validated and then used for a high throughput screening (HTS) campaign for drug discovery. Taking functionally active molecules or ” hits” from either HTS or HCS campaigns to lead drug candidate requires substantial process of characterization and refinement. From a chemistry perspective, the “hit to lead” paradigm includes improving potency and a host of pharmaceutical and physiochemical properties to produce compounds with drug-like properties. Optimal compounds (leads) can then be used in subsequent studies (i.e., in vivo testing). Very few lead compounds become drugs, but elaboration of drug-like properties into leads is an imperative step towards identifying a viable drug candidate. Compounds showing significant potency in screening assays (e.g., >50% inhibition at 10 μM or Z scores >10 in phenotypic assays) are judged to be sufficiently potent to be rescreened in a dose–response fashion in the same assay to validate their “hit” status and more accurately assess their potency. Compounds that show dose-dependent potency are labeled as validated “hits” and ranked in order of potency and drug-like properties for future development. Identification of screening “hits” is then verified with chemical counter tests. This involves confirming the chemical purity and integrity of the “hits” by analytical techniques and resynthesis.
The next (sometimes lengthy) recursive series of steps involves optimization of the “hits” for potency and drug-likeness properties. Because an optimization campaign is long and expensive, it is important to select screening “hit(s) ” with the greatest potential as lead candidates. Selection considerations include: potency and the drug-like nature of the template based on the lead-like properties described above. Additional important criteria include: ease of synthesis (<10 steps) and ease of synthesis of analogs (using a common core structure), cost-effectiveness and availability of diverse starting material for synthesis of analogs (diversity in the functional groups to be considered using a Craig-diagram [78], availability of SAR information, and promiscuity of the template and patent landscape. Christopher Lipinski’s rule-of-five analysis gives guidelines about properties and structural features that make molecules more or less drug-like with regard to their pharmaceutical and pharmacological profile including absorption, distribution, metabolism, excretion, and toxicology (ADMET properties) [79]. For development of new intellectual property, the most powerful patents include new composition of matter. Accordingly, synthesis of novel analogs of “hits” is desirable. For purposes of obtaining drug-like cardiomyocyte differentiation agents, the biological timing of a “hit” in the cardiac differentiation program is an important criterion as molecules working in early time windows of differentiation yield little to no benefits for stimulating endogenous stem cells and hence are of minimal value as drug candidates.
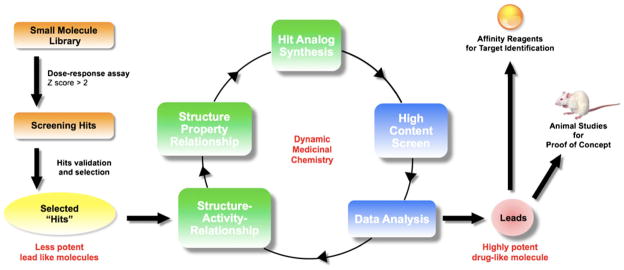
In summary, selected “hit(s) ” are optimized using a recursive approach including (1) purchasing analogs for testing (i.e., analog by catalog) and (2) synthesis of analogs and in vitro testing. Over the years, we have developed an approach called dynamic medicinal chemistry that takes into consideration potency and drug-likeness (e.g., ADMET properties) and involves medicinal chemical intuition and experience of the chemists [80] (Fig. 4). SAR and pharmaceutical structure–property relationships emerge from the data and guide the exploration, refinement, and optimization of potency and pharmaceutical property chemistry.
Fig. 4.
Overview of “hit” to drug refinement. Once identified by screening, a “hit” is selected for optimization based on its pharmacological and pharmaceutical prospects, and improved through several rounds of dynamic medicinal chemistry. Ultimately this should result in drug-like leads that can be used for testing in animal models and as the basis for affinity reagents to study mechanism of action
Prospects and Conclusions
The rationale of cardiac regeneration as a therapeutic target is predicated on the recognition that new myocytes can be created from multiple sources, including pre-existing myocytes and stem/progenitor populations. Since the signaling pathways that could enhance the regenerative potential of the adult heart are poorly understood, adopting a large-scale phenotypic screening approach to discovering probes and toolbox reagents may lead to the identification of novel drug targets that should eventually contribute to the development of pharmaceutical therapies to stimulate endogenous stem or progenitor cells to regenerate myocytes (and other cells) needed to restore the heart’s function as a biomechanical pump.
