Abstract
Triple negative breast cancers are an aggressive subtype of breast cancer with poor survival, but there remains little known about the etiological factors which promote its initiation and development. Commonly inherited breast cancer risk factors identified through genome wide association studies (GWAS) display heterogeneity of effect among breast cancer subtypes as defined by estrogen receptor (ER) and progesterone receptor (PR) status. In the Triple Negative Breast Cancer Consortium (TNBCC), 22 common breast cancer susceptibility variants were investigated in 2,980 Caucasian women with triple negative breast cancer and 4,978 healthy controls. We identified six single nucleotide polymorphisms (SNPs) significantly associated with risk of triple negative breast cancer, including rs2046210 (ESR1), rs12662670 (ESR1), rs3803662 (TOX3), rs999737 (RAD51L1), rs8170 (19p13.11) and rs8100241 (19p13.11). Together, our results provide convincing evidence of genetic susceptibility for triple negative breast cancer.
Keywords: genetic susceptibility, neoplasms, association study, subtypes, common variant
Introduction
Triple negative (TN) breast cancers are a biologically and clinically distinct subtype of breast cancer, defined as tumors that exhibit low or no expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2) (1). Women with TN disease account for approximately 15% of all invasive breast cancers and are more likely to be younger, African American, have an earlier age at menarche, higher body mass index during premenopausal years, higher parity, and a lower lifetime duration of breast feeding (2-4). In addition, TN tumors are typically of higher histologic grade and are associated with more aggressive disease and poorer survival (1, 5, 6). These differences in tumor pathology, non-genetic risk factors, and survival among women with TN disease suggest that the etiology of these tumors may differ from other breast cancer subtypes.
Genome wide association studies (GWAS) have recently identified common, low-penetrance susceptibility variants that are associated with risk of breast cancer (7-16). Growing evidence suggests substantial heterogeneity by tumor subtype, defined by hormone receptor status, for associations with these SNPs. In particular, variants in 5p12, FGFR2, 8q24, 1p11.2, 9p21.3, 10q21.2, and 11q13 are associated with risk of developing ER-positive tumors (9-12, 14, 17, 18) but not ER-negative tumors, whereas variants in 2q35, TOX3, LSP1, MAP3K1 TGFB1 and RAD51L1 are associated with both ER-positive and ER-negative disease (19). To date, no variants have been specifically associated with ER-negative or TN disease. However, variants at TOX3, 2q35, and two distinct signals at 19p13.1 have been associated with breast cancer risk in BRCA1 mutation carriers, who predominantly develop tumors displaying an ER-negative and TN phenotype (15, 20, 21). Thus, additional studies specifically investigating ER-negative and TN disease are necessary to understand genetic susceptibility to these breast cancer subtypes.
Here we report on the first TNBCC study of genetic susceptibility to TN breast cancer in which associations between 22 common breast cancer susceptibility loci and risk among 2,980 cases and 4,978 controls were evaluated. This comprehensive study included 21 common variants from all known susceptibility loci identified through currently published breast cancer GWAS (1p11.2, 2q35, 3p24/NEK10, 5p12/MRPS30, MAP3K1, ESR1, 8q24, 9p21.3, 9q31.2, 10p15.1, 10q21.2/ZNF365, 10q22.3/ZMIZ1, FGFR2, LSP1, 11q13, RAD51L1, TOX3, 17q23/COX11, 19p13.1) and a SNP from CASP8 identified in a candidate-gene study of CASP8 (22, 23). We show that SNPs from four of these loci are strongly associated with risk of TN breast cancer.
Materials and Methods
Ethics Statement
Study subjects were recruited on protocols approved by the Institutional Review Boards at each participating institution, and all subjects provided written informed consent.
Study populations
Samples from several TN breast cancer case-control series, including 2,778 TN breast cancer cases and 1,406 unaffected controls, were genotyped on the iPLEX platform. These subjects were ascertained by 22 studies in 10 different countries: United States, Australia, Great Britain, Finland, Germany, Netherlands, Greece, Ireland, and Sweden. These included cases from the KBCP and POSH cohort studies, cases and controls from the MCCS cohort study, and cases and controls from established population-based breast cancer case-control studies (BBCS, GENICA, MARIE, SEARCH), hospital or clinic based case-control studies (ABCS, BIGGS, LMBC, MCBCS, OBCS, SBCS, and RPCI), case-only studies with geographically matched controls (BBCC, KARBAC, SKKDKFZS, FCCC), and unselected cases identified in tumor collections (DFCI, ABCTB, DEMOKRITOS). Data from an ongoing GWAS of TN breast cancer, including cases and controls from several of the studies described above, and the TN cases from the HEBCS GWAS along with population control data (n=273) were also included (24). In addition, data from four publicly available control GWAS data sets (Wellcome Trust Case Control Consortium UK 1958 Birth Cohort (WTCCC), National Cancer Institute's Cancer Genetic Markers of Susceptibility (CGEMS) project, Cooperative Health Research in the Region of Augsburg (KORA) study, and the Australian Twin Cohort study from the Queensland Institute of Medical Research (QIMR)) (n=3,593) were utilized. Age distributions and years of diagnosis for individual study sites are provided in Supplementary Table 1, and these studies are described in more detail in Supplementary Material.
Pathology and tumor markers
A TN breast cancer case was defined as an individual with an ER–negative, PR–negative and HER2–negative (0 or 1 by immunohistochemical staining (IHC)) breast cancer diagnosed after age 18. Criteria used for defining ER, PR, and HER2 status varied by study. These are described in detail in Supplementary Table 2. CK5/6 and EGFR IHC data for identification of basal tumors were not available.
