P90 RSK, but not Akt/PKB, facilitates nuclear retention of Chk1 through Chk1–Ser-280 phosphorylation in response to serum stimulation. Chk1–Ser-280 phosphorylation is also elevated in a p90 RSK–dependent manner after UV irradiation and accelerates the Chk1 activation process (Ser-345 and Ser-296 phosphorylation on Chk1) after UV irradiation.
Abstract
The ataxia telangiectasia mutated- and rad3-related kinase (ATR)/Chk1 pathway is a sentinel of cell cycle progression. On the other hand, the Ras/mitogen-activated protein kinase/90-kDa ribosomal S6 kinase (p90 RSK) pathway is a central node in cell signaling downstream of growth factors. These pathways are closely correlated in cell proliferation, but their interaction is largely unknown. Here we show that Chk1 is phosphorylated predominantly at Ser-280 and translocated from cytoplasm to nucleus in response to serum stimulation. Nonphosphorylated Chk1–Ser-280 mutation attenuates nuclear Chk1 accumulation, whereas the phosphomimic mutation has a reverse effect on the localization. Treatment with p90 RSK inhibitor impairs Chk1 phosphorylation at Ser-280 and accumulation at the nucleus after serum stimulation, whereas these two phenomena are induced by the expression of the constitutively active mutant of p90 RSK in serum-starved cells. In vitro analyses indicate that p90 RSK stoichiometrically phosphorylates Ser-280 on Chk1. Together with Chk1 phosphorylation at Ser-345 by ATR and its autophosphorylation at Ser-296, which are critical for checkpoint signaling, Chk1–Ser-280 phosphorylation is elevated in a p90 RSK–dependent manner after UV irradiation. In addition, Chk1 phosphorylation at Ser-345 and Ser-296 after UV irradiation is also attenuated by the treatment with p90 RSK inhibitor or by Ser-280 mutation to Ala. These results suggest that p90 RSK facilitates nuclear Chk1 accumulation through Chk1–Ser-280 phosphorylation and that this pathway plays an important role in the preparation for monitoring genetic stability during cell proliferation.
INTRODUCTION
Cell proliferation requires timely signals from extracellular growth factors. Two core-signaling pathways exist downstream of receptor tyrosine kinases (RTKs). One is a pathway from Ras to the mitogen-activated protein kinase (MAPK) cascade, consisting of Raf–MAPK kinase (MEK) 1/2–extracellular signal-regulated kinase (ERK) 1/2 (Lewis et al., 1998; Nishimoto and Nishida, 2006; Cargnello and Roux, 2011). The 90-kDa ribosomal S6 kinase (p90 RSK; also called MAPK-activated protein kinase-1) is a Ser/Thr kinase that lies downstream of the Ras-MAPK pathway. Following the stimulation of cells with growth factors, p90 RSK is phosphorylated at multiple residues by several kinases and then activated; these phosphorylation events are triggered by ERK1/2-induced phosphorylation of Thr-573 in the C-terminal kinase domain of p90 RSK (Anjum and Blenis, 2008; Pearce et al., 2010). The other is a pathway from phosphatidylinositol 3-kinase (PI3-K) to Akt/protein kinase B (PKB). PI3-K is activated downstream of RTKs and then synthesizes phosphatidylinositol(3,4,5)phosphate (PIP3). Akt/PKB activation is triggered by recruitment to the plasma membrane through direct interaction of its pleckstrin homology (PH) domain with PIP3, which induces Akt/PKB phosphorylation at Thr-308 and Thr-473, critical sites for its kinase activation (Vivanco and Sawyers, 2002; Engelman, 2009; Pearce et al., 2010; Wong et al., 2010). PTEN, a potent tumor suppressor, antagonizes PI3-K–Akt/PKB function through PIP3 dephosphorylation (Vivanco and Sawyers, 2002; Engelman, 2009; Wong et al., 2010). Ras-MAPK and PI3-K–Akt pathways were reported to be up-regulated in a wide spectrum of human cancers through mutations in or deregulation of their components (Vivanco and Sawyers, 2002; Anjum and Blenis, 2008; Engelman, 2009; Wong et al., 2010; Cargnello and Roux, 2011).
