An essential role for hnRNP-Q1 in regulating dendritic development and focal adhesions is identified. This function for hnRNP-Q1 involves repression of RhoA mRNA translation. These findings suggest that negative regulation of RhoA translation is required for proper cell development and morphogenesis.
Abstract
The small GTPase RhoA has critical functions in regulating actin dynamics affecting cellular morphogenesis through the RhoA/Rho kinase (ROCK) signaling cascade. RhoA signaling controls stress fiber and focal adhesion formation and cell motility in fibroblasts. RhoA signaling is involved in several aspects of neuronal development, including neuronal migration, growth cone collapse, dendrite branching, and spine growth. Altered RhoA signaling is implicated in cancer and neurodegenerative disease and is linked to inherited intellectual disabilities. Although much is known about factors regulating RhoA activity and/or degradation, little is known about molecular mechanisms regulating RhoA expression and the subsequent effects on RhoA signaling. We hypothesized that posttranscriptional control of RhoA expression may provide a mechanism to regulate RhoA signaling and downstream effects on cell morphology. Here we uncover a cellular function for the mRNA-binding protein heterogeneous nuclear ribonucleoprotein (hnRNP) Q1 in the control of dendritic development and focal adhesion formation that involves the negative regulation of RhoA synthesis and signaling. We show that hnRNP-Q1 represses RhoA translation and knockdown of hnRNP-Q1 induced phenotypes associated with elevated RhoA protein levels and RhoA/ROCK signaling. These morphological changes were rescued by ROCK inhibition and/or RhoA knockdown. These findings further suggest that negative modulation of RhoA mRNA translation can provide control over downstream signaling and cellular morphogenesis.
INTRODUCTION
The small GTPase RhoA has critical functions in regulating actin dynamics affecting cellular morphogenesis through the RhoA/Rho kinase (ROCK) signaling cascade (Maekawa et al., 1999; Govek et al., 2005). RhoA signaling controls stress fiber and focal adhesion formation and cell motility (Nobes and Hall, 1995; Narumiya et al., 2009), and altered RhoA expression and signaling contribute to tumor cell invasion and metastasis (Narumiya et al., 2009). RhoA signaling is involved in several aspects of neuronal development, including neuronal migration (Govek et al., 2011), growth cone collapse (Swiercz et al., 2002; Wu et al., 2005), dendrite branching, and spine growth (Nakayama et al., 2000; Tashiro and Yuste, 2008). Specific mutations affecting RhoA signaling have been linked to inherited intellectual disability and autism (Govek et al., 2004; Jiang et al., 2010). RhoA signaling also mediates a local inhibitory effect on nerve regeneration following injury in the CNS, which can be overridden by genetic and pharmacological inhibition of the RhoA signaling pathway (Kubo et al., 2007; Duffy et al., 2009).
Considering the importance of RhoA signaling in health and disease, it becomes critical to understand mechanisms involved in the regulation of both RhoA expression and signaling. Like many other small GTPases, RhoA cycles between the GDP-bound inactive form and GTP-bound active form. GTP-bound RhoA interacts with and activates downstream effectors, such as ROCK (Maekawa et al., 1999). Levels of GTP-bound active RhoA are tightly controlled by RhoA GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs; Sit and Manser, 2011). Besides this conventional regulatory mechanism, recent studies suggest that the regulation of RhoA signaling can also be achieved by modulating RhoA protein levels through specific protein degradation (Wang et al., 2003), miRNA-mediated translational repression (Kong et al., 2008; Chiba et al., 2009), and extracellular signaling-triggered RhoA synthesis (Wu et al., 2005). However, trans-acting protein factors regulating RhoA translation remain unidentified. In light of the broad functions of RNA-binding proteins in the posttranscriptional regulation of gene expression (Anderson, 2008; Besse and Ephrussi, 2008), we sought to identify a possible role for a specific RNA-binding protein in RhoA synthesis and signaling.
The mRNA-binding protein heterogeneous nuclear ribonucleoprotein (hnRNP) Q1 is the cytoplasmic isoform of hnRNP-Q proteins derived by alternative splicing, distinguished by a unique carboxy terminus that has one nuclear localization sequence instead of two (Mourelatos et al., 2001). hnRNP-Q1 is ubiquitously expressed and was previously identified as NS1-associating protein-1 (Nsap1; Harris et al., 1999) and synaptotagmin-binding, cytoplasmic RNA-interacting protein (Syncrip; Mizutani et al., 2000). At the molecular level, hnRNP-Q1 has been shown to bind to cis-acting mRNA sequences of its target mRNAs and play roles in mRNA editing (Blanc et al., 2001), activation of internal ribosome entry site (IRES)–mediated translation (Cho et al., 2007; Kim et al., 2007), and coding region determinant–mediated mRNA decay (Grosset et al., 2000). However, the cellular functions of hnRNP-Q1 are not characterized. Previous studies revealed that hnRNP-Qs exhibit high expression in hippocampal neurons (Bannai et al., 2004) and that the expression of hnRNP-Qs in the CNS is developmentally regulated (Mizutani et al., 2000; Rossoll et al., 2002). Thus, we hypothesized that hnRNP-Q–mediated regulation of target mRNA expression may play an important role in neuronal development. By analyzing the cellular and molecular effects of hnRNP-Q1 silencing in mature hippocampal neurons and C2C12 myoblastoma cells, our results revealed an essential role for hnRNP-Q1 in regulating dendritic development and focal adhesions, which are mediated by negatively regulating RhoA protein synthesis and signaling. These findings provide new insight into the posttranscriptional regulation of RhoA signaling and mechanisms underlying cellular morphogenesis.
RESULTS
Reduced dendritic complexity and density of dendritic protrusions in hippocampal neurons upon hnRNP-Q1 knockdown
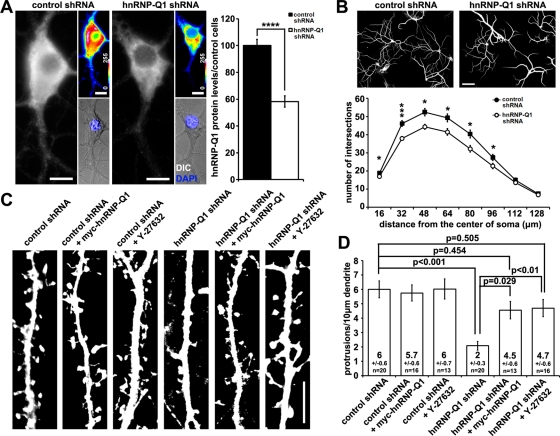
Previous studies suggested a function for hnRNP-Q in neuronal development (Mizutani et al., 2000; Bannai et al., 2004). Therefore, we examined the expression of hnRNP-Q isoforms in both embryonic and postnatal mouse brain regions and various tissues by Western blot analysis with antibodies recognizing all hnRNP-Q isoforms (Mizutani et al., 2000). We found that hnRNP-Q1 was detected as the major protein isoform of hnRNP-Q in all cells and tissues examined (Supplemental Figure S1A). Previous studies described the differential subcellular distribution of hnRNP-Q isoforms, of which hnRNP-Q1 is mostly cytoplasmic, compared with hnRNP-Q2/3, which are mostly nuclear (Chen et al., 2008). This suggests that hnRNP-Q1 is likely involved in posttranscriptional regulation in the cytoplasm. We assessed the effect of hnRNP-Q1–specific knockdown in mouse hippocampal cultures to uncover a possible function of hnRNP-Q1 in neuronal development. To achieve hnRNP-Q1–specific knockdown, we generated lentiviruses encoding short hairpin RNA (shRNA) targeting the 3′ untranslated region (UTR), which is unique to the mouse hnRNP-Q1 transcript (hnRNP-Q1 shRNA). Scrambled shRNA was used as a control. Western blot analysis of transduced neuronal cultures revealed that protein levels of hnRNP-Q1, but not hnRNP-Q3 or hnRNP-R, were specifically reduced in cells expressing hnRNP-Q1 shRNA (Supplemental Figure S2A). Immunofluorescence staining with an hnRNP-Q1–specific antibody generated for this study (Supplemental Figure S1B) showed that lentivirus-mediated hnRNP-Q1 shRNA expression led to >40% reduction of hnRNP-Q1 fluorescence intensity in the cell body (Figure 1A). We used the Sholl analysis to examine possible effects of hnRNP-Q1 knockdown on the development of dendrites. The Sholl analysis involves drawing concentric circles centered at the cell bodies with increased radius and measuring the number of intersections of each circle with dendrites (Sholl, 1953). The number of intersections in the Sholl analysis reflects the complexity of dendritic arbor, which is an overall effect of dendritic branching, outgrowth, and retraction (Miller, 1981). We found that after hnRNP-Q1 knockdown, hippocampal neurons exhibited reduced dendritic complexity (Figure 1B).
