Abstract
Apoptosis is a mode of programmed cell death in multicellular organisms and plays a central role in controlling embryonic development, growth and differentiation and monitoring the induction of tumor cell death through anticancer therapy. Since the most effective chemotherapeutics rely on apoptosis, imaging apoptotic processes can be an invaluable tool to monitor therapeutic intervention and discover new drugs modulating apoptosis. The most attractive target for developing specific apoptosis imaging probes is caspases, crucial mediators of apoptosis. Up to now, various optical imaging strategies for apoptosis have been developed as an easy and economical modality. However, current optical applications are limited by poor sensitivity and specificity. A subset of molecular imaging contrast agents known as “activatable” or “smart” molecular probes allow for very high signal-to-background ratios compared to conventional targeted contrast agents and open up the possibility of imaging intracellular targets. In this review, we will discuss the unique design strategies and applications of activatable probes recently developed for fluorescence and bioluminescence imaging of caspase activity.
Keywords: Activatable probes, apoptosis, bioluminescence, caspases, optical imaging
Introduction
Apoptosis is a highly organized mode of cell death that plays an essential role in regulating growth, development and immune response, and clearing redundant or abnormal cells in organisms [1]. Apoptotic cell death occurs through the intrinsic pathway or the extrinsic pathway [2]. The intrinsic apoptotic pathway can be stimulated by chemotherapeutics, mitochondrial damage or ionizing radiation. The extrinsic apoptotic pathway is activated in response to ligation of death receptors of the tumour necrosis factor receptor type I family. Caspases, or cysteine aspartyl-specific proteases, are a family of cysteine proteases that cleave at an aspartate residue and are recognized as essential mediators of apoptosis. There are a total of 14 caspases identified so far, which play distinct roles in inflammation and apoptosis [3]. Among these caspases, caspases-2, -3, -6, -7, -8, -9 and -10 are involved in the apoptotic pathway, either as upstream initiators of the proteolytic cascade (caspases-2, -8, -9 and -10), or as downstream effectors (caspase-3, -6 and -7) that cleave cellular proteins [4]. Many reports have indicated that the dysregulation of apoptosis can lead to the destruction of normal tissues in a variety of disorders [5-7]. In particular, successful tumor therapeutic treatments, such as radiation [8], chemotherapy [9], thermal therapy [10] and photodynamic therapy [11], require the iatrogenic induction of apoptosis. Given the central role of apoptosis, it would be desirable to have a promising imaging method to detect and monitor this process.
In view of the specific biochemical changes in cells undergoing apoptosis, three major imaging mechanisms have been identified [12], including i) exposure of phosphatidylserine on the extracellular face of the plasma membrane which can be detected by dye-labeled phospholipid-binding proteins such as annexin V [13], ii) intracellular caspase activation, imaged by using molecular beacons comprised of dye-labeled caspase substrates [14] and iii) mitochondrial membrane potential collapse, monitored by reduced levels of phosphoniumcations that normally accumulate in healthy mitochondria [7].
Various targeted imaging strategies were developed using several imaging modalities, such as optical, MR, ultrasound and radionuclide imaging. Several comprehensive reviews have summarized and discussed these recent applications [15-16]. Among these diverse strategies, more and more investigators focus on the development of optical molecular probes targeting caspase activation; because, activated caspases represent obvious target molecules for apoptosis and optical imaging is a highly sensitive and easy imaging modality. Very recently, “activatable probes” or “smart probes” have become a particularly attractive platform for targeted optical imaging. These probes allow the researchers to control and manipulate the outputs of the maximized target signal and the minimized background signal by altering the chemical environment. Therefore, they have higher target-to-background ratios than conventional “always on” imaging agents. In the current review, we will focus on research progress on activatable imaging probes targeting apoptosis based on fluorescent amplification platforms for fluorescence and bioluminescence imaging (BLI). Given the increasing attention paid to nanotechnology, various types of nanoparticle-based activatable probes will be discussed.
Design of activatable optical imaging probes
Design of activatable fluorescent imaging probes
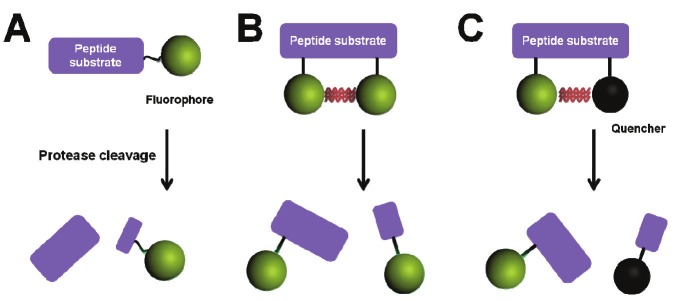
In vivo fluorescence imaging has added an easy and economical modality to the rapidly growing field of molecular imaging. Currently, several types of activatable fluorescent imaging probes have been developed including auto-quenched probes (Figure 1A), dye-dye self-quenched probes (Figure 1B), and fluorophore-quencher pair labeled probes (Figure 1C). Since the auto-quenched and self-quenched probes do not efficiently minimize background signals, we will focus on fluorophore-quencher pair probes. As shown in Figure 2A, activatable probes are composed of two or three basic parts: i) a peptide substrate which can be recognized and cleaved by targeted enzymes, ii) a silent or quenching moiety matched with a fluorophore which generates a strongly activated fluorescence signal after substrate cleavage, and iii) a carrier that optimizes pharmacokinetics or improves the cellular uptake of the probe [17-20]. These components must be integrated into one probe to increase the target-to-background ratio. In this system, the targeting substrate must be highly specific to the target tissues in the heterogeneous environment. The reactions between enzymes and substrates are usually very specific because of their complementary shape, charge and hydrophilic/hydrophobic characteristics [21]. There are a variety of highly, specific peptide-based protease substrates that can be the ideal candidates for a targeting ligand of activatable probes.
Figure 1.
Schematic diagrams of activatable fluorescent imaging probes. A) Auto-quenched probe, B) self-quenched probe and c) fluorophore/quencher pair-labeled probe.
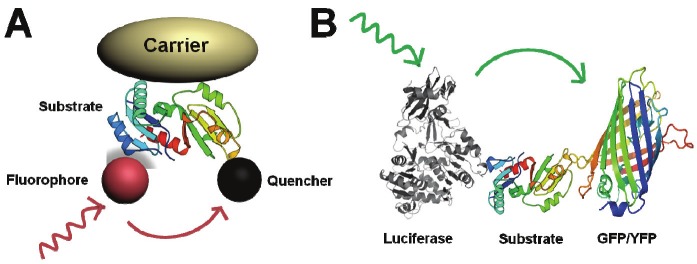
Figure 2.
Design of activatable probes based on A) Fluorescence Resonance Energy Transfer (FRET) and B) Bioluminescence Resonance Energy Transfer (BRET).
