The N-terminal region is the lipid raft–targeting determinant. A novel regulatory mechanism is found by which Rnd proteins function as RhoA antagonists, as is a striking mechanism by which differential membrane targeting governs activities of G proteins having similarities in effector interaction.
Abstract
The Rnd proteins Rnd1, Rnd2, and Rnd3/RhoE are well known as key regulators of the actin cytoskeleton in various cell types, but they comprise a distinct subgroup of the Rho family in that they are GTP bound and constitutively active. Functional differences of the Rnd proteins in RhoA inhibition signaling have been reported in various cell types. Rnd1 and Rnd3 antagonize RhoA signaling by activating p190 RhoGAP, whereas Rnd2 does not. However, all the members of the Rnd family have been reported to bind directly to p190 RhoGAP and equally induce activation of p190 RhoGAP in vitro, and there is no evidence that accounts for the functional difference of the Rnd proteins in RhoA inhibition signaling. Here we report the role of the N-terminal region in signaling. Rnd1 and Rnd3, but not Rnd2, have a KERRA (Lys-Glu-Arg-Arg-Ala) sequence of amino acids in their N-terminus, which functions as the lipid raft-targeting determinant. The sequence mediates the lipid raft targeting of p190 RhoGAP correlated with its activation. Overall, our results demonstrate a novel regulatory mechanism by which differential membrane targeting governs activities of Rnd proteins to function as RhoA antagonists.
INTRODUCTION
Most small G proteins function as molecular switches by cycling between GDP-bound inactive and GTP-bound active states. Their activation is controlled by guanine nucleotide-exchange factors (GEFs) and GTPase-activating proteins (GAPs). For most Rho family proteins, the GDP-bound form is predominant at the resting state and interacts with a guanine nucleotide dissociation inhibitor (GDI) protein that covers the C-terminal geranylgeranyl moiety and stabilizes them as a cytosolic Rho-GDI complex (Sasaki and Takai, 1998). By contrast, the Rnd proteins Rnd1, Rnd2, and Rnd3/RhoE comprise a distinct branch of Rho family GTPases in that they have a low affinity for GDP and very low intrinsic GTPase activities (Foster et al., 1996; Guasch et al., 1998; Nobes et al., 1998). Rnd3 has been reported to be farnesylated (Foster et al., 1996), and Rnd1 and Rnd2 C-terminal sequences are supposed to be farnesylated and not associated with a GDI. This suggests that the regulatory mechanism of Rnd protein localization is likely to be different from those of other Rho family members. In addition, considering constitutive activities of Rnd GTPases, GTP/GDP-cycling subcellular trafficking is excluded from regulatory mechanism for the localization.
The functions of Rnd proteins have been studied in many cell types, and a clear role for these proteins in the actin cytoskeleton has emerged. Expression of Rnd1 or Rnd3 induces loss of focal adhesions that anchor the stress fibers to the extracellular matrix, and cells losing their adhesions cannot spread and tend to round (Nobes et al., 1998). The morphological effects of Rnd1 or Rnd3 expression that have been described so far are related to RhoA inhibition. It was recently shown that Rnd proteins interact with p190 RhoGAP, the most abundant GAP for RhoA in cells (Wennerberg et al., 2003). Rnd proteins bind directly to the middle domain of p190 RhoGAP, and this interaction promotes p190 RhoGAP–mediated GAP activity toward RhoA and is critical to the cellular effects elicited by expression of Rnd1 or Rnd3 in fibroblasts. Apart from p190 RhoGAP, Rnd3 directly interacts with and inhibits a RhoA effector, Rho-associated kinase (ROCK) I (Riento et al., 2003), which is known to stimulate actin reorganization by phosphorylating a number of actin-associated proteins (Riento and Ridley, 2003). Rnd1, which does not bind to ROCK I, is efficient at inducing the loss of actin stress fibers. Thus stimulation of p190 RhoGAP activity is believed to be the most universal explanation for the effects of Rnd1 and Rnd3 (Chardin, 2006). However, all the members of the Rnd subfamily—Rnd1, Rnd2, and Rnd3—can interact with and activate p190 RhoGAP in vitro (Wennerberg et al., 2003), although Rnd2 has little or no inhibitory effect on RhoA in cells (Nobes et al., 1998; Tanaka et al., 2006). There has been no evidence that accounts for the functional difference of the Rnd proteins in RhoA inhibition signaling.
Subcellular localizations of the Rnd proteins have been reported to be different; Rnd1 and Rnd3 are plasma membrane associated, whereas Rnd2 lacks this association (Roberts et al., 2008). The plasma membrane of most cell types contains specialized subdomains with distinct lipid and protein compositions, referred to as lipid raft microdomains (Brown and London, 1998; Kurzchalia and Parton, 1999; Simons and Toomre, 2000). Lipid rafts are cholesterol- and sphingolipid-rich lipid microdomains, and one of the most important properties of lipid rafts is that they can include or exclude proteins to variable extents. Rafts function in cellular signaling by concentrating or separating specific molecules in a unique lipid environment in many cell types. Among the Rho family of small GTPases, RhoA, Rac1, and TC10 are known to be concentrated and function in lipid rafts and caveolae, and their localization is crucial for the activation of many signal transduction pathways (Watson et al., 2001; Palazzo et al., 2004; del Pozo et al., 2004). Here we show that the N-terminal KERRA sequence of Rnd1 functions as the lipid raft–targeting determinant and its raft targeting is required for p190 RhoGAP–mediated RhoA inhibition signaling.
RESULTS
Rnd1 is lipid raft dependently localized to the plasma membrane
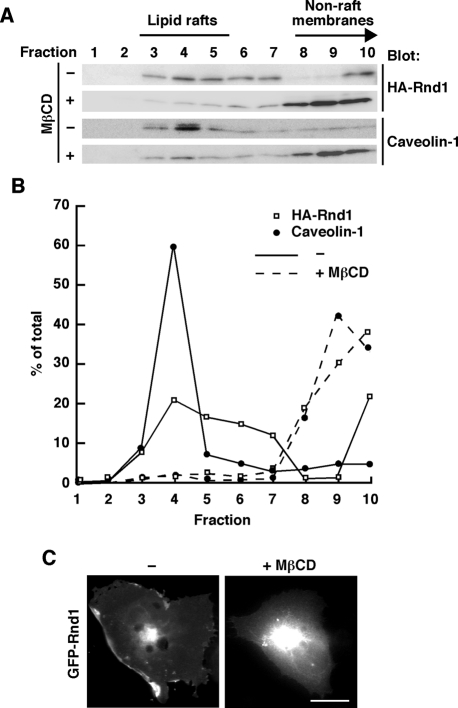
Previous reports showed that Rnd1 localizes to the plasma membrane and is concentrated at the cell periphery (Nobes et al., 1998; Roberts et al., 2008). The plasma membrane of cells contains specialized subdomains with distinct lipid and protein compositions, referred to as lipid raft microdomains (Brown and London, 1998; Kurzchalia et al., 1999; Simons and Toomre, 2000). To determine whether Rnd1 resides in lipid rafts, we performed sucrose density gradient fractionation assay of COS-7 cells transiently transfected with hemagglutinin (HA)-tagged Rnd1. In our fractionation procedure, an endogenous raft marker protein, caveolin-1 (Palazzo et al., 2004), was present mainly in fractions 3–5. Gα12 and Gα13 are known to be concentrated at lipid raft and nonraft, respectively (Waheed and Jones, 2002), and they were found in 3–5 (Gα12) and 9 and10 (Gα13) in the fractionation (unpublished data). Thus, in our experiments, we defined fractions 3–5 as lipid raft fractions. Rnd1 resided in the raft fractions (Figure 1, A and B), and ectopically expressed green fluorescent protein (GFP)-tagged Rnd1 showed plasma membrane localization and concentrated at the cell periphery in COS-7 cells (Figure 1C). Lipid raft localization of Rnd1 and caveolin-1 was decreased (Figure 1A) and plasma membrane localization of GFP-Rnd1 was abolished (Figure 1C) by treatment of the cells with methyl-β-cyclodextrin (MβCD), which depletes cellular cholesterol and disrupts lipid rafts (del Pozo et al., 2004). These data demonstrate lipid raft targeting of Rnd1 and raft-dependent localization of Rnd1 to the plasma membrane.