FIGURE 1:
Morphological analysis of cultured hippocampal neurons upon hnRNP-Q1 knockdown. (A) shRNA-mediated hnRNP-Q1 knockdown in hippocampal neurons. hnRNP-Q1 fluorescence intensity of the soma was reduced by >40% in neurons treated with hnRNP-Q1 shRNA lentiviral particles. t test, ****p < 0.001. The difference of hnRNP-Q1 fluorescence intensity was displayed as converted 16-color coded intensity graphs. Virus treatment did not affect the health of neurons, as demonstrated by differential interference contrast images and DAPI staining, as shown here. Scale bar, 10 μm. (B) hnRNP-Q1 knockdown reduced dendritic complexity. Dendrites were identified by Map2 staining and traced using NeuronJ for Sholl analysis. Representative images are shown on the top. Scale bar, 50 μm. Totals of 125 and 145 cells from four experiments were analyzed for control and hnRNP-Q1–knockdown cells, respectively. The numbers on the y-axis represent the sum of intersections within every 16 μm of dendrites as measure of distance from soma on the x-axis. Kruskal–Wallis analysis followed by Mann–Whitney U-test, *p < 0.05, ***p < 0.001. (C, D) hnRNP-Q1 knockdown reduced the density of dendritic protrusions, which was rescued by expression of shRNA-insensitive myc-hnRNP-Q1 or ROCK inhibitor, Y-27632. Neurons were transfected at 7DIV and fixed at 12DIV. (C) Representative images of GFP-Lifeact–positive neurons from each condition as indicated. Scale bar, 10 μm. (D) Quantification revealed a 60% reduction of dendritic protrusions in hnRNP-Q1–depleted neurons. Average densities of protrusions and number of cells analyzed for each condition are listed. One-way analysis of variance (ANOVA) post hoc Tukey Honestly Significant Difference (HSD).
Next we examined the effect of hnRNP-Q1 knockdown on the development of dendritic protrusions in cultured hippocampal neurons. In these experiments, neurons were cotransfected with shRNA plasmids (the same ones used to make lentiviruses) and a green fluorescent protein (GFP)-Lifeact–expressing plasmid to label dendritic protrusions (Figure 1C). GFP-Lifeact binds to actin filaments with low affinity without affecting actin filament dynamics (Riedl et al., 2008), and we used this method previously to label and analyze dendritic protrusions in cultured neurons (Gross et al., 2010). Neurons were cotransfected at day in vitro (DIV) 7 and fixed at DIV12 for image acquisition and analysis. Neurons that express hnRNP-Q1 shRNA were compared with scrambled shRNA as a control. hnRNP-Q1 knockdown was again confirmed by hnRNP-Q1–specific immunofluorescence staining (Supplemental Figure S3). Owing to the relative paucity of mature spines at this developmental stage and the prevalence of both thin spines and filopodia, we quantified the density of total protrusions, which included spines and filopodial protrusions. We observed that the protrusion density on primary and secondary dendrites of hnRNP-Q1–depleted neurons was reduced by >60% compared with control neurons (Figure 1, C and D, and Supplemental Figure S3). The reduction in protrusion density was rescued by reintroducing shRNA-insensitive myc-hnRNP-Q1 (lacking the 3′ UTR) (Figure 1, C and D, and Supplemental Figure S3). hnRNP-Q1 overexpression in control cells had no effect on protrusion densities (Figure 1, C and D, and Supplemental Figure S3). This suggests that either the endogenous hnRNP-Q1–mediated function on dendritic morphology was saturated or the levels of overexpression were insufficient to exert further effects. Nonetheless, the morphological effects obtained with hnRNP-Q1 knockdown and rescue experiments implicate a role for hnRNP-Q1 in dendritic and possibly synaptic development.
ROCK inhibition rescued the dendritic protrusion phenotype induced by hnRNP-Q1 knockdown
The reduced dendritic complexity and protrusion density in hnRNP-Q1-depleted neurons are reminiscent of phenotypes observed when RhoA/ROCK signaling is elevated (Govek et al., 2005; Elia et al., 2006; Zhang and Macara, 2008). We hypothesized that the effect of hnRNP-Q1 knockdown on dendritic morphogenesis may be caused by excess RhoA/ROCK signaling. We then examined the effects of a ROCK inhibitor, Y-27632, on protrusion density. We found that Y-27632 treatment of hnRNP-Q1–depleted neurons restored the density of dendritic protrusions to a level similar to that of control neurons (Figure 1, C and D). Consistent with a previous report (Nakayama et al., 2000), Y-27632 treatment had no effect on control cells (Figure 1, C and D). A similar dendritic protrusion phenotype was observed after hnRNP-Q1 knockdown with small interfering RNA (siRNA) targeting a different region of the hnRNP-Q1 3′ UTR (Supplemental Figure S2B), and this phenotype was also rescued by Y-27632 treatment (Supplemental Figures S4A and S4B). Taken together, our results suggest that hnRNP-Q1 silencing leads to excess RhoA/ROCK signaling, and the possible effectors of hnRNP-Q1 are upstream of ROCK.