In order to design a robust activatable imaging probe, it is important to choose suitable fluorophore-quencher pairs based on resonance energy transfer (RET). RET was first described in the 1940’s by Förster and is characterized by the radiationless transfer of excited state energy from a donor to an acceptor molecule. Förster and others have shown that energy transfer efficiency is highly dependent on the distance between the donor and acceptor moieties and their relative orientation with respect to each other. In most RET-based assays, the typical effective distance between the donor and acceptor is 1 to 10 nm [22]. This range correlates well with most biological interactions thus making RET an excellent tool for monitoring macromolecular interactions. Examples of RET-based technologies are Fluorescence Resonance Energy Transfer (FRET) and Bioluminescence Resonance Energy Transfer (BRET) [23]. Typically, fluorophore-quencher pairs abide by FRET with few exceptions, such as a non-FRET mechanism called static quenching [24-25]. Based on this FRET mechanism, various self-quenched and fluorophore-quencher pair labeled activatable probes has been developed for imaging different target cells or tissues.
To improve the pharmacokinetics of probes in vivo, biocompatible polymer backbones and nanoparticles (NPs) are considered ideal carriers. Many reports have demonstrated that conjugating poly (ethylene glycol) (PEG), PEGylation, reduces the rate of mononuclear phagocyte system uptake and increases circulation half-life, improving the targeting efficiency of probes [26]. The possible reason is that PEG creates a steric shield around the probe, effectively preventing plasma proteins from adhering to the particle surface and thus avoiding subsequent uptake by mononuclear phagocytes. In our previous study, we synthesized a series of PEGylated activatable probes and assessed the expression of matrix metalloproteinases (MMPs) in a tumor-bearing mouse mode [27]. The results demonstrate that a probe modified with an optimized molecular weight of PEG could significantly affect the probe pharmacokinetics and enhance the tumor-to-background ratio in vivo. NPs used as carriers show excellent pharmacokinetics owing to their small size and large surface area. Notably, NPs can accumulate at tumor sites by the enhanced permeability and retention (EPR) effect. Some polymeric and inorganic NPs that combine the fluorophore with quenching moiety provide the potential development of activatable nanoprobes in tumor imaging [28-29]. General applications of polymer and NP-associated protease activatable probes for optical imaging have been reviewed elsewhere [17,19-20].
Design of activatable bioluminescence imaging probes
BLI of luciferase reporters is a relatively simple, robust, cost-effective and extremely sensitive means to image fundamental biological processes, that have lead to new advances and discoveries in life sciences. During the last decade, BLI has become an indispensable tool for visualizing molecular events at a cellular level both in vivo and in vitro, owing to exceptionally high signal-to-noise levels. BLI is based on the expression of luciferase proteins, which comprise a family of photoproteins that emit detectable photons in the presence of oxygen and ATP during metabolism of substrates such as luciferin into oxyluciferin. There are a variety of luciferase enzymes from other organisms that possess unique spectral characteristics and substrate requirements. Frequently-used luciferases include Firefly and Renilla [30], generating light upon their interaction with the substrates luciferin and coelenterazine, respectively. A central limitation of BLI reporter-gene strategies is the need to introduce the reporter construct into the cell populations of interest. Gene transfection using viral or non-viral vectors have proved useful in delivering reporter genes; however, they suffer from variable efficiency, especially when attempting to introduce genes into certain primary cells. These techniques also often require cell activation and culturing for varying periods of time, which might alter the biological activity of the cells. In addition, sensitivity of BLI is dependent on various factors, such as luciferase expression levels in the target cells, oxygenation and viability of the cells or tissues, sensitivity and settings of the camera, signal quenching by tissues, and numerous other factors. However, it is no doubt that BLI is a powerful tool to study biological processes such as gene-expression patterns [31], gene transfer efficiency [32], tumor growth and response to therapy [33-35], protein–protein interactions [36-41], and the location and proliferation of stem cells as well as in drug discovery and development [42-43].
There are two kinds of inactivatable/activatable BLI apoptosis probes reported so far: i) luciferin loses its bioluminescence activity by changing structure and ii) the luminescence is quenched by forming luminescent donor and fluorescent acceptor protein pairs based on Bioluminescence Resonance Energy Transfer (BRET). BRET is a naturally occurring phenomenon resulting from the nonradiative transfer of energy between luminescent donor and fluorescent acceptor proteins [23]. In the sea pansy Renilla reniformis, the luminescence resulting from the catalytic degradation of coelenterazine by luciferase (Rluc) is transferred to the fluorescent protein, which in turn, emits fluorescence upon dimerization of the two proteins. This BRET phenomenon between energy donors (luciferase) and acceptors (fluorescent protein) makes it an ideal system of choice to monitor protein–protein interactions in living cells. Utilizing this system, the substrate of luciferase, coelenterazine and D-luciferin, and yellow fluorescent protein (YFP), green fluorescent protein (GFP) and fluorophore can be used as a donor and acceptor, respectively. By combining the donor-acceptor pairs based on BRET, different activatable BLI probes are established to study protein-protein interactions and monitor the protease expression of cells. The remaining sections discuss recently reported highly sensitive activatable probes that combine various fluorophores with substrate, polymers, nanoparticles, and reporter genes to image apoptosis.
Optical imaging of caspase activities
Optical imaging is less expensive and more convenient than other imaging modalities such as MRI and PET. As such, fluorescent imaging and BLI are the most widely used techniques for imaging protease activity. With the first demonstration of in vivo optical imaging of protease activity by Weissleder et al. [44], various activatable imaging probes have been studied, such as peptide-based and NP-based activatable probes. Herein we mainly focus on the imaging of caspase activity of these probes. For an efficient activatable system, the following three aspects should be considered: how to efficiently i) improve specific interactions between enzymes and peptide substrates, ii) decrease background signals, and iii) increase activated fluorescent output signals. Since enzyme activity and substrate specificity are generally constant in the same reaction conditions, the development of highly sensitive, activatable probes will be governed by combining decreased background with increased output signals.
Fluorescent imaging of caspase activity: peptide-based fluorescent probes
Typical peptide-based probes detect caspase activities by restoring fluorescence signals following caspase-dependent cleavage that is quenched in the native state. The simplest and most conventional of these peptide-based probes is the fluorophore and quencher pair connected with a caspase-specific peptide substrate. This probe is optically silent in its quenched (native) state, but it becomes highly fluorescent following proteolysis of the caspase substrate linker by the target enzymes. Pham et al. synthesized a nonfluorogenic near-infrared quencher (NIRQ) that has a broad absorption peak from 550 nm to 800 nm and the maximum absorption is in the range of 700 to 800 [45]. Then they designed an activatable caspase-3 probe by directly anchoring NIRQ to the N-terminus of a caspase-3 cleavable peptide substrate (-DEVD-) labeled with Alexa Fluor 680. The fluorescent signal of this activatable probe was increased about four-fold, while no activation was observed when the caspase-3 specific inhibitor was present. Lee et al. fabricated a complementary imaging probe, LS498, consisting of tetraazacyclododecanetetraacetic acid (DOTA) chelating the radionuclide 64Cu, a NIR dye LS-276, a caspase-3 substrate and a quencher dye IRdye QC-1. The fluorescent signal is activated by cleaving the peptide substrate for caspase-3 [46]. In vivo experiments demonstrate a time-dependent five-fold NIR fluorescence enhancement in an artificial model of subcutaneous tumor in mice but radioactivity was the same in caspase-3 positive and negative controls. This study indicates it is feasible to use radionuclide imaging to localize and quantify the distribution of molecular probes and optical imaging to report the functional status of diagnostic enzymes.