Genotyping
The following 22 SNPs were genotyped on the iPLEX platform: rs11249433 (1p11.2), rs13387042 (2q35), rs4973768 (3p24), rs10941679 (5p12), rs889312 (MAP3K1), rs2046210 (ESR1), rs12662670 (ESR1, surrogate for rs9397435), rs13281615 (8q24), rs1011970 (9p21.3), rs865686 (9q31.2), rs2380205 (10p15.1), rs10509168 (10q21.2, surrogate for rs10995190), rs704010 (10q21.2), rs2981582 (FGFR2), rs3817198 (LSP1), rs614367 (11q13), rs999737 (RAD51L1), rs3803662 (TOX3), rs6504950 (17q23), rs8170 (19p13.11), rs8100241 (19p13.11), and rs17468277 (tagSNP for CASP8 D302H). For 10q21.2, rs10509168 was genotyped as a surrogate for rs10995190 (14).
Genotype data for 22 SNPs were generated for 2,778 cases and 1,406 controls using a single multiplex on the iPLEX Mass Array platform (Sequenom). Samples were plated by study as random mixtures of cases and controls with no-template and CEPH controls in every plate. Genotyping quality for SNPs and samples was evaluated using an iterative quality control (QC) process. SNPs and samples were excluded based on the following criteria: SNP call rate <95%, Hardy-Weinberg equilibrium (HWE) p-value <0.01 among controls, and sample call rate <95%. The final dataset of 2707 cases and 1385 controls exhibited SNP call rates >99%, HWE p-value >0.01, and sample call rates >95%.
In addition, genotype data from cases and controls included in a TN GWAS were available to supplement the iPLEX genotypes. Cases from 10 study sites (ABCTB, BBCC, DFCI, FCCC, GENICA, MARIE, MCBCS, MCCS, POSH, SBCS) were genotyped using the Illumina 660-Quad SNP array. A subset of MARIE cases were genotyped using the Illumina CNV370 SNP array. HEBCS cases and controls were genotyped using the Illumina 550-Duo SNP array. GWAS data for public controls were generated using the following arrays: Illumina 660-Quad (QIMR), Illumina 550(v1) (CGEMS), Illumina 550 (KORA), and Illumina 1.2M (WTCCC). For HEBCS, population allele and genotype frequencies on 221 healthy population controls genotyped on Illumina HumanHap 370CNV in the NordicDB, a Nordic pool and portal for genome-wide control data, were obtained from the Finnish Genome Center (25). These GWAS data were independently evaluated by an iterative QC process with the following exclusion criteria: minor allele frequency (MAF) <0.01, call rate <95%, HWE p-value <1×10-7 among controls and sample call rate <98%. When DNA was available (n=1,402), we re-genotyped samples from the TN GWAS as part of the iPLEX study in an effort to obtain as much data as possible from a single platform. Therefore, following preferential selection of data from the iPLEX study, genotypes for an additional 273 cases and 3,593 controls were included from the GWAS data (Table 1). No GWAS genotype data were available for rs10941679 (5p12), rs2046210 (ESR1), rs6504950 (17q23) and only partial data were available for five other SNPs because of the absence of these SNPs from some or all of the GWAS genotyping platforms (Table 1). As a further measure of genotype quality, genotype concordance was evaluated for the 1,402 samples included in both the iPLEX and GWAS. Eighteen of 19 SNPs, had concordance rates >98% and rs8100241 showed concordance of 96.3% .
Table 1.
Subjects by country and genotyping platform (iPLEX, GWAS)
| Country | No. of studies | Age range (mean)a |
Years of diagnosisa | iPLEX |
GWAS |
Combined |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Total | Cases | Controls | Total | Cases | Controls | Total | |||
| U.S.A | 5 | 25 - 92 (52) | 24 - 92 (62) | 1990 - 2010 | 711 | 448 | 1159 | 35 | 1126 | 1161 | 746 | 1574 | 2320 |
| Australia | 3 | 25 - 91 (56) | 29 - 72 (46) | 1990 - 2009 | 186 | 59 | 245 | 21 | 657 | 678 | 207 | 716 | 923 |
| U.K. | 5 | 22 - 93 (45) | 42 - 81 (53) | 1971 - 2010 | 573 | 111 | 684 | 6 | 1374 | 1380 | 579 | 1485 | 2064 |
| Finland | 3 | 27 - 90 (55) | 18 - 80 (57) | 1990 - 2004 | 101 | 88 | 189 | 85 | 221 | 306 | 186 | 309 | 495 |
| Germany | 6 | 22 - 88 (57) | 24 - 81 (58) | 1993 - 2008 | 740 | 501 | 1241 | 126 | 215 | 341 | 866 | 716 | 1582 |
| Greece | 1 | 21 - 79 (53) | 34 - 82 (50) | 1997 - 2010 | 273 | 85 | 358 | 0 | 0 | 0 | 273 | 85 | 358 |
| Netherlands | 1 | 26 - 62 (39) | NA | 1995 - 2007 | 67 | 0 | 67 | 0 | 0 | 0 | 67 | 0 | 67 |
| Sweden | 1 | 48 - 88 (62) | 48 - 85 (62) | 1998 - 2000 | 27 | 26 | 53 | 0 | 0 | 0 | 27 | 26 | 53 |
| Total | 25 | 21 - 93 (52) | 18 - 92 (56) | 1971 - 2010 | 2707 | 1385 | 4092 | 273 | 3593 | 3866 | 2980 | 4978 | 7958 |
Study-specific distributions shown in Supplementary Table 1
Statistical methods
Allele frequencies for each of the 22 SNPs included in these analyses were estimated using the iPLEX genotype data and the combined GWAS and iPLEX data for cases, controls, and all subjects (Supplementary Table 3). Associations for TN breast cancer were estimated using unconditional logistic regression adjusted for country of residence. The sites were categorized by country of origin (American, Australian, British, Finnish, German, Greek, Irish, and Swedish) (Table 1). SNPs were coded for a gene-dose effect by assigning a three-level (0, 1, 2) variable to each genotype (log-additive model). We calculated p-values, odds ratios (ORs) and 95% confidence intervals from these logistic regressions. Pair-wise interactions were tested by including multiplicative interaction terms in logistic regression models. Homogeneity of ORs by country was tested using the Q statistic (26) and the extent of heterogeneity was estimated by the I2 statistic (27). All analyses were conducted using SAS version 9.2, R version 2.11.0, or Plink version 1.07.