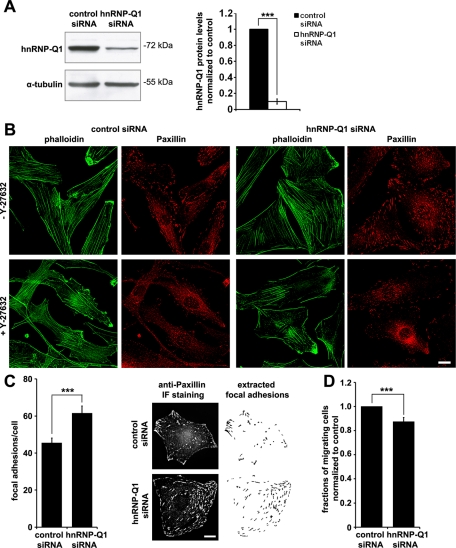
Knockdown of hnRNP-Q1 enhanced focal adhesion and stress fiber formation in C2C12 cells
The ubiquitous expression of hnRNP-Q1 (Supplemental Figure S1A) suggests that its functions are likely conserved among different cell types. On the basis of our observations in hippocampal neurons, we surveyed the effects of hnRNP-Q1 knockdown on actin filaments and focal adhesions in C2C12 myoblastoma cells. A critical role of RhoA/ROCK signaling is to promote stress fiber and focal adhesion formation (Hall, 1998). Immunoblot analysis showed that hnRNP-Q1 was depleted by 90% in C2C12 cells after 72 h of hnRNP-Q1 siRNA transfection, as determined by quantitative Western blot (Figure 2A). Knockdown of hnRNP-Q1 resulted in increased cell spreading, as evident by increases in cell area and number of focal adhesions (Figure 2B). Quantitative analysis revealed a 38% increase of focal adhesions in hnRNP-Q1 siRNA–treated C2C12 cells compared with control cells (Figure 2C). In addition, hnRNP-Q1–knockdown cells exhibited a pronounced increase of stress fibers (Figure 2B). Treatment with Y-27632 dramatically reduced focal adhesions and stress fibers in both control and hnRNP-Q1 siRNA–transfected cells (Figure 2B), suggesting the involvement of the RhoA/ROCK signaling pathway in focal adhesion and stress fiber formation induced by hnRNP-Q1 depletion. Furthermore, hnRNP-Q1 knockdown reduced C2C12 cell motility (Figure 2D), another phenotype associated with elevated RhoA/ROCK signaling (Besson et al., 2004; Mohseni and Chishti, 2008). Collectively, these observations illustrate a conserved function of hnRNP-Q1 in regulating cellular morphogenesis in different cell types, and this function is mediated by the RhoA/ROCK signaling pathway.
FIGURE 2:
hnRNP-Q1 knockdown enhances focal adhesion and stress fiber numbers in C2C12 cells and reduces cell motility. (A) Western blot analysis of hnRNP-Q1 knockdown in C2C12 cells. hnRNP-Q1 protein levels were reduced by 90% relative to control cells 3 d post hnRNP-Q1 siRNA transfection, normalized to tubulin. t test, ***p < 0.001. (B) Representative images of paxillin immunofluorescence staining for focal adhesions and phalloidin staining for actin filaments in C2C12 cells. Cells were treated with or without Y-27632 for 16 h before fixation. n = 4; t test, ***p < 0.001. Scale bar, 10 μm. (C) Quantitative analysis revealed a 38% increase of focal adhesion numbers in hnRNP-Q1 siRNA–transfected cells (three experiments; 80 and 90 cells were analyzed for control and hnRNP-Q1 knockdown cells, respectively). t test, ***p < 0.005. Representative images and corresponding graphs showing extracted focal adhesions for quantification are shown on the right. Scale bar, 10 μm. (D) Transwell migration assay shows reduced motility of C2C12 cells after hnRNP-Q1 knockdown. n = 4; t test, ***p < 0.005.
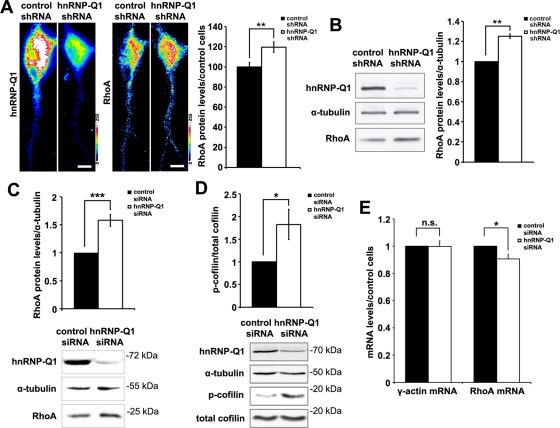
hnRNP-Q1 knockdown up-regulated RhoA protein expression and cofilin phosphorylation
Recent studies suggest that posttranscriptional regulation of RhoA translation and protein levels may affect the RhoA/ROCK signaling pathway (Wu et al., 2005; Kong et al., 2008; Chiba et al., 2009). Our observed effects of hnRNP-Q1 knockdown on RhoA signaling motivated us to investigate whether hnRNP-Q1 regulates the expression of RhoA by a posttranscriptional mechanism. Hence we first examined RhoA protein levels upon hnRNP-Q1 knockdown in hippocampal neurons using a monoclonal antibody against RhoA protein. The specificity of this antibody was validated by examining its ability to detect modulated RhoA expression changes that were achieved by overexpression of RhoA protein (Supplemental Figure S5A) or RhoA knockdown using siRNA specifically against RhoA mRNA (Supplemental Figure S5B). Our results revealed a 20% increase of RhoA protein in hippocampal neurons upon hnRNP-Q1 knockdown, as determined by quantitative immunofluorescence staining (Figure 3A), and that a similar up-regulation of RhoA protein (>20%) in hippocampal neurons was detected by Western blot (Figure 3B). Western blot analysis also showed a 50% increase of RhoA protein in both C2C12 cells (Figure 3C) and primary mouse embryonic fibroblast cells (Supplemental Figure S4C) following hnRNP-Q1 knockdown. These results indicate that hnRNP-Q1 negatively regulates RhoA protein expression in neuronal and nonneuronal cells.
FIGURE 3:
hnRNP-Q1 knockdown up-regulates RhoA protein expression and cofilin phosphorylation. (A) Significant up-regulation of RhoA protein expression in hippocampal neurons following hnRNP-Q1 knockdown. hnRNP-Q1 and RhoA proteins in hippocampal neurons from both conditions were detected by quantitative immunofluorescence staining with rabbit hnRNP-Q1 and mouse RhoA-specific antibodies. Insets are converted 16-color coded intensity graphs for visualization. Scale bar, 10 μm. RhoA fluorescent signals in proximal primary dendrites of 103 neurons from four experiments were measured for both conditions. t test, **p < 0.01. (B) Western blot analysis of RhoA protein expression in hippocampal neurons following hnRNP-Q1 knockdown. t test, **p < 0.01. (C) Up-regulation of RhoA protein by 50% in C2C12 cells following hnRNP-Q1 knockdown as detected by Western blot. Five experiments, t test, ***p < 0.005. Representative Western blots are shown at the bottom. (D) Increased cofilin phosphorylation in C2C12 cells after hnRNP-Q1 knockdown. n = 9; t test, *p < 0.05. (E) Reduced RhoA mRNA levels in hnRNP-Q1–knockdown C2C12 cells. RNA was extracted 72 h post siRNA transfection. An equal volume of cDNA reverse transcribed from 1 μg of total RNA was used for qRT-PCR analysis. A 10% reduction of RhoA mRNA was detected in hnRNP-Q1–knockdown cells compared with control. n = 10, t test, *p < 0.05.