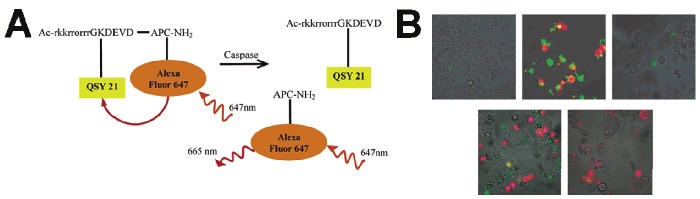
Unlike other extracellular-proteases such as MMP and urokinase plasminogen activator (uPA) which are excreted in the extracellular environment, caspases are intracellular-proteases. Therefore to target caspases, the probes need to be delivered into the cytoplasm. Many peptide probes do not possess permeability into the cell membrane; so, one prerequisite for imaging caspase activity is to efficiently deliver peptide probes into the cells. Bullok et al. prepared a small, membrane-permeable probe, TcapQ647, comprising of a Tat permeation peptide sequence (Ac-rkkrrorrr), an effector caspase recognition sequence (DEVD), and an activatable dye pair of QSY 21/Alexa Fluor 647 (Figure 3) [47]. TcapQ647 show high quenching efficiencies and a low fluorescent background signal. TcapQ647 is specifically cleaved by caspase-3 and 7 as shown by in vitro enzyme assays, thereby releasing the fluorophore and enabling apoptosis imaging. Cellular apoptosis imaging in HeLa and KB 3-1 cells treated with anticancer drugs demonstrated TcapQ647 is a sensitive, effector caspase-specific “smart” probe to monitor apoptosis in cells [48]. In addition, TcapQ647 was able to detect and monitor single-cell apoptosis in a mouse glaucoma model [49]. A similar probe design with a different cell-penetrating peptide sequence KKKRKV resulted in improved signal sensitivity with less cellular toxicity [50]. Although cell-permeable peptide-mediated delivery of activatable probes can improve the overall efficiency of activation, it can also result in non-specific activation of the probe in vivo due to non-specific accumulation in tissues.
Figure 3.
A membrane-permeable, caspase-activatable fluorescent probe, TcapQ647, for imaging apoptosis. A) Schematic of TcapQ647 activation following cleavage by effector caspase. B) Intracellular activation of TcapQ647 during apoptosis. Modified with permission from ref [47]. Copyright 2005 American Chemical Society.
NPs-based fluorescent probes
A significant number of peptide-based imaging probes have been used to image protease activity in vitro and in vivo; however, these small-molecules are limited by several aspects, such as they are i) unstable and can be rapidly cleared from the body and ii) difficult to alter while maintaining their biological activities due to the lack of functional sites for chemical modification. In contrast, polymers or NPs provide many functional groups with large surface areas for efficient modification with a broad range of molecules including substrates, fluorophores and quenchers. More importantly, the quencher-fluorophore pairs of peptide-based probes are one-to-one, resulting in low quenching efficiency and high background signal. NP-based probes provide a platform for different quencher-fluorophore combinations such as multiple-to-one or multiple-to-multiple pairs. Very recently, polymeric and inorganic metal NP-based activatable probes have successfully been developed.
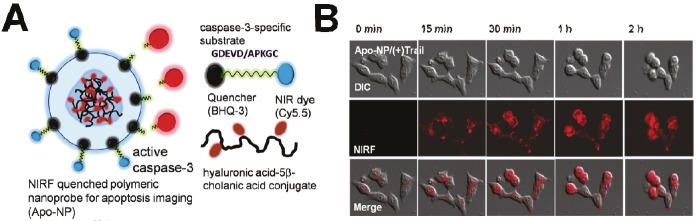
Lee et al. reported a new activatable nanoprobe designed on the basis of a polymer nanoparticle platform [51]. This nanoprobe consists of strongly dual-quenched (dye-dark quencher and dye-dye quenching mechanisms), caspase-3-specific, and NIR fluorogenic peptides on the surface of hyaluronic acid-based, self-assembled polymeric nanoparticles (HA-NPs) that serve as carriers (Figure 4). The results indicate that the activatable nanoprobe efficiently delivers dual-quenched caspase-3-sensitive fluorogenic peptides into cells, allowing caspase-3-dependent strong fluorescence amplification to be imaged in apoptotic cells in real-time and at high resolution. To demonstrate the utility of this nanoprobe to image apoptosis in vivo, nanoprobes were intratumorally injected into tumor-bearing mice. Nanoprobes demonstrate high NIR fluorescence signal activation in mice treated with doxorubicin, enabling clear visualization of apoptosis in tumors. In contrast, the fluorescence signal from the vehicle-treated tumors and normal tissues were substantially weaker.
Figure 4.
Real-time imaging of apoptosis in single cells with a polymeric nanoprobe. A) Schematic diagram of the caspase-3 activatable nanoprobe (Apo-NP). B) Activity of caspase-3 in HCT116 cells were video imaged using the Apo-NPs. Modified with permission from ref [51]. Copyright 2011 American Chemical Society.
In another embodiment, various types of gold NP-based activatable probes were designed along with a caspase-3 substrate, DEVD. Sun et al. developed a simple and efficient apoptosis imaging probe with gold NPs [52]. A near-infrared fluorescence dye was attached to the gold NP surface through the bridge of caspase-3 substrate, DEVD. The fluorescence was quenched in physiological conditions due to the high quenching efficiency of gold NPs [53], and the quenched fluorescence was recovered after the DEVD was cleaved by caspase-3. The results indicate that the substrate cleavage and the dequenching process are very fast, reducing the need to fix the cells for imaging. In addition, real-time monitoring of caspase activity is achieved in live cells, which enables early detection of apoptosis compared to a conventional apoptosis kit such as Annexin V-FITC. To provide a very clear signal at the single-molecule level in the highly heterogeneous and high-background scattering environment of live cells, Jun et al. designed a series of crown NP plasmon rulers for caspase-3 activation (Figure 5) [54]. These assemblies were linked together by peptides (DEVD) via NeutrAvidin-biotin and Au-thiol chemistries. It was expected that these crown NPs could result in an immediate reduction in light-scattering intensity by caspase-3 cleavage of DEVD peptides. As expected, the results show that these crown NP plasmon rulers can be used for continuous observation of caspase-3 activity and analysis of caspase-3 activation kinetics on the single-molecule level. Lin et al. prepared gold quantum dots (GQDs) as nucleus shuttles and functionalized their surfaces with peptides that recognize activated caspase-3, to enable the real-time monitoring of cellular apoptosis [55]. As shown in Figure 6, GQDs were firstly functionalized with a peptide moiety that contains both nuclear export signal (NES) and nuclear localization signal (NLS) sequences, which are used to mimic the actions of nucleus shuttle proteins. To monitor cell apoptosis, the proteolytic moiety DEVD-peptide sequence was placed between the NES and NLS to form the apoptotic sensing probe, GQD-NES-linker-DEVD-linker-NLS. During apoptosis, the activated caspase-3 proteolytically cleaved the DEVD-linker-NLS moiety from the remaining GQDs-NES, enabling the GQDs exported from the nucleus to remain in the cytoplasm. Consequently, the photoluminescence of GQDs within the nucleus gradually diminished during apoptosis from its preapoptotic maximum at equilibrium. As such, the NES-linker-DEVD-linker-NLS peptide enables the GQDs to function as molecular probes for real-time monitoring of cellular apoptosis (Figure 6).