FIGURE 1:
Lipid raft localization of Rnd1. (A) Rnd1-transfected or nontransfected COS-7 cells treated with (+) or without (–) MβCD were subjected to the sucrose density gradient fractionation assay. Each fraction (sampled and numbered from the top of the gradient) was analyzed by immunoblotting using anti-HA antibody (top) or anti–caveolin-1 antibody (bottom). (B) Densitometric analysis of the bands on the immunoblots in A. The amount of the protein in each fraction was expressed as a percentage of the total. (C) COS-7 cells were transiently transfected with GFP-Rnd1 and treated with (+) or without (–) MβCD (20 mM) before fixation. Scale bar, 20 μm.
Rnd1 and Rnd3, but not Rnd2, reside in lipid rafts
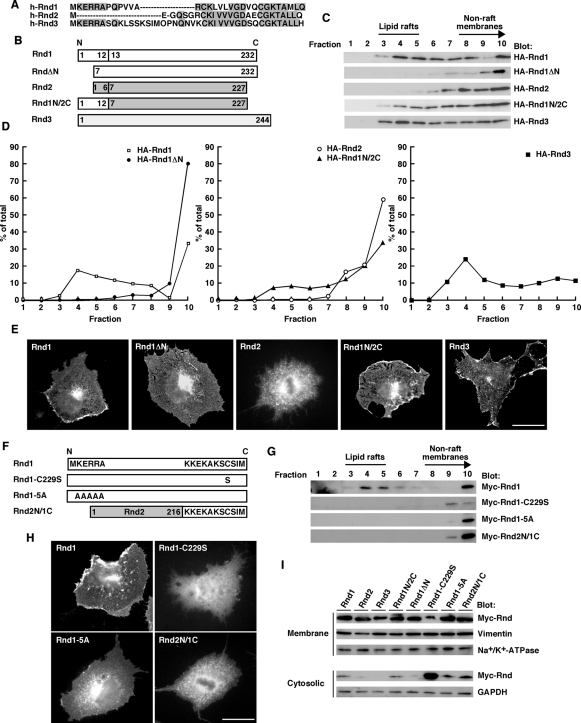
Subcellular localization of the Rnd proteins has been reported to be different. Rnd1 is predominantly plasma membrane associated, Rnd2 is not plasma membrane associated and is localized to early endosomes, and Rnd3 is found in the Golgi complex, as well as in the plasma membrane (Tanaka et al., 2006; Roberts et al., 2008). Sequence alignment of the Rnd proteins revealed that Rnd1 and Rnd3, but not Rnd2, have a Lys-Glu-Arg-Arg-Ala (KERRA) sequence of amino acids in their N-terminus (Figure 2A). As shown in Figure 2C, Rnd1 and Rnd3, having the KERRA sequence of amino acids in their N-terminus, were localized to lipid rafts. By contrast, Rnd2, lacking the sequence, could not localize to lipid rafts. To examine the role of the KERRA sequence in lipid raft targeting, we prepared Rnd1ΔN, a deletion mutant of Rnd1 that lacks the first seven N-terminal amino acids, and Rnd1N/2C, a chimeric construct in which the first six N-terminal amino acids of Rnd2 is replaced by the first 12 N-terminal amino acids of Rnd1 (Figure 2B). Lipid raft localization of Rnd1 was decreased by the deletion of the N-terminal KERRA sequence of Rnd1, whereas addition of the N-terminal KERRA sequence of Rnd1 to Rnd2 backbone sequences induced lipid raft localization (Figure 2, C and D). The morphologies of COS-7 cells are not affected by the expression of Rnd1 or Rnd3 (Katoh et al., 2002), and immunofluorescence microscopy analysis revealed that Rnd1, Rnd1N/2C, and Rnd3, containing the KERRA sequence, were localized in the cell periphery. By contrast, Rnd1ΔN or Rnd2, lacking the sequence, could not localize to the cell periphery (Figure 2E). To further confirm the role of the N-terminal KERRA sequence in lipid raft targeting, we prepared Rnd1-5A, in which the N-terminal five residues of Rnd1 have each been changed to alanine (Figure 2F). Alanine substitution for the N-terminal KERRA sequence completely suppressed lipid raft localization (Figure 2G) and plasma membrane localization (Figure 2H) of Rnd1. These results suggest that the N-terminal KERRA sequence of Rnd1 is the lipid raft–targeting determinant.
FIGURE 2:
The N-terminal KERRA sequence of Rnd1 mediates lipid raft targeting and plasma membrane localization. (A) The amino-terminal amino acid sequences of human Rnd1, Rnd2, and Rnd3. Identical amino acids are boxed in gray. (B) Schematic representation of the constructs used in this study. Numbers indicate amino acid positions within the sequence. (C) COS-7 cells transfected with the indicated expression plasmids were subjected to the sucrose density gradient fractionation assay. (D) Densitometric analysis of HA-Rnd bands on the immunoblots in C. The amount of the protein in each fraction was expressed as a percentage of the total. (E) COS-7 cells were transiently transfected with the Myc-tagged expression plasmids of Rnd proteins and stained with anti-Myc monoclonal antibody. Scale bar, 20 μm. (F) Schematic representation of the constructs. (G) COS-7 cells transfected with the indicated expression plasmids were subjected to the sucrose density gradient fractionation assay. (H) COS-7 cells were transiently transfected with the Myc-tagged expression plasmids of Rnd proteins and stained with anti-Myc monoclonal antibody. Scale bar, 20 μm. (I) Cellular homogenates from COS-7 cells transiently transfected with the indicated plasmids were separated into the crude membrane and the cytosolic fractions and then analyzed by immunoblotting. The fractions were immunoblotted with anti-vimentin and anti–Na+/K+-ATPase antibodies for the control membrane proteins and with anti-GAPDH antibody for the control soluble protein.