Such oncogenic changes often accompany stalled DNA replication and DNA damage, which activates DNA replication/damage checkpoints (Bartek et al., 2007; Halazonetis et al., 2008; Jackson and Bartek, 2009). The checkpoint activation facilitates the elimination of transformed cells from the proliferation cell pool through the induction of cellular senescence or death, which works as a carcinogenesis barrier (Bartek et al., 2007; Jackson and Bartek, 2009; Ciccia and Elledge, 2010; Ma et al., 2011). In the center of checkpoint signaling pathways, there exists a protein kinase cascade from ataxia telangiectasia mutated- and rad3-related kinase (ATR) to Chk1 (McGowan and Russell, 2004; Reinhardt and Yaffe, 2009). ATR is activated in response to stalled DNA replication or damaged DNA induced by genotoxic stimuli such as UV, ionizing radiation (IR), and DNA-damaging agents (Cimprich and Cortez, 2008; Flynn and Zou, 2011). The activated ATR phosphorylates Chk1 at Ser-317 and Ser-345 (Zhao and Piwnica-Worms, 2001; Polo and Jackson, 2011), which then induces functionally essential Chk1–Ser-296 autophosphorylation (Kasahara et al., 2010). A series of Chk1 phosphorylation events is indispensable for cell cycle arrest (Niida and Nakanishi, 2006; Kasahara et al., 2010), which provides time to repair damaged DNA lesions (Branzei and Foiani, 2008).
Several groups reported that the PI3-K–Akt/PKB pathway overrides DNA damage–induced G2 arrest (Henry et al., 2001; Shtivelman et al., 2002; Nimbalkar and Quelle, 2008; Xu et al., 2010). Chk1 had been considered to be a likely candidate of Akt/PKB substrate for the suppression of G2/M checkpoint. Akt/PKB was reported to induce Chk1 phosphorylation at Ser-280 (Shtivelman et al., 2002; King et al., 2004) and to reduce nuclear localization of Chk1 (Puc et al., 2005). However, recent studies revealed that Chk1–Ser-280 mutants behaved like Chk1 wild type (WT) in the G2/M checkpoint (Tonic et al., 2010; Xu et al., 2010). Thus the role of Chk1–Ser-280 phosphorylation remains controversial. Here we show that p90 RSK, but not Akt/PKB, facilitates nuclear retention of Chk1 through Chk1–Ser-280 phosphorylation in response to serum stimulation. Chk1–Ser-280 phosphorylation is also elevated in a p90 RSK–dependent manner after UV irradiation and accelerates the Chk1 activation process (Ser-345 and Ser-296 phosphorylation on Chk1) after UV irradiation.
RESULTS
Chk1 is phosphorylated at Ser-280 and translocated from cytoplasm to nucleus in response to serum stimulation
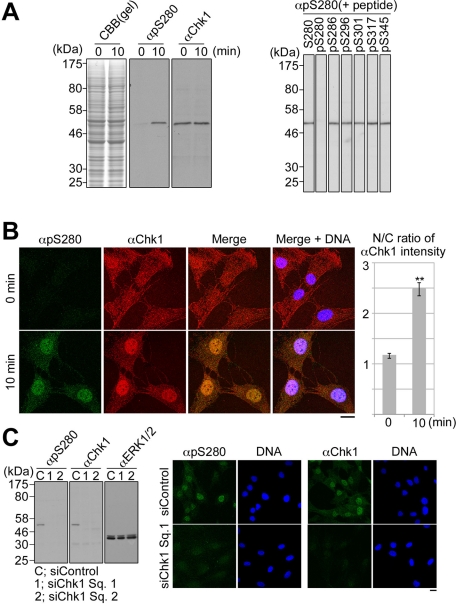
To analyze Chk1–Ser-280 phosphorylation in cells, we first characterized anti–phospho-Ser-280 on Chk1 (αpS280). As shown in Figure 1A, αpS280 specifically immunoreacted with a ∼54-kDa band corresponding to Chk1 in the lysate of h-TERT-immortalized retinal pigment epithelia (RPE1) cells stimulated with serum (in which growth factors are rich) for 10 min. This immunoreactivity was impaired specifically by preincubation with a phosphopeptide pS280 corresponding to Ser-280–phosphorylated Chk1 but not with nonphosphorylated peptide S280 and phosphopeptides for other sites within Chk1 (Figure 1A, right). Following the stimulation of cells with serum, αpS280-immunocytochemical signals emerged mainly in the nucleus and colocalized with αChk1 signals (Figure 1B). As shown in Figure 1C, Chk1 depletion by Chk1-specific small interfering RNA (siRNA) reduced αpS280-immunoreactive signals not only in the immunoblotting, but also in the immunocytochemistry.