Figure 5.
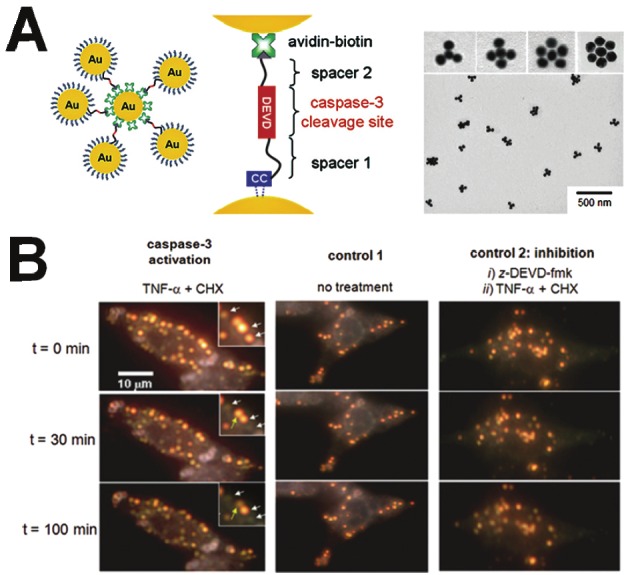
A) Schematic diagram (left) and TEM images (right) of Au-based nanoprobes, which are composed of a NeutrAvidin-coated gold-core nanoparticle with multiple biotinylated gold satellite nanoparticles. B) Cellular delivery of gold nanoprobes in live cells and single-molecule imaging of caspase-3 activation in apoptotic cells. Modified with permission from ref [54]. Copyright 2009 National Academy of Sciences, USA.
Figure 6.
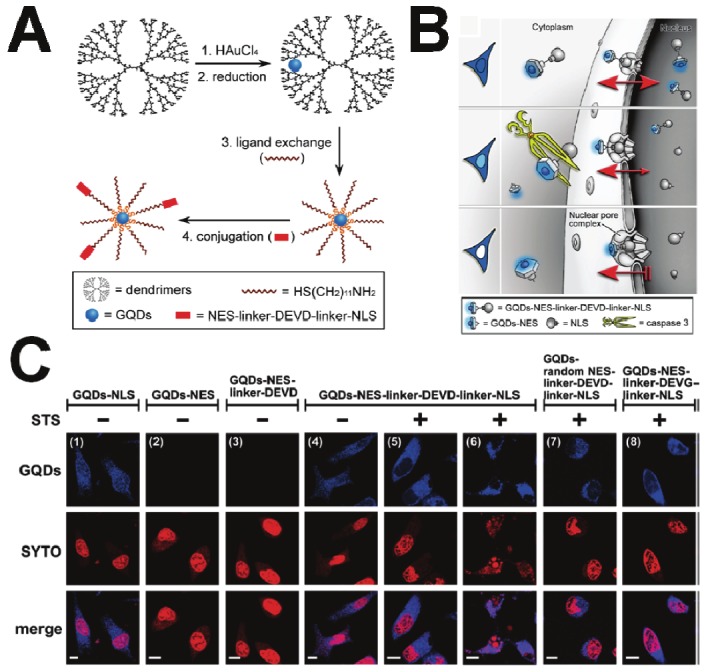
Photoluminescent gold quantum dots (GQDs) for real-time monitoring of cell apoptosis. A) Illustration of GQD synthesis and derivatization. B) Schematic representation of the nucleus shuttle of GQDs functionalized with the peptide moiety containing NLS, NES and the caspase-3 responsive DEVD that allow monitoring of cell apoptotic progression. C) Confocal microscopic images showing the GQDs’ clearance from the nucleus in response to the STS-induced cell apoptosis. Modified with permission from ref [55]. Copyright 2010 American Chemical Society.
BLI of caspase activity
Most activatable probes utilize fluorescence as a reporter signal, but bioluminescence is also another promising modality for optically imaging cancer and its biomarkers. BLI is more sensitive than fluorescence imaging because it does not require an external excitation light source [56-57], which can detect as few as 10 cells in vitro and 100–1,000 cells in vivo [58]. BLI is performed in absolute darkness, thus avoiding the interference with background light and autofluorescence, making the measurement of small lesions more reliable [59-60]. Based on BLI techniques, many strategies exist for genetically engineered bioluminescent proteins for apoptosis imaging, such as structural changes of substrate-based and BRET-based probes. However, up to now, there have been no report on BLI for imaging apoptosis using NP-based activatable probes.
Structure changes of substrate-based BLI activatable probes
Laxman et al. constructed a series of hybrid recombinant reporter molecules wherein the activity of the reporter can be silenced by fusion with the estrogen receptor regulatory domain (ER) [61]. Once adequate silencing of the reporter’s enzymatic activity was achieved, the inclusion of a protease cleavage site for caspase-3 (DEVD) between these two domains allowed for protease-mediated activation of the reporter molecule after separation from the silencing domain (ER). Stable human glioma cell lines were constructed expressing luciferase (Luc) with single or double ER fusions. In vitro studies revealed that the double ER fusion molecule had the greatest attenuation of Luc activity that could be reactivated by caspase-3 induction. Furthermore, in vivo studies also demonstrated that caspase-3 could be imaged noninvasively by BLI upon activation by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) treatment.
The Promega Corporation developed and commercialized Z-DEVD-aminoluciferin soluble substrate (VivoGloTM Caspase-3/7 Substrate), an activatable luciferin substrate targeting apoptosis for BLI. The substrate is a circularly-permuted Firefly luciferase. They joined the original termini of the Firefly luciferase with peptide substrate sequence, thereby locking the luciferase in an inactive state. When Z-DEVD-aminoluciferin is still linked to DEVD peptide, it cannot interact with luciferase because of steric hindrance whereas, after caspase cleavage, the released aminoluciferin is used by luciferase, together with ATP and O2, to induce light. Using this Z-DEVD-aminoluciferin substrate system, several laboratories have evaluated the feasibility of caspase-3/7 activity with BLI [62-67].
The common recombinant bioluminescent probes are composed of a “sandwich” linear structure, Luc-DEVD-receptor. However, their molecular size is quite large and the intensity of the luminescence signals is relatively low because of insufficient digestion of the DEVD sequence during apoptosis. To overcome this limitation, Kanno et al. engineered a genetically encoded cyclic luciferase to detect protease activities in living cells and animals [68]. Two fragments of DnaE intein, producing a high yield of cyclic pepetide or protein, were fused to neighboring ends of Fluc connected with a substrate sequence for the protease (Figure 7). After translation into a single polypeptide in living cells, the amino (N) and carboxy (C) terminals of the luciferase were ligated by protein splicing, resulting in a closed circular polypeptide chain. Since the structure of the cyclic luciferase was distorted, the luciferase loses its bioluminescence activity. If the substrate sequence was digested by a protease, the luciferase transforms into an active form and restores its activity. This cyclic luciferase was used to quantitatively sense caspase-3 activity in living cells upon extracellular stimuli and to noninvasively image the time-dependent caspase activity in living mice. The results demonstrated that the response of cyclic Fluc upon caspase-3 activation was very fast and can be used for quantitative detection of caspase-3 activity in living cells and animals [69].