We further examined whether Rnd1 tail contributes to localization. We prepared Rnd2N/1C, a chimeric construct in which the last 11 C-terminal amino acids of Rnd2 is replaced by the last 11 C-terminal amino acids of Rnd1 (Figure 2F). As shown in Figure 2, G and H, Rnd2N/1C was localized in nonraft membranes and could not localize to plasma membrane. The CAAX (where C represents cysteine, A is an aliphatic amino acid, and X is a terminal amino acid) motif-signaled posttranslational modification has been reported to regulate subcellular localization of Rnd proteins (Foster et al., 1996; Roberts et al., 2008). The C-terminal sequence of Rnd proteins has been supposed to be farnesylated, and this modification is required for association with the plasma membrane (Foster et al., 1996). To test the role of the C-terminal CAAX motif of Rnd1 in lipid raft localization, we prepared Rnd1-C229S, in which the CAAX motif of Rnd1 was inactivated by a change of Cys to Ser (Figure 2F). As shown in Figure 2G, Rnd1-C229S could not localize to the lipid raft. Moreover, we observed a very little amount of Rnd1-C229S in nonraft membranes. Our additional biochemical fractionation analysis using crude membrane and cytosolic fractions revealed that membrane localization is severely impaired (Figure 2I), and Rnd1-C229S was observed diffusely throughout the cytoplasm and nucleus in cells (Figure 2H). These data suggest that although the C-terminal CAAX motif of Rnd1 is required for membrane association, it is by itself insufficient to induce lipid raft localization.
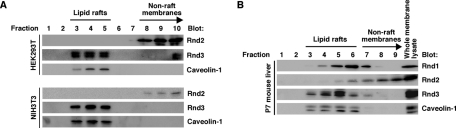
To further examine the localization of endogenous Rnd proteins, we performed sucrose density gradient fractionation assay of a human epithelial cell line, HEK293T, and a mouse fibroblast cell line, NIH3T3, where we could detect endogenous Rnd2 and Rnd3. In both cell types, an endogenous raft marker protein, caveolin-1, was present mainly in fractions 3–5, and endogenous Rnd3 protein resided in the raft fractions. We further performed lipid raft fractionation analysis of perfused liver dissected from postnatal day 7 (P7) mouse, where we could detect all the Rnd subfamilies. As shown in Figure 3B, the endogenous raft marker protein caveolin-1 was present mainly in fractions 3–6, and Rnd1 and Rnd3 were localized in the caveolin-rich fractions of lipid rafts in mouse liver. By contrast, Rnd2 was predominant in the fractions of nonraft membranes in cell culture (Figure 3A) and tissue (Figure 3B). These data suggest that Rnd proteins display differential membrane localization in vivo.
FIGURE 3:
Rnd1 and Rnd3, but not Rnd2, reside in lipid rafts in vivo. (A) HEK293T cells (top) and NIH3T3 cells (bottom) were subjected to the sucrose density gradient fractionation assay. Ten fractions were sampled and numbered from the top of the gradient, and each fraction was analyzed by immunoblotting. (B) Liver tissue homogenate prepared from postnatal day 7 mouse was subjected to the sucrose density gradient fractionation assay. Nine fractions were sampled and numbered from the top of the gradient, and each fraction and the whole membrane lysate were analyzed by immunoblotting.
The N-terminal KERRA sequence is required for RhoA inhibition
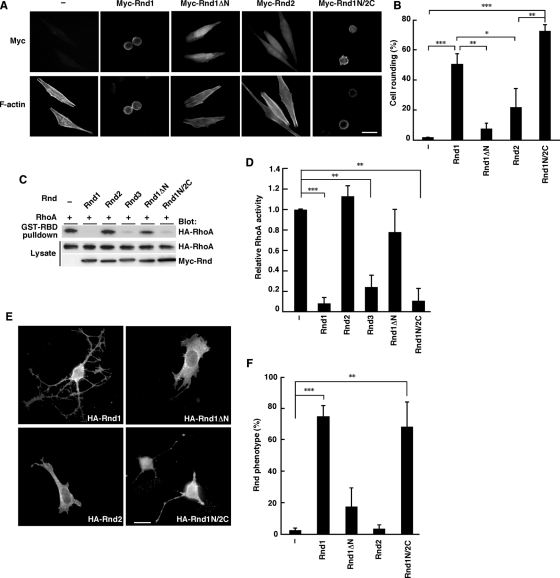
Rnd1 and Rnd3 have been reported to induce the loss of focal adhesions, and cells losing their adhesions cannot spread and tend to round (Nobes et al., 1998). We next examined the role of the N-terminal KERRA sequence of Rnd1 in the signaling. We performed immunofluorescence microscopy of the HeLa cells expressing various Rnd constructs with anti-Myc monoclonal antibody. As shown in Figure 4, A and B, Rnd1 induced strong cell rounding, but Rnd2 did not. Furthermore, like Rnd1, transient expression of Rnd1N/2C induced cell rounding, even though its backbone is Rnd2. Rnd1ΔN could not induce cell rounding. These data demonstrate that the N-terminal KERRA sequence is required for inducing cell rounding.
FIGURE 4:
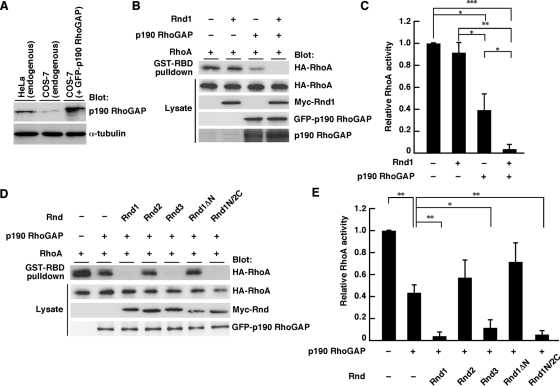
The N-terminal sequence of Rnd1 is required for RhoA inhibition signaling. (A) HeLa cells were transiently transfected with the indicated expression plasmids and stained with anti-Myc monoclonal antibody and with phalloidin to visualize F-actin. (B) Myc-staining–positive cells with an area <300 μm2 were scored as a percentage of the total number of transfected cells (n = 50/sample). (C) HeLa cells were transfected with the indicated expression plasmids, and the cell lysates were incubated with GST-fused, Rho-binding domain of rhotekin (GST-RBD). Bound RhoA and total cell lysates were analyzed by immunoblotting. (D) Relative RhoA activity was determined by the amount of RhoA bound to GST-RBD normalized to the amount of RhoA in cell lysates. The results are the mean ± SEM of three independent experiments. (E) Swiss 3T3 fibroblasts were microinjected with the indicated expression plasmids and stained with anti-HA monoclonal antibody. (F) Cells exhibiting the Rnd phenotype were scored as a percentage of the total number of microinjected cells. The results are the mean ± SEM of three independent experiments (n = 30/sample). Scale bars, 25 μm. ***p < 0.001, **p < 0.01, *p < 0.05 (one-way ANOVA).
Rnd1 and Rnd3 have been reported to possess antagonistic effects on the action of RhoA in various cell types (Chardin, 2006). We next examined the role of the N-terminal sequence of Rnd1 in the RhoA inhibition signaling. HeLa cells were transfected with an expression vector encoding HA-tagged wild-type RhoA, together with Myc-tagged Rnd GTPases, and RhoA activity was determined by pulldown assay with glutathione S-transferase–fused Rho-binding domain (GST-RBD) of a RhoA effector, rhotekin. Expression of Rnd1 decreased RhoA activity, but expression of Rnd1ΔN, which lacks the N-terminal region of Rnd1 and cannot localize to lipid rafts (Figure 2C) and cell periphery (Figure 2E), showed no effect on RhoA activity (Figure 4, C and D). Moreover, expression of Rnd2 could not inhibit RhoA activity, whereas expression of Rnd1N/Rnd2C, which contains the N-terminal KERRA sequence of Rnd1 and localizes to lipid rafts (Figure 2C) and cell periphery (Figure 2E), could inhibit RhoA activity (Figure 4, C and D).