Results
We evaluated 22 breast cancer susceptibility SNPs identified in breast cancer GWAS for associations with TN disease using genotype data from an iPLEX study of the 22 SNPs supplemented with data from a TN GWAS. The combined data resulted in a case-control study of 2,980 cases and 4,978 controls from 25 studies in eight countries (Table 1). All 22 SNPs were in Hardy-Weinberg equilibrium among controls at p>0.01. Only rs17468277 and rs1011970 showed evidence of heterogeneity by country (rs17468277: p=0.047, I2=50.8%; rs1011970: p=0.093, I2=42.8%). Of the 22 SNPs from 20 loci, eight were significantly associated with risk of TN breast cancer (p<0.05) (Table 2). Six SNPs from four loci, rs2046210 (p=4.38 × 10-7), rs12662670 (p=1.13 × 10-4), rs999737 (p=2.96 × 10-4), rs3803662 (p=3.66 × 10-5), rs8170 (p=2.25 × 10-8), and rs8100241 (p=8.66 × 10-7), remained significant after correction for multiple testing (p<2.27 × 10-3). Adjustment for age did not change the magnitude or significance of our results. In addition, we did not find evidence of significant interactions with age for any of the 22 SNPs.
Table 2.
Breast cancer susceptibility SNP (n=22) associations with TN breast cancer in a log-additive model
| SNP | Gene/Locus | Chr | Tested (Minor) Allele | Overall | iPLEX | Published OR (95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | P-trend | OR (95% CI) | Cases | Controls | P-trend | OR (95% CI) | |||||
| rs11249433 | 1p11.2 | 1p11.2 | G | 2976 | 4968 | 0.27 | 0.96 (0.90-1.03) | 2707 | 1385 | 0.54 | 0.97 (0.88-1.07) | 1.16 (1.09-1.24) (12) |
| rs17468277a | CASP8 | 2q33.1 | T | 2979 | 4977 | 0.005 | 0.87 (0.78-0.96) | 2707 | 1385 | 0.16 | 0.90 (0.78-1.04) | 0.88 (0.84-0.92) (22) |
| rs13387042 | 2q35 | 2q35 | G | 2977 | 4976 | 0.26 | 0.96 (0.90-1.03) | 2705 | 1384 | 0.92 | 0.99 (0.91-1.09) | 1.20 (1.14-1.26) (9) |
| rs4973768 | SLC4A7:NEK10 | 3p24 | T | 2960 | 4974 | 0.24 | 1.04 (0.97-1.12) | 2688 | 1382 | 0.21 | 1.06 (0.97-1.17) | 1.11 (1.08-1.13) (11) |
| rs10941679 | MRPS30:FGF10 | 5p12 | G | 2705 | 1385 | 0.43 | 1.04 (0.94-1.16) | 2705 | 1385 | 0.43b | 1.04 (0.94-1.16) | 1.19 (1.11-1.28) (10) |
| rs889312 | MAP3K1 | 5q11.2 | C | 2844 | 2757 | 0.13 | 1.07 (0.98-1.17) | 2707 | 1385 | 0.20 | 1.07 (0.97-1.19) | 1.12 (1.08-1.16) (7) |
| rs2046210 | ESR1 | 6q25.1 | A | 2707 | 1385 | 4.38 × 10-7 | 1.29 (1.17-1.42) | 2707 | 1385 | 4.38 × 10-7b | 1.29 (1.17-1.42) | 1.15c (1.03-1.28) (13) |
| rs12662670 | ESR1 | 6q25.1 | G | 2707 | 2759 | 1.13 × 10-4 | 1.33 (1.15-1.53) | 2707 | 1385 | 3.52 × 10-4 | 1.37 (1.15-1.62) | 1.18 (1.10-1.26) (28) |
| rs13281615 | 8q24 | 8q24.21 | G | 2841 | 3413 | 0.79 | 0.99 (0.92-1.07) | 2707 | 1385 | 0.70 | 0.98 (0.89-1.08) | 1.08 (1.05-1.12) (7) |
| rs1011970a | CDKN2BAS:CDKN2A:CDKN2B | 9p21.3 | T | 2979 | 4977 | 0.13 | 1.07 (0.98-1.17) | 2707 | 1385 | 0.02 | 1.16 (1.02-1.31) | 1.09 (1.04-1.14) (14) |
| rs865686 | LOC100128657 | 9q31.2 | G | 2979 | 4971 | 0.65 | 1.02 (0.95-1.09) | 2707 | 1385 | 0.96 | 1.00 (0.91-1.1) | 0.89 (0.85-0.92) (16) |
| rs2380205 | ANKRD16:FBXO18 | 10p15.1 | T | 2979 | 4974 | 0.71 | 0.99 (0.92-1.06) | 2707 | 1385 | 0.94 | 1.00 (0.91-1.1) | 0.94 (0.91-0.89) (14) |
| rs10509168 | ZNF365 | 10q21.2 | T | 2980 | 4976 | 0.79 | 1.01 (0.94-1.08) | 2707 | 1385 | 0.88 | 0.99 (0.90-1.09) | 0.86 (0.82-0.91) (14) |
| rs704010 | ZMIZ1 | 10q22.3 | T | 2964 | 4963 | 0.80 | 0.99 (0.93-1.06) | 2692 | 1370 | 0.99 | 1.00 (0.91-1.1) | 1.07 (1.03-1.11) (14) |
| rs2981582 | FGFR2 | 10q26 | A | 2707 | 2756 | 0.24 | 0.95 (0.88-1.03) | 2707 | 1385 | 0.64 | 0.98 (0.89-1.08) | 1.26 (1.22-1.29) (7) |
| rs3817198 | LSP1 | 11p15.5 | C | 2929 | 4756 | 0.49 | 1.03 (0.95-1.10) | 2707 | 1385 | 0.68 | 1.02 (0.92-1.13) | 1.07 (1.04-1.11) (7) |