Figure 7.
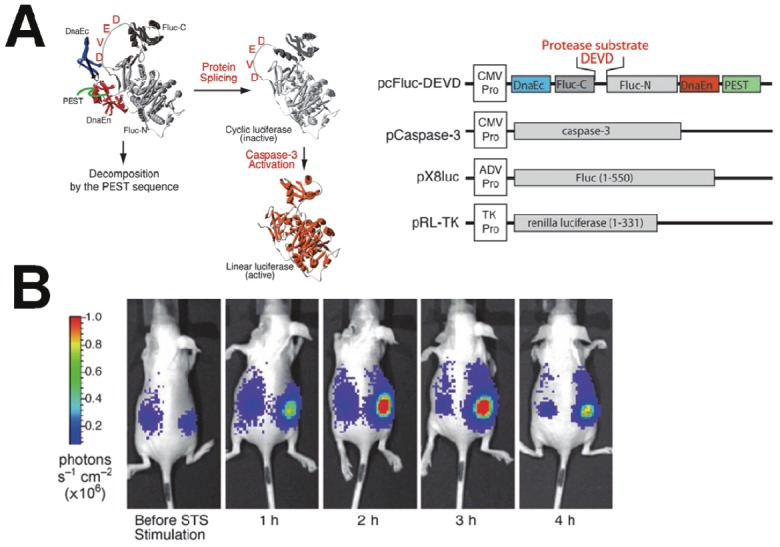
A genetically encoded cyclic luciferase for the detection of caspase-3 activities. (A) Schematic diagram of monitoring caspase-3 activation and structure of cyclic Firefly luciferase. (B) Time-dependent caspase-3 activity in living mice carrying transiently transfected HeLa cells expressing cyclic Fluc. Images were taken at the indicated times after intraperitoneal injection of staurosporine. Modified with permission from ref [68]. Copyright 2007 John Wiley and Sons.
Coppola et al. developed a structure change/reassembly activatable BLI system [70]. In this system, the reporter constituted of a fusion of small interacting peptides, peptide A and peptide B, with the NLuc and CLuc fragments of luciferase with a caspase-3 cleavage site (DEVD) between pepANLuc (ANLuc) and pepBCLuc (BCLuc). During apoptosis, caspase-3 cleaves the reporter, enabling separation of ANLuc from BCLuc. A high-affinity interaction between peptide A and peptide B restores luciferase activity by NLuc and CLuc complementation. Treatment of live cells and mice carrying D54 tumor xenografts with chemotherapeutic agents and radiation resulted in increased bioluminescence activity due to enhanced apoptosis. Renilla and Firefly luciferases, having substrates of coelenterazine and D-luciferin, respectively, have been used to monitor different biological processes in a single animal [30,71]. The ability to image two or more biological processes greatly increases the utility of luciferase imaging by using two different luciferases or activatable luciferases. Shah et al. utilized dual substrate/reporter bioluminescence imaging (Fluc: Firefly luciferase–luciferin and Rluc: Renilla luciferase–coelenterazine) to test the efficacy of TRAIL using replication-deficient herpes simplex virus (HSV) type 1 amplicon vectors in gliomas [71]. Growth of tumors stably transfected with Fluc (Gli36fluc+) was readily monitored in vivo by BLI following luciferin administration. HSV amplicon vectors bearing the genes for TRAIL and Rluc injected directly into Gli36fluc+-expressing subcutaneous gliomas revealed peak Rluc activity 36 h after intratumoral injection as determined by coelenterazine injection followed by imaging. This strategy offers a unique way to monitor both gene delivery and efficacy of TRAIL-induced apoptosis in tumors in vivo in real time by dual enzyme substrate (Rluc/Fluc) imaging. Following this example, a secreted version of TRAIL was used as a therapeutic protein to treat gliomas and tumor apoptosis was successfully monitored and imaged by dual-substrate bioluminescence [72].
In another example, a more complicated BLI system to monitor caspase activity was developed. Ray et al. constructed a fusion protein, combining three different reporter proteins, red fluorescent protein (mRFP1), Firefly luciferase (FL), and HSV1-sr39 truncated thymidine kinase (TK), linked through a caspase-3 recognizable peptide linker [73]. In the fused form, all three reporter proteins had markedly attenuated activity. However, a significant signal increase of mRFP1, FL, and TK activity was observed upon cleavage by activated caspase-3. The results demonstrate that upon induction with staurosporine, a general drug to induce caspase-3 activation, the fusion protein showed a 2.8-fold increase in FL, a 1.5-fold increase in TK, and a 2-fold increase in mRFP1 activity in 293T cells. Furthermore, bioluminescence and micropositron emission tomography imaging of the apoptotic B16F10-mtf-hrl melanoma showed a 2-fold higher FL activity and a 2-fold higher TK activity than control tumors when normalized with RL activity.
BRET-based BLI activatable probes
The original BRET system used Rluc as the donor, the derivative of coelenterazine as its substrate and a yellow fluorescent protein (YFP) as the acceptor. In recent years, some GFPs and fluorophores were introduced as acceptor along with luciferin as the donor. In 2000, Angers et al. first reported the detection of caspase-3 by BRET technique in the study of G protein-coupled receptor (GPCR) homodimers at the surface of living cells [36]. The Rluc-DEVD-YFP fusion gene was subcloned from the pT7 plasmid into the mammalian expression vector. The stimulation of caspase-3 activity by a treatment of the cells with staurosporine promoted a marked decrease in the BRET ratio, indicating that a proportion of the fusion protein was cleaved, resulting in the physical separation of the BRET partners. The staurosporine-induced reduction in BRET was blocked by a pretreatment with the caspase-3 inhibitor, confirming the specificity of the effect. After this report, several groups used BRET techniques to study protein-protein interactions in live cells and some pathophysiological cellular functions [37,74-77]. However, few impressive studies were reported to monitor the protease expression based on a BRET system, especially for caspase activity imaging. In 2009, Gammon et al. chose a series of BRET pairs based on luciferases and tdTomato (acceptor) for caspase-3 activity imaging to predict optimal luciferase-fluorophore pairing [78]. The results indicated that one should choose a peptide-linker length that is as short as can be allowed by available cloning strategies or consensus sequences to achieve maximal change in BRET signal upon protease cleavage. Thus, it is important to keep the fluorescent protein as close to the active site of the luciferase as possible. However, this strategy may change the structures of luciferase and fluorescent protein and decrease protease activation.
Conclusion and perspective
In many cases, poor specificity and sensitivity of current optical imaging probes limit their imaging ability and applications. From this point of view, the emergence of activatable molecular imaging probes based on signal amplification strategies are a promising set of imaging tools which can overcome these limitations. They offer several distinct advantages over conventional dye-labeled imaging probes: i) multiple fluorophores are able to be cleaved by a single enzyme resulting in an intrinsic signal amplification, ii) quenching of the probe before activation results in significantly low background fluorescence and iii) the activation efficiency strongly depends on the protease expression of target cells resulting in highly specific detection of target molecules. The combined effect results in high signal-to-background ratios and improved detection sensitivity and specificity.