Previous studies showed that expression of Rnd1 or Rnd3 induces the cell body retraction to produce a dendritic phenotype in Swiss 3T3 cells (Guasch et al., 1998; Nobes et al., 1998). We microinjected HA-tagged Rnd expression plasmids into the nuclei of Swiss 3T3 fibroblasts and observed the morphological effects. As shown in Figure 4E, microinjection of Rnd1 or Rnd1N/2C, but not Rnd2 or Rnd1ΔN, induced the rounding of the cell body. These data suggest that the N-terminal KERRA sequence of Rnd1 is required for RhoA inhibition signaling and serves as the determinant for the Rnd proteins to function as antagonists of RhoA.
RhoA inhibition by RhoGAP requires the N-terminal KERRA sequence
Rnd proteins have been reported to function as RhoA antagonists through p190 RhoGAP (Wennerberg et al., 2003). We next examined the role of the N-terminal sequence in p190 RhoGAP–mediated RhoA inhibition signaling. COS-7 cells were used to observe p190 RhoGAP–dependent RhoA inhibition signaling. The endogenous expression level of p190 RhoGAP in COS-7 cells was relatively low compared with that in HeLa cells, which was significantly heightened by ectopic expression of GFP-p190 RhoGAP (Figure 5A). Expression of Rnd1 alone did not affect RhoA activity, but Rnd1 decreased RhoA activity when coexpressed with GFP-p190 RhoGAP (Figure 5, B and C). This decrease was also observed with Rnd3 or Rnd1N/2C but not with Rnd2 or Rnd1ΔN (Figure 5, D and E). These data suggest that the N-terminal KERRA sequence is required for p190 RhoGAP-dependent RhoA inhibition.
FIGURE 5:
Role of the N-terminal sequence of Rnd1 in p190 RhoGAP-mediated RhoA inhibition. (A) Whole-cell lysates from untransfected or GFP-p190 RhoGAP–transfected cells were analyzed by immunoblotting with anti–p190 RhoGAP and anti–α-tubulin antibodies. (B–E) COS-7 cells were transfected with the indicated expression plasmids, and relative RhoA activity was measured as described in the legend to Figure 4, C and D. The results are the mean ± SEM of three independent experiments. ***p < 0.001, **p < 0.01, *p < 0.05 (one-way ANOVA).
Lipid raft targeting enables efficient binding of Rnd proteins to p190 RhoGAP in cells
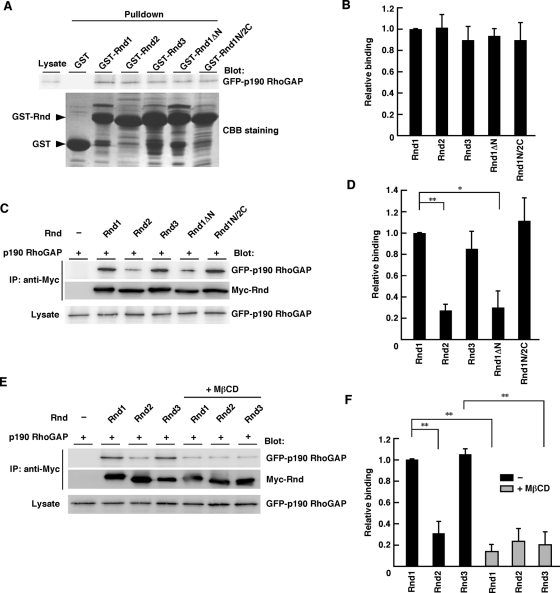
It has been reported that all the Rnd proteins—Rnd1, Rnd2, and Rnd3—bind directly to p190 RhoGAP and their interaction equally induces activation of p190 RhoGAP in vitro (Wennerberg et al., 2003). Our GST-pulldown assays showed that all the Rnd proteins, including Rnd1N/2C and Rnd1ΔN, could interact equally with p190 RhoGAP in vitro (Figure 6, A and B). In contrast, coimmunoprecipitation analysis in COS-7 cells revealed that p190 RhoGAP preferentially bound to Rnd1, Rnd3, or Rnd1N/2C rather than Rnd2 or Rnd1ΔN (Figure 6, C and D), suggesting that the N-terminal KERRA sequence enables efficient binding of Rnd proteins to p190 RhoGAP in cells. We further examined whether lipid rafts mediate the enhancement of the interaction of Rnd1 and Rnd3 with p190 RhoGAP. As shown in Figure 6, E and F, cholesterol depletion with MβCD treatment suppressed the interaction of Rnd1 and Rnd3 with p190 RhoGAP in COS-7 cells. These data demonstrate the lipid raft–dependent interaction of Rnd1 and Rnd3 with p190 RhoGAP in cells.
FIGURE 6:
Lipid raft enables efficient binding of Rnd1 to p190 RhoGAP. (A) COS-7 cells were transfected with an expression vector encoding GFP-tagged p190 RhoGAP, and the cell lysate was incubated with GST-fused Rnd proteins. GST proteins used in the pulldown assay were verified by CBB staining. (C, E) Lysates from transiently transfected COS-7 cells with or without MβCD treatment (20 mM, 20 min) were immunoprecipitated with an antibody against Myc. Bound and total proteins were analyzed by immunoblotting with the indicated antibodies. (B, D, F) Relative binding was determined by the amount of bound p190 RhoGAP normalized to the amount of p190 RhoGAP in cell lysates. The results are the mean ± SEM of three independent experiments. **p < 0.01, *p < 0.05 (one-way ANOVA).
Rnd1 induces lipid raft targeting of p190 RhoGAP
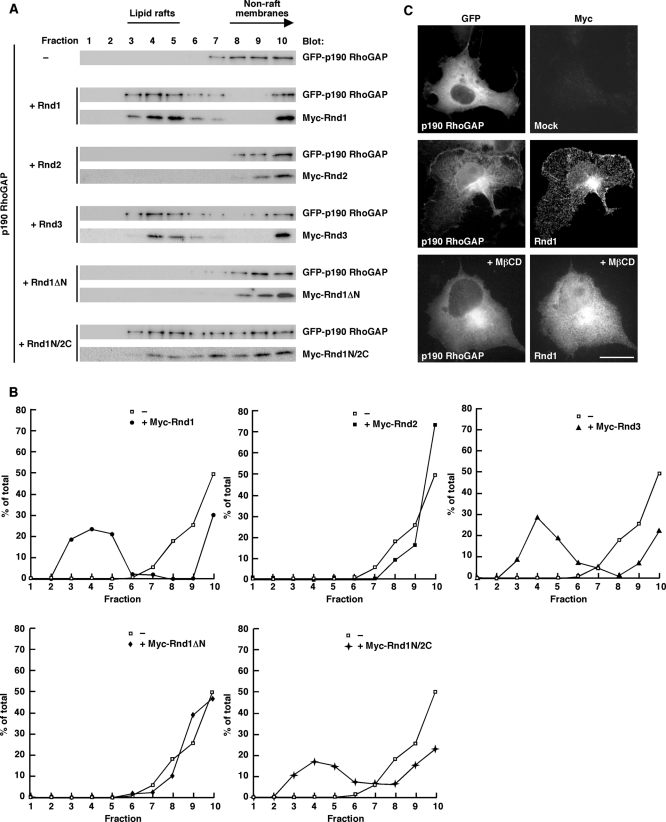
p190 RhoGAP must be recruited to lipid rafts for inactivation of RhoA, since active RhoA is concentrated and functions in lipid rafts (Palazzo et al., 2004). We examined whether Rnd proteins regulate localization of p190 RhoGAP. p190 RhoGAP in nonraft membranes was localized to lipid rafts when coexpressed with Rnd1. This change in p190 RhoGAP localization was also observed with Rnd3 or Rnd1N/2C but not with Rnd2 or Rnd1ΔN (Figure 7, A and B). These results suggest that the N-terminal KERRA sequence is required for lipid raft targeting of p190 RhoGAP. We also performed immunofluorescence microscopy analysis. To observe subcellular localization of transiently transfected p190 RhoGAP in COS-7 cells, we used poly-l-lysine at 20 μg/ml to prevent cell detachment and cell rounding. We found that p190 RhoGAP was localized to the cell periphery when coexpressed with Rnd1, and this localization was suppressed by MβCD treatment (Figure 7C). These data indicate that Rnd1 mediates lipid raft-dependent plasma membrane localization of p190 RhoGAP.