| rs614367 | MYEOV:CCND1 | 11q13 | T | 2926 | 4749 | 0.17 | 1.07 (0.97-1.18) | 2707 | 1385 | 0.12 | 1.12 (0.97-1.28) | 1.15 (1.10-1.20) (14) |
| rs999737 | RAD51L1 | 14q24.1 | T | 2978 | 4977 | 2.96 × 10-4 | 0.86 (0.80-0.93) | 2706 | 1385 | 0.05 | 0.90 (0.80-1.00) | 0.94 (0.88-0.99) (12) |
| rs3803662 | TOX3 | 16q12.1 | A | 2980 | 4973 | 3.66 × 10-5 | 1.17 (1.09-1.26) | 2707 | 1385 | 8.25 × 10-4 | 1.20 (1.08-1.33) | 1.19 (1.15-1.23) (7) |
| rs6504950 | COX11 | 17q23.2 | A | 2707 | 1385 | 0.54 | 0.97 (0.87-1.07) | 2707 | 1385 | 0.54b | 0.97 (0.87-1.07) | 0.95 (0.92-0.97) (11) |
| rs8170 | C19orf62:ANKLE1 | 19p13.1 | T | 2979 | 4978 | 2.25 × 10-8 | 1.27 (1.17-1.38) | 2707 | 1385 | 7.30 × 10-8 | 1.40 (1.24-1.58) | 1.26 (1.17-1.35) (21) |
| rs8100241 | C19orf62:ANKLE1 | 19p13.1 | A | 2980 | 4320 | 8.66 × 10-7 | 0.84 (0.78-0.90) | 2707 | 1385 | 1.81 × 10-6 | 0.79 (0.71-0.87) | 0.84 (0.80-0.89) (21) |
These SNPs showed evidence of country-based heterogeneity.
No additional samples included in overall analysis compared to iPLEX-only
Estimated OR in Europeans
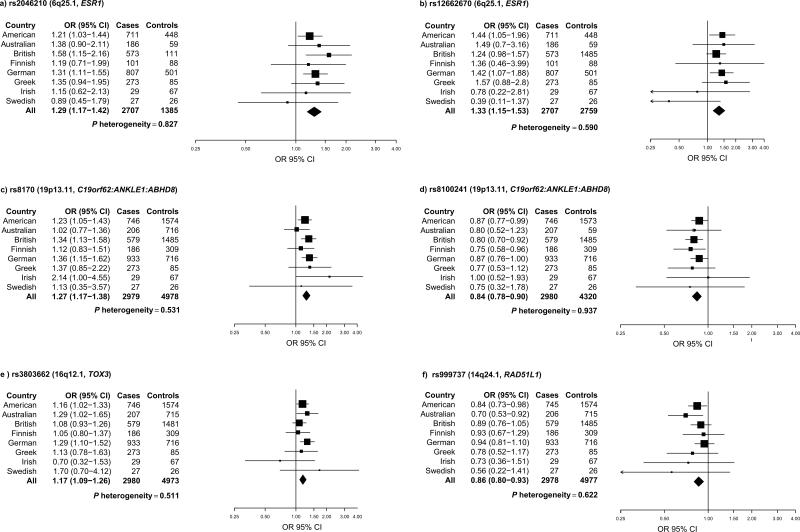
Rs2046210, located upstream of ESR1 on chromosome 6q25.1, exhibited a strong association with TN disease [odds ratio (OR)=1.29, 95% Confidence Interval (CI) 1.17 – 1.42; p=4.38 × 10-7] (Figure 1a), whereas rs12662670, located further upstream of ESR1, displayed a similar effect but slightly less significant association with TN disease [OR=1.33-fold, 95% CI 1.15 – 1.53; p=1.13 × 10-4] (Figure 1 b). To assess the independence of these two ESR1 SNPs, which are not correlated in HapMap subjects of European ancestry (r2=0.09), we included both SNPs in a multivariate model. Rs2046210 was more strongly associated with TN risk than rs12662670 [rs2046210 OR=1.24, 95% CI 1.12 – 1.38; p=5.64 × 10-5; rs12662670 OR=1.20, 95% CI 1.00 – 1.44; p=0.053] in this model, suggesting that rs2046210 may account in part for these two associations. In addition, two SNPs at 19p13.1 shown to have genome wide significant associations with breast cancer in BRCA1 mutation carriers, were highly significantly associated with TN breast cancer [rs8170: OR=1.27, 95% CI 1.17 – 1.38; p=2.25 × 10-8] [rs8100241: OR=0.84, 95% CI 0.78 – 0.90; p=8.66 × 10-7] (Figure 1 c,d). Multivariate modeling of these two SNPs, which are moderately correlated in HapMap subjects of European ancestry (r2=0.74), showed that rs8170 is more strongly associated with TN breast cancer risk [rs8170: OR=1.22, 95% CI 1.10 – 1.34; p=7.56 × 10-5; rs8100241: OR=0.90, 95% CI 0.83 – 0.98; p=0.014] although both variants are retained in the model. Additionally, rs3803662 (TOX3), which has been strongly associated with risk of ER-negative breast cancer (OR=1.15, p=2.1 × 10-10) (19), was associated with a 1.17-fold increase in risk of TN disease [OR=1.17, 95% CI 1.09 – 1.26; p=3.66 × 10-5] (Figure 1e). Likewise, the rs999737 (RAD51L1) SNP was significantly associated with risk of TN breast cancer [rs999737 OR=0.86, 95% CI 0.80 – 0.93; p=2.96 × 10-4] (Figure 1f). In contrast, rs17468277 (ALS2CR12/CASP8) (p=0.005) was not significantly associated with TN breast cancer risk after correction for multiple testing, suggesting that this result should be interpreted with caution. None of these six SNPs showed evidence of heterogeneity by country (Figure 1). To further understand the influence of variants in the 6q25.1 and 19p13.11 loci on TN risk, we looked for statistical interactions between the SNPs in these regions. While there was no evidence for a statistical interaction between rs2046210 and rs1266270 (p=0.820) at 6q25.1, we found strong evidence of an interaction (p=0.004) between rs8170 and rs8100241 from 19p13.1, in a multiplicative model.
