T1736 is a novel phosphorylation site on the integrin β4 subunit that is phosphorylated downstream of protein kinase C and EGF receptor activation and is a substrate for protein kinase D1 in vitro and in cells. It contributes to the regulation of HD dynamics through modulating the association of β4 with plectin.
Abstract
During wound healing, hemidesmome disassembly enables keratinocyte migration and proliferation. Hemidesmosome dynamics are altered downstream of epidermal growth factor (EGF) receptor activation, following the phosphorylation of integrin β4 residues S1356 and S1364, which reduces the interaction with plectin; however, this event is insufficient to drive complete hemidesmome disassembly. In the studies reported here, we used a fluorescence resonance energy transfer–based assay to demonstrate that the connecting segment and carboxy-terminal tail of the β4 cytoplasmic domain interact, which facilitates the formation of a binding platform for plectin. In addition, analysis of a β4 mutant containing a phosphomimicking aspartic acid residue at T1736 in the C-tail suggests that phosphorylation of this residue regulates the interaction with the plectin plakin domain. The aspartic acid mutation of β4 T1736 impaired hemidesmosome formation in junctional epidermolysis associated with pyloric atresia/β4 keratinocytes. Furthermore, we show that T1736 is phosphorylated downstream of protein kinase C and EGF receptor activation and is a substrate for protein kinase D1 in vitro and in cells, which requires its translocation to the plasma membrane and subsequent activation. In conclusion, we identify T1736 as a novel phosphorylation site that contributes to the regulation of hemidesmome disassembly, a dynamically regulated process involving the concerted phosphorylation of multiple β4 residues.
INTRODUCTION
Hemidesmosomes (HDs) are junctional protein complexes that maintain epithelial tissue integrity. HDs mediate the stable adhesion of epithelial cells to the underlying basement membrane by linking the extracellular matrix to the intermediate filament system. In simple epithelia this link is formed by type II HDs, which consist of integrin α6β4 and plectin, the latter of which binds directly to the keratin filament system. In squamous and complex epithelia more stable type I HDs occur (Green and Jones, 1996; Borradori and Sonnenberg, 1999). Type 1 HDs are formed via interaction between integrin α6β4 and plectin, with further stabilization occurring through interaction with bullous pemphigoid antigens 180 (BP180) and 230 (BP230) (Litjens et al., 2006). Genetic mutations that alter the expression and/or function of any of these proteins result in tissue integrity defects known as epidermolysis bullosa (EB; Pulkkinen and Uitto, 1999).
Two known sites of interaction exist between integrin α6β4 and plectin. The primary site is formed by the first pair of fibronectin type III (FnIII) domains and a small part of the connecting segment (CS) of β4 that binds to the actin-binding domain (ABD) of plectin (Geerts et al., 1999; de Pereda et al., 2009). This interaction is sufficient for the formation of HDs in cultured keratinocytes (Niessen et al., 1997; Schaapveld et al., 1998). Moreover, HD formation was not impaired in mice carrying a targeted deletion of the C-terminal cytoplasmic tail β4 up to residue 1355, which leaves the primary binding site for the plectin-ABD intact (Nikolopoulos et al., 2005). Further support for the importance of this primary interaction site in regulating HD dynamics comes from the occurrence of two missense mutations in the second fibronectin type III (FNIII) domain of β4, identified in patients with nonlethal forms of junctional EB (Pulkkinen and Uitto, 1999; Nakano et al., 2001). Both mutations were shown to inhibit interaction with plectin-ABD (Geerts et al., 1999; Koster et al., 2001; de Pereda et al., 2009). The secondary binding site is formed between the carboxy terminus of the CS, the carboxy-terminal tail (C-tail) of β4, and the plakin domain of plectin (Rezniczek et al., 1998; Koster et al., 2004). A nonsense mutation (Q1767X) in the C-tail of β4 was identified in an EB patient displaying a relatively mild, trauma-induced blistering of the skin (Nakano et al., 2001). This secondary interaction is predicted to stabilize the β4 and plectin-ABD complex, but its exact role in HD dynamics is not known.
During the migration and proliferation of keratinocytes, HD assembly/disassembly and adhesion to the extracellular matrix must be dynamically regulated. Several growth factors have been implicated in regulating HD disassembly, such as epidermal growth factor (EGF), hepatocyte growth factor, and macrophage-stimulating protein (Santoro et al., 2003; Werner and Grose, 2003; Litjens et al., 2006; Barrientos et al., 2008). These factors collectively induce the Ras/MAPK signaling pathway and stimulate β4 phosphorylation on serine residues in the CS (Rabinovitz et al., 2004; Wilhelmsen et al., 2007). Recently we showed that ERK1/2 and its downstream effector kinase p90RSK1/2 phosphorylate β4 on S1356 and S1364, respectively, destabilizing the interaction between the FNIII domains of β4 and the ABD of plectin (Frijns et al., 2010), but the signaling mechanisms that regulate the interaction between the C-tail of β4 and the plectin plakin domain remain unknown. Protein kinase D1 (PKD1) is an important regulator of proliferation, apoptosis, and tumor cell invasion (Van Lint et al., 2002; Rozengurt et al., 2005), processes that require the (partial) disassembly of HDs. Here we set out to determine whether PKD1 has a putative role in regulating HD (dis)assembly and interaction between the C-tail of β4 and plectin.
We demonstrate that the C-tail is positioned in close proximity to the CS of β4 and that a β4 T1736 phosphorylation mimic prevents interaction between the C-tail of β4 and the plectin plakin domain. Furthermore, PKD1 directly phosphorylated T1736 in vitro, and T1736 was phosphorylated after phorbol-12-myristate-13-acetate (PMA)– or EGF-induced activation and membrane translocation of PKD1 in junctional epidermolysis associated with pyloric atresia (PA-JEB)/β4 keratinocytes. In summary, our results suggest that multiple phosphorylation events are necessary to regulate the interaction of plectin with β4.
RESULTS
Mimicking phosphorylation of T1727 or T1736 prevents interaction between the C-terminal tail of β4 and the plectin plakin domain
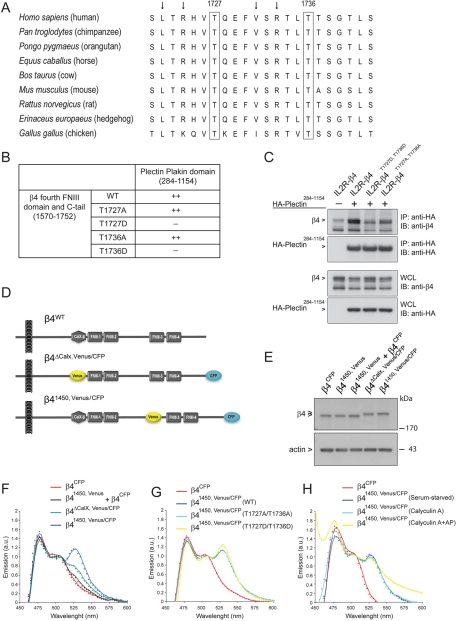
We previously showed that the C-tail (amino acids 1667–1752) of the β4 cytoplasmic domain is important for binding to the plakin domain of plectin, and that phosphorylation of S1356 and S1364 in the CS disrupts the interaction with the ABD of plectin (Wilhelmsen et al., 2007; Frijns et al., 2010). To determine whether phosphorylation events can also potentially regulate the interaction of the C-tail with the plakin domain of plectin, putative phosphorylation sites in the C-tail of β4 were screened using the Netphos program. Threonines 1727 and 1736 were identified to contain consensus sequences for PKD1-mediated phosphorylation (L/V-X-R-X-X-pS/T). Of importance, both threonines are evolutionarily conserved among several species (Figure 1A).