The major hurdle of the chemical biology approach—screening phenotypic assays as a means to identify targets—is that the current state-of-the-art approaches such as pull down and systems biology technologies lack the throughput needed to comprehensively evaluate even a fraction of the druggable targets within a cell. This issue makes the case for using focused libraries of molecules that have characterized targets, such as GPCR, kinase inhibitor, and known drug collections, or even siRNA and microRNA libraries that present a more straightforward path to new target information. Whether from unbiased or focused libraries, tool molecules should be useful to probe pathways involved in regeneration, test delivery modalities, and as reagents to enhance production of myocardial cells from ESCs and iPSCs for research and tissue engineering applications [81].
Developing high throughput in vitro assays that mimic complex in vivo biology represents a merger of stem cell and chemical biology technologies and should increase our knowledge of the pathways and proteins that control regeneration. Using chemical biology to expose the logic of regeneration should lead to a new generation of drug therapies for the treatment of heart disease.
Acknowledgments
This work was supported by grants from the NIH (R37HL59502, R33HL088266) and California Institute for Regenerative Medicine (CIRM) (RC1001321) to MM; CIRM (SEED RS1001691) and T Foundation to JRC; and CIRM Training Grant T2-00004 and American Heart Association for postdoctoral grant to EW.
Contributor Information
Erik Willems, Sanford-Burnham Medical Research Institute, 10901 N. Torrey Pines Rd., La Jolla, CA 92037, USA. ChemRegen Inc., 11171 Corte Cangrejo, San Diego, CA 92130, USA.
Marion Lanier, Human BioMolecular Research Institute, 5310 Eastgate Mall, San Diego, CA 92121, USA. ChemRegen Inc., 11171 Corte Cangrejo, San Diego, CA 92130, USA.
Elvira Forte, Sanford-Burnham Medical Research Institute, 10901 N. Torrey Pines Rd., La Jolla, CA 92037, USA.
Frederick Lo, Department of Bioengineering, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA.
John Cashman, Human BioMolecular Research Institute, 5310 Eastgate Mall, San Diego, CA 92121, USA. ChemRegen Inc., 11171 Corte Cangrejo, San Diego, CA 92130, USA.
Mark Mercola, Email: mmercola@sanfordburnham.org, Sanford-Burnham Medical Research Institute, 10901 N. Torrey Pines Rd., La Jolla, CA 92037, USA. ChemRegen Inc., 11171 Corte Cangrejo, San Diego, CA 92130, USA.
References
- 1.AHA. AHA Update 2010. 2010 http://www.aha.org.
- 2.Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes & Development. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 3.Kehat I, Gepstein A, Spira A, Itskovitz-Eldor J, Gepstein L. High-resolution electrophysiological assessment of human embryonic stem cell-derived cardiomyocytes: a novel in vitro model for the study of conduction. Circulation Research. 2002;91:659–661. doi: 10.1161/01.res.0000039084.30342.9b. [DOI] [PubMed] [Google Scholar]
- 4.Binah O, et al. Functional and developmental properties of human embryonic stem cells-derived cardiomyocytes. Journal of Electrocardiology. 2007;40:S192–S196. doi: 10.1016/j.jelectrocard.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 5.Kita-Matsuo H, et al. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS ONE. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Fu JD, Siu CW, Li RA. Functional sarcoplasmic reticulum for calcium handling of human embryonic stem cell-derived cardiomyocytes: insights for driven maturation. Stem cells (Dayton, Ohio) 2007;25:3038–3044. doi: 10.1634/stemcells.2007-0549. [DOI] [PubMed] [Google Scholar]
- 7.Dolnikov K, et al. Functional properties of human embryonic stem cell-derived cardiomyocytes: intracellular Ca2+ handling and the role of sarcoplasmic reticulum in the contraction. Stem cells (Dayton, Ohio) 2006;24:236–245. doi: 10.1634/stemcells.2005-0036. [DOI] [PubMed] [Google Scholar]
- 8.Laflamme MA, et al. Formation of human myocardium in the rat heart from human embryonic stem cells. The American Journal of Pathology. 2005;167:663–671. doi: 10.1016/S0002-9440(10)62041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germanguz I, et al. Molecular characterization and functional properties of cardiomyocytes derived from human inducible pluripotent stem cells. J Cell Mol Med. 2011;15:38–51. doi: 10.1111/j.1582-4934.2009.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laflamme MA, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nature Biotechnology. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 11.van Laake LW, Passier R, Doevendans PA, Mummery CL. Human embryonic stem cell-derived cardiomyocytes and cardiac repair in rodents. Circulation Research. 2008;102:1008–1010. doi: 10.1161/CIRCRESAHA.108.175505. [DOI] [PubMed] [Google Scholar]