Over the past decade, activatable fluorescent probes underwent major improvements, but the increased fluorescent signal outputs are not exciting, especially for in vivo application. Besides the high biological tissue absorption of fluorescent signals, fluorescent activatable probes need to prevent additional, equally important, limitations like efficiently decreasing background signal and increasing fluorescent output signal. As mentioned in the previous section, the quencher-fluorophore pairs of peptide probes are generally one-to-one format, limiting improvement in the signal-to-background ratio. In addition, the efficient targeted intracellular delivery of activatable probes limits the development of imaging caspase activity. Combining NPs with activatable probes can ensure high payload delivery of these probes to the target site, penetrating the cytoplasm by a different cellular uptake mechanism. With the incorporation of polymer NPs and novel metals such as gold NPs into one platform, use of complementary imaging modalities like PET, SPECT, MRI and CT is conceivable and is no longer confined to depth penetration concerns of optical imaging techniques.
The largest limitation of BLI is that many clinical trials using viral vectors for transfection have been terminated since the application of these vectors have induced unexpected adverse effects such as toxicity, immunogenicity and oncogenicity. Nanomaterials are gaining more interest as alternatives to viral vectors for gene delivery, but it is hard to achieve a major breakthrough in a short term [79]. However, it is emerging as a powerful technology to study viral pathogenesis, immune responses to infection, and quantifying effects of therapy in living animals. Although the outlook of activatable probes is clear and exciting, most platforms are in the proof-of-concept stage. Many obstacles, such as pharmacokinetics, non-specific degradation and aggregation, and potential toxicity of the probe, should be considered.
Despite the application of these technologies for optical imaging, “smart probes” for apoptosis imaging is still in its infancy and has a long way to go. The ability to utilize these aforementioned technologies in vivo will surely lead to advancements in early detection, improved diagnosis and therapeutic treatments tailored for individual patients.
Acknowledgement
This work was supported by the Intramural Research Program (IRP) of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH. We thank Maggie Swierczewska for proofreading the manuscript.
References
- 1.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–54. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 2.Putcha GV, Harris CA, Moulder KL, Easton RM, Johnson EM, Jr, Thompson CB. Intrinsic and extrinsic pathway signaling during neuronal apoptosis: lessons from the analysis of mutant mice. J Cell Biol. 2002;157:441–53. doi: 10.1083/jcb.200110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Logue SE, Martin SJ. Caspase activation cascades in apoptosis. Biochem Soc Trans. 2008;36:1–9. doi: 10.1042/BST0360001. [DOI] [PubMed] [Google Scholar]
- 4.Howley B, Fearnhead HO. Caspases as therapeutic targets. J Cell Mol Med. 2008;12:1502–16. doi: 10.1111/j.1582-4934.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–16. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–9. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 8.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Ren X, Alt E, Bai X, Huang S, Xu Z, Lynch PM, Moyer MP, Wen XF, Wu X. Chemoprevention of colorectal cancer by targeting APC-deficient cells for apoptosis. Nature. 2010;464:1058–61. doi: 10.1038/nature08871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukami T, Nakasu S, Baba K, Nakajima M, Matsuda M. Hyperthermia induces translocation of apoptosis-inducing factor (AIF) and apoptosis in human glioma cell lines. J Neurooncol. 2004;70:319–31. doi: 10.1007/s11060-004-9168-0. [DOI] [PubMed] [Google Scholar]
- 11.Starkey JR, Rebane AK, Drobizhev MA, Meng F, Gong A, Elliott A, McInnerney K, Spangler CW. New two-photon activated photodynamic therapy sensitizers induce xenograft tumor regressions after near-IR laser treatment through the body of the host mouse. Clin Cancer Res. 2008;14:6564–73. doi: 10.1158/1078-0432.CCR-07-4162. [DOI] [PubMed] [Google Scholar]
- 12.Tait JF. Imaging of apoptosis. J Nucl Med. 2008;49:1573–6. doi: 10.2967/jnumed.108.052803. [DOI] [PubMed] [Google Scholar]
- 13.Boersma HH, Kietselaer BL, Stolk LM, Bennaghmouch A, Hofstra L, Narula J, Heidendal GA, Reutelingsperger CP. Past, present, and future of annexin A5: from protein discovery to clinical applications. J Nucl Med. 2005;46:2035–50. [PubMed] [Google Scholar]
- 14.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 15.Niu G, Chen X. Apoptosis imaging: beyond annexin V. J Nucl Med. 2010;51:1659–62. doi: 10.2967/jnumed.110.078584. [DOI] [PubMed] [Google Scholar]
- 16.Vangestel C, Peeters M, Mees G, Oltenfreiter R, Boersma HH, Elsinga PH, Reutelingsperger C, Van Damme N, De Spiegeleer B, Van de Wiele C. In Vivo Imaging of Apoptosis in Oncology: An Update. Mol Imaging. doi: 10.2310/7290.2010.00058. [DOI] [PubMed] [Google Scholar]
- 17.Lee S, Xie J, Chen X. Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem Rev. 110:3087–111. doi: 10.1021/cr900361p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi H, Choyke PL. Target-cancer-cell-specific activatable fluorescence imaging probes: rational design and in vivo applications. Acc Chem Res. 44:83–90. doi: 10.1021/ar1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Park K, Kim K, Choi K, Kwon IC. Activatable imaging probes with amplified fluorescent signals. Chem Commun (Camb) 2008:4250–60. doi: 10.1039/b806854m. [DOI] [PubMed] [Google Scholar]
- 20.Lee S, Xie J, Chen X. Activatable molecular probes for cancer imaging. Curr Top Med Chem. 10:1135–44. doi: 10.2174/156802610791384270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha Y. Structure and mechanism of intramembrane protease. Semin Cell Dev Biol. 2009;20:240–50. doi: 10.1016/j.semcdb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Lee CJ, Simeone DM. Identification of human pancreatic cancer stem cells. Methods in molecular biology. 2009;568:161–73. doi: 10.1007/978-1-59745-280-9_10. [DOI] [PubMed] [Google Scholar]
- 23.Xia Z, Rao J. Biosensing and imaging based on bioluminescence resonance energy transfer. Curr Opin Biotechnol. 2009;20:37–44. doi: 10.1016/j.copbio.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson MK. Choosing reporter-quencher pairs for efficient quenching through formation of intramolecular dimers. Methods Mol Biol. 2006;335:17–29. doi: 10.1385/1-59745-069-3:17. [DOI] [PubMed] [Google Scholar]