The RhoA/ROCK/LIM kinase signaling pathway has been demonstrated to be a main signaling cascade by which RhoA regulates actin filament dynamics (Maekawa et al., 1999; Amano et al., 2001; Sumi et al., 2001). Activation of LIM kinase by ROCK following RhoA activation leads to cofilin phosphorylation, which inactivates the function of cofilin as an actin filament destabilizer (Sumi et al., 1999). To investigate whether hnRNP-Q1 knockdown up-regulates RhoA downstream signaling, we looked for a possible increase in cofilin phosphorylation. We found that following hnRNP-Q1 knockdown in C2C12 cells, levels of phospho-cofilin were increased by >80% over control cells (Figure 3D), suggesting that hnRNP-Q1 knockdown in C2C12 cells led to enhanced RhoA signaling, possibly mediated by the up-regulation of RhoA protein levels.
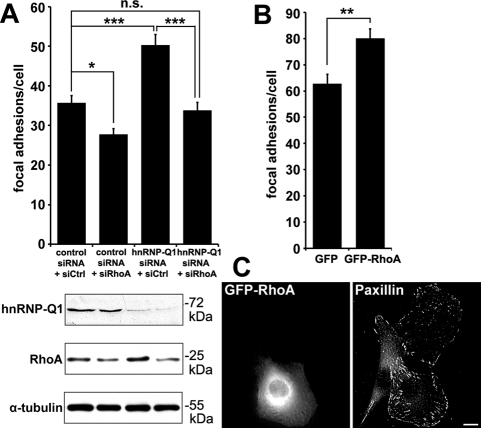
RhoA knockdown abolished the effect of hnRNP-Q1 depletion on focal adhesion formation in C2C12 cells
To investigate whether the cellular phenotypes from hnRNP-Q1 were partly due to elevated RhoA expression and signaling, we investigated whether RhoA knockdown can possibly rescue the morphological changes incurred by hnRNP-Q1 knockdown. For this purpose, scrambled control (siCtrl) or RhoA siRNA (siRhoA) was mixed with hnRNP-Q1 or control siRNA at a ratio of 1:2 and cotransfected into C2C12 cells. Under this condition, siRhoA effectively abolished the effects of hnRNP-Q1 silencing on RhoA protein expression, as well as focal adhesion formation in C2C12 cells (Figure 4A). When cotransfected with control siRNA for hnRNP-Q1, siRhoA had only a modest effect on RhoA protein level and slightly reduced focal adhesion numbers (Figure 4A). The low efficiency of RhoA knockdown was intentionally achieved by using one-third of the recommended amount of RhoA siRNA. To ensure that we indeed depleted RhoA specifically but not other Rho homologues, we examined the effects of siRhoA transfection on mRNA levels for several small GTPases, including RhoA, RhoB, RhoC, Rac1, and Cdc42, and found that RhoA mRNA is the only target specifically affected by siRhoA (Supplemental Figure S6). In addition, elevation of RhoA protein levels by transient expression of GFP-RhoA(wt) was sufficient to increase focal adhesion numbers (Figure 4B). Such an increase of focal adhesion numbers was evident in cells expressing GFP-RhoA when compared with adjacent nontransfected cells (Figure 4C). Taken together, these data suggest that the enhanced focal adhesion formation caused by hnRNP-Q1 knockdown is mediated at least in part through the up-regulation of RhoA protein levels. Our results further suggest that RhoA signaling can be regulated by RhoA protein levels. In line with our observations, Sawada et al. (2009) showed that modest up-regulation of RhoA protein levels by transgenic expression could significantly increase RhoA downstream signaling. In addition, in DRG neurons, the function for semaphorin signaling to induce growth cone collapse is mediated by up-regulation of RhoA protein levels in growth cones through localized RhoA mRNA translation (Wu et al., 2005). Here we identify hnRNP-Q1 as a modulator of RhoA expression that is sufficient to affect downstream signaling and cell morphogenesis.
FIGURE 4:
Up-regulation of RhoA protein levels mediates the effect of hnRNP-Q1 silencing to induce focal adhesions. (A) siRhoA transfection abolished the up-regulation of RhoA protein levels and focal adhesion formation induced by hnRNP-Q1 knockdown in C2C12 cells. RhoA knockdown alone modestly reduces RhoA protein levels and focal adhesion number. Representative Western blots demonstrating hnRNP-Q1 and RhoA knockdown are shown at the bottom. One-way ANOVA post hoc Tukey HSD, *p < 0.05, ***p < 0.001; total 85–98 cells from three experiments were analyzed. (B) Transient expression of GFP-RhoA(wt) in C2C12 cells increases focal adhesions by ∼30% over GFP control vector-transfected cells. t test, ***p < 0.001, 58 (GFP), and 67 (GFP-RhoA) cells from two experiments were quantified. (C) Representative images showing GFP-RhoA expression induced focal adhesion formation in C2C12 cells. Focal adhesion complexes were identified by anti-paxillin immunofluorescence staining. Scale bar, 10 μm.
The 3′ UTR of RhoA mRNA mediates the function of hnRNP-Q1 to regulate RhoA expression
To analyze whether elevated RhoA protein levels in hnRNP-Q1–depleted cells were possibly due to increased mRNA expression, we assessed RhoA mRNA levels in both control and hnRNP-Q1–knockdown cells. It is surprising that, in contrast to the increase of RhoA protein expression shown earlier, a slight reduction (∼10%) of RhoA mRNA levels was detected in C2C12 cells after hnRNP-Q1 knockdown (Figure 3E). Such a reduction was not observed for γ-actin mRNA (Figure 3E). These data suggest that up-regulated RhoA protein expression in hnRNP-Q1–depleted cells was not caused by increased RhoA mRNA stabilization or transcription. These results led us to study a possible direct role for hnRNP-Q1 on RhoA expression by negatively regulating RhoA mRNA translation.
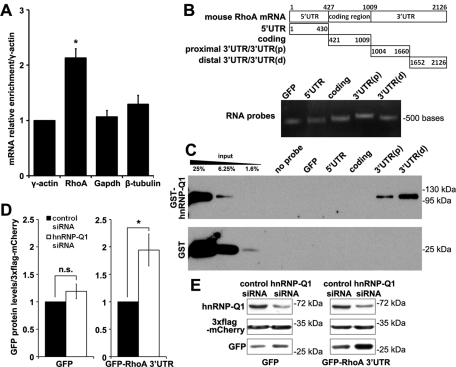
We investigated the association of hnRNP-Q1 with RhoA mRNA. Coimmunoprecipitation (coIP) followed by quantitative real-time (qRT)–PCR analysis showed that RhoA mRNA, but not glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-tubulin mRNA, was selectively enriched in FLAG-mCherry-hnRNP-Q1 precipitates from Neuro2a neuroblastoma cells (Figure 5A). Furthermore, an in vitro RNA pulldown assay using biotinylated RNA probes (Figure 5B) and purified recombinant glutathione S-transferase (GST)–hnRNP-Q1 demonstrated that hnRNP-Q1 directly interacts with RhoA mRNA via the 3′ UTR since only probes corresponding to the proximal and distal sequences of RhoA mRNA 3′ UTR, rather than coding and 5′ UTR sequences, efficiently pulled down GST-hnRNP-Q1 but not control GST protein (Figure 5C). This result further suggests that the 3′ UTR of RhoA mRNA contains multiple binding elements for hnRNP-Q1 and might mediate the inhibitory function of hnRNP-Q1 in regulating RhoA protein expression through translation. We then examined the effects of hnRNP-Q1 knockdown on the expression of a GFP reporter construct fused with the 3′ UTR of mouse RhoA mRNA (pGFP-RhoA-3′ UTR) in Neuro2a cells. Similar to the up-regulation of endogenous RhoA protein, hnRNP-Q1 knockdown also up-regulated GFP expression from the pGFP-RhoA-3′ UTR reporter by ∼90% but not the control vector (Figure 5, D and 5E). A similar up-regulation, although toward a lesser extent, of firefly luciferase RhoA-3′ UTR mRNA reporter was also observed in primary cortical neurons following hnRNP-Q1 knockdown (Supplemental Figure S7A). In addition, the up-regulation of firefly luciferase expression from the RhoA 3′ UTR mRNA reporter was not associated with an up-regulation of firefly luciferase mRNA levels (Supplemental Figure S7B). Taken together, our results suggest an essential role for the RhoA 3′ UTR in hnRNP-Q1–mediated repression of RhoA expression. Because hnRNP-Q1 knockdown did not affect RhoA or its reporter mRNA levels, yet expression of the RhoA-3′ UTR reporters was up-regulated, our results suggest a function for hnRNP-Q1 in the negative regulation of RhoA mRNA translation that is mediated through the 3′ UTR, and this function for hnRNP-Q1 is conserved between neurons and nonneuronal cells.