- 12.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circulation Research. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 14.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh PC, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nature Medicine. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn B, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Natural Medicines. 2007;13:962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Dedifferentiation and proliferation of mammalian cardiomyocytes. PLoS One. 2010;5:e12559. doi: 10.1371/journal.pone.0012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojakowski W, et al. Mobilization of CD34/CXCR4+, CD34/CD117+, c-met+stem cells, and mononuclear cells expressing early cardiac, muscle, and endothelial markers into peripheral blood in patients with acute myocardial infarction. Circulation. 2004;110:3213–3220. doi: 10.1161/01.CIR.0000147609.39780.02. [DOI] [PubMed] [Google Scholar]
- 20.Limana F, et al. Myocardial infarction induces embryonic reprogramming of epicardial c-kit(+) cells: role of the pericardial fluid. J Mol Cell Cardiol. 2010;48:609–618. doi: 10.1016/j.yjmcc.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Di Meglio F, et al. Epicardial cells are missing from the surface of hearts with ischemic cardiomyopathy: a useful clue about the self-renewal potential of the adult human heart? Int J Cardiol. 2010;145(2):e44–e46. doi: 10.1016/j.ijcard.2008.12.137. [DOI] [PubMed] [Google Scholar]
- 22.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 23.Oh H, et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laugwitz KL, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hierlihy AM, Seale P, Lobe CG, Rudnicki MA, Megeney LA. The post-natal heart contains a myocardial stem cell population. FEBS Letters. 2002;530:239–243. doi: 10.1016/s0014-5793(02)03477-4. [DOI] [PubMed] [Google Scholar]
- 26.Messina E, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circulation Research. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 27.Smith RR, et al. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- 28.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, et al. Cardiomyocyte grafting for cardiac repair: graft cell death and anti-death strategies. Journal of Molecular and Cellular Cardiology. 2001;33:907–921. doi: 10.1006/jmcc.2001.1367. [DOI] [PubMed] [Google Scholar]
- 30.Fransioli J, et al. Evolution of the c-kit-positive cell response to pathological challenge in the myocardium. Stem Cells. 2008;26:1315–1324. doi: 10.1634/stemcells.2007-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubo H, et al. Increased cardiac myocyte progenitors in failing human hearts. Circulation. 2008;118:649–657. doi: 10.1161/CIRCULATIONAHA.107.761031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raya A, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(Suppl 1):11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacLellan WR, Schneider MD. Genetic dissection of cardiac growth control pathways. Annual Review of Physiology. 2000;62:289–320. doi: 10.1146/annurev.physiol.62.1.289. [DOI] [PubMed] [Google Scholar]
- 36.Rubart M, Field LJ. Cardiac regeneration: repopulating the heart. Annual Review of Physiology. 2006;68:29–49. doi: 10.1146/annurev.physiol.68.040104.124530. [DOI] [PubMed] [Google Scholar]
- 37.Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-{beta} signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci USA. 2006;103(45):16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsley RC, Gill JG, Kyba M, Murphy TL, Murphy KM. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 39.D’Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nature Biotechnology. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 40.Yasunaga M, et al. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nature Biotechnology. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- 41.Willems E, Leyns L. Patterning of mouse embryonic stem cell-derived pan-mesoderm by Activin A/Nodal and Bmp4 signaling requires Fibroblast Growth Factor activity. Differentiation. 2008;76:745–759. doi: 10.1111/j.1432-0436.2007.00257.x. [DOI] [PubMed] [Google Scholar]
- 42.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes & Development. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes & Development. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foley AC, Korol O, Timmer AM, Mercola M. Multiple functions of Cerberus cooperate to induce heart downstream of Nodal. Developmental Biology. 2007;303:57–65. doi: 10.1016/j.ydbio.2006.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foley AC, Mercola M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes & Development. 2005;19:387–396. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naito AT, et al. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueno S, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitamura R, et al. Stage-specific role of endogenous Smad2 activation in cardiomyogenesis of embryonic stem cells. Circulation Research. 2007;101:78–87. doi: 10.1161/CIRCRESAHA.106.147264. [DOI] [PubMed] [Google Scholar]
- 49.Chen VC, Stull R, Joo D, Cheng X, Keller G. Notch signaling respecifies the hemangioblast to a cardiac fate. Nature Biotechnology. 2008;26:1169–1178. doi: 10.1038/nbt.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qyang Y, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 51.Kwon C, et al. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10894–10899. doi: 10.1073/pnas.0704044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirosh-Finkel L, et al. BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development. 2010;137:2989–3000. doi: 10.1242/dev.051649. [DOI] [PubMed] [Google Scholar]