FIGURE 1:
Mimicking phosphorylation of β4 on T1727 or T1736 prevents binding of the C-tail of β4 to the plakin domain of plectin. (A) Amino acid sequence (1721–1742) alignment of β4 from different species using ClustalW. Besides the residues T1727 and T1736, the leucine, valine, and arginines (arrows) are highly conserved. (B) Substitution of T1727 or T1736 on β4 by a phosphomimicking aspartic acid residue resulted in loss of binding of the fourth FNIII domain and C-tail of β4 to the plectin plakin domain in a yeast two-hybrid interaction assay. (C) Coimmunoprecipitation from lysates of COS-7 cells cotransfected with different IL2R/β4 chimeric and HA-tagged plectin plakin-domain constructs showed association of the plakin domain of plectin with the cytoplasmic domains of wild-type β4 and mutant β4 in which T1736 had been replaced by alanine. Replacement of T1736 by aspartic acid eliminated binding. (D) Two β4 Venus-CFP fusion constructs were generated in which a CFP fluorophore was fused to the C-terminus and a Venus fluorophore, which either replaced the nonfunctional CalX domain (β4ΔCalX,Venus/CFP) or was inserted in the CS at position 1450 (β41450,Venus/CFP), where in the β4B variant an additional 53 amino acids are located. (E) COS-7 cells transiently transfected with β4CFP, β41450,Venus, a combination of β4CFP and β41450,Venus, β4ΔCalX, Venus/CFP, or β41450,Venus/CFP were lysed, and the levels of the different β4 Venus-CFP fusion proteins were analyzed by immunoblotting with polyclonal antibodies against β4. (F) COS-7 cells transiently expressing the indicated β4 Venus-CFP fusion proteins were serum starved overnight and lysed. The fluorescence emission spectra of the cell lysates upon excitation at 390 nm were recorded using a spectrophotometer and plotted. The expression of β41450,Venus/CFP resulted in a FRET signal at 527 nm, indicating that the C-tail is in close proximity to the CS of β4. (G, H) Serum-starved COS-7 cells transiently expressing the indicated β4 Venus-CFP fusion proteins were either left untreated or treated with calyculin A (50 nM) in growth medium (DMEM + 10% FCS) for 25 min and lysed. An aliquot of the lysate from the calyculin A–treated cells was incubated with alkaline phosphatases (AP; 60 U/ml) for 30 min at 37°C. The emission spectra of the cell lysates after excitation at 390 nm were analyzed and plotted. Substitution of T1727 and T1736 by alanine or aspartic acid, or treatment of lysates with calyculin A or AP, did not influence the emission spectra significantly.
To investigate whether phosphorylation of T1727 and T1736 regulates the interaction of β4 with the plectin plakin domain, yeast two-hybrid and coimmunoprecipitation (coIP) assays were performed. In agreement with previous studies, interaction of a fragment of β4 comprising the fourth FNIII domain and C-tail (amino acids 1570–1752) with the plakin domain of plectin (amino acids 284–1154, spectrin repeats 1–6; Sonnenberg et al., 2007) was observed in yeast (Figure 1B; Rezniczek et al., 1998; Koster et al., 2004). This interaction was prevented by the substitution of either T1727 or T1736 by a phosphomimicking aspartic acid, but not an alanine. In agreement, coIP experiments from transiently transfected cells confirmed the binding of the plectin plakin domain to the β4 cytoplasmic domain (Figure 1C). Furthermore, as observed in yeast, binding of the plakin domain to the β4 cytoplasmic domain was prevented by the substitution of T1727 and T1736 by aspartic acid, but not alanine. Taken together, these data suggest that the interaction between β4 and the plakin domain of plectin is regulated by the phosphorylation of T1727 and/or T1736.
Analysis of intermolecular interaction of the β4 cytoplasmic domain by fluorescence resonance energy transfer
Interaction between β4 and the plectin plakin domain depends on the C-tail of β4 and a specific region within the CS (1382–1436), which yeast two-hybrid and blot overlay assays suggest interact with each other (Rezniczek et al., 1998; Koster et al., 2004). It is not known, however, whether the interaction of the C-tail and the CS occurs within the full-length β4 molecule. Fluorescence resonance energy transfer (FRET), in which the excited cyan fluorescent protein (CFP) fluorophore (donor) transfers energy to Venus fluorophore (acceptor) when in close proximity (<10 nm) and at the right dipole orientation, was used to study the β4 C-tail/CS tertiary structure further. Two β4 Venus-CFP fusion proteins were generated. The CFP fluorophore was fused to the C-terminus, and the Venus fluorophore either replaced the nonfunctional CalX domain (β4ΔCalX, Venus/CFP) or was placed in the CS at position 1450 (β41450,Venus/CFP; Figure 1D). Rationale for Venus fluorophore positioning within the CalX domain of β4 was based on previous findings that deletion of this domain did not alter HD dynamics (Niessen et al., 1997). Consistent with these findings, introduction of CFP and/or Venus at these different positions did not affect the function of β4: the two β4 fusion proteins were efficiently coprecipitated with the ABD of plectin (Supplemental Figure S1A). β4 Venus-CFP fusion proteins were transfected into COS-7 cells, and FRET was measured by recording the fluorescence emission spectra of lysates after excitation of CFP. Differences in emission spectra could not be ascribed to any variations in protein expression, as the expression levels of all β4 Venus-CFP fusion proteins were comparable (Figure 1E and Supplemental Figure S1B). Excitation of CFP in COS-7 cell lysates expressing β4CFP resulted in an emission peak at 475 nm, corresponding to CFP emission (Figure 1F-H). A FRET signal was detected after excitation of CFP in lysates of COS-7 cells expressing β41450, Venus/CFP, as sensitized Venus fluorescence emission at 527 nm was detected in addition to the CFP emission at 475 nm (Figure 1F). Minimal 527-nm signal was detected in the lysates of COS-7 cells expressing β4ΔCalX,Venus/CFP. These results indicate that the C-tail is in close proximity to the CS of β4 but not to the CalX domain. In addition, coexpression of β4CFP and β41450,Venus did not result in a FRET signal, indicating that intramolecular rather than intermolecular FRET is detected. To confirm the existence of intramolecular FRET between the C-tail and the CS of β4 in live cells, we applied fluorescence lifetime imaging microscopy (FLIM) to PA-JEB keratinocytes transiently expressing β41450,Venus/CFP or β4ΔCalX,Venus/CFP. PA-JEB keratinocytes that express β4CFP showed an average lifetime value of 2.44 ± 0.0019 ns. A significant decrease in donor chromophore (CFP) lifetime values, indicative of FRET, occurred in cells expressing β41450,Venus/CFP or β4ΔCalX,Venus/CFP (Supplemental Figure S2). Similar to the results of the FRET determinations in cell lysates, a higher FRET efficiency was observed in cells expressing β41450,Venus/CFP than in β4ΔCalX,Venus/CFP cells. These data suggest that the β4 cytoplasmic domain supports intramolecular association between the C-tail and the CS, not only in cell lysates, but also in living cells.
Substitution of T1727 and T1736 by phosphomimicking aspartic acids does not alter the conformation of the β4 cytoplasmic domain
To investigate whether intramolecular conformation is regulated by phosphorylation, aspartic acids or alanines were introduced into the putative phosphorylation sites T1727 and T1736 in β4 Venus-CFP. Both sets of amino acid substitutions (i.e., T1727A/T1736A and T1727D/T1736D) had no effect on FRET efficiency (Figure 1G). Moreover, no change in FRET signal was detected for β41450,Venus/CFP in lysates from cells pretreated with calyculin A, an inhibitor of protein phosphatases-1 and -2A, and incubated with or without alkaline phosphatase (Figure 1H and Supplemental Figure S3). The same results were obtained in a different cell line, HEK293T, and using FLIM to measure FRET (unpublished observations). Together the data show that residues T1727 and T1736, which prevent interaction of β4 with the plectin plakin domain, do not disrupt the intramolecular interaction between the CS and the fourth FnIII domain/C-tail of β4 integrin.
Integrin β4 is phosphorylated on T1736 by PKD1 in vitro
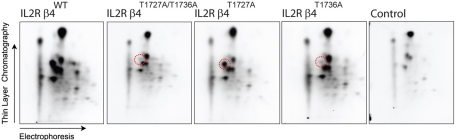
With the knowledge that aspartic acid substitution of the PKD1-consensus phosphorylation sites T1727 and T1736 prevented the interaction between β4 and the plectin plakin domain, we next wanted to determine whether PKD1 could phosphorylate β4. To this end, an in vitro kinase assay was performed in which PKD1 was incubated with either wild-type or mutant β4 proteins, in which one or both threonine residues were replaced by alanine. Subsequent phosphopeptide mapping showed that wild-type β4 is phosphorylated on several residues, whereas substitution of T1736, but not of T1727, resulted in the loss of a single phosphopeptide (Figure 2). This indicates that T1736, but not T1727, is a PKD1 phosphorylation site in vitro.