FIGURE 7:
Rnd-dependent raft targeting of p190 RhoGAP. (A) COS-7 cells transiently transfected with the indicated plasmids were subjected to the sucrose density gradient fractionation assay. (B) Densitometric analysis of the GFP-p190 RhoGAP bands on the immunoblots in A. The amount of the GFPxp190 RhoGAP protein in each fraction was expressed as a percentage of the total. –, in the absence of Rnd1. (C) COS-7 cells were transiently transfected with GFP–p190 RhoGAP or GFP–p190 RhoGAP plus Myc-tagged Rnd1 and stained with anti-Myc monoclonal antibody. MβCD (20 mM) was treated for 20 min before fixation. Scale bar, 20 μm.
Rnd1 induces lipid raft–dependent activation of p190 RhoGAP
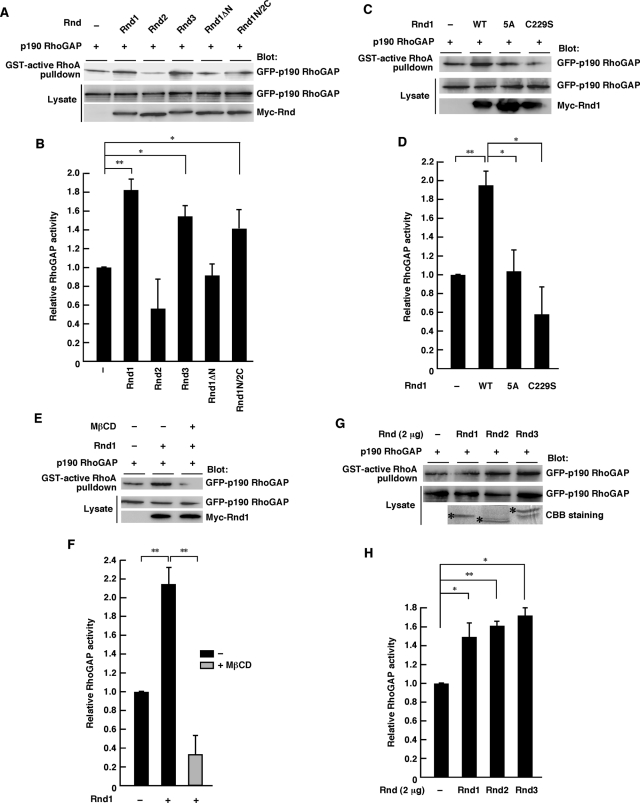
Functional activation of p190 RhoGAP can be measured by the increased ability to associate with its substrate, active RhoA (Noren et al., 2003; Barberis et al., 2005). We used the GST-fused, constitutively active form of RhoA (RhoA G14V) to pull down functionally activated p190 RhoGAP from COS-7 cell lysates expressing the indicated Rnd proteins. As shown in Figure 8A, activity of p190 RhoGAP in COS-7 cells was increased by the expression of Rnd1, Rnd3, or Rnd1N/2C but not by Rnd2 or Rnd1ΔN. p190 RhoGAP activation by Rnd1 was abolished by the 5A mutation (alanine substitution for the N-terminal KERRA sequence) or C229S mutation (inactivation of the CAAX motif by a change of Cys to Ser). These data suggest that both the N-terminal KERRA sequence and the C-terminal CAAX motif are required for activation of p190 RhoGAP in cells. We further found that the Rnd1-mediated p190 RhoGAP activation was suppressed by MβCD treatment (Figure 8, E and F). In contrast, all the Rnd proteins—Rnd1, Rnd2, and Rnd3—which could equally interact with p190 RhoGAP in vitro (Figure 6, A and B), could equally activate p190 RhoGAP in an in vitro setting using purified Rnd proteins (Figure 8, G and H). These results suggest that although all the Rnd proteins can potentially activate p190 RhoGAP in vitro, differential membrane targeting governs p190 RhoGAP interaction (Figure 6, C–F) and activation (Figure 8, A–F) in cells.
FIGURE 8:
Rnd1 induces lipid raft–dependent p190 RhoGAP activation. (A, C, E) Functionally active p190 RhoGAP in COS-7 lysates expressing the indicated Rnd proteins was pulled down by GST-fused active RhoA. MβCD treatment (20 mM) was performed for 20 min before cell lysis. (G) COS-7 cells were transfected with an expression vector encoding GFP-tagged p190 RhoGAP, and the cell lysate was incubated with 2 μg of nonfused recombinant Rnd proteins. Rnd proteins used in the assay were verified by CBB staining (bottom, indicated by asterisks). (B, D, F, H) Relative RhoGAP activity was determined by the amount of bound p190 RhoGAP normalized to the amount of p190 RhoGAP in cell lysates. The results are the mean ± SEM of three independent experiments. **p < 0.01, *p < 0.05 (one-way ANOVA).
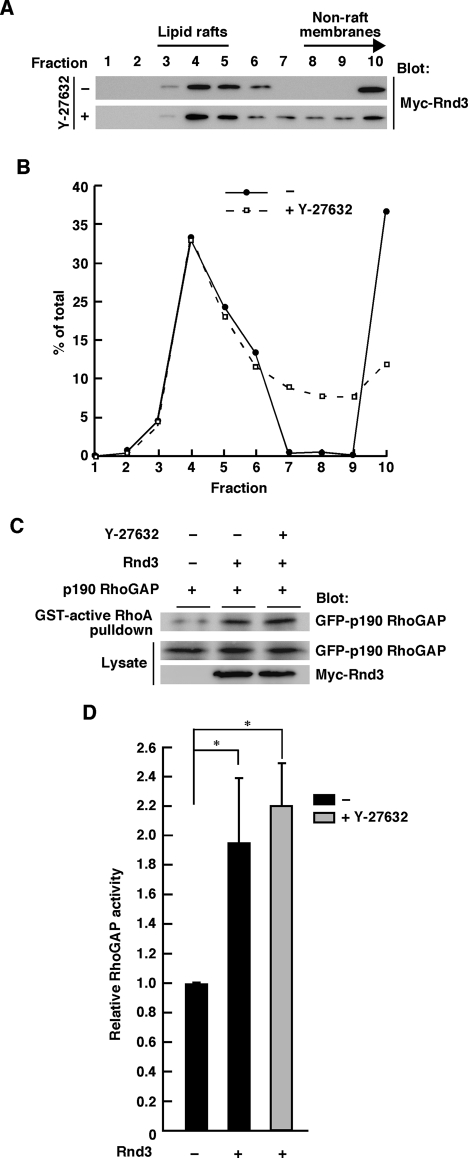
Rnd3 displays ROCK-independent lipid raft localization and activation of p190 RhoGAP
Rnd3 has been reported to be phosphorylated at the N-terminal region by a RhoA effector, ROCK I (Riento et al., 2003). We investigated whether ROCK-mediated phosphorylation contributes to Rnd3 localization. As shown in Figure 9, A and B, although shift in localization within nonraft membrane fractions was observed, lipid raft localization of Rnd3 was not affected by treatment of the cells with a specific ROCK inhibitor, Y-27632. Moreover, Y-27632 treatment did not affect the Rnd3-mediated p190 RhoGAP activation in COS-7 cells (Figure 9, C and D). These results suggest that Rnd3 displays ROCK-independent lipid raft localization and activation of p190 RhoGAP.