- 53.Campa VM, et al. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. The Journal of Cell Biology. 2008;183:129–141. doi: 10.1083/jcb.200806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collesi C, Zentilin L, Sinagra G, Giacca M. Notch1 signaling stimulates proliferation of immature cardiomyocytes. The Journal of Cell Biology. 2008;183:117–128. doi: 10.1083/jcb.200806091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Shi Y, Ding S. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 56.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 57.Zeineddine D, et al. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Developmental Cell. 2006;11:535–546. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi T, et al. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation. 2003;107:1912–1916. doi: 10.1161/01.CIR.0000064899.53876.A3. [DOI] [PubMed] [Google Scholar]
- 59.Sadek H, et al. Cardiogenic small molecules that enhance myocardial repair by stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6063–6068. doi: 10.1073/pnas.0711507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bushway PJ, Mercola M, Price JH. A comparative analysis of standard microtiter plate reading versus imaging in cellular assays. Assay and Drug Development Technologies. 2008;6:557–567. doi: 10.1089/adt.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu X, Ding S, Ding Q, Gray NS, Schultz PG. Small molecules that induce cardiomyogenesis in embryonic stem cells. Journal of the American Chemical Society. 2004;126:1590–1591. doi: 10.1021/ja038950i. [DOI] [PubMed] [Google Scholar]
- 62.Wei ZL, et al. Isoxazolyl-serine-based agonists of peroxisome proliferator-activated receptor: design, synthesis, and effects on cardiomyocyte differentiation. Journal of the American Chemical Society. 2004;126:16714–16715. doi: 10.1021/ja046386l. [DOI] [PubMed] [Google Scholar]
- 63.Dinsmore J, et al. Embryonic stem cells differentiated in vitro as a novel source of cells for transplantation. Cell Transplantation. 1996;5:131–143. doi: 10.1177/096368979600500205. [DOI] [PubMed] [Google Scholar]
- 64.De Tullio MC, Arrigoni O. Hopes, disillusions and more hopes from vitamin C. Cellular and Molecular Life Sciences. 2004;61:209–219. doi: 10.1007/s00018-003-3203-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bushway PJ, Mercola M. High-throughput screening for modulators of stem cell differentiation. Methods in Enzymology. 2006;414:300–316. doi: 10.1016/S0076-6879(06)14017-3. [DOI] [PubMed] [Google Scholar]
- 66.Frormann S, Jas G. Natural Products and Combinatorial Chemistry: the comeback of nature in drug discovery. Business Briefing Future Drug Discovery. 2002:84–90. [Google Scholar]
- 67.Rishton GM. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discovery Today. 2003;8:86–96. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]
- 68.Feher M, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. Journal of Chemical Information and Computer Sciences. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 69.Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. Journal of Chemical Information and Computer Sciences. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- 70.Teague SJ, Davis AM, Leeson PD, Oprea T. The design of leadlike combinatorial libraries. Angewandte Chemie (International Ed in English) 1999;38:3743–3748. doi: 10.1002/(SICI)1521-3773(19991216)38:24<3743::AID-ANIE3743>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 71.Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. Journal of Natural Products. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 72.Gupta S, Maurya MR, Subramaniam S. Identification of crosstalk between phosphoprotein signaling pathways in RAW 264.7 macrophage cells. PLoS Comput Biol. 2010;6:e1000654. doi: 10.1371/journal.pcbi.1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pradervand S, Maurya MR, Subramaniam S. Identification of signaling components required for the prediction of cytokine release in RAW 264.7 macrophages. Genome Biology. 2006;7:R11. doi: 10.1186/gb-2006-7-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brill LM, et al. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Hoof D, et al. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 76.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nature Chemical Biology. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chi X, et al. Expression of Nkx2-5-GFP bacterial artificial chromosome transgenic mice closely resembles endogenous Nkx2-5 gene activity. Genesis. 2003;35:220–226. doi: 10.1002/gene.10181. [DOI] [PubMed] [Google Scholar]
- 78.Craig PN. Interdependence between physical parameters and selection of substituent groups for correlation studies. Journal of Medicinal Chemistry. 1971;14:680–684. doi: 10.1021/jm00290a004. [DOI] [PubMed] [Google Scholar]
- 79.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 80.Cashman JR, MacDougall JM. Dynamic medicinal chemistry in the elaboration of morphine-6-glucuronide analogs. Current Topics in Medicinal Chemistry. 2005;5:585–594. doi: 10.2174/1568026054367647. [DOI] [PubMed] [Google Scholar]
- 81.Domian IJ, et al. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]