FIGURE 2:
The integrin β4 subunit is phosphorylated on T1736 by PKD1 in vitro. Phosphopeptide maps of chimeric proteins consisting of the extracellular and transmembrane domains of the IL2R fused to the cytoplasmic domain of β4WT, β4T1727A/T1736A, β4T1727A, and β4T1736A, isolated from transfected COS7 cells and phosphorylated in vitro by PKD1. As a control, precipitates prepared with anti-β4 mAb 450-11A from untransfected COS-7 cells were incubated with PKD1 and analyzed by gel electrophoresis, and an equivalent area of the gel where IL2R/β4 would run was excised and subjected to phosphopeptide analysis. Substitution of T1736 by an alanine resulted in the loss of a single phosphopeptide (encircled in red), indicating that T1736 is a phosphorylation site for PKD1 in vitro.
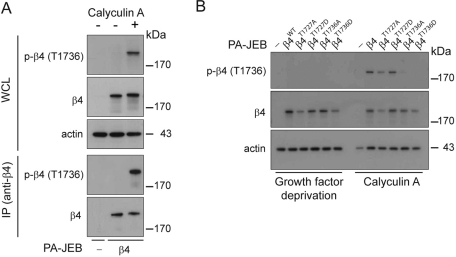
Generation and characterization of phosphorylated T1736-specific antibodies
To evaluate whether T1736 is phosphorylated in keratinocytes, we generated a rabbit polyclonal antibody against a phospho-T1736 peptide and characterized the specificity of the antibody by immunoblot (IB). Lysates from PA-JEB and PA-JEB/β4 keratinocytes treated with or without calyculin A were analyzed by T1736 phosphospecific antibody IBs (Figure 3A, top). Antibody specificity for β4 was determined by IB analysis of β4 IPs (Figure 3, fourth panel) and was confirmed on lysates from PA-JEB keratinocytes expressing either wild-type or mutant β4 subunits. No reaction was obtained with β4 carrying a T1736A or T1736D mutation, whereas a reaction was observed with β4 carrying a T1727A or T1727D mutation (Figure 3B). These results show that our antibody specifically recognizes β4 when phosphorylated on T1736.
FIGURE 3:
The integrin β4 subunit is phosphorylated on T1736 in PA-JEB/β4 keratinocytes. (A) PA-JEB and PA-JEB/β4 keratinocytes were deprived of growth factors overnight and incubated with keratinocyte medium with or without bovine pituitary extract, EGF, and 50 nM calyculin A for 25 min. The cells were lysed, and β4 was immunoprecipitated using mAb 450-11A. Phosphorylation of β4 was detected by immunoblotting using polyclonal antibodies specifically recognizing β4 phosphorylated at T1736. The total levels of β4 were verified using an antibody against β4. (B) PA-JEB keratinocytes reconstituted with wild-type or mutant β4 in which T1727 or T1736 were substituted by an alanine or aspartic acid were deprived of growth factors overnight and incubated with keratinocyte medium with or without bovine pituitary extract, EGF, and 50 nM calyculin A for 25 min. The cells were lysed in 1% NP-40 buffer and analyzed by immunoblotting using an antibody directed against phosphorylated β4 (T1736).
PKD1 mediates PMA-stimulated phosphorylation of T1736 on β4
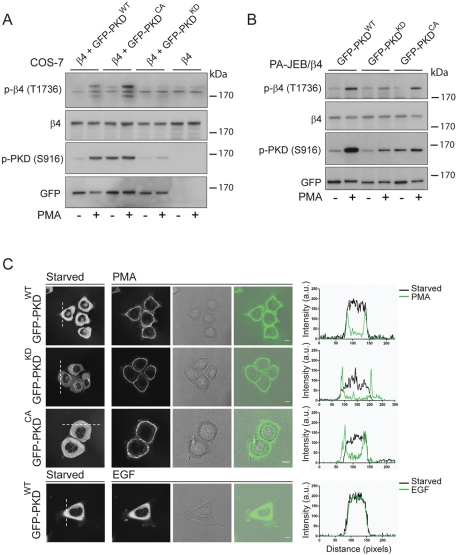
To further address the involvement of PKD1 in the phosphorylation of β4 on T1736, we coexpressed β4 with wild-type, constitutively active (S744E/S748E), or kinase-dead GFP-PKD1 (D733A) fusion proteins. PKD1 is activated by diacylglycerol (DAG) produced by Gβγ and/or growth factor–stimulated activation of phospholipase C (PLC), as well as artificially by phorbol esters that mimic DAG (Van Lint et al., 1995; Matthews et al., 2000). Treatment of transiently transfected cells with PMA resulted in the phosphorylation of β4 on T1736 in cells expressing wild-type or active PKD1 but not kinase-dead PKD1 (Figure 4A). Compared to wild-type PKD1, coexpression of β4 with the active mutant resulted in a more pronounced increase in phosphorylation of β4 on T1736. Of interest, despite the fact that the constitutively active PKD1 is kept in an active conformation, as shown by the phosphorylation of S961, phosphorylation of β4 on T1736 was minimal in the absence of PMA. Next we investigated whether β4 is phosphorylated by PKD1 in keratinocytes. To this end, we generated PA-JEB/β4 keratinocytes stably expressing wild-type, constitutively active, or kinase-dead GFP-PKD1 fusion proteins. Similar to the results obtained in transiently transfected COS-7 cells, PMA-stimulation increased T1736 phosphorylation on β4 in cells that express wild-type or constitutive active but not kinase-dead PKD1 (Figure 4B). Thus, in coexpression experiments using COS-7 cells and PA-JEB keratinocytes, PKD1 mediates the phosphorylation of β4 on T1736 in a PMA-dependent manner.
FIGURE 4:
PMA-induced plasma membrane translocation and activation of PKD1 result in the phosphorylation of β4 on T1736 in transfected COS-7 and PA-JEB keratinocytes. (A) COS-7 cells transiently cotransfected with plasmids expressing β4 and wild-type, GFP-tagged PKD1WT, β4, and constitutively active GFP-PKD1CA or β4 and kinase-dead GFP-PKD1KD, or transfected with the β4 expression plasmid alone were starved overnight and left unstimulated or stimulated with 100 ng/ml PMA for 15 min. Cells were lysed in 1% NP40 lysis buffer supplemented with protease and phosphatase (50 nM calyculin A) inhibitors and analyzed by immunoblotting using antibodies against phosphorylated β4 (T1736). Activation of PKD1 was verified using antibodies specific for phosphorylated PKD1 (S916), whereas the total levels of PKD1 were verified using anti-GFP. (B) PA-JEB/β4 keratinocytes stably expressing wild-type GFP-PKD1WT, kinase-dead GFP-PKD1KD, or constitutively active GFP-PKD1CA were deprived of growth factors and were left unstimulated or were stimulated with 100 ng/ml PMA for 15 min. Cells were lysed and analyzed by immunoblotting with antibodies against phosphorylated β4 (T1736), total β4, phosphorylated PKD1 (S916), or total PKD1 (anti-GFP). (C) The localization of GFP-PKD1WT, GFP-PKD1KD, and GFP-PKD1CA in PA-JEB/β4 keratinocytes that were deprived of growth factors overnight was analyzed using confocal microscopy in a fluorescence, a transmission, and an overlay image. Stimulation with 100 ng/ml PMA induced a rapid translocation of PKD1 from the cytoplasm and TGN to the plasma membrane, whereas stimulation with 100 ng/ml EGF did not. The translocation of PKD1 is visualized by plotting the fluorescent intensity of the dotted white line before (black curve) and after (green curve) stimulation.