FIGURE 5:
hnRNP-Q1 directly interacts with the 3′ UTR of RhoA mRNA and regulates RhoA translation. (A) Selective enrichment of endogenous RhoA mRNA in FLAG-mCherry-hnRNP-Q1 precipitates from Neuro2a cells. n = 3; one-way ANOVA post hoc Tukey HSD, *p < 0.05. (B) RNA probes corresponding to the 5′ UTR, coding region and the proximal and distal 3′ UTR of mouse RhoA mRNA and GFP RNA probes were generated by in vitro transcription with biotin–11-CTP. Quality of RNA probes was examined by RNA gel as shown. (C) Representative Western blot showing GST–hnRNP-Q1 pulled down by biotinylated RNA probes in vitro. Only 3′ UTR(p) and 3′ UTR(d) probes pulled down GST-hnRNP-Q1. No probe pulled down control GST protein. (D) hnRNP-Q1 knockdown increased GFP protein expression from a GFP reporter bearing the 3′ UTR of mouse RhoA mRNA in Neuro2a cells. Triple FLAG-tagged mCherry (3XFLAG-mCherry) was cotransfected and used for normalization. A 90% up-regulation of GFP expression from the reporter construct was observed in hnRNP-Q1–knockdown cells. n = 5, t test, *p < 0.05; n.s., not significant. (E) Representative Western blots.
DISCUSSION
Here we report a mechanistic and functional link between the mRNA-binding protein hnRNP-Q1 and the small GTPase RhoA. Our results demonstrate a cellular function for hnRNP-Q1 in RhoA-dependent cellular morphogenesis and a molecular function for hnRNP-Q1 as a RhoA mRNA translation repressor. Neither of these functions for hnRNP-Q1 has been reported previously.
The human hnRNP-Q protein family, also known as SYNCRIP and NSAP1, is composed of three isoforms, hnRNP-Q1, Q2, and Q3, which exist due to alternative splicing of human hnRNP-Q transcripts (Mourelatos et al., 2001). In mouse cells, only two splicing variants encoding hnRNP-Q1 (NCBI accession number NM_019796) and Q3 (NM_019666), respectively, have been identified. Previous studies suggested that isoforms of hnRNP-Q play roles in pre-mRNA splicing, RNA editing, cytoplasmic mRNA transport, IRES-mediated translational activation, and mRNA degradation (Grosset et al., 2000; Blanc et al., 2001; Mourelatos et al., 2001; Bannai et al., 2004; Kanai et al., 2004; Chen et al., 2008). hnRNP-Q3 is mostly nuclear, whereas hnRNP-Q1 is enriched in the cytoplasm (Chen et al., 2008). hnRNP-Q3 in involved in RNA editing in the nucleus, where it interacts with APOBEC1 to catalyze a C-to-U ribonucleotide transition in apolipoprotein B mRNA (Blanc et al., 2001). hnRNP-Q1 plays a role in coding region instability determinant–mediated and AU-rich element–mediated mRNA degradation (Grosset et al., 2000). One well-documented function for hnRNP-Q1 is that its direct interaction with the IRESs of BiP and serotonin N-acetyltransferase mRNAs can activate translation (Cho et al., 2007; Kim et al., 2007). In addition, hnRNP-Q1 was also identified as component of transport mRNP granules in neurons, as it was copurified with the KIF5 kinesin cargo-binding domain (Kanai et al., 2004) and colocalized with staufen and inositol 1,4,5-trisphosphate receptor type 1 reporter mRNA in neuronal processes (Bannai et al., 2004).
In this study, we report a novel function for hnRNP-Q1 as a translational repressor that negatively regulates RhoA protein expression in a RhoA mRNA 3′ UTR-dependent manner in neurons and nonneuronal cells. Knockdown of hnRNP-Q1 up-regulated endogenous RhoA protein expression in neurons and nonneuronal cells, which was not attributed to changes in RhoA mRNA and suggests a translational mechanism. However, our results do not exclude the possibility that hnRNP-Q1 indirectly regulates RhoA protein stability, independent of mRNA regulation, since RhoA protein levels can be regulated through proteosome-mediated RhoA protein degradation (Wang et al., 2003). However, this is unlikely to explain hnRNP-Q1 knockdown–induced up-regulation of RhoA protein expression. We consistently observed that hnRNP-Q1 knockdown up-regulated the expression of both GFP and firefly luciferase protein from the corresponding reporter constructs fused with the RhoA 3′ UTR; although robust in Neuro2a cells, the extent of up-regulation was more moderate in neurons, perhaps due to technical limitations with cotransfection of multiple constructs. The up-regulation of GFP protein levels from the 3′ UTR reporter in Neuro2a cells was not a result of GFP protein stabilization, since a similar change was not observed with the GFP control vector. Of importance, our results in neurons and nonneuronal cells suggest that the 3′ UTR of RhoA mRNA plays an essential cis-acting role in hnRNP-Q1–mediated regulation of RhoA expression, which can be conferred to a heterologous reporter. Our results further suggest that hnRNP-Q1 functions as a translational repressor that negatively regulates RhoA protein expression. The derepression of RhoA mRNA translation by physiological signals may stimulate up-regulation of RhoA protein and signaling, analogous to that induced by hnRNP-Q1 depletion. Future work may uncover how extracellular signaling pathways may trigger the degradation of hnRNP-Q1 or reduce its affinity for its target mRNA, perhaps by phosphorylation, as a physiological means to derepress RhoA mRNA translational inhibition and result in activation of RhoA signaling.