FIGURE 1:
The specificity of anti–phospho-Ser-280 on Chk1 (αpS280) or anti-Chk1 (αChk1). (A) After 48 h of serum starvation, RPE1 cells were incubated with the fresh growing medium for 0 or 10 min as described in Materials and Methods. Specificity of each antibody was analyzed by immunoblotting. Amounts of loading cell lysates were determined by staining of SDS–PAGE gel with Coomassie brilliant blue (CBB; left). In the competition assay, αpS280 was preincubated with 50 ng/ml nonphosphopeptide S280 or each phosphopeptide (pS), and then the serum-stimulated cell lysate was immunoblotted (right). (B) Cells were stained with αpS280 (green), αChk1 (red), and 4′,6-diamidino-2-phenylindole (DAPI; blue; left). The N/C ratio of αChk1 intensity is shown. Data represent mean ± SEM for at least 20 cells in each cell group, **p < 0.01 using Student's t test (right). (C) RPE1 cells were transfected with the mixture of the indicated siRNA and Lipofectamine RNAiMAX reagent according to the reverse transfection procedures (Invitrogen). At 16 h after the transfection, the cells were cultured in the serum-free medium for 48 h and then incubated in the growing medium for 10 min. Similar diminishment of the antibody signals in the immunocytochemistry was observed in cells transfected with siChk1 Sq. 2 (unpublished data). Scale bar, 10 μm (B, C).
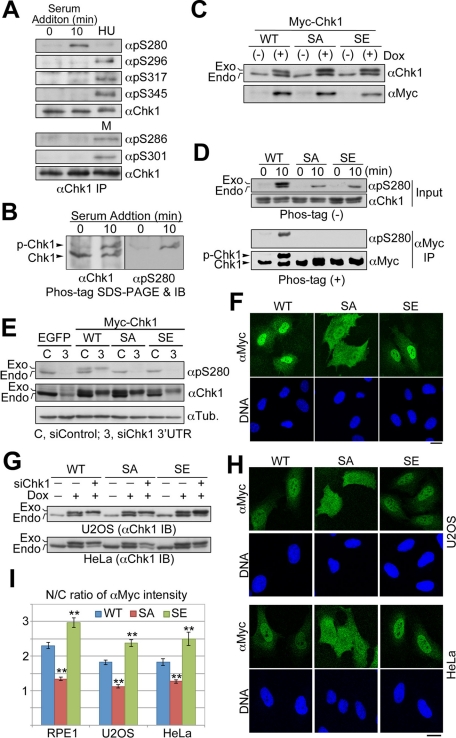
In response to serum stimulation, Chk1 was phosphorylated at Ser-280 but not at Ser-296 (a Chk1 site; Kasahara et al., 2010), at Ser-317 and Ser-345 (ATR sites; Zhao and Piwnica-Worms, 2001), or at Ser-286 and Ser-301 (Cdk1 sites; Shiromizu et al., 2006; Figure 2A). For the estimation of the extent of Chk1 phosphorylation in cells, the αChk1 immunoprecipitates were subjected to Mn2+-Phos-tag SDS–PAGE (Kinoshita et al., 2006; Kinoshita-Kikuta et al., 2007) and then analyzed by immunoblotting. Owing to the interaction of a phosphate group with Mn2+-Phos-tag–modified polyacrylamide, phosphorylated Chk1 (p-Chk1) migrated more slowly than Chk1 without phosphorylation; about half of Chk1 molecules were estimated to be phosphorylated in cells stimulated by serum for 10 min (Figure 2B). To confirm that Ser-280 is only one major phosphorylation site after serum stimulation, we mutated Chk1–Ser-280 to Ala (SA) or Glu (SE) and then established Tet-On RPE1 cells in which each Myc-tagged Chk1 is expressed in a doxycycline (Dox)-dependent manner (Figure 2C). As shown in Figure 2D, the mobility shift in Mn2+-Phos-tag–modified polyacrylamide was completely diminished by Chk1 mutation at Ser-280 (Figure 2D). These results suggested that Chk1 is phosphorylated predominantly at Ser-280 after serum stimulation.