FIGURE 9:
Rnd3 displays ROCK-independent lipid raft localization and activation of p190 RhoGAP. (A) Rnd3-transfected COS-7 cells treated with (+) or without (–) Y-27632 (25 μM) treatment were subjected to the sucrose density gradient fractionation assay. (B) Densitometric analysis of the bands on the immunoblots in A. The amount of the protein in each fraction was expressed as a percentage of the total. (C) Functionally active p190 RhoGAP in Rnd3-transfected COS-7 lysates with or without Y-27632 (25 μM) treatment was pulled down by GST-fused active RhoA. (D) Relative RhoGAP activity was determined by the amount of bound p190 RhoGAP normalized to the amount of p190 RhoGAP in cell lysates. The results are the mean ± SEM of three independent experiments. *p < 0.05 (one-way ANOVA).
DISCUSSION
It has been shown that Rnd1 is predominantly plasma membrane associated, Rnd2 is not plasma membrane associated and localized to early endosomes, and Rnd3/RhoE is found in the Golgi complex, as well as in the plasma membrane (Chardin, 2006; Tanaka et al., 2006). Rnd1 and Rnd3 show inhibitory effects on RhoA in cells but Rnd2 does not (Nobes et al., 1998; Tanaka et al., 2006). The Rnd proteins Rnd1, Rnd2, and Rnd3 have been considered constitutively active, and how their activity and localization are regulated has been obscure. Our results identify the N-terminal region of Rnd1 or Rnd3 as the lipid raft–targeting determinant and demonstrate a novel regulatory mechanism by which Rnd proteins function as antagonists of RhoA.
The plasma membrane of most cell types contains specialized subdomains with distinct lipid and protein compositions, referred to as lipid raft microdomains (Brown et al., 1998; Kurzchalia et al., 1999; Simons and Toomre, 2000). Lipid rafts are cholesterol- and sphingolipid-rich lipid microdomains and function in cellular signaling by concentrating or separating specific molecules in a unique lipid environment in many cell types. It is well known that members of the Rho family GTPases are key regulators of the actin cytoskeleton in various cell types (Hall, 1998). Among the Rho family of small GTPases, RhoA, Rac1, and TC10 are known to be concentrated and function in lipid rafts and caveolae, and their localization is crucial for the activation of many signal transduction pathways (Watson et al., 2001; del Pozo et al., 2004; Palazzo et al., 2004). For most Rho family proteins, this GDP-bound form is predominant at the resting state and interacts with the GDI protein that covers the C-terminal geranylgeranyl moiety and stabilizes them as a cytosolic Rho-GDI complex (Sasaki and Takai, 1998). By contrast, Rnd proteins Rnd1, Rnd2, and Rnd3/RhoE comprise a distinct branch of Rho family GTPases, in that they have a low affinity for GDP and very low intrinsic GTPase activities (Foster et al., 1996; Guasch et al., 1998; Nobes et al., 1998). Previous work showed that Rnd proteins are farnesylated and do not seem to be associated with a GDI (Roberts et al., 2008). Thus it is presumable that the regulatory mechanism of Rnd protein localization and activation is likely to be different from those of other Rho family members. In this article, we found that the N-terminal KERRA sequence of Rnd1 is a determinant for its subcellular localization.
Basic amino acid residues have been known to present in a multitude of plasma membrane–targeted proteins. For example, although H-Ras and N-Ras are farnesylated and palmitoylated, K-Ras has a cluster of basic residues in its C-terminal hypervariable region as its additional plasma membrane–targeting signal (Hancock et al., 1989). Basic residues have also been implicated in the plasma membrane localization of GAP 43, Src, myristoylated alanine-rich C kinase substrate, G protein–coupled receptor kinases, and other signaling proteins (McLaughlin and Aderem, 1995; McLaughlin and Murray, 2005; Murray et al., 1998; Thiyagarajan et al., 2004). Regions of positive charges on the surface of a protein function as membrane-binding signals via electrostatic interactions with negatively charged head groups of membrane lipids (McLaughlin and Aderem, 1995; McLaughlin and Murray, 2005; Murray et al., 1998). Potential lipid-binding partners for basic clusters could be the monovalent acidic phosphatidylserine or the more negatively charged phosphatidylinositol-4,5-bisphosphate (Ghomashchi et al., 1995). Moreover, these acidic lipids are concentrated at the plasma membrane compared with other intracellular membranes (McLaughlin and Murray, 2005). This may provide a simple mechanism for the N-terminal KERRA sequence to direct Rnd1 or Rnd3 more preferentially to the inner surface of the plasma membrane. However, the sequence is rather small and contains an acidic residue. It will be important in future work to define which membrane lipids or molecules are interacting with the N-terminal region of Rnd1 and Rnd3.
Despite the functional difference in RhoA inhibition signaling in cells, all the members of the Rnd subfamily—Rnd1, Rnd2, and Rnd3—have been considered constitutively active (Foster et al., 1996; Guasch et al., 1998; Nobes et al., 1998), and all the members have been reported to bind directly to p190 RhoGAP and equally induce activation of p190 RhoGAP in vitro (Wennerberg et al., 2003). In this study, we found that Rnd2 or Rnd1ΔN bind to p190 RhoGAP less effectively than Rnd1 or Rnd3 in cells, even though they show almost the same affinities in vitro. We further found that lipid rafts mediate the enhancement of the interaction of Rnd1 and Rnd3 with p190 RhoGAP. We suppose that lipid raft targeting of p190 RhoGAP by Rnd1 or Rnd3 may result in concentration of the molecule in a specified lipid compartment, inducing more efficient interaction with Rnd1 or Rnd3 in cells.
Overall, our study sheds light on how constitutively active small GTPases are functionally regulated, presenting a striking mechanism by which differential membrane targeting governs the activities of G proteins having similarities in effector interaction.