PMA-induced plasma membrane translocation of PKD1 is required for phosphorylation of β4
The finding that phosphorylation of β4 by active PKD1 requires PMA stimulation suggests that, in addition to protein kinase C (PKC)–mediated phosphorylation of S744/748 in the activation loop of PKD1, its membrane translocation is also necessary to stimulate phosphorylation of T1736 on β4. To visualize a possible translocation, we performed immunofluorescence analysis of the localization of GFP-PKD1 fusion proteins in PA-JEB/β4 keratinocytes. In unstimulated cells, wild-type and active PKD1 are primarily localized in the cytoplasm, with a small fraction of them being associated with the Golgi apparatus. The kinase-dead mutant, however, was predominantly found at the Golgi apparatus, as judged by immunostaining of the trans-Golgi network (TGN; Supplemental Figure S4A). Stimulation with PMA induced a rapid and near-complete translocation of all three PKD1 fusion proteins to the plasma membrane (Figure 4C; Matthews et al., 1999a, 2000). Quantification of the fluorescence intensity of the cytoplasm over time revealed that the translocation of will-type PKD1 is completed within 2 min after PMA-stimulation (Supplemental Figure S4B). In contrast, EGF was considerably less potent in stimulating the translocation of wild-type PKD1, and only a modest translocation of PKD1 to the plasma membrane was observed (Figure 4C, bottom). Moreover, this translocation was highly transient and seemed to be associated with membrane ruffling induced by EGF. This suggests that EGF receptor (EGFR) signaling in PA-JEB/β4 cells does not lead to a sufficiently strong activation of PLCγ and production of DAG to mediate membrane translocation of PKD1. In conclusion, the phosphorylation of β4 on T1736 requires both the activation of PKD1 and its translocation to plasma membrane bound β4.
EGF-stimulated phosphorylation of T1736 on β4 is not mediated by PKD1
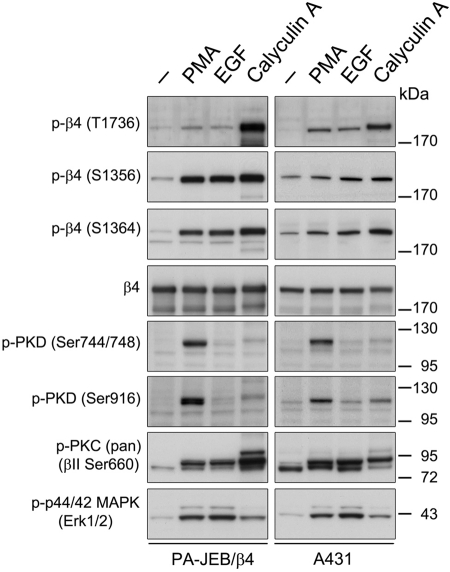
Next we determined whether endogenous PKD1 mediates phosphorylation of β4 in PA-JEB/β4 cells. Cells were stimulated with PMA or EGF, the latter known to activate the PLCγ-DAG-PKC signaling pathway. Analysis of the phosphorylation state of PKD1 showed that only PMA stimulated PKD1 activation (Figure 5). This result is in line with our findings that PMA, but not EGF, induces translocation of PKD1 to the plasma membrane, where it is phosphorylated and activated by PKC. As with EGF-stimulated phosphorylation of T1736, the calyculin A–inhibited dephosphorylation of this residue is not dependent on the activation of PKD1. Thus, although PKD1 might be involved in the phosphorylation of β4 on T1736 after PMA stimulation, another kinase may phosphorylate T1736 in response to EGF and calyculin A. Similar results were obtained using A431 epidermoid carcinoma cells, although PMA and EGF induced a stronger stimulation of phosphorylation on T1736 in these cells than in PA-JEB/β4 keratinocytes (Figure 5). PMA and EGF also induced ERK1/2 activation and β4 phosphorylation on S1356 and S1364 in PA-JEB/β4 keratinocytes, consistent with previous results (Frijns et al., 2010). In contrast, A431 cells showed only a modest increase in β4 phosphorylation at S1356 and S1364 following PMA and EGF stimulation. A431 cells also demonstrate constitutive β4 phosphorylation, which is likely due to the high basal level of EGFR expression and activation in these cells. Thus stimulation of PA-JEB/β4 keratinocytes and A431 epidermoid carcinoma cells with both PMA and EGF induces T1736 phosphorylation, but kinases in addition to PKD1 may be involved.
FIGURE 5:
EGF and PMA stimulate phosphorylation of β4 at T1736 in PA-JEB/β4 keratinocytes and A431 epidermoid carcinoma cells. PA-JEB/β4 (left) and A431 (right) cells were deprived of growth factors overnight and left untreated or were stimulated with 100 ng/ml PMA or 50 ng/ml EGF for 15 min. The cells were lysed in RIPA buffer and analyzed for the phosphorylation of β4 at T1736, S1356, and S1364, PKD1 (S744/748 or S916), p44/p42 (T202/Y204), and PKC (βII S660) and for total levels of β4 using immunoblotting.
The PKD inhibitor kb-NB 142-70 suppresses PMA- and EGF-induced phosphorylation of β4 on T1736
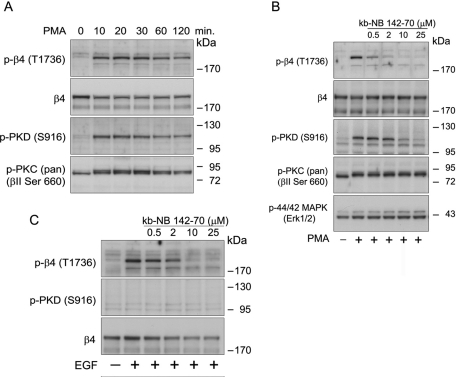
To further investigate the role of PKD1 in PMA-induced phosphorylation of T1736 of β4, we compared the relationship between the activation of PKD1 and the phosphorylation of β4 in A431 cells treated with PMA (Figure 6A). The kinetics of PKD1 (S916) phosphorylation, a measure used to indicate activity, was similar to that of β4 phosphorylation. Maximal phosphorylation of both PKD1 and β4 was observed at 20–30 min and sustained for at least 2 h following stimulation. Next we examined the effect of kb-NB 142-70, a potent and selective PKD inhibitor, on T1736 phosphorylation (LaValle et al., 2010). PMA-induced phosphorylation of β4 was reduced in a dose-dependent manner, and significant inhibition was detectable at 0.5 and 2 μM kb-NB 142-70, concentrations that had no noticeable effect on PKD1 (S916) phosphorylation (Figure 6B). Maximum inhibition of PKD1 and β4 phosphorylation was seen at 25 and 10 μM kb-NB 142-70, respectively. The compound kb-NB 142-70 also inhibited EGF-induced β4 phosphorylation of T1736 only slightly less efficiently than it inhibited PMA-induced phosphorylation of β4 (Figure 6C). This inhibition was unexpected since PKD1 is not obviously activated downstream of EGFR and suggests that kinases other then PKD1 may also be inhibited by the compound. These kinases are likely to be structurally related to PKD1 and to belong to the CAMK family; nonetheless, to further control for off-target effects, the compound was shown to not interfere with the activation and phosphorylation of PKC and ERK1/2 (Figure 6B).
FIGURE 6:
Time course of PMA-induced phosphorylation of β4 at T1736 and inhibition of T1736 phosphorylation by the PKD inhibitor kb-NB 142-70. (A) Time courses of PMA-stimulated phosphorylation of β4 and PKD1 in A431 cells. Growth factor–starved A431 cells were stimulated with PMA for the indicated times. Cell lysates were analyzed by immunoblotting with antibodies specific for phosphorylated β4 (T1736) and PKD1 (S916). (B) Inhibition of PMA-induced β4 phosphorylation at T1736 in A431 cells. A431 cells were pretreated with the indicated concentrations of kb-NB 142-70 for 30 min and then stimulated with 100 ng/ml PMA for 15 min. Cell lysates were analyzed by immunoblotting with antibodies specific for phosphorylated β4 (T1736), PKD (S916), PKC (βII S660), and p44/p42 MAPK. Immunoblotting for total β4 and PKD1 verified that equal amounts of these proteins were evaluated in the A431 lanes. (C) Inhibition of EGF-induced β4 phosphorylation at T1736 in A431 cells. A431 cells were pretreated with the indicated concentrations of kb-NB 142-70 for 30 min and then stimulated with 50 ng/ml EGF for 15 min. Cell lysates were analyzed by immunoblotting with antibodies specific for phosphorylated β4 (T1736), PKD (S916), β4, and PKD1.