Figure 1. Breast cancer susceptibility loci and risk of TN breast cancer.
Forest plots for six breast cancer susceptibility loci and risk of TN breast cancer are shown by country. Country-specific odds ratios (95% CIs) are denoted by black boxes (black lines). Overall OR estimates are represented by black diamonds, where diamond width corresponds to 95% CI bounds. Box and diamond heights are inversely proportional to precision of the OR estimate. I2 values were 0 for each of these 6 SNPs, indicating no heterogeneity by country.
Next we performed a subset analysis using the iPLEX data alone (2,707 cases, 1,385 controls) for the 19 SNPs with both iPLEX and GWAS genotypes to assess the consistency of our results. Analysis of associations with TN disease in the iPLEX-only dataset showed that odds ratios for the 19 SNPs were consistent in both direction and magnitude of effect compared to the analysis using all available genotype data, although some variation in the significance of the associations was observed (Table 2). Four of the SNPs significantly associated with TN breast cancer in the overall analysis retained statistical significance in the iPLEX-only analysis (rs12662670 p=3.52 × 10-4; rs3803662 p=8.25 × 10-4; rs8170 p=7.30 × 10-8; rs8100241 p=1.81 × 10-6) after correction for multiple testing. Results were unchanged for rs2046210 from the ESR1 locus, because the overall analysis was restricted to iPLEX data as a result of missing GWAS data for this variant. Finally, while the rs999737 (RAD51L1) SNP was only marginally associated with TN breast cancer risk in the iPLEX-only analysis (rs999737 p=0.053), the estimate of effect for this SNP was consistent with the effect observed in the overall analysis.
Importantly, genotype data from a subset of these cases and controls have previously been used in association studies involving a number of these SNPs by the Breast Cancer Association Consortium (BCAC). To avoid duplication and to assess the degree to which these BCAC samples influenced our results, we also performed a subset analysis in which we excluded all cases and controls used in the BCAC studies (n=1,819 cases; n=4,038 controls) (Supplementary Table 4). The effect estimates and significance of associations with TN disease in either the iPLEX or combined analyses were not substantially modified following the removal of these cases and controls (Supplementary Table 5).
Discussion
Here we report on the first study by the TNBCC and the largest study to date of genetic susceptibility to TN breast cancer, which is comprised 2,980 cases and 4,978 controls from 25 studies in eight countries. We show that a subset of breast cancer susceptibility SNPs identified through GWAS are also associated with risk of TN breast cancer. Specifically, we determined that six breast cancer susceptibility SNPs from four loci- rs2046210 (ESR1), rs12662670 (ESR1), rs999737 (RAD51L1), rs3803662 (TOX3), rs8170 (19p13.1) and rs8100241 (19p13.1)- are associated with risk of TN breast cancer. Of these, rs8170 (19p13.1) achieved genome-wide significance (p=2.25 × 10-8). Overall, these findings provide strong evidence of genetic susceptibility to triple negative breast cancer.
We identified highly significant associations between SNPs at 6q25.1 and risk of TN breast cancer, including rs12662670 (p=1.13 × 10-4) and rs2046210, which reached near genome-wide significance (p=4.38 × 10-7). These variants are located approximately 30kb and 60kb upstream of the first untranslated exon and 180kb and 210kb upstream of the first coding exon of ESR1, which encodes the estrogen receptor-α protein.
The rs2046210 SNP was originally reported in a breast cancer GWAS in Chinese women (13) where a stronger association among ER-negative than ER-positive breast cancers was observed. Importantly, the magnitude of effect in this TN study [OR=1.29, 95% CI 1.17 – 1.42] was identical to that reported for ER-negative breast cancer in the Chinese study [OR=1.29, 95% CI 1.21-1.37]. In contrast, a study of women of European ancestry did not observe an association with breast cancer, although analyses were not stratified by ER status (28). When combined with our results the suggestion is that this SNP may be specifically associated with TN or ER-negative disease. The second variant in the ESR1 locus, rs12662670, was originally associated with breast cancer in the same study of women of European ancestry [OR=1.12, 95% CI 1.03 – 1.21] and was used as a surrogate for rs9397435 , which is associated with breast cancer risk [OR=1.15, 95% CI 1.06 – 1.25] independently of rs2046210 (28). Here rs12662670 showed a strong influence on TN breast cancer risk [OR=1.33, 95% CI 1.15 – 1.53] again suggesting that variation in the ESR1 loci is specifically associated with risk of ER-negative and/or TN breast cancer. It remains to be determined whether a single locus represented by rs2046210 or two loci accounted for by rs2046210 and rs9397435, are associated with ER-negative and TN breast cancer at chromosome 6q25.
Since TN breast cancer is defined in part by the absence of expression of estrogen receptors, we can speculate that inherited variation may down-regulate ESR1 expression and promote formation of ERα negative tumors. However, recent studies in mice have shown that the mammary stem cell compartment can be regulated by 17β-estradiol and progesterone through a paracrine-signalling mechanism from steroid receptor-positive luminal cells to steroid receptor-negative stem cells (29, 30). Thus, SNPs in the ESR1 locus may promote expansion of receptor negative precursors and subsequent development of TN tumors. Interestingly, variation in the 5’ region of ESR1 has been associated with an increased risk of breast cancer relapse in a British prospective cohort study (31), which was accounted for by including tumor grade and nodal status in multivariate models. Thus the causal SNPs in this area may be associated with a more aggressive tumor phenotype.