FIGURE 2:
Chk1 is phosphorylated specifically at Ser-280 in response to serum stimulation. (A, B) Endogenous Chk1 was immunoprecipitated from cells stimulated by 10% serum for 0 or 10 min, HU-treated or mitotic (M) cells. Each immunoprecipitate was subjected to the SDS–PAGE with (B) or without (A) Mn2+-Phos-tag, followed by immunoblotting with the indicated antibody. (C) Establishment of each Tet-On RPE1 cell line. Cells were treated with (+) or without (−) 2 ng/ml doxycycline (Dox) for 48 h. SA or SE indicates Myc-tagged Chk1 mutated at Ser-280 to Ala or Glu, respectively. (D) Tet-On RPE1 cell line was cultured in the serum-free medium containing 5 ng/ml Dox for 48 h. After serum starvation, cells were incubated in the growing medium for 0 or 10 min. After treatment, cells were subjected to αMyc immunoprecipitation. The immunoprecipitate (αMyc IP) or a fraction of each cell extract (Input) was subjected to the SDS–PAGE with (+) or without (−) Mn2+-Phos-tag, followed by immunoblotting, respectively. (E–I) Each Tet-On cell line was transfected with control or Chk1 3′ UTR siRNA according to the forward transfection procedures (Invitrogen). At 4 h after transfection, the medium was replaced with the fresh growing medium containing Dox. At 24 h after transfection, cells were analyzed by immunoblotting (E, G) or immunocytochemistry (F, H, I). In E, we used Tet-On RPE1 cell line expressing EGFP as a negative control. In G, each Tet-On cell line was also incubated with or without Dox for 24 h in order to evaluate inducible expression of each Myc-Chk1. The N/C ratio of αMyc intensity is shown. Data represent mean ± SEM for at least 20 cells in each cell group, **p < 0.01 vs. WT-replacing cells (I). Similar results were obtained using another Chk1 3′UTR sequence (unpublished data). Scale bar, 10 μm (F, H).
In RPE1 Tet-On cell lines, endogenous Chk1 was replaced with exogenous Chk1 mutant under the cultivation with the growing medium by the induction of Myc-tagged Chk1 in combination with RNA interference–mediated depletion of endogenous Chk1 (Figure 2E). Compared with WT protein, a nonphosphorylated mutant of Ser-280 (SA) failed to localize to the nucleus, although a phosphomimic mutant (SE) had a reverse effect on the localization (Figure 2, F and I). Similar results were obtained using other Tet-On cell lines (U2OS and HeLa; Figure 2, G–I). These results suggest that nuclear accumulation of Chk1 is mediated through Chk1–Ser-280 phosphorylation after serum stimulation.
MAPK cascade–p90 RSK pathway controls Chk1–Ser-280 phosphorylation and nuclear Chk1 accumulation after serum stimulation
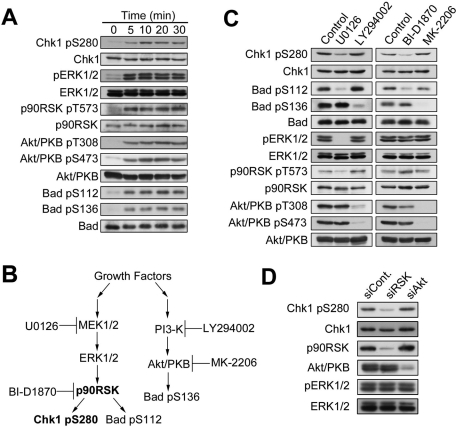
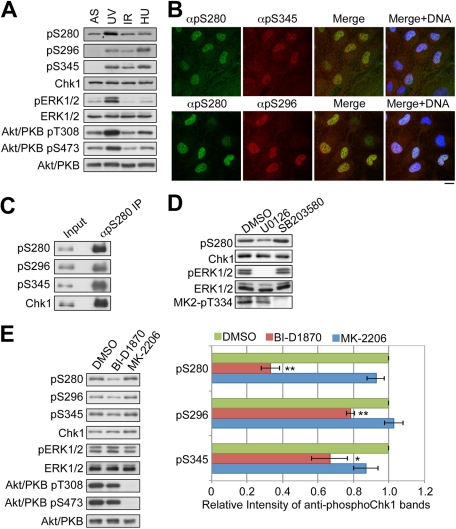