- 25.Lee S, Park K, Lee SY, Ryu JH, Park JW, Ahn HJ, Kwon IC, Youn IC, Kim K, Choi K. Dark quenched matrix metalloproteinase fluorogenic probe for imaging osteoarthritis development in vivo. Bioconjug Chem. 2008;19:1743–7. doi: 10.1021/bc800264z. [DOI] [PubMed] [Google Scholar]
- 26.Kang JS, Deluca PP, Lee KC. Emerging PEGylated drugs. Expert Opin Emerg Drugs. 2009;14:363–80. doi: 10.1517/14728210902907847. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L, Xie J, Swierczewska M, Zhang F, Quan Q, Ma Y, Fang X, Kim K, Lee S, Chen X. Real-Time Video Imaging of Protease Expression In Vivo. Theranostics. 1:18–27. doi: 10.7150/thno/v01p0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Ryu JH, Park K, Lee A, Lee SY, Youn IC, Ahn CH, Yoon SM, Myung SJ, Moon DH, Chen X, Choi K, Kwon IC, Kim K. Polymeric nanoparticle-based activatable near-infrared nanosensor for protease determination in vivo. Nano Lett. 2009;9:4412–6. doi: 10.1021/nl902709m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Cha EJ, Park K, Lee SY, Hong JK, Sun IC, Kim SY, Choi K, Kwon IC, Kim K, Ahn CH. A near-infrared-fluorescence-quenched gold-nanoparticle imaging probe for in vivo drug screening and protease activity determination. Angew Chem Int Ed Engl. 2008;47:2804–7. doi: 10.1002/anie.200705240. [DOI] [PubMed] [Google Scholar]
- 30.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci U S A. 2002;99:377–82. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raty JK, Liimatainen T, Unelma Kaikkonen M, Grohn O, Airenne KJ, Yla-Herttuala S. Noninvasive Imaging in Gene Therapy. Mol Ther. 2007;15:1579–86. doi: 10.1038/sj.mt.6300233. [DOI] [PubMed] [Google Scholar]
- 32.Cao F, Xie X, Gollan T, Zhao L, Narsinh K, Lee RJ, Wu JC. Comparison of gene-transfer efficiency in human embryonic stem cells. Mol Imaging Biol. 12:15–24. doi: 10.1007/s11307-009-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeamari S, Rumping G, Floot B, Lyons S, Stewart FA. In vivo bioluminescence imaging of locally disseminated colon carcinoma in rats. Br J Cancer. 2004;90:1259–64. doi: 10.1038/sj.bjc.6601637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shan L, Wang S, Korotcov A, Sridhar R, Wang PC. Bioluminescent animal models of human breast cancer for tumor biomass evaluation and metastasis detection. Ethn Dis. 2008;18:S2-65–9. [PubMed] [Google Scholar]
- 35.Ozawa T, James CD. Establishing intracranial brain tumor xenografts with subsequent analysis of tumor growth and response to therapy using bioluminescence imaging. J Vis Exp. doi: 10.3791/1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc Natl Acad Sci U S A. 2000;97:3684–9. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfleger KD, Dromey JR, Dalrymple MB, Lim EM, Thomas WG, Eidne KA. Extended bioluminescence resonance energy transfer (eBRET) for monitoring prolonged protein-protein interactions in live cells. Cell Signal. 2006;18:1664–70. doi: 10.1016/j.cellsig.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Pfleger KD, Seeber RM, Eidne KA. Bioluminescence resonance energy transfer (BRET) for the real-time detection of protein-protein interactions. Nat Protoc. 2006;1:337–45. doi: 10.1038/nprot.2006.52. [DOI] [PubMed] [Google Scholar]
- 39.De A, Loening AM, Gambhir SS. An improved bioluminescence resonance energy transfer strategy for imaging intracellular events in single cells and living subjects. Cancer Res. 2007;67:7175–83. doi: 10.1158/0008-5472.CAN-06-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De A, Ray P, Loening AM, Gambhir SS. BRET3: a red-shifted bioluminescence resonance energy transfer (BRET)-based integrated platform for imaging protein-protein interactions from single live cells and living animals. Faseb J. 2009;23:2702–9. doi: 10.1096/fj.08-118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazars A, Fahraeus R. Using BRET to study chemical compound-induced disruptions of the p53-HDM2 interactions in live cells. Biotechnol J. 5:377–84. doi: 10.1002/biot.200900272. [DOI] [PubMed] [Google Scholar]
- 42.Compte M, Cuesta AM, Sanchez-Martin D, Alonso-Camino V, Vicario JL, Sanz L, Alvarez-Vallina L. Tumor immunotherapy using genemodified human mesenchymal stem cells loaded into synthetic extracellular matrix scaffolds. Stem Cells. 2009;27:753–60. doi: 10.1634/stemcells.2008-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Suzuki Y, Huang M, Cao F, Xie X, Connolly AJ, Yang PC, Wu JC. Comparison of reporter gene and iron particle labeling for tracking fate of human embryonic stem cells and differentiated endothelial cells in living subjects. Stem Cells. 2008;26:864–73. doi: 10.1634/stemcells.2007-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weissleder R, Tung CH, Bogdanov A, Jr, Mahmood U. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–8. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 45.Pham W, Weissleder R, Tung CH. An azulene dimer as a near-infrared quencher. Angew Chem Int Ed Engl. 2002;41:3659–62. doi: 10.1002/1521-3773(20021004)41:19<3659::AID-ANIE3659>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 46.Lee H, Akers WJ, Cheney PP, Edwards WB, Liang K, Culver JP, Achilefu S. Complementary optical and nuclear imaging of caspase-3 activity using combined activatable and radiolabeled multimodality molecular probe. J Biomed Opt. 2009;14:040507. doi: 10.1117/1.3207156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bullok K, Piwnica-Worms D. Synthesis and characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. J Med Chem. 2005;48:5404–7. doi: 10.1021/jm050008p. [DOI] [PubMed] [Google Scholar]
- 48.Bullok KE, Maxwell D, Kesarwala AH, Gammon S, Prior JL, Snow M, Stanley S, Piwnica-Worms D. Biochemical and in vivo characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. Biochemistry. 2007;46:4055–65. doi: 10.1021/bi061959n. [DOI] [PubMed] [Google Scholar]
- 49.Barnett EM, Zhang X, Maxwell D, Chang Q, Piwnica-Worms D. Single-cell imaging of retinal ganglion cell apoptosis with a cell-penetrating, activatable peptide probe in an in vivo glaucoma model. Proc Natl Acad Sci U S A. 2009;106:9391–6. doi: 10.1073/pnas.0812884106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maxwell D, Chang Q, Zhang X, Barnett EM, Piwnica-Worms D. An improved cell-penetrating, caspase-activatable, near-infrared fluorescent peptide for apoptosis imaging. Bioconjug Chem. 2009;20:702–9. doi: 10.1021/bc800516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S, Choi KY, Chung H, Ryu JH, Lee A, Koo H, Youn IC, Park JH, Kim IS, Kim SY, Chen X, Jeong SY, Kwon IC, Kim K, Choi K. Real time, high resolution video imaging of apoptosis in single cells with a polymeric nanoprobe. Bioconjug Chem. 22:125–31. doi: 10.1021/bc1004119. [DOI] [PubMed] [Google Scholar]
- 52.Sun IC, Lee S, Koo H, Kwon IC, Choi K, Ahn CH, Kim K. Caspase sensitive gold nanoparticle for apoptosis imaging in live cells. Bioconjug Chem. 21:1939–42. doi: 10.1021/bc1003026. [DOI] [PubMed] [Google Scholar]