MATERIALS AND METHODS
DNA constructs and mutagenesis
GFP-tagged human p190 RhoGAP was from H. Sabe (Hokkaido University, Sapporo, Japan). Rnd and RhoA constructs were described previously (Katoh et al., 2002; Oinuma et al., 2003). Rnd1ΔN (lacking the first seven N-terminal amino acids), Rnd1-5A (alanine substitution for the N-terminal KERRA sequence), Rnd1-C229S (mutation in the C-terminal CAAX motif by a change of Cys to Ser), chimeric Rnd1N/2C (replacing the first seven N-terminal amino acids of Rnd2 by the first 12 N-terminal amino acids of Rnd1), and chimeric Rnd2N/1C (replacing the last 11 C-terminal amino acids of Rnd2 by the last 11 C-terminal amino acids of Rnd1) were generated by PCR amplification.
Antibodies and reagents
We used the following antibodies: mouse monoclonal antibodies against GFP, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and RhoA; rabbit polyclonal antibody against HA (Santa Cruz Biotechnology, Santa Cruz, CA); a rat monoclonal antibody against HA (Roche, Indianapolis, IN); a rabbit polyclonal antibody against Myc (MBL, Woburn, MA); a mouse monoclonal antibody against caveolin-1 (BD Transduction Laboratories, Lexington, KY); mouse monoclonal antibodies against Myc, Na+/K+-ATPase α subunit, p190 RhoGAP, and Rnd3 (Millipore, Billerica, MA); mouse monoclonal antibodies against α-tubulin and vimentin (Sigma-Aldrich, St. Louis, MO); horseradish peroxidase–conjugated secondary antibodies (DakoCytomation, Hamburg, Germany); and Alexa Fluor–conjugated secondary antibodies (Molecular Probes, Invitrogen, Carlsbad, CA). Rabbit polyclonal antibodies against Rnd1 and Rnd2 were described previously (Fujita et al., 2002; Ishikawa et al., 2003; Tanaka et al., 2006). We used the following reagents: Y-27632 (Wako, Osaka, Japan); poly-l-lysine, lysophosphatidic acid (LPA), and MβCD (Sigma-Aldrich).
Cell culture and transfection
COS-7, HeLa, HEK293T, NIH3T3, and Swiss3T3 cells were cultured and transfected as described previously (Katoh et al., 2002; Oinuma et al., 2003; Hiramoto et al., 2006; Tanaka et al., 2006). In cholesterol depletion experiments, COS-7 cells were incubated with 20 mM MβCD at 37°C for 20 min. Y-27632 (25 μM) was treated for 1 h prior to cell collection.
Immunoblotting
Immunoblotting was performed as described previously (Oinuma et al., 2003). Images were captured using an LAS 1000 analyzer equipped with Image Gauge 4.0 software (Fuji, Tokyo, Japan), and densitometric analysis of the immunoblots was performed with ImageJ software (National Institutes of Health, Bethesda, MD).
Purification of recombinant proteins
All GST-fused proteins were purified from Escherichia coli as described previously (Katoh et al., 2002, Tanaka et al., 2006). For purification of Rnd proteins, GTP (1 mM) was added throughout the procedure. Nonfused Rnd proteins were recovered by incubation with 10 U of thrombin (Sigma-Aldrich) for 12 h at 4°C, and then thrombin was removed by absorption to p-amino-benzamidine–agarose beads (Sigma-Aldrich). Protein concentration was determined by comparing with bovine serum albumin standards after SDS–PAGE and Coomassie brilliant blue (CBB) staining.
Binding assay
GST-pulldown assays using purified Rnd GTPases and immunoprecipitation analysis have been described (Oinuma et al., 2003, Tanaka et al., 2006).
Immunofluorescence microscopy
Immunofluorescence staining was performed as described previously (Katoh et al., 2002). Images were captured using a microscope (Eclipse E800; Nikon, Melville, NY) and a 40× 0.75 objective or a 100× 1.25 objective (Nikon) equipped with a digital camera (DC350F; Leica, Wetzlar, Germany). To observe subcellular localization of transiently expressed p190 RhoGAP, poly-l-lysine was used at 20 μg/ml to prevent cell detachment and cell rounding.
Cell rounding assay
HeLa cell rounding assay was performed as described (Tanaka et al., 2006). Myc-staining–positive cells with an area <300 μm2 were determined as contracted cells and were scored as the total number of transfected cells.
Measurement of RhoA activity
Measurement of RhoA activity in COS-7 cells and HeLa cells was performed as described previously (Oinuma et al., 2003; Tanaka et al., 2006).
Measurement of p190 RhoGAP activity
Measurement of p190 RhoGAP activity was performed as described previously (Barberis et al., 2005). Twelve micrograms of the GST-fused, constitutively active form of RhoA (RhoA G14V) was used to pull down functionally activated p190 RhoGAP.
Microinjection
Swiss 3T3 fibroblasts (2 × 104) were seeded onto round, 13-mm glass coverslips. Then cells were serum starved for 16 h, and the indicated plasmids were microinjected into the nucleus as described previously (Katoh et al., 2002). Two hours after injection, cells were stimulated with 1 μM LPA for 30 min and fixed. Cells exhibiting the Rnd phenotype were defined as cells comprising <1000 μm2 of the cell body with cellular processes >50 μm.
Sucrose density gradient fractionation
Cells were lysed by sonication with the sonication buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 250 mM sucrose). The lysates were then centrifuged for 30 min at 100,000 × g at 4°C, and the supernatants were removed. The pellets were resuspended and homogenized with the ice-cold homogenization buffer (500 mM Na2CO3, 10 mM MgCl2, and 1 mM PMSF) using a Potter-Elvehjem homogenizer. The homogenates were adjusted to 45% sucrose by adding the equal volume of 90% sucrose in ice-cold homogenization buffer, then placed in an S55S ultracentrifuge tube (Hitachi High-Technologies, Tokyo, Japan) and overlaid with 35, 30, 25, and 5% sucrose solution in 2-(N-morpholino)ethanesulfonic acid (MES)–buffered saline (25 mM MES-HCl, pH 6.5, 150 mM NaCl, 250 mM Na2CO3, and 10 mM MgCl2). All of these steps were performed on ice. After centrifugation (12 h, 167,000 × g, 4°C), 10 aliquot fractions were collected from the top.
Livers were dissected from a P7 mice perfused transcardially with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4), and sucrose density gradient fractionation of tissue was performed as described previously (Kim et al., 2008). Briefly, 700 μl of 1% Triton X-100 in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (25 mM HEPES-HCl, pH 6.5, 150 mM NaCl, 1 mM ethylene glycol tetraacetic acid, 1 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin) was added to 300 μl of tissue homogenate and suspended well by gentle pipetting. Then the lysate was homogenized (30 strokes) and incubated on ice for 30 min. After incubation, the homogenate was adjusted to 40% sucrose by adding the equal volume of 80% sucrose in ice-cold HEPES buffer and then placed in an S55S ultracentrifuge tube and overlaid with 30 and 5% sucrose solution in HEPES buffer. All of these steps were performed on ice. After centrifugation (20 h, 167,000 × g, 4°C), nine aliquot fractions were collected from the top. The fractioned samples were boiled for 20 min with SDS-sample buffer containing 50 mM dithiothreitol (DTT) and then analyzed by immunoblotting.