Mimicking phosphorylation of T1736 induces HD disassembly
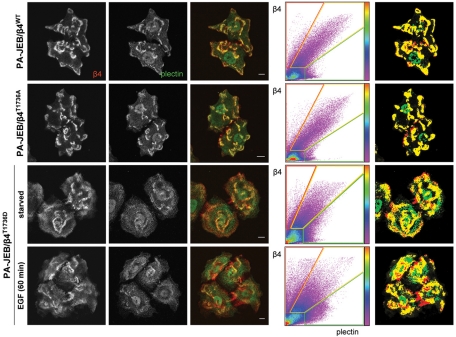
The data thus far demonstrate that phosphorylation of β4 on T1736 prevents the binding of β4 to the plectin plakin domain, suggesting a role of T1736 phosphorylation in the regulation of HD (dis)assembly. To investigate whether this interaction in necessary for HD formation in cells, we performed immunofluorescence microscopy of PA-JEB keratinocytes stably expressing wild-type or mutant β4 constructs in which T1736 was substituted by either aspartic acid or alanine. As shown in Figure 7, all three PA-JEB/β4 cell lines were able to assemble HDs as demonstrated by the colocalization of β4 and plectin. However, when PA-JEB keratinocytes expressing β4T1736D are deprived of growth factors to reduce basal β4 phosphorylation, fewer HDs are present in comparison to PA-JEB keratinocytes expressing wild-type β4 or β4T1736A. A further reduction in the number of HDs in PA-JEB keratinocytes expressing β4T1736D is observed when the cells are also stimulated with EGF, which induces the phosphorylation of β4 on S1356 and S1364 and disrupts the interaction between the ABD of plectin and β4 (Figure 5; Wilhelmsen et al., 2007; Frijns et al., 2010). Together these data show that HD (dis)assembly is a tightly regulated process involving the phosphorylation of several residues on β4, including T1736, S1356, and S1364.
FIGURE 7:
The assembly of HDs in PA-JEB/β4 keratinocytes is prevented by mimicking the phosphorylation of β4 at T1736, which contributes to the EGF-induced HD disassembly. PA-JEB keratinocytes expressing wild-type or mutant β4 in which T1736 is substituted by alanine or aspartic acid were starved overnight, stimulated with or without 50 ng/ml EGF for 60 min, and fixed for immunolabeling of β4 (red) and plectin (green). The degree of colocalization of β4 and plectin is visualized in the overlay image (yellow) and using a scatter plot in which the intensity of β4 (y-axis) and plectin (x-axis) for each pixel is plotted. The color code is a measure for the number of pixels with similar β4/plectin intensity. Right, pixels in which β4 and plectin are strongly colocalized (yellow) and β4 (red) and plectin (green) are not colocalized.
DISCUSSION
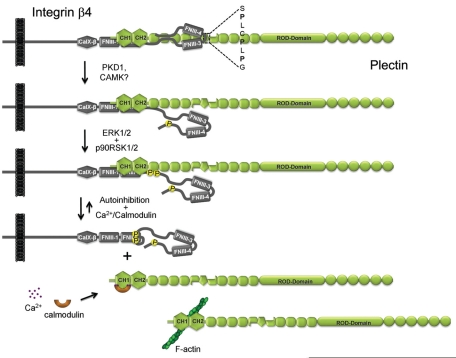
In this study we show that the phosphorylation of T1736 in the C-tail of β4 augments HD disassembly. Aspartic acid mutation of T1736 prevented the interaction between the C-tail of β4 and the plectin plakin domain. We present evidence that β4 can be phosphorylated on T1736 by PKD1, an event that requires both activation and membrane translocation of PKD1 following PMA stimulation in cells. Previous studies showed enhanced HD dynamics and β4 phosphorylation on residues S1356 and S1364 downstream of activated EGFR (Rabinovitz et al., 2004; Wilhelmsen et al., 2007; Frijns et al., 2010). S1356 and S1364 phosphorylation prevents the binding of a region of β4 comprising the first pair of FNIII domains and a small N-terminal fragment of the CS (1115–1355) to plectin-ABD. Here, the T1736D mutation augmented disassembly of HDs when PA-JEB/β4 keratinocytes were stimulated with EGF, suggesting phosphorylation of this particular amino acid in conjunction with the phosphorylation of S1356 and S1364 regulates interaction with plectin through a series of posttranslational modifications.
Two other regions on β4 that mediate binding to the plakin domain of plectin are located in the C-terminal part of the CS (1382–1436) and the C-tail (1667–1752). These two β4 fragments are separated in the primary structure by a stretch of amino acids that forms the second pair of FNIII domains. Using FRET-based assays, we demonstrated that in the tertiary structure these two regions are in close proximity, forming a binding platform for the plakin domain. Three proline residues present in the relatively large segment that connects the third and fourth FNIII domains may be important for conferring a bent conformation on this region of the β4 cytoplasmic domain, thereby bringing the CS (1382–1436) and C-terminal tail (1667–1752) of β4 closely together (Figure 8). Although the two regions are dispensable for the formation of HDs in vitro, their role in stabilizing the binding of β4 to the plectin-ABD is suggested because a nonsense mutation Q1767X (Q1714X in the mature protein) was identified in an EB patient with a mild skin-blistering condition (Nakano et al., 2001). This particular mutation is interesting because it introduces a premature stop codon resulting in the deletion of 39 amino acids from the C-terminus of β4. T1727 and T1736, the threonine residues that modulated interaction with the plakin domain of plectin, are located in this 39–amino acid stretch. However, because the binding site for the ABD of plectin remains, we predict that HDs are still formed with this truncated variant of integrin β4, resulting in only a relatively mild skin-blistering condition.
FIGURE 8.
Model for phosphorylation-induced dissociation of the β4-plectin complex. Phosphorylation of β4 on T1736 by PKD1 or other CAMK-like kinases results in the dissociation of the C-tail and the plakin domain of plectin, whereas phosphorylation of β4 at S1356 and S1364 by ERK1/2 and p90RSK1/2 causes a loss of plectin-ABD binding. Subsequent binding of plectin to F-actin or Ca2+/calmodulin may shift the equilibrium toward complete disassociation of the β4-plectin complex.
The same regions involved in the binding of β4 to the plakin domain have also been identified in yeast two-hybrid and dot blot assays to mediate intramolecular and/or intermolecular interaction (Rezniczek et al., 1998; Koster et al., 2004). Our FRET data are in agreement with a model that predicts these two regions interact in the β4 molecule. However, because no FRET signal was obtained when the Venus and CFP fluorophores were presented in separate β4 molecules (β41450,Venus and β4CFP), we have no evidence yet that these regions are also involved in self-association of β4 molecules. An inherent low affinity between the CS and C-tail regions of β4 and a relatively low protein concentration is likely responsible for the fact that intermolecular interaction could not be demonstrated by FRET in cell lysates. Because the substitution of T1736 by aspartic acid does not affect the intramolecular interaction of β4, the binding site on the C-terminal region for the CS might be different from that for the plakin domain of plectin. However, it should be realized that the results on the intramolecular interaction and the binding of the β4 to the plakin domain to plectin were obtained by different assays and that an effect of the T1736D mutation on the interaction between β4 and the plakin domain of plectin may remain undetected in the FRET-based assay.
In the C-tail of β4, T1736 resides in a highly conserved consensus sequence for PKD1 (L/V-X-R-X-X-S/Tp). PKD1 is a member of the PKD family of Ca2+/calmodulin-dependent kinase (CAMK)–related protein kinases and is activated in response to multiple stimuli, including growth factors, phorbol esters, and G protein–coupled receptors (Rozengurt et al., 2005). Our data show that PKD1 phosphorylates β4 on T1736 in vitro and when it is (over)expressed together with β4 in COS-7 or PA-JEB cells. In both cell types, PMA stimulation was required for the phosphorylation of β4 on T1736 by PKD1. PMA appeared to be essential not only for PKD1 activation, but also for its translocation to the plasma membrane. Thus PKD1 might contribute to HD disassembly at the plasma membrane. This is in agreement with results of previous studies in which PKD1 expression levels were correlated with the proliferation of keratinocytes, a process that requires HD disassembly to occur (Rennecke et al., 1999; Geuijen and Sonnenberg, 2002).