This present study also reveals a novel cellular function for hnRNP-Q1 in modulating cellular morphogenesis. Previously, the cellular functions of hnRNP-Q1 were unknown, although it has been shown to be ubiquitously expressed in mammalian tissues. Here we report that hnRNP-Q1 knockdown reduced dendrite complexity and spine density in mature hippocampal neurons and enhanced cell spreading, stress fiber, and focal adhesion number in C2C12 cells. These morphological changes may largely be caused by the up-regulation of RhoA proteins levels and downstream signaling. This conclusion is supported by two pieces of evidence. First, these morphological phenotypes can be rescued by inhibition of RhoA/ROCK signaling with the ROCK inhibitor Y-27632. Second, analysis of C2C12 cells showed that the effect of hnRNP-Q1 knockdown on focal adhesion number was rescued by RNA interference–induced RhoA silencing or mimicked by transient expression of wild-type RhoA protein. These results have important implications, as they suggest that control of RhoA expression by hnRNP-Q1 can be a critical factor to mediate RhoA signaling and modulate cellular morphogenesis. Given that hnRNP-Q1 may also regulate other, currently unidentified mRNA targets and perhaps the levels of other RhoA modulators, including GAPs and GEFs, their expression may also play critical roles in the regulation of RhoA function. It is likely that the phenotypes from hnRNP-Q1 knockdown are not entirely dependent on elevated RhoA, but other hnRNP-Q1 target mRNAs may contribute to the observed effects on cell morphogenesis. Identification of these target mRNAs and the specific role(s) of hnRNP-Q1 on such targets will be essential to generate a complete picture for the function of hnRNP-Q1 in development and cellular morphogenesis.
Alterations in the RhoA signaling pathway are associated with intellectual disabilities and autism spectrum disorders that present with abnormal dendritic morphology. For instance, the loss of function of a Rho GTPase-activating protein, oligophrenin-1, is associated with an inherited form of X-linked intellectual disability (Govek et al., 2004). In addition, increased RhoA signaling was observed in a mouse model of Smith–Lemli–Opitz syndrome (Jiang et al., 2010). The typical features in human patients with this syndrome include cognitive deficits, behavioral abnormality, and autism. In the future, it will be interesting to investigate the possible function of hnRNP-Q1 in synaptic plasticity and learning that rely on morphological modulation of dendritic spines (Segal, 2005). Beyond the role of hnRNP-Q1 in regulation of dendritic and spine development, our findings motivate inquiries into its possible role in other cellular activities known to depend on RhoA signaling, such as cancer cell invasion and metastasis (Narumiya et al., 2009), axon regeneration (Gross et al., 2007; Kubo et al., 2007; Duffy et al., 2009), and axon guidance (Wu et al., 2005).
An interesting area for future research will be to assess whether hnRNP-Q1 regulates RhoA mRNA localization in dendrites and axons, since hnRNP-Q1 has been observed in transport mRNP granules (Bannai et al., 2004; Kanai et al., 2004). RhoA mRNA has been shown to be localized to axons, where its translation is induced by semaphorin signaling (Wu et al., 2005). Axonal synthesis of RhoA is dependent on the 3′ UTR and is necessary for semaphorin-induced growth cone collapse (Wu et al., 2005). RhoA mRNA was also detected in purified synaptic compartments, where RhoA translation can be activated by brain-derived neurotrophic factor (Troca-Marin et al., 2010). In breast cancer cells, RhoA translation can be activated by semaphorin signaling as well (Pan and Bachelder, 2010). Because we show that the 3′ UTR of RhoA mRNA mediates the inhibitory effect of hnRNP-Q1 on RhoA protein expression, future studies should examine a possible function for hnRNP-Q1 in extracellular signal-stimulated RhoA synthesis; such an event may take place in growth cones and dendritic compartments of neuronal cells locally. In addition, whether hnRNP-Q1 is involved in RhoA mRNA transport and localization will also need to be addressed in future studies.
MATERIALS AND METHODS
Plasmids, siRNA, shRNA, and lentivirus production
Full-length cDNA of human hnRNP-Q1 was obtained by RT-PCR from total RNA extracted from HEK293 cells and inserted into pEGFPC1 and p3Xflag-mCherryC1 vectors via the XhoI site. To express recombinant GST-hnRNP-Q1, hnRNP-Q1 coding sequences were cut from pEGFP-hnRNP-Q1 and inserted into pGEX-2T (GE Healthcare Bio-Sciences, Piscataway, NJ) by BamH1 and EcoRI sites. To generate a GFP reporter construct, the mouse RhoA mRNA 3′ UTR was amplified from a pCMV6-msRhoA mammalian expressing plasmid (OriGene, Rockville, MD, accession number NM_016802) and inserted into pEGFPC1. To generate lentiviral transfer plasmid encoding hnRNP-Q1 shRNA, DNA oligos with sequences 5′-CCGGAAGCTTGCAGTGGAGTAATGGCTCGAGCCATTACTCCACTGCAAGCTTTTTTTG-3′ and 5′-AATTCAAAAAAAGCTTGCAGTGGAGTAATGGCTCGAGCCATTACTCCACTGCAAGCTT-3′ were annealed into double-strand DNA and inserted into a pLKO.1-cloning vector (plasmid 10878; Addgene, Cambridge, MA) using AgeI and EcoRI sites. A control plasmid bearing scrambled shRNA sequence (plasmid 1864) was from Addgene. hnRNP-Q1 siRNA (sense, GAUGCAGUUUCAGGUGUAAUCUCA; antisense, UGAUGAUUACACCUGAAACUGCAUC) and control siRNA (sense, GAUUUGAGACUUGUGCUAAACGUCA; antisense, UGACGUUUAGCACAAGUCUCAAAUC) were from Invitrogen. GFP-RhoA–expressing plasmid (plasmid 12965) was from Addgene. RhoA siRNA (sc-29471) and scrambled control siRNA (sc-37007) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Lentiviruses were packaged in HEK293 cells according to Addgene's pLKO.1 protocol (www.addgene.org/plko). Lentiviral particles in cell culture medium were enriched and purified by ultracentrifugation.
Cell cultures and transfection
Neuro2a and C2C12 myoblastoma (American Type Culture Collection, Manassas, VA) cells were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) with 10% fetal bovine serum (Sigma-Aldrich), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen, Carlsbad, CA) at 5% CO2 and 37°C. Primary mouse hippocampal neurons were cultured from E16 mouse embryos as described previously (Gross et al., 2010).
All cells were transfected with Lipofectamine 2000 (Invitrogen) transfection reagent according to the manufacturer's protocol. For mouse hippocampal neuron transfection, shRNA, GFP-Lifeact, and myc-hnRNP-Q1(human) expressing plasmid or pcDNA3 were mixed with a ratio of 6:1:3. For siRNA transfection, freshly trypsinized C2C12 and Neuro2a cells were incubated with siRNA–Lipofectamine 2000 mixture at room temperature for 30 min and then plated into cell culture plates with normal culture medium. To examine the expression of GFP-reporter for RhoA 3′ UTR, Neuro2a cells pretreated with siRNA for 48 h were transfected with GFP-expressing plasmid, p3Xflag-mCherry, and pcDNA3 with a ratio of 1:1:10, and proteins were expressed for 16 h.