Separation of crude membrane and cytosolic fractions
Transiently transfected COS-7 cells were suspended in homogenization buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM PMSF, 10 mM MgCl2, 1 mM DTT, and 250 mM sucrose). The cell suspensions were then lysed by sonication (five times at 5 s each), and the lysates were centrifuged at 9300 × g for 5 min at 4°C to remove the unbroken cells and nuclear fractions. The supernatants were further fractioned at 100,000 × g for 1 h at 4°C. The particle pellet was resuspended in the same volume as the cytosolic fraction, and equal volumes of each were analyzed by SDS–PAGE and immunoblotting.
Statistical analyses
All data are reported as mean ± SEM of three independent experiments. Statistical significance of intergroup differences was determined by one-way analysis of variance (ANOVA) with Tukey's post hoc test using GraphPad Prism, version 5.0, statistical software (GraphPad Software, La Jolla, CA). Differences at the level of p < 0.05 were considered statistically significant.
Acknowledgments
We thank H. Sabe for providing a plasmid for p190 RhoGAP. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (Challenging Exploratory Research 23657127 to I.O. and Scientific Research (B) 23390019 to M.N.).
Abbreviations used:
- ANOVA
analysis of variance
- CBB
Coomassie brilliant blue
- GAP
GTPase-activating protein
- GDI
guanine nucleotide dissociation inhibitor
- GEF
guanine nucleotide-exchange factor
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HA
hemagglutinin, LPA, lysophosphatidic acid
- MβCD
methyl-β-cyclodextrin
- PMSF
phenylmethylsulfonyl fluoride
- RBD
Rho-binding domain
- ROCK
Rho-associated kinase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0900) on February 22, 2012.
REFERENCES
- Barberis D, Casazza A, Sordella R, Corso S, Artigiani S, Settleman J, Comoglio PM, Tamagnone L. p190 Rho-GTPase activating protein associates with plexins and it is required for semaphoring signalling. J Cell Sci. 2005;118:4689–4700. doi: 10.1242/jcs.02590. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Chardin P. Function and regulation of Rnd proteins. Nat Rev Mol Cell Biol. 2006;7:54–62. doi: 10.1038/nrm1788. [DOI] [PubMed] [Google Scholar]
- del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RG, Schwartz MA. Integrins regulate Rac targeting by internalization of membrane domains. Science. 2004;303:839–842. doi: 10.1126/science.1092571. [DOI] [PubMed] [Google Scholar]
- Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J, Settleman J. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol Cell Biol. 1996;16:2689–2699. doi: 10.1128/mcb.16.6.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Katoh H, Ishikawa Y, Mori K, Negishi M. Rapostlin is a novel effector of Rnd2 GTPase inducing neurite branching. J Biol Chem. 2002;277:45428–45434. doi: 10.1074/jbc.M208090200. [DOI] [PubMed] [Google Scholar]
- Ghomashchi F, Zhang X, Liu L, Gelb MH. Binding of prenylated and polybasic peptides to membranes: affinities and intervesicle exchange. Biochemistry. 1995;34:11910–11918. doi: 10.1021/bi00037a032. [DOI] [PubMed] [Google Scholar]
- Guasch RM, Scambler P, Jones GE, Ridley AJ. RhoE regulates actin cytoskeleton organization and cell migration. Mol Cell Biol. 1998;18:4761–4771. doi: 10.1128/mcb.18.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPase and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- Hiramoto K, Negishi M, Katoh H. Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exp Cell Res. 2006;312:4205–4216. doi: 10.1016/j.yexcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Katoh H, Negishi M. A role of Rnd1 GTPase in dendritic spine formation in hippocampal neurons. J Neurosci. 2003;23:11065–11072. doi: 10.1523/JNEUROSCI.23-35-11065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Harada A, Mori K, Negishi M. Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol Cell Biol. 2002;22:2952–2964. doi: 10.1128/MCB.22.9.2952-2964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KB, Lee JS, Ko YG. The isolation of detergent-resistant lipid rafts for two-dimensional electrophoresis. Methods Mol Biol. 2008;424:413–422. doi: 10.1007/978-1-60327-064-9_32. [DOI] [PubMed] [Google Scholar]
- Kurzchalia TV, Parton RG. Membrane microdomains and caveolae. Curr Opin Cell Biol. 1999;11:424–431. doi: 10.1016/s0955-0674(99)80061-1. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- Murray D, Hermida-Matsumoto L, Buser CA, Tsang J, Sigal CT, Ben-Tal N, Honig B, Resh MD, McLaughlin S. Electrostatics and the membrane association of Src: theory and experiment. Biochemistry. 1998;37:2145–2159. doi: 10.1021/bi972012b. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Lauritzen I, Mattei MG, Paris S, Hall A, Chardin P. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J Cell Biol. 1998;141:187–197. doi: 10.1083/jcb.141.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Arthur WT, Burridge K. Cadherin engagement inhibits RhoA via p190RhoGAP. J Biol Chem. 2003;278:13615–13618. doi: 10.1074/jbc.C200657200. [DOI] [PubMed] [Google Scholar]
- Oinuma I, Katoh H, Harada A, Negishi M. Direct interaction of Rnd1 with Plexin-B1 regulates PDZ-RhoGEF-mediated Rho activation by Plexin-B1 and induces cell contraction in COS-7 cells. J Biol Chem. 2003;278:25671–25677. doi: 10.1074/jbc.M303047200. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Eng CH, Schlaepfer DD, Marcantonio EE, Gundersen GG. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science. 2004;303:836–839. doi: 10.1126/science.1091325. [DOI] [PubMed] [Google Scholar]
- Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol. 2003;23:4219–4229. doi: 10.1128/MCB.23.12.4219-4229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behavior. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- Roberts PJ, Mitin N, Keller PJ, Chenette EJ, Madigan JP, Currin RO, Cox AD, Wilson O, Kirschmeier P, Der CJ. Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J Biol Chem. 2008;283:25150–25163. doi: 10.1074/jbc.M800882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Takai Y. The Rho small G protein family-Rho GDI system as a temporal and spatial determinant for cytoskeletal control. Biochem Biophys Res Commun. 1998;245:641–645. doi: 10.1006/bbrc.1998.8253. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Katoh H, Negishi M. Pragmin, a novel effector of Rnd2 GTPase, stimulates RhoA activity. J Biol Chem. 2006;281:10355–10364. doi: 10.1074/jbc.M511314200. [DOI] [PubMed] [Google Scholar]
- Thiyagarajan MM, Stracquatanio RP, Pronin AN, Evanko DS, Benovic JL, Wedegaertner PB. A predicted amphipathic helix mediates plasma membrane localization of GRK5. J Biol Chem. 2004;279:17989–17995. doi: 10.1074/jbc.M310738200. [DOI] [PubMed] [Google Scholar]
- Waheed AA, Jones TL. Hsp90 interactions and acylation target the G protein Ga12 but not Ga13 to lipid rafts. J Biol Chem. 2002;277:32409–32412. doi: 10.1074/jbc.C200383200. [DOI] [PubMed] [Google Scholar]
- Watson RT, Shigematsu S, Chiang SH, Mora S, Kanzaki M, Macara IG, Saltiel AR, Pessin JE. Lipid raft microdomain compartmentalization of TC10 is required for insulin signaling and GLUT4 translocation. J Cell Biol. 2001;154:829–840. doi: 10.1083/jcb.200102078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K, Forget MA, Ellerbroek SM, Arthur WT, Burridge K, Settleman J, Der CJ, Hansen SH. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr Biol. 2003;13:1106–1115. doi: 10.1016/s0960-9822(03)00418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]