The SNPs rs8170 (p=2.25 × 10-8) and rs8100241 (p=8.66 × 10-7) located at 19p13.1 were first identified as modifiers of breast cancer risk in BRCA1 carriers (15) and as risk factors for ovarian cancer (32) and were also shown to be significantly associated with ER-negative breast cancer (15). In this study we showed that rs8170 displayed a genome wide significant association with TN breast cancer, suggesting that we can now identify variation in the 19p13.1 locus as a risk factor for TN disease. Interestingly, rs8170 attenuated the significance of rs8100241 when the SNPs were included in a multivariate regression model for breast cancer, whereas these both SNPs retained significance in multivariate models evaluating effects on BRCA1 associated breast cancer and ER-negative breast cancer (15). In addition, our data suggest that these SNPs have a multiplicative effect on TN breast cancer risk. Further studies are required to determine whether these SNPs represent independent signals in the 19p13.1 locus. Additional studies are also needed to identify the underlying causative genetic events in this locus and to determine if the causative events for BRCA1, ER-negative, and TN breast cancer as well as ovarian cancer are in common.
These 19p13.1 variants are located in a cluster of genes including C19orf62, ANKLE1, and ABHD8. ABHD8 encodes the abhydrolase domain containing 8 protein, which is a gene of uncharacterized function, and is located about 13 kb downstream of both rs8170 and rs8100241. The SNP rs8170 is located within C19orf62, which encodes the MERIT40 protein, while rs8100241 is located within ANKLE1, a protein of unknown function which encodes ankyrin repeat and LEM domains. MERIT40 is the most plausible candidate in this region for breast cancer susceptibility because it is a component of the BRCA1-A complex and is required to ensure the integrity and localization of this complex during the repair of DNA double-strand breaks, specifically through the recruitment and retention of the BRCA1-BARD1 ubiquitin ligase and the BRCC36 deubiquitination enzyme (33-35). However, it remains to be determined whether the causal variants at 19p13.1 alter MERIT40 expression or function or influence other genes in the region such as ANKLE1 or ABHD8.
We also found that variants in RAD51L1 (rs999737, p=2.96 × 10-4) and TOX3 (rs3803662, p=3.66 × 10-5) were strongly associated with risk of TN breast cancer. Rs999737 (RAD51L1) was originally identified in a recent breast cancer GWAS of women of European ancestry (12). Detailed studies of breast tumors have suggested that rs999737 is associated with both ER-positive and ER-negative breast cancer, which is consistent with our findings. RAD51L1 is a member of the Rad51-like family and functions in the double-strand break repair and homologous recombination pathway (36). When coupled with the association of the 19p13.1/MERIT40 locus with TN, the suggestion is that modification of DNA repair genes is an important mechanism involved in predisposition to TN breast cancer. The SNP rs3803662, located telomeric to the gene TOX3, was also strongly associated with TN breast cancer in our study (p=3.66 × 10-5). This SNP was originally identified in two GWAS of breast cancer (7, 9) and has been associated with risk of developing both ER-positive and ER-negative tumors (9). The SNP is also associated with risk of BRCA1 related breast cancers (15), which are primarily ER-negative or TN. TOX3 encodes a protein containing an HMG-box that is speculated to be involved in the modification of DNA and chromatin structure (37).
Only a subset of the 22 susceptibility loci were associated with TN disease in this study. This suggests that there may be heterogeneity in the predisposition loci associated with different breast tumor subtypes. However, it is important to consider whether limited statistical power may have influenced our results. Among the 16 SNPs that did not reach statistical significance in this study, the effect estimates for variants at 1p11.2, 2q35, 8q24, 9q31.2, 10p15.1, 10q21.2/ZNF365, 10q22.3/ZMIZ1, and FGFR2 showed either no evidence for association or were in the opposite direction compared to the original GWAS findings. Interestingly, 2q35 has been associated with both ER-negative (19) and BRCA1-related breast cancer (21), and was marginally significant in a smaller set of TN breast cancer (19). However, we found no evidence for association at 2q35 among TN breast cancer, indicating that risk for this locus may be limited to non-TN, ER-negative breast cancer. In contrast, the ORs for SNPs at CASP8, 9p21.3, and COX11 were comparable in magnitude to the original GWAS findings, while the ORs for variants at 3p24/NEK10, 5p12, MAP3K1, LSP1, and 11q13 had only mildly attenuated effects. Our results are also consistent with a recent study reporting associations between MAP3K1, 3p24/NEK10, COX11, and CASP8 and ER-negative breast cancer (19). These results suggest that we may have had insufficient power to detect significant associations for these SNPs among TN breast cancers.
Several limitations should be considered when interpreting these results. First, different ascertainment criteria were used among the contributing breast cancer studies with cases being ascertained from population-based or hospital-based case-control studies. Importantly, genetic main effects models in other large breast cancer consortia such as BCAC have provided stable risk estimates for SNPs across a wide range of study designs. This would suggest that in the case of these genetic variants, ascertainment and study design issues had limited influence on the results of genetic association studies for breast cancer. The consistency in effect estimates among BRCA1-related breast cancers, ER negative breast cancer, and now triple negative breast cancer for variants at 19p13.1, 6q25, and TOX3 provide additional evidence that these estimates are robust to variability in study design. Further, our evaluation of interactions with age was underpowered, and unavailability of family history on the majority of studies precluded investigations of interactions by family history. There is also variability in the criteria used to define ER, PR, and HER2 status of cases between studies (Supplementary Table 2). For HER2, cases with scores of 0 or 1 by IHC were defined as HER2 negative. Cases with IHC of 2+ were not included in order to minimize erroneous inclusion of HER2 positive cases. In general, cases were considered ER or PR negative based on IHC of tumors using thresholds of <1% of cells stained, <10% of cells stained, or an Allred score of 0-2, which incorporates both intensity and percentage of staining in tumor cells. In addition to variability in thresholds for positivity, factors such as tissue fixation, antibody choice, and interpretation of positive immunostaining may also affect the definition or ER or PR status across study sites (38, 39). The resulting heterogeneity in the definition of triple negative breast cancer may influence our ability to detect associations with susceptibility loci that are specific to triple negative or ER negative disease. However, we did successfully identify six genetic loci associated with triple negative disease, and the lack of heterogeneity in effect estimates across study sites in this analysis (Figure 1) would suggest that our findings are generally robust to the differences noted above. Additionally, in a sensitivity analysis including only cases from studies with the most stringent criteria for defining TN cases (<1% of cells stained positive for ER and PR, HER2 0 or 1+ on IHC), the effect estimates were very similar to those from the complete analysis for the six SNPs in ESR1, 19p13.11, TOX3, and RAD51L1, with some attenuation of significance. Finally, it is important to note that the results of this study are specific to Caucasian women. While greater proportions of African Americans and Latinas than Caucasians develop TN breast cancer, it is not known whether similar associations with the SNPs described here exist in these populations. Further studies are needed to address this question.