Antibodies and immunofluorescence
The following antibodies were used in this study: rabbit anti-Syncrip (1:10,000; kindly provided by Katsuhiko Mikoshiba; Mizutani et al., 2000); mouse anti-Map2 (1:1500; Sigma-Aldrich); mouse anti–α-tubulin (1:40,000; Sigma-Aldrich); mouse anti-paxillin (1:150; BD Biosciences, San Diego, CA), mouse anti-RhoA (1:200; Cytoskeleton, Denver, CO); rabbit anti–phospho-cofilin (1:5000, Cell Signaling, Technology, Beverly, MA), and rabbit anti-cofilin (1:10,000, Sigma-Aldrich). Rabbit anti–hnRNP-Q1 was produced by immunizing rabbits with a keyhole limpet hemocyanin-conjugated synthetic peptide corresponding to the C-terminal sequence of hnRNP-Q1 (KGVEAGPDLLQ, through Sigma-Genosys, The Woodlands, TX). The hnRNP-Q1 antibody was purified using an affinity column covalently coupled with the same peptide. Secondary antibodies conjugated with cyanine dyes were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA), and Alexa 488–conjugated phalloidin was from Invitrogen.
For immunofluorescence staining, all cells were fixed with 4% paraformaldehyde in PBS/MgCl2 for 18 min and washed with phosphate-buffered saline (PBS)/MgCl2 three times. Cells were permeabilized with PBS/0.3% Triton X-100 and incubated with blocking buffer containing 5% bovine serum albumin (BSA) in PBS/0.1% Triton X-100 (PBST). Primary and secondary antibodies were diluted in blocking buffer. Cells were incubated with primary antibodies for 1 h at room temperature or overnight at 4°C and incubated in secondary antibody for 1 h at room temperature, followed by three washes with PBST. Coverslips were mounted in mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) and propyl gallate.
Fluorescence microscopy and digital imaging
Fixed cells were visualized using a Nikon Eclipse inverted microscopes (TE300 or Ti) equipped with a 60× Plan-Neofluar objective (Nikon, Melville, NY). Images were captured with a cooled charge-coupled device camera (Quantix; Photometrics, Tucson, AZ) and processed using IP Lab Spectrum (Scanalytics, Rockville, MD) or Nikon Elements. For quantitative analysis, exposure time was kept constant and below grayscale saturation. Images were deconvolved using AutoQuant X (Media Cybernetics, Bethesda, MD).
Densities of dendritic protrusion were analyzed as described previously (Gross et al., 2010). Dendritic protrusions quantified in this study include all filopodia and mushroom-like protrusions. For Sholl analysis, dendrites were manually traced with the NeuronJ plug-in in ImageJ (National Institutes of Health, Bethesda, MD), and the tracing result was saved as a single image using the snapshot function (Meijering et al., 2004). Sholl analysis was performed using the Sholl analysis plug-in developed for the ImageJ program by the Ghosh lab at the University of California, San Diego. To quantify focal adhesions, deconvolved images were processed using the rolling ball background subtraction plug-in for ImageJ and subjected to autothreshold. Numbers of focal adhesions were quantified in ImageJ using particle analysis function. Focal adhesions were defined as continuous paxillin puncta occupying >64 pixel units, except for GFP and GFP-RhoA overexpression experiments, in which 32 pixel units was set as the minimal size for focal adhesions.
Transwell migration experiment
Transwell cell migration assay was performed using 8-μm-pore, poly(ethylene terephthalate), track-etched membrane cell culture inserts (BD Biosciences, San Diego, CA) coated with fibronectin (Sigma-Aldrich) on the bottom surface. At 72 h after siRNA transfection, C2C12 cells were trypsinized and plated on the top surface of inserts. Cells were fixed with 4% paraformaldehyde 8 h after plating and stained with DAPI. Ten random optical fields from each insert were imaged with a 4× objective, and the average number of DAPI foci was quantified as the total number of cells. Then cells on the top surface were removed and remaining cells were counted as migrating cells. Cell migration efficiency was measured as the ratio of migrating cells to total cells.
Immunoprecipitation and qRT-PCR analysis
Neuro2a cell lysates were prepared with cell lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, and 1% NP-40, pH 7.5) supplemented with complete protease inhibitors (Roche, Indianapolis, IN) and RNase inhibitor (SUPERase-In; Applied Biosystems/Ambion, Austin, TX). FLAG-tagged proteins were immunoprecipitated with anti-FLAG agarose (Sigma-Aldrich), followed by extensive washes with cell lysis buffer. After the final wash, RNA was extracted from IP pellets with TRIzol reagent (Invitrogen). Total RNAs were reverse transcribed with SSIII reverse transcriptase (Invitrogen) according to the manual.
Real-time PCR and analysis was performed in a LightCycler real-time PCR system with LightCycler SYBR Green I reagent (Roche). Gene-specific primers were used as follows: CATTGACAGCCCTGATAGTT and TCGTCATTCCGAAGGTCCTT for RhoA; CTGGTGGATCTCTGTGAGCAC and AAACGTTCCCAACTCAAGGC for γ-actin; GAGTCTACTGGTGTCTTCAC and CCACAATGCCAAAGTTGTCAT for GAPDH; and TCGTGGAATGGATCCCAAC and TCCATCTCTGTCCTGCCT for β-tubulin. CoIP efficiency of γ-actin, RhoA, GAPDH, and β-tubulin was quantified by normalizing levels of each mRNA in IP pellets to inputs. To assess mRNA-selective enrichment, coIP efficiency was normalized to γ-actin mRNA.
Recombinant protein purification
GST and GST-hnRNP-Q1 were purified from Rosetta2 (DE3) bacteria (EMD Millipore, Merck, Darmstadt, Germany) transformed with pGEX-2T vector or pGEX-hnRNP-Q1 using a GST protein purification system (Novagen, EMD4Biosciences, Gibbstown, NJ) according to the manual. Expression of recombinant proteins was induced at 16°C for 4 h with 0.1 M isopropyl-β-d-thiogalactoside.
RNA probe labeling and RNA-based affinity purification
DNA sequences of interest were amplified by PCR and inserted into pGEM T or T-easy vectors (Promega, Madison, WI), except for the 5′ UTR. To generate a DNA template for the 5′ UTR, pCMV6-msRhoA was linearized by EcoRV. Biotinylated RNA probes were generated by in vitro transcription using T7/Sp6 Maxiscript kit (Ambion) with Biotin-11-cytidine-5′-triphosphate (biotin-11-CTP; Roche). Biotinylated CTP and normal CTP were used at a ratio of 1:4 to ensure transcription yield and sufficient labeling. Nonincorporated nucleotides were removed with a G-25 column (Amersham-Pharmacia Biotech, GE Healthcare Bio-Sciences, Piscataway, NJ), followed by ethanol precipitation. RNA concentration was determined by A260 absorption, and the quality was examined by denatured RNA electrophoresis. NeutrAvidin agarose (Thermo Scientific, Waltham, MA) preblocked with BSA was used to precipitate biotinylated RNA probes. Recombinant GST-hnRNP-Q1 or GST (10 ng) was incubated with 200 ng of indicated RNA probes in the presence of yeast tRNA (Roche) for 20 min at room temperature, followed by incubation with NeutrAvidin beads and extensive washes. Proteins were detected by Western blot against GST.
Western blot and quantification
Western blot was performed according to standard procedure. Protein-specific signal was quantified using ImageJ. RhoA protein levels were normalized to α-tubulin. To measure levels of phospho-cofilin, membranes previously blotted with phospho-cofilin antibody were stripped with Restore stripping buffer (Thermo Scientific) and reprobed with cofilin antibodies. Levels of phospho-cofilin were normalized to total cofilin. For the GFP reporter assay, GFP levels were normalized to 3XFLAG-mCherry.