The time-course experiment revealed that the level of Chk1–Ser-280 phosphorylation was elevated in a time-dependent manner, peaked around 10 min after serum stimulation, and was then maintained thereafter (Figure 3A). Similarly, we observed the elevation in the level of ERK1/2 phosphorylated at Thr-202 and Tyr-204 (pERK1/2), p90 RSK phosphorylated at Thr-573, Akt/PKB phosphorylated at Thr-308 and at Ser-473 (also see Introduction), and Bad phosphorylated at Ser112 by p90 RSK (Bonni et al., 1999; Tan et al., 1999) and at Ser-136 by Akt/PKB (Mabuchi et al., 2002; Tran et al., 2002). This suggested that both the MAPK cascade–p90 RSK and PI3-KxAkt/PKB pathways were activated in RPE1 cells after serum stimulation (Figure 3A). To examine which pathway participates in serum-induced Chk1xSer-280 phosphorylation, we used U0126 (MEK1/2 inhibitor), BI-D1870 (p90 RSK inhibitor; Sapkota et al., 2007), LY294002 (PI3-K inhibitor), or MK-2206 (an allosteric inhibitor that blocks Akt/PKB activation process through the inhibition of the binding between PIP3 and Akt/PKB-PH domain; Lindsley, 2010; Meuillet et al., 2010). As shown in Figure 3, B and C, U0126 specifically inhibited the MAPK cascade–p90 RSK pathway from ERK1/2 phosphorylation (pERK1/2) to Bad–Ser-112 phosphorylation by p90 RSK. BI-D1870 specifically decreased the level of Bad–Ser-112 phosphorylation, suggesting successful inhibition of p90 RSK. On the other hand, LY294002 or MK-2206 specifically inhibited Akt/PKB activation pathway, as judged by specific reduction of Akt–Thr-308/Ser-473 phosphorylation and Bad–Ser-136 phosphorylation. Under these conditions, U0126 or BI-D1870 inhibited Chk1–Ser-280 phosphorylation, although LY294002 or MK-2206 had no significant effects. As shown in Figure 3D, the depletion of p90 RSK 1/2/3, but not of Akt1/2, by transfection with specific siRNAs decreased the level of Chk1 phosphorylation at Ser-280.
FIGURE 3:
Chk1–Ser-280 is phosphorylated by p90 RSK in response to serum stimulation. (A) After 48 h of serum starvation, RPE1 cells were incubated with the growing medium for the indicated time. (B) Schema shows a kinase that each chemical agent inhibits in MAP kinase and PI3-K–Akt/PKB pathways. (C) RPE1 cells were treated with 10 μM U0126, 10 μM LY294002, 10 μM BI-D1870, 1 μM MK-2206, or equal volume of DMSO (control) as described in Materials and Methods. At 5 min after serum addition, cells were analyzed by immunoblotting. (D) HeLa cells were transfected with p90 RSK1/2/3 or Akt1/2 (5 nM of Dharmacon ON-TARGETplus SMARTpool per each protein) according to the reverse transfection procedures (Invitrogen). As a negative control, we used 15 nM of Dharmacon ON-TARGETplus siCONTROL. At 16 h after transfection, cells were cultured in DMEM medium containing 0.5% FBS for 48 h and then incubated in the medium containing 10% FBS for 5 min.
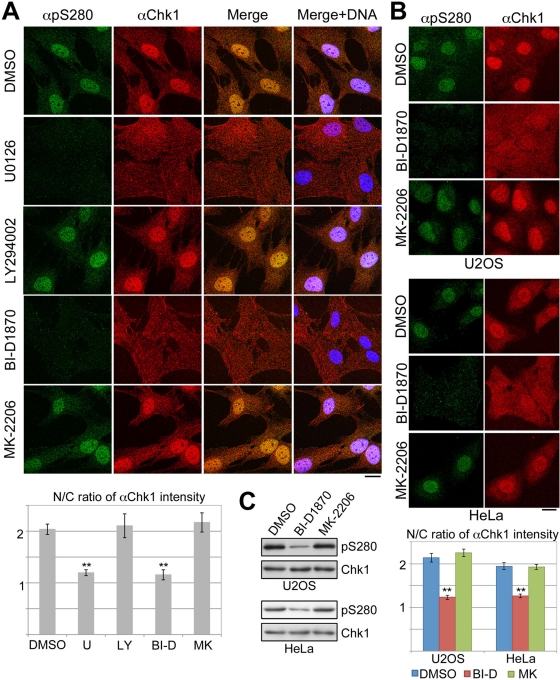
We next examined the effects of the foregoing inhibitors on Chk1 phosphorylation and localization. U0126 or BI-D1870, but not LY294002 or MK-2206, inhibited Chk1–Ser-280 phosphorylation and nuclear accumulation of Chk1 after serum stimulation in RPE1 cells (Figure 4A). In U2OS and HeLa cells, the treatment with BI-D1870 also reduced Chk1–Ser-280 phosphorylation and attenuated nuclear Chk1 accumulation, whereas the treatment with MK-2206 had almost no effect (Figure 4, B and C). All these results suggest that p90 RSK regulates both Chk1–Ser-280 phosphorylation and Chk1 translocation to the nucleus.