In conclusion, our study provides convincing evidence for genetic susceptibility to TN breast cancer and suggests that susceptibility loci may differ by histological breast tumor subtype, defined by ER, PR and HER2 status. These findings add to the evidence suggesting that these subtypes likely arise through distinct etiologic pathways. Additional studies, such as those from the Breast Cancer Association Consortium, will be important for determining whether these SNPs are exclusively associated with ER-negative, TN disease, or even basal breast cancer, a more refined subgroup of TN tumors. Fine mapping and functional analyses of these susceptibility loci are needed to identify the casual variants and mechanisms underlying the associations with TN breast cancer risk.
Supplementary Material
Acknowledgements
Mammary Carcinoma Risk Factor Investigation (MARIE)
MARIE would like to thank Tracy Slanger and Elke Mutschelknauss for their valuable contributions, and S. Behrens, R. Birr, W. Busch, U. Eilber, B. Kaspereit, N. Knese, K. Smit, for their excellent technical assistance.
Melbourne Collaborative Cohort Study (MCCS)
We acknowledge the contribution of the MCCS investigators John L Hopper, Dallas R English, and Melissa C Southey.
Sheffield Breast Cancer Study (SBCS)
We thank Helen Cramp, Dan Connley and Ian Brock for patient recruitment, database management and DNA preparation respectively.
Prospective Study of Outcomes in Sporadic Versus Hereditary Breast Cancer (POSH)
We thank the 126 participating investigators who recruited cases to the study and the NCRN for supporting recruitment to the study.
Leuven Multidisciplinary Breast Centre (LMBC)
LMBC thanks Gilian Peuteman, Dominiek Smeets and Sofie van Soest for Technical assistance.
Mayo Clinic Breast Cancer Study (MCBCS)
We would like to thank Georgia Chenevix-Trench for her valuable contributions.
Helsinki Breast Cancer Study (HEBCS)
HEBCS thanks RN Hanna Jäntti and Irja Erkkilä for their help with the patient data and samples and Drs. Päivi Heikkilä, Ari Ristimäki, Tuomas Heikkinen, Mira Heinonen and Laura Hautala for their help with the tumor marker and pathology information, and gratefully acknowledges the Finnish Cancer Registry for the cancer data. The population allele and genotype frequencies were obtained from the data source funded by the Nodic Center of Excellence in Disease Genetics based on samples regionally selected from Finland, Sweden and Denmark.
Breast Cancer in Galway Genetic Study (BIGGS)
Thanks are given to Drs Gabrielle Colleran, Niall McInerney, Nicola Miller and Professor Michael Kerin, University Hospital Galway, for their help collecting patient data and samples.
Amsterdam Breast Cancer Study (ABCS)
We acknowledge ABCS/BOSOM study collaborators, among others LJ Van't Veer, FE van Leeuwen, R van Hien, S Cornelissen, A Broeks and AJ van den Broek, and the NKI-AVL Family Cancer Clinic, especially FB Hogervorst.
Australian Breast Cancer Tissue Bank (ABCTB)
RLB is a Cancer Institute New South Wales Fellow.
Oulu Breast Cancer Study (OBCS)
We wish to thank Mervi Grip and Kari Mononen for their help with patient contacts and sample and data collection, and Meeri Otsukka for assistance with sample and data handling.
Kuopio Breast Cancer Project (KBCP)
KBCP is grateful to Mrs Eija Myöhänen and Mrs Helena Kemiläinen for their skillful assistance.
Grant Support
Mammary Carcinoma Risk Factor Investigation (MARIE)
The MARIE study was supported by the Deutsche Krebshilfe e.V., grant number 70-2892-BR I, the Hamburg Cancer Society, the German Cancer Research Center (DKFZ) and the DNA extraction and genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany grant 01KH0402.
Gene Environment Interaction and Breast Cancer in Germany (GENICA)
The GENICA Network was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation of Medical Research, Stuttgart, Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University Tübingen,Germany (HB, Christina JustenhovenJ); Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany (UH); Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany (YDK, Christian Baisch); Institute of Pathology, Medical Faculty of the University of Bonn, Germany (Hans-Peter Fischer); Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, Germany (Thomas Brüning, Beate Pesch, Volker Harth, Sylvia Rabstein)
Melbourne Collaborative Cohort Study (MCCS)
The MCCS was supported by Australian NHMRC grants 209057, 251553 and 504711 and infrastructure provided by the Cancer Council Victoria.
Sheffield Breast Cancer Study (SBCS)
The SBCS was supported by the Breast Cancer Campaign (grant 2004Nov49 to AC), and by Yorkshire Cancer Research core funding.
Dana Farber Cancer Institute (DFCI)
This work was supported in part by the DFCI Breast Cancer SPORE NIH P50 CA089393.
Prospective Study of Outcomes in Sporadic Versus Hereditary Breast Cancer (POSH)
The POSH study (CI DM Eccles) was funded by Cancer Research UK. Blood samples were collected by the University of Southampton Cancer Sciences Human Tissue Bank (HTA license 12009).