Statistical analysis
Specific statistical analyses are described in figure legends and were performed in SSPS 17.0. All data were tested for normality and homogeneity of variances. The analysis for Figure 4A was performed on square root–transformed data since heterogeneity of variances for original data was observed. Nonparametric testes were used for data from Sholl analysis. All error bars represent SEM.
Supplementary Material
Acknowledgments
We thank Christina Gross and Sharon Swanger for helpful discussions and critical reading of the manuscript. We also thank Wilfried Rossoll for sharing p3XFLAG-mCherry and GFP–hnRNP-Q3 plasmids and Katsuhiko Mikoshiba for providing pan–hnRNP-Q antibodies. This work was supported by aid from the Muscular Dystrophy Association (to G.J.B.) and National Institutes of Health (NIH) Training Grant T32GM008367-21 (for K.R.W.). This research project was supported in part by Viral Vector Core and Microscopy Core of the Emory Neuroscience NINDS (NIH) Core Facilities grant P30NS055077.
Abbreviations used:
- DIV
day in vitro
- hnRNP
heterogeneous nuclear ribonucleoprotein
- IRES
internal ribosome entry site
- ROCK
Rho kinase
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-10-0867) on February 22, 2012.
REFERENCES
- Amano T, Tanabe K, Eto T, Narumiya S, Mizuno K. LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem J. 2001;354:149–159. doi: 10.1042/0264-6021:3540149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- Bannai H, Fukatsu K, Mizutani A, Natsume T, Iemura S, Ikegami T, Inoue T, Mikoshiba K. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J Biol Chem. 2004;279:53427–53434. doi: 10.1074/jbc.M409732200. [DOI] [PubMed] [Google Scholar]
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Besson A, Gurian-West M, Schmidt A, Hall A, Roberts JM. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 2004;18:862–876. doi: 10.1101/gad.1185504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Navaratnam N, Henderson JO, Anant S, Kennedy S, Jarmuz A, Scott J, Davidson NO. Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing. J Biol Chem. 2001;276:10272–10283. doi: 10.1074/jbc.M006435200. [DOI] [PubMed] [Google Scholar]
- Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol Cell Biol. 2008;28:6929–6938. doi: 10.1128/MCB.01332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of RhoA in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180:713–719. doi: 10.1164/rccm.200903-0325OC. [DOI] [PubMed] [Google Scholar]
- Cho S, Park SM, Kim TD, Kim JH, Kim KT, Jang SK. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol Cell Biol. 2007;27:368–383. doi: 10.1128/MCB.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy P, Schmandke A, Sigworth J, Narumiya S, Cafferty WB, Strittmatter SM. Rho-associated kinase II (ROCKII) limits axonal growth after trauma within the adult mouse spinal cord. J Neurosci. 2009;29:15266–15276. doi: 10.1523/JNEUROSCI.4650-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia LP, Yamamoto M, Zang K, Reichardt LF. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 2006;51:43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Hatten ME, Van Aelst L. The role of Rho GTPase proteins in CNS neuronal migration. Dev Neurobiol. 2011;71:528–553. doi: 10.1002/dneu.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Akerman CJ, Cross JR, Van der Veken L, Van Aelst L. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat Neurosci. 2004;7:364–372. doi: 10.1038/nn1210. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Gross C, Nakamoto M, Yao X, Chan CB, Yim SY, Ye K, Warren ST, Bassell GJ. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross RE, Mei Q, Gutekunst CA, Torre E. The pivotal role of RhoA GTPase in the molecular signaling of axon growth inhibition after CNS injury and targeted therapeutic strategies. Cell Transplant. 2007;16:245–262. doi: 10.3727/000000007783464740. [DOI] [PubMed] [Google Scholar]
- Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Harris CE, Boden RA, Astell CR. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J Virol. 1999;73:72–80. doi: 10.1128/jvi.73.1.72-80.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XS, Wassif CA, Backlund PS, Song L, Holtzclaw LA, Li Z, Yergey AL, Porter FD. Activation of Rho GTPases in Smith-Lemli-Opitz syndrome: pathophysiological and clinical implications. Hum Mol Genet. 2010;19:1347–1357. doi: 10.1093/hmg/ddq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Hata K, Yamaguchi A, Yamashita T. Rho-ROCK inhibitors as emerging strategies to promote nerve regeneration. Curr Pharm Des. 2007;13:2493–2499. doi: 10.2174/138161207781368657. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Miller M. Maturation of rat visual cortex. I. A quantitative study of Golgi-impregnated pyramidal neurons. J Neurocytol. 1981;10:859–878. doi: 10.1007/BF01262658. [DOI] [PubMed] [Google Scholar]
- Mizutani A, Fukuda M, Ibata K, Shiraishi Y, Mikoshiba K. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J Biol Chem. 2000;275:9823–9831. doi: 10.1074/jbc.275.13.9823. [DOI] [PubMed] [Google Scholar]
- Mohseni M, Chishti AH. The headpiece domain of dematin regulates cell shape, motility, and wound healing by modulating RhoA activation. Mol Cell Biol. 2008;28:4712–4718. doi: 10.1128/MCB.00237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 2001;20:5443–5452. doi: 10.1093/emboj/20.19.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Pan H, Bachelder RE. Autocrine semaphorin3A stimulates eukaryotic initiation factor 4E-dependent RhoA translation in breast tumor cells. Exp Cell Res. 2010;316:2825–2832. doi: 10.1016/j.yexcr.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W, Kroning AK, Ohndorf UM, Steegborn C, Jablonka S, Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum Mol Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- Sawada N, Itoh H, Miyashita K, Tsujimoto H, Sone M, Yamahara K, Arany ZP, Hofmann F, Nakao K. Cyclic GMP kinase and RhoA Ser188 phosphorylation integrate pro- and antifibrotic signals in blood vessels. Mol Cell Biol. 2009;29:6018–6032. doi: 10.1128/MCB.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M. Dendritic spines and long-term plasticity. Nat Rev Neurosci. 2005;6:277–284. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124:679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- Sumi T, Matsumoto K, Nakamura T. Specific activation of LIM kinase 2 via phosphorylation of threonine 505 by ROCK, a Rho-dependent protein kinase. J Biol Chem. 2001;276:670–676. doi: 10.1074/jbc.M007074200. [DOI] [PubMed] [Google Scholar]
- Sumi T, Matsumoto K, Takai Y, Nakamura T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase 2. J Cell Biol. 1999;147:1519–1532. doi: 10.1083/jcb.147.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz JM, Kuner R, Behrens J, Offermanns S. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 2002;35:51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Yuste R. Role of Rho GTPases in the morphogenesis and motility of dendritic spines. Methods Enzymol. 2008;439:285–302. doi: 10.1016/S0076-6879(07)00421-1. [DOI] [PubMed] [Google Scholar]
- Troca-Marin JA, Alves-Sampaio A, Tejedor FJ, Montesinos ML. Local translation of dendritic RhoA revealed by an improved synaptoneurosome preparation. Mol Cell Neurosci. 2010;43:308–314. doi: 10.1016/j.mcn.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.