Together the correlation between PKD activation and β4 phosphorylation kinetics and the inhibition of PMA-induced phosphorylation of β4 by the PKD inhibitor kb-NB 142-70 implies that PKD1 is also the kinase that endogenously regulates β4 phosphorylation at T1736. However, the finding that β4 phosphorylation is suppressed already at concentrations that do not inhibit PKD1 activity (as judged by the phosphorylation of PKD1 at S916) suggests that not PKD1 but another kinase phosphorylates β4 at T1736. Moreover, the finding that kb-NB 142-70 inhibited EGF-induced phosphorylation, which is independent of PKD1 activity, indicates that the drug inhibits more targets than previously reported (LaValle et al., 2010). Thus it remains possible that both PMA- and EGF-stimulated β4 phosphorylation at T1736 is mediated by the same kinase and that this kinase could differ from PKD1. Consistent with the fact that PKD1 was not activated downstream of EGFR activation, this kinase was also not or only minimally translocated to the plasma membrane upon EGF stimulation of PA-JEB/β4 keratinocytes.
The substitution of T1736 by a phosphomimicking aspartic acid results in a relatively small effect on the formation and stability of HDs in PA-JEB/β4 keratinocytes. This observation is consistent with previous findings that show HD formation is primarily regulated through the interaction of β4 with the ABD of plectin (Schaapveld et al., 1998; Geerts et al., 1999). Concomitantly, EGF-mediated phosphorylation of S1356 and S1364 on β4 further enhanced the T1736D-mediated dissociation from plectin. Thus we propose a model in which several kinases cooperate to regulate the interaction between the two binding sites of β4 and plectin: phosphorylation of T1736 by PKD1, or another CAMK-like kinase, initiates β4 C-tail dissociation from the plectin plakin domain, and ERK1/2- and p90RSK1/2-mediated phosphorylation of S1356 and S1364, through a proposed mechanism of phosphorylation-dependent auto-inhibition, disrupts the interaction between the first pair of FNIII domains and the plectin ABD (Litjens et al., 2006). Phosphorylation enables dynamic interaction between β4 and plectin; however, for complete disassociation, binding of either β4 or plectin to other molecules may still be required. Indeed, the plectin-ABD can bind directly to F-actin, and binding of β4 and F-actin to plectin was found to be mutually exclusive (Geerts et al., 1999; Garcia-Alvarez et al., 2003). In addition, there is evidence that calmodulin binds to the ABD of plectin in a calcium-dependent manner and inhibits the association of plectin with β4 (Kostan et al., 2009). Thus binding of the plectin-ABD to either F-actin or calmodulin can shift the equilibrium toward a more complete dissociation of the two molecules (Figure 7). Recent data suggest that other mechanisms may also control HD disassembly. For example Germain et al. (2009) showed that a novel β4 phosphorylation site, S1424, regulates HD disassembly in the trailing edge of migrating keratinocytes. In addition, T1727 might be another residue that is involved in the regulation of HDs (this study). Although there is no evidence yet that T1727 is phosphorylated, the finding that a phosphomimetic, but not an unphosphorylatable, amino acid substitution prevented the binding of β4 to the plakin domain of plectin makes this residue an interesting target for further investigation.
MATERIALS AND METHODS
Cell culture
The immortalized PA-JEB keratinocyte cell line derived from a PA-JEB patient was described previously (Schaapveld et al., 1998). These keratinocytes were cultured in serum-free keratinocyte medium (SFM; Invitrogen, Rockville, MD) supplemented with 50 μg/ml bovine pituitary gland extract, 5 ng/ml EGF, 100 U/ml penicillin, and 100 U/ml streptomycin. PA-JEB/β4 and PA-JEB/β4+GFP-PKD1 keratinocytes were generated by retroviral transduction, as described previously (Geuijen and Sonnenberg, 2002). A431, COS-7, and HEK 293 cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 U/ml streptomycin. COS-7 and HEK293 cells were transiently transfected with 10 μg of cDNA, using the DEAE-dextran method or FuGENE transfection agents, respectively (Seed and Aruffo, 1987).
Antibodies
Polyclonal rabbit antibodies specific for β4 phosphorylated at residue T1736 were raised against a synthetic peptide, CTQEFVSRTLTTSGTLSTHM, in which the underlined threonine residue contained a phosphate group. To prevent recognition of unphosphorylated β4 in immunoblotting, the antiserum was used in combination with 30 μM of the synthetic peptide lacking the phosphate group. The generation and specificity of the rabbit polyclonal antibodies that recognize phosphorylated S1356 or S1364 on the integrin β4 subunit have been described previously (Frijns et al., 2010). The rabbit polyclonal antibodies recognizing the first pair of FNIII domains (residues 1115–1355) of β4 were generated as described previously (Wilhelmsen et al., 2007). The rabbit polyclonal antibodies against phosphorylated PKD (S916), PKD (S744/S748), and PKC (βII S660) were purchased from Cell Signaling (Beverly, MA), and the rabbit polyclonal antibody against TGN46 was purchased from Novus Biologicals (Huissen, Netherlands). The mouse monoclonal antibodies (mAbs) against plectin (clone 31) were obtained from BD Biosciences (San Jose, CA), actin (clone C4) from Chemicon International (Temecula, CA), GFP (B34) from Covance (Princeton, NJ), anti-hemagglutinin (HA) epitope 12CA5 from Santa Cruz Biotechnology (Santa Cruz, CA), and β4 (clone 450-11A) from BD Biosciences (San Diego, CA). The secondary antibodies conjugated to Texas red goat anti-mouse immunoglobulin G (IgG) antibody (T-862) was from Invitrogen, and goat anti-rabbit IgG conjugated to Cy5 649 was from Jackson ImmunoResearch Laboratories (West Grove, PA). Secondary antibodies linked to horseradish peroxidase (HRP) were purchased from GE Healthcare UK (Little Chalfont, United Kingdom).
cDNA constructs
The construction of expression vectors encoding full-length β4 and the chimeric protein containing the extracellular and transmembrane domains of the IL2R fused to the intracellular domain of the IL2R/β4cyto has been described (Niessen et al., 1997; Nievers et al., 2000). Point mutants of β4 T1727 and/or T1736 were generated by site-directed mutagenesis with the PCR-based overlap extension method using Pwo DNA polymerase (Roche Molecular Biochemicals, Indianapolis, IN), and fragments containing the different mutations were exchanged with corresponding fragments in the β4 pcDNA3 or IL2R/β4cyto vectors. Retroviral vectors containing mutant β4 cDNAs were generated by subcloning the mutant β4 cDNAs into the EcoRI restriction site of the LZRS-MS-IRES-ZEO vector (Geuijen and Sonnenberg, 2002). The plectin-1C ABD-plakin (1–1154) and plakin domain (284–1154) constructs were cloned into the pcDNA3-HA vector using EcoRI restriction sites (Koster et al., 2004). Wild-type GFP-PKD1, kinase-dead GFP-PKD (D733A) and constitutively active GFP-PKD (S744E/S748E) were cloned in the pEF-plink2-GFPC3 expression vector (Matthews et al., 1999a, 1999b).
The β4 Venus-CFP recombinant fusion constructs used in this study, shown in Figure 1C, were cloned into pcDNA3 (Invitrogen) in five steps. First, the coding region for CFP with a stop codon was placed downstream of a sequence encoding five glycine residues and the complete coding sequence for the β4 subunit, using the PCR-based overlap extension method. Subsequently, the resulting β4-CFP fragment was cloned into the EcoRV/EcoRI restriction sites of pcDNA3. Second, a PCR product containing the Venus (A206K) coding sequence flanked by XhoI restriction sites was generated and ligated into the pGEM-T Easy Vector (Promega, Madison, WI). Third, cDNA fragments of β4 with an XhoI restriction site either at the position of the CalX domain or in the CS at amino acid position 1450 were generated by the PCR-based overlap extension method, using the Pwo DNA polymerase (Roche Molecular Biochemicals). The two β4 PCR fragments were then exchanged with the corresponding fragments in full-length β4 cDNA in pUC18. Fourth, the Venus coding sequence containing the point mutation A206K and flanked by XhoI restriction sites was inserted into full-length β4 using the XhoI restriction sites to generate the β4ΔCalX,Venus and β41450,Venus constructs. Finally, the SfiI fragment of β4-CFP was replaced by the corresponding fragments of β4ΔCalX,Venus or β41450,Venus, yielding β4ΔCalX,Venus/CFP or β41450,Venus/CFP, respectively.