- 53.Swierczewska M, Lee S, Chen X. The design and application of fluorophore-gold nanoparticle activatable probes. Phys Chem Chem Phys. 13:9929–41. doi: 10.1039/c0cp02967j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jun YW, Sheikholeslami S, Hostetter DR, Tajon C, Craik CS, Alivisatos AP. Continuous imaging of plasmon rulers in live cells reveals early-stage caspase-3 activation at the single-molecule level. Proc Natl Acad Sci U S A. 2009;106:17735–40. doi: 10.1073/pnas.0907367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin SY, Chen NT, Sun SP, Chang JC, Wang YC, Yang CS, Lo LW. The protease-mediated nucleus shuttles of subnanometer gold quantum dots for real-time monitoring of apoptotic cell death. J Am Chem Soc. 132:8309–15. doi: 10.1021/ja100561k. [DOI] [PubMed] [Google Scholar]
- 56.El Hilali N, Rubio N, Martinez-Villacampa M, Blanco J. Combined noninvasive imaging and luminometric quantification of luciferase-labeled human prostate tumors and metastases. Lab Invest. 2002;82:1563–71. doi: 10.1097/01.lab.0000036877.36379.1f. [DOI] [PubMed] [Google Scholar]
- 57.Ray P, De A, Min JJ, Tsien RY, Gambhir SS. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 2004;64:1323–30. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edinger M, Cao YA, Verneris MR, Bachmann MH, Contag CH, Negrin RS. Revealing lymphoma growth and the efficacy of immune cell therapies using in vivo bioluminescence imaging. Blood. 2003;101:640–8. doi: 10.1182/blood-2002-06-1751. [DOI] [PubMed] [Google Scholar]
- 59.Ponomarev V, Doubrovin M, Serganova I, Vider J, Shavrin A, Beresten T, Ivanova A, Ageyeva L, Tourkova V, Balatoni J, Bornmann W, Blasberg R, Gelovani Tjuvajev J. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging. 2004;31:740–51. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 60.Troy T, Jekic-McMullen D, Sambucetti L, Rice B. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol Imaging. 2004;3:9–23. doi: 10.1162/15353500200403196. [DOI] [PubMed] [Google Scholar]
- 61.Laxman B, Hall DE, Bhojani MS, Hamstra DA, Chenevert TL, Ross BD, Rehemtulla A. Noninvasive real-time imaging of apoptosis. Proc Natl Acad Sci U S A. 2002;99:16551–5. doi: 10.1073/pnas.252644499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scabini M, Stellari F, Cappella P, Rizzitano S, Texido G, Pesenti E. In vivo imaging of early stage apoptosis by measuring real-time caspase-3/7 activation. Apoptosis. 16:198–207. doi: 10.1007/s10495-010-0553-1. [DOI] [PubMed] [Google Scholar]
- 63.Liu JJ, Wang W, Dicker DT, El-Deiry WS. Bioluminescent imaging of TRAIL-induced apoptosis through detection of caspase activation following cleavage of DEVD-aminoluciferin. Cancer Biol Ther. 2005;4:885–92. doi: 10.4161/cbt.4.8.2133. [DOI] [PubMed] [Google Scholar]
- 64.Kizaka-Kondoh S, Itasaka S, Zeng L, Tanaka S, Zhao T, Takahashi Y, Shibuya K, Hirota K, Semenza GL, Hiraoka M. Selective killing of hypoxia-inducible factor-1-active cells improves survival in a mouse model of invasive and metastatic pancreatic cancer. Clin Cancer Res. 2009;15:3433–41. doi: 10.1158/1078-0432.CCR-08-2267. [DOI] [PubMed] [Google Scholar]
- 65.O'Brien MA, Daily WJ, Hesselberth PE, Moravec RA, Scurria MA, Klaubert DH, Bulleit RF, Wood KV. Homogeneous, bioluminescent protease assays: caspase-3 as a model. J Biomol Screen. 2005;10:137–48. doi: 10.1177/1087057104271865. [DOI] [PubMed] [Google Scholar]
- 66.Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC, Randi AM. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hickson J, Ackler S, Klaubert D, Bouska J, Ellis P, Foster K, Oleksijew A, Rodriguez L, Schlessinger S, Wang B, Frost D. Noninvasive molecular imaging of apoptosis in vivo using a modified firefly luciferase substrate, Z-DEVD-aminoluciferin. Cell Death Differ. 17:1003–10. doi: 10.1038/cdd.2009.205. [DOI] [PubMed] [Google Scholar]
- 68.Kanno A, Yamanaka Y, Hirano H, Umezawa Y, Ozawa T. Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew Chem Int Ed Engl. 2007;46:7595–9. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- 69.Zhang F, Zhu L, Liu G, Hida N, Lu G, Eden HS, Niu G, Chen X. Multimodality Imaging of Tumor Response to Doxil. Theranostics. 2011;1 doi: 10.7150/thno/v01p0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coppola JM, Ross BD, Rehemtulla A. Noninvasive imaging of apoptosis and its application in cancer therapeutics. Clin Cancer Res. 2008;14:2492–501. doi: 10.1158/1078-0432.CCR-07-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shah K, Tang Y, Breakefield X, Weissleder R. Real-time imaging of TRAIL-induced apoptosis of glioma tumors in vivo. Oncogene. 2003;22:6865–72. doi: 10.1038/sj.onc.1206748. [DOI] [PubMed] [Google Scholar]
- 72.Shah K, Tung CH, Breakefield XO, Weissleder R. In vivo imaging of S-TRAIL-mediated tumor regression and apoptosis. Mol Ther. 2005;11:926–31. doi: 10.1016/j.ymthe.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 73.Ray P, De A, Patel M, Gambhir SS. Monitoring caspase-3 activation with a multimodality imaging sensor in living subjects. Clin Cancer Res. 2008;14:5801–9. doi: 10.1158/1078-0432.CCR-07-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Otsuji T, Okuda-Ashitaka E, Kojima S, Akiyama H, Ito S, Ohmiya Y. Monitoring for dynamic biological processing by intramolecular bioluminescence resonance energy transfer system using secreted luciferase. Anal Biochem. 2004;329:230–7. doi: 10.1016/j.ab.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 75.Arai R, Nakagawa H, Kitayama A, Ueda H, Nagamune T. Detection of protein-protein interaction by bioluminescence resonance energy transfer from firefly luciferase to red fluorescent protein. J Biosci Bioeng. 2002;94:362–4. doi: 10.1263/jbb.94.362. [DOI] [PubMed] [Google Scholar]
- 76.Lechardeur D, Dougaparsad S, Nemes C, Lukacs GL. Oligomerization state of the DNA fragmentation factor in normal and apoptotic cells. J Biol Chem. 2005;280:40216–25. doi: 10.1074/jbc.M502220200. [DOI] [PubMed] [Google Scholar]
- 77.De A, Gambhir SS. Noninvasive imaging of protein-protein interactions from live cells and living subjects using bioluminescence resonance energy transfer. Faseb J. 2005;19:2017–9. doi: 10.1096/fj.05-4628fje. [DOI] [PubMed] [Google Scholar]
- 78.Gammon ST, Villalobos VM, Roshal M, Samrakandi M, Piwnica-Worms D. Rational design of novel red-shifted BRET pairs: Platforms for real-time single-chain protease biosensors. Biotechnol Prog. 2009;25:559–69. doi: 10.1002/btpr.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu G, Swierczewska M, Lee S, Chen X. Functional nanoparticles for molecular imaging guided gene delivery. Nano Today. 5:524–539. doi: 10.1016/j.nantod.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]