FIGURE 4:
Chk1 is translocated from cytoplasm to nucleus in a p90 RSK–dependent manner. RPE1 (A), U2OS, or HeLa (B, C) cells were treated with each chemical agent as described in Materials and Methods. At 5 (A) or 10 (B) min after serum addition, cells were stained with αpS280 (green), αChk1 (red), and DAPI (blue). In C, treated cells were also analyzed by immunoblotting. The N/C ratio of αChk1 intensity is also shown. Data represent mean ± SEM for at least 20 cells in each cell group, **p < 0.01 vs. cells treated with DMSO (A, B). Scale bar, 10 μm (A, B).
P90 RSK directly phosphorylates Ser-280 on Chk1
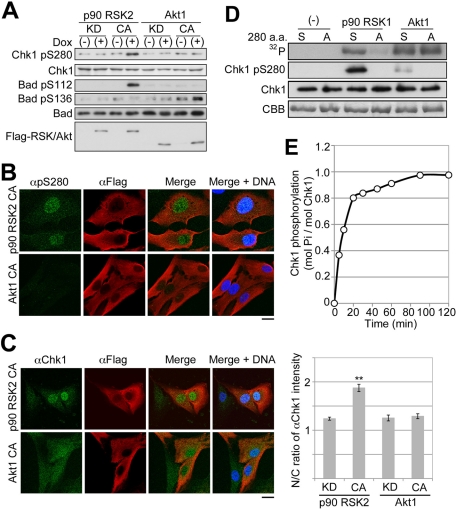
Using each Tet-On RPE1 cell expressing a constitutively active (CA) or kinase-dead (KD) mutant of p90 RSK2 or Akt1 in a Dox-dependent manner, we examined the effect of each mutant expression under the serum-starved condition (Figure 5, A–C, and Supplemental Figure S1). Each CA mutant remained active in cells without serum stimulation because the induction of p90 RSK2 CA or Akt1 CA enhanced Bad phosphorylation at Ser-112 or Ser-136, respectively (Figure 5A). The expression of p90 RSK CA mutant but not of Akt1 CA induced Chk1 phosphorylation at Ser-280 (Figure 5, A and B) and nuclear Chk1 accumulation (Figure 5C). Since these Chk1 phenomena were not observed in the case of KD induction (Figure 5A and Supplemental Figure S1), p90 RSK catalytic activity was required for these phenomena in the cells.
FIGURE 5:
P90 RSK phosphorylates Ser-280 on Chk1. (A–C) Tet-On RPE1 cell line was cultured in the serum-free medium for 48 h. After treatment, cells were cultured in serum-free medium with (+) or without (−) Dox for 6 h. Cells were analyzed by immunoblotting (A) or immunocytochemistry (B, C). CA or KD indicates a constitutively active or kinase-dead mutant, respectively. The N/C ratio of αChk1 intensity is shown. Data represent mean ± SEM for at least 20 cells in each cell group, **p < 0.01 vs. RSK2 KD-expressing cells (C; also see Supplemental Figure S1). Bar, 10 μm (B, C). (D) Purified Chk1 mutant KD (S) or KD/S280A (A) was incubated with or without p90 RSK1 or Akt1 for 20 min as described in Materials and Methods. The reaction mixture was analyzed by SDS–PAGE or immunoblotting with αpS280 (Chk1–pSer-280) or αChk1 (Chk1). After SDS–PAGE, bands of Chk1 or radioactive Chk1 were visualized by staining with CBB or autoradiography (32P), respectively. (E) Time course of Chk1 KD phosphorylation by p90 RSK1.
Next we performed in vitro kinase assays using purified proteins. As shown in Figure 5D, p90 RSK1 and Akt1 can phosphorylate Chk1 to a similar extent in vitro. However, Ser-280 mutation to Ala diminished Chk1 phosphorylation by p90 RSK1 but not by Akt1. The immunoblotting with αpS280 also revealed that p90 RSK1 phosphorylates Ser-280 on Chk1 more preferably than Akt1 (Figure 5D). The