Immunofluorescence
PA-JEB/β4 keratinocytes grown on glass coverslips were deprived of growth factors for 16 h. Cells were then stimulated with 50 ng/ml EGF (Sigma-Aldrich, St. Louis, MO) for 1 h, fixed with 1% paraformaldehyde, and permeabilized with 0.5% Triton X-100. Cells were blocked with PBS containing 2.5% bovine serum albumin (Sigma-Aldrich) for 1 h and incubated with the appropriate primary and secondary antibodies for 45 min each. Coverslips were mounted onto glass slides in Mowiol-DAPCO and visualized using an AOBS confocal microscope (Leica, Mannheim, Germany). Colocalization between β4 and plectin was analyzed in sequentially acquired images using a custom-made Visual Basic (version 6.0) program that depicted results in a scatter plot, as described previously (Wilhelmsen et al., 2007).
PA-JEB/β4 keratinocytes expressing GFP-PKD1 were grown on glass coverslips and deprived of growth factors overnight. Cells were stimulated with 100 ng/ml PMA for 10 min, and slides were prepared and visualized as described. The trans-Golgi apparatus and GFP-PKD1 were visualized by a maximum-intensity projection.
PA-JEB/β4+GFP-PKD1 was grown on glass coverslips and serum starved overnight. Fluorescence and transmission images were taken before and after stimulation with 100 ng/ml PMA or 50 ng/ml EGF. GFP-PKD1 translocation was analyzed using ImageJ (National Institutes of Health, Bethesda, MD).
Immunoblotting and coimmunoprecipitation assays
PA-JEB/β4 and PA-JEB/β4+GFP-PKD1 keratinocytes were starved overnight in growth factor–free keratinocyte-SFM, and COS-7 and A431 cells were starved overnight in DMEM without fetal calf serum (FCS). Cells were left untreated or treated with 50 ng/ml EGF, 100 ng/ml PMA, or 50 nM calyculin A (Cell Signaling, Beverly, MA) for 15 min. In some experiments growth factor–deprived PA-JEB keratinocytes were challenged by replacing the keratinocyte-SFM medium with complete medium containing bovine pituitary extract, EGF, and 50 nM calyculin A and serum-starved COS-7 cells by replacing DMEM with DMEM supplemented with 10% FCS and 50 nM calyculin A. For studies with the PKD inhibitor kb-NB 142-70 (TOCRIS Biosciences, Bristol, United Kingdom), A431 cells were pretreated with the compound for 30 min before they were stimulated with PMA. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with 1.5 mM Na3VO4, 15 mM NaF, 50 nM calyculin A, and protease inhibitor cocktail (Sigma-Aldrich). The cell lysates were cleared by centrifugation at 20,000 × g for 60 min at 4°C. Proteins were separated using 4–12% NuPAGE Novex Bis-Tris gels (Invitrogen) and transferred to Immobilon-P transfer membrane (Millipore, Billerica, MA) for immunoblot analysis.
COS-7 cells cotransfected with the HA-tagged plectin1-1154 and β4 CFP-Venus cDNAs or the HA-tagged plectin254-1154 plectin and IL2R/β4cyto cDNAs were lysed in 1% NP-40 lysis buffer supplemented with protease inhibitors. Cell lysates were cleared by centrifugation and incubated for 4 h with the mouse mAb 12CA5 and subsequently incubated with GammaBind G-Sepharose (Amersham-Pharmacia Biotech, GE Healthcare Bio-Sciences, Piscataway, NJ) to precipitate HA-tagged plectin1-1154. The immunoblots were analyzed with antibodies against integrin β4 and HA and secondary antibodies linked to HRP. Results were visualized by chemiluminescence (GE Healthcare UK).
Fluorescence resonance energy transfer
COS-7 cells transfected with cDNAs encoding β4 CFP-Venus recombinant fusions were serum starved in DMEM overnight and treated or not with 50 nm calyculin A for 25 min. Cells were lysed in 1% NP-40 lysis buffer supplemented with protease inhibitors and cleared. Phosphate groups were removed by incubating cell lysates with alkaline phosphatase (60 U/ml; Roche, Mannheim, Germany) at 37°C for different periods of time. CFP was excited in the whole-cell lysates at 390-nm wavelength, and the emission spectrum was collected between 450 and 600 nm with a 3-nm step size and a 2-s integration time, using the spectrofluorimeter (PTI Quantamaster, MD-5020). The data were normalized for 508 nm after subtraction of the background emission signal.
Frequency-domain FLIM measurements were obtained with Li-FLIM hardware and software (Lambert Instruments, Roden, Netherlands) with a Il18MD MCP and a Vosskühler (CCD-1300D) camera coupled to the microscope (Leica DMIRE2; Leica Microsystems, Heidelberg, Germany) with a 63× objective (numerical aperture 1.3, glycerin). A 1-W, 442-nm LED was modulated at 36 MHz, and emitted light (480 ± 15 nm) was collected from transiently transfected PA-JEB keratinocytes on 24-mm coverslips. The keratinocytes were placed in preheated (37°C) 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–buffered saline (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM glucose, and 10 mM HEPES, pH 7.4). All the experiments were done at 37°C.
Yeast two-hybrid assay
Yeast two-hybrid interaction assays were performed as described previously (Geerts et al., 1999; Koster et al., 2004). Constructs were generated using standard cloning techniques in which plasmid inserts were generated using restriction enzyme digestion or PCR using the Pwo DNA polymerase (Roche Molecular Biochemicals). For the yeast GAL4 BD or GAL4 AD fusion proteins, the pAS2.1 or pACT2 expression vector was used, respectively (Clontech, Mountain View, CA).
Phosphopeptide mapping
From COS-7 cell lysates expressing IL2R/β4, β4 was immunoprecipitated using mAb 450-11A and subsequently incubated in vitro with purified PKD1 (Upstate, Millipore, Billerica, MA) in the presence of 50 μCi of [γ-32P]orthophosphate, 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 20 μg/ml phosphatidyl serine, 8 μg/ml diacylglycerol, and 0.5 mM ethylene glycol tetraacetic acid for 1 h at 30°C. The samples were subjected to SDS–PAGE, and the gel was dried. The film was exposed for 10 min at room temperature. Radioactive β4 was isolated from the gel and digested with trypsin. Phosphopeptide mapping was performed as described previously (van der Geer et al., 1993; Wilhelmsen et al., 2007).
Supplementary Material
Acknowledgments
We thank D. Cantrell for providing the different cDNAs for PKD1. This work was supported by grants from the Dutch Cancer Society and the Netherlands Science Organization (NWO/ALW).
Abbreviations used:
- ABD
actin-binding domain
- CS
connecting segment
- DAG
diacylglycerol
- EGF
epidermal growth factor
- EGFR
EGF receptor
- FLIM
fluorescence lifetime imaging microscopy
- FRET
fluorescence resonance energy transfer
- FNIII
fibronectin type III
- HD
hemidesmosome
- PA-JEB
junctional epidermolysis associated with pyloric atresia
- PKC
protein kinase C
- PKD1
protein kinase D1;
- PLC
phospholipase C
- PMA
phorbol-12-myristate-13-acetate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0957) on February 22, 2012.