Molecular Diagnostics Laboratory IRRP, National Centre for Scientific Research (DEMOKRITOS)
This work was supported by the Hellenic Cooperative Oncology Group research grant (HR R_BG/04) and the Greek General Secretary for Research and Technology (GSRT) Program, Research Excellence II, funded at 75% by the European Union.
Bavarian Breast Cancer Cases and Controls (BBCC)
Peter Fasching was partly funded by the Dr. Mildred Scheel Stiftung of the Deutsche Krebshilfe e.V.
British Breast Cancer Study (BBCS)
The BBC NCRN study is funded by Cancer Research UK and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR biomedical Research Centre and the National Cancer Research Network (NCRN).
Leuven Multidisciplinary Breast Centre (LMBC)
LMBC is supported by European Union Framework Programme 6 Project LSHC-CT-2003-503297 (the Cancerdegradome) and by the ‘Stichting tegen Kanker’ (232-2008).
Oulu Breast Cancer Study (OBCS)
OBCS was supported by grants and other funding from the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the Academy of Finland, the University of Finland, and Oulu University Hospital.
Mayo Clinic Breast Cancer Study (MCBCS)
MCBCS was supported by NIH Grants CA122340 and a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), and grants from the Komen Foundation for the Cure and the Breast Cancer Research Foundation (BCRF).
Study of Epidemiology and Risk factors in Cancer Heredity (SEARCH)
SEARCH was supported by Cancer Research UK grants C1287/A7497, C490/A11021, C1287/A10118 and C1287/A5260.
Helsinki Breast Cancer Study (HEBCS)
The HEBCS study has been financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society, and the Sigrid Juselius Foundation.
Fox Chase Cancer Center (FCCC)
A.K.G. was funded by SPORE P-50CA83638, U01CA69631, 5U01CA113916, and the Eileen Stein Jacoby Fund.
Roswell Park Cancer Institute (RPCI)
Data and samples were obtained from the RPCI DataBank and BioRepository (DBBR) (40), a Cancer Center Support Grant Shared Resource (P30 CA016056-32).
Städtisches Klinikum Karlsruhe and Deutsches Krebsforschungszentrum Breast Cancer Study (SKKDKFZS)
The SKKDKFZS study was supported by the Deutsches Krebsforschungszentrum.
Breast Cancer in Galway Genetic Study (BIGGS)
ES is funded by the National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centre, Guy's & St. Thomas’ NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust.
Australian Breast Cancer Tissue Bank (ABCTB)
The ABCTB is generously supported by the National Health and Medical Research Council of Australia, The Cancer Institute NSW and the National Breast Cancer Foundation.
Amsterdam Breast Cancer Study (ABCS)
MKS was funded by the Dutch Cancer Society grant number 2009-4363.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007;16:439–43. doi: 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 3.Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 4.Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109:123–39. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–8. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 6.Irvin WJ, Jr., Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44:2799–805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 7.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 10.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 11.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–92. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, et al. Novel Breast Cancer Susceptibility Locus at 9q31.2: Results of a Genome-Wide Association Study. J Natl Cancer Inst. 2011 doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Closas M, Chanock S. Genetic susceptibility loci for breast cancer by estrogen receptor status. Clin Cancer Res. 2008;14:8000–9. doi: 10.1158/1078-0432.CCR-08-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antoniou AC, Spurdle AB, Sinilnikova OM, Healey S, Pooley KA, Schmutzler RK, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82:937–48. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoniou AC, Beesley J, McGuffog L, Sinilnikova OM, Healey S, Neuhausen SL, et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742–54. doi: 10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–8. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 23.Milne RL, Gaudet MM, Spurdle AB, Fasching PA, Couch FJ, Benitez J, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the Breast Cancer Association Consortium: a combined case-control study. Breast Cancer Res. 2010;12:R110. doi: 10.1186/bcr2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Humphreys K, Darabi H, Rosin G, Hannelius U, Heikkinen T, et al. A genome-wide association scan on estrogen receptor-negative breast cancer. Breast Cancer Res. 2010;12:R93. doi: 10.1186/bcr2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leu M, Humphreys K, Surakka I, Rehnberg E, Muilu J, Rosenstrom P, et al. NordicDB: a Nordic pool and portal for genome-wide control data. Eur J Hum Genet. 2010;18:1322–6. doi: 10.1038/ejhg.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacey SN, Sulem P, Zanon C, Gudjonsson SA, Thorleifsson G, Helgason A, et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asselin-Labat ML, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 30.Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–7. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 31.Tapper W, Hammond V, Gerty S, Ennis S, Simmonds P, Collins A, et al. The influence of genetic variation in 30 selected genes on the clinical characteristics of early onset breast cancer. Breast Cancer Res. 2008;10:R108. doi: 10.1186/bcr2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–4. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–28. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–54. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–39. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lio YC, Mazin AV, Kowalczykowski SC, Chen DJ. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J Biol Chem. 2003;278:2469–78. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- 37.O'Flaherty E, Kaye J. TOX defines a conserved subfamily of HMG-box proteins. BMC Genomics. 2003;4:13. doi: 10.1186/1471-2164-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gown AM. Current issues in ER and HER2 testing by IHC in breast cancer. Mod Pathol. 2008;21(Suppl 2):S8–S15. doi: 10.1038/modpathol.2008.34. [DOI] [PubMed] [Google Scholar]
- 39.Allred DC, Carlson RW, Berry DA, Burstein HJ, Edge SB, Goldstein LJ, et al. NCCN Task Force Report: Estrogen Receptor and Progesterone Receptor Testing in Breast Cancer by Immunohistochemistry. J Natl Compr Canc Netw. 2009;7(Suppl 6):S1–S21. doi: 10.6004/jnccn.2009.0079. quiz S2-3. [DOI] [PubMed] [Google Scholar]
- 40.Ambrosone CB, Nesline MK, Davis W. Establishing a cancer center data bank and biorepository for multidisciplinary research. Cancer Epidemiol Biomarkers Prev. 2006;15:1575–7. doi: 10.1158/1055-9965.EPI-06-0628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.