REFERENCES
- Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- Borradori L, Sonnenberg A. Structure and function of hemidesmosomes: more than simple adhesion complexes. J Invest Dermatol. 1999;112:411–418. doi: 10.1046/j.1523-1747.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- de Pereda JM, Lillo MP, Sonnenberg A. Structural basis of the interaction between integrin alpha6beta4 and plectin at the hemidesmosomes. EMBO J. 2009;28:1180–1190. doi: 10.1038/emboj.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frijns E, Sachs N, Kreft M, Wilhelmsen K, Sonnenberg A. EGF-induced MAPK signaling inhibits hemidesmosome formation through phosphorylation of the integrin beta4. J Biol Chem. 2010;285:37650–37662. doi: 10.1074/jbc.M110.138818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez B, Bobkov A, Sonnenberg A, de Pereda JM. Structural and functional analysis of the actin binding domain of plectin suggests alternative mechanisms for binding to F-actin and integrin beta4. Structure. 2003;11:615–625. doi: 10.1016/s0969-2126(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Geerts D, Fontao L, Nievers MG, Schaapveld RQ, Purkis PE, Wheeler GN, Lane EB, Leigh IM, Sonnenberg A. Binding of integrin alpha6beta4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J Cell Biol. 1999;147:417–434. doi: 10.1083/jcb.147.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain EC, Santos TM, Rabinovitz I. Phosphorylation of a novel site on the beta4 integrin at the trailing edge of migrating cells promotes hemidesmosome disassembly. Mol Biol Cell. 2009;20:56–67. doi: 10.1091/mbc.E08-06-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuijen CA, Sonnenberg A. Dynamics of the alpha6beta4 integrin in keratinocytes. Mol Biol Cell. 2002;13:3845–3858. doi: 10.1091/mbc.02-01-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Jones JC. Desmosomes and hemidesmosomes: structure and function of molecular components. FASEB J. 1996;10:871–881. doi: 10.1096/fasebj.10.8.8666164. [DOI] [PubMed] [Google Scholar]
- Kostan J, Gregor M, Walko G, Wiche G. Plectin isoform-dependent regulation of keratin-integrin alpha6beta4 anchorage via Ca2+/calmodulin. J Biol Chem. 2009;284:18525–18536. doi: 10.1074/jbc.M109.008474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J, Kuikman I, Kreft M, Sonnenberg A. Two different mutations in the cytoplasmic domain of the integrin beta4 subunit in nonlethal forms of epidermolysis bullosa prevent interaction of beta4 with plectin. J Invest Dermatol. 2001;117:1405–1411. doi: 10.1046/j.0022-202x.2001.01567.x. [DOI] [PubMed] [Google Scholar]
- Koster J, van Wilpe S, Kuikman I, Litjens SH, Sonnenberg A. Role of binding of plectin to the integrin beta4 subunit in the assembly of hemidesmosomes. Mol Biol Cell. 2004;15:1211–1223. doi: 10.1091/mbc.E03-09-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaValle CR, Bravo-Altamirano K, Giridhar KV, Chen J, Sharlow E, Lazo JS, Wipf P, Wang QJ. Novel protein kinase D inhibitors cause potent arrest in prostate cancer cell growth and motility. BMC Chem Biol. 2010;10:5. doi: 10.1186/1472-6769-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens SH, de Pereda JM, Sonnenberg A. Current insights into the formation and breakdown of hemidesmosomes. Trends Cell Biol. 2006;16:376–383. doi: 10.1016/j.tcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Matthews S, Iglesias T, Cantrell D, Rozengurt E. Dynamic re-distribution of protein kinase D (PKD) as revealed by a GFP-PKD fusion protein: dissociation from PKD activation. FEBS Lett. 1999a;457:515–521. doi: 10.1016/s0014-5793(99)01090-x. [DOI] [PubMed] [Google Scholar]
- Matthews SA, Iglesias T, Rozengurt E, Cantrell D. Spatial and temporal regulation of protein kinase D (PKD) EMBO J. 2000;19:2935–2945. doi: 10.1093/emboj/19.12.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews SA, Rozengurt E, Cantrell D. Characterization of serine 916 as an in vivo autophosphorylation site for protein kinase D/protein kinase Cmu. J Biol Chem. 1999b;274:26543–26549. doi: 10.1074/jbc.274.37.26543. [DOI] [PubMed] [Google Scholar]
- Nakano A, Pulkkinen L, Murrell D, Rico J, Lucky AW, Garzon M, Stevens CA, Robertson S, Pfendner E, Uitto J. Epidermolysis bullosa with congenital pyloric atresia: novel mutations in the beta 4 integrin gene (ITGB4) and genotype/phenotype correlations. Pediatr Res. 2001;49:618–626. doi: 10.1203/00006450-200105000-00003. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Hulsman EH, Oomen LC, Kuikman I, Sonnenberg A. A minimal region on the integrin beta4 subunit that is critical to its localization in hemidesmosomes regulates the distribution of HD1/plectin in COS-7 cells. J Cell Sci. 1997;110:1705–1716. doi: 10.1242/jcs.110.15.1705. [DOI] [PubMed] [Google Scholar]
- Nievers MG, Kuikman I, Geerts D, Leigh IM, Sonnenberg A. Formation of hemidesmosome-like structures in the absence of ligand binding by the (alpha)6(beta)4 integrin requires binding of HD1/plectin to the cytoplasmic domain of the (beta)4 integrin subunit. J Cell Sci. 2000;113:963–973. doi: 10.1242/jcs.113.6.963. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos SN, Blaikie P, Yoshioka T, Guo W, Puri C, Tacchetti C, Giancotti FG. Targeted deletion of the integrin beta4 signaling domain suppresses laminin-5-dependent nuclear entry of mitogen-activated protein kinases and NF-kappaB, causing defects in epidermal growth and migration. Mol Cell Biol. 2005;25:6090–6102. doi: 10.1128/MCB.25.14.6090-6102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen L, Uitto J. Mutation analysis and molecular genetics of epidermolysis bullosa. Matrix Biol. 1999;18:29–42. doi: 10.1016/s0945-053x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- Rabinovitz I, Tsomo L, Mercurio AM. Protein kinase C-alpha phosphorylation of specific serines in the connecting segment of the beta 4 integrin regulates the dynamics of type II hemidesmosomes. Mol Cell Biol. 2004;24:4351–4360. doi: 10.1128/MCB.24.10.4351-4360.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennecke J, Rehberger PA, Furstenberger G, Johannes FJ, Stohr M, Marks F, Richter KH. Protein-kinase-Cmu expression correlates with enhanced keratinocyte proliferation in normal and neoplastic mouse epidermis and in cell culture. Int J Cancer. 1999;80:98–103. doi: 10.1002/(sici)1097-0215(19990105)80:1<98::aid-ijc19>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Rezniczek GA, de Pereda JM, Reipert S, Wiche G. Linking integrin alpha6beta4-based cell adhesion to the intermediate filament cytoskeleton: direct interaction between the beta4 subunit and plectin at multiple molecular sites. J Cell Biol. 1998;141:209–225. doi: 10.1083/jcb.141.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozengurt E, Rey O, Waldron RT. Protein kinase D signaling. J Biol Chem. 2005;280:13205–13208. doi: 10.1074/jbc.R500002200. [DOI] [PubMed] [Google Scholar]
- Santoro MM, Gaudino G, Marchisio PC. The MSP receptor regulates alpha6beta4 and alpha3beta1 integrins via 14-3-3 proteins in keratinocyte migration. Dev Cell. 2003;5:257–271. doi: 10.1016/s1534-5807(03)00201-6. [DOI] [PubMed] [Google Scholar]
- Schaapveld RQ, Borradori L, Geerts D, van Leusden MR, Kuikman I, Nievers MG, Niessen CM, Steenbergen RD, Snijders PJ, Sonnenberg A. Hemidesmosome formation is initiated by the beta4 integrin subunit, requires complex formation of beta4 and HD1/plectin, and involves a direct interaction between beta4 and the bullous pemphigoid antigen 180. J Cell Biol. 1998;142:271–284. doi: 10.1083/jcb.142.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed B, Aruffo A. Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc Natl Acad Sci USA. 1987;84:3365–3369. doi: 10.1073/pnas.84.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Rojas AM, de Pereda JM. The structure of a tandem pair of spectrin repeats of plectin reveals a modular organization of the plakin domain. J Mol Biol. 2007;368:1379–1391. doi: 10.1016/j.jmb.2007.02.090. [DOI] [PubMed] [Google Scholar]
- Van der Geer P, Luo K, Sefton BM, Hunter T. 1993. pp. 31–59. Phosphopeptide mapping and phosphoamino acid analysis on cellulose thin-layer plates. In: Protein Phosphorylation; a Practical Approach, ed. DG Hardie, Oxford: IRL Press.
- Van Lint J, Rykx A, Maeda Y, Vantus T, Sturany S, Malhotra V, Vandenheede JR, Seufferlein T. Protein kinase D: an intracellular traffic regulator on the move. Trends Cell Biol. 2002;12:193–200. doi: 10.1016/s0962-8924(02)02262-6. [DOI] [PubMed] [Google Scholar]
- Van Lint JV, Sinnett-Smith J, Rozengurt E. Expression and characterization of PKD, a phorbol ester and diacylglycerol-stimulated serine protein kinase. J Biol Chem. 1995;270:1455–1461. doi: 10.1074/jbc.270.3.1455. [DOI] [PubMed] [Google Scholar]
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Kuikman I, Margadant C, van Rheenen J, Sonnenberg A. Serine phosphorylation of the integrin beta4 subunit is necessary for epidermal growth factor receptor induced hemidesmosome disruption. Mol Biol Cell. 2007;18:3512–3522. doi: 10.1091/mbc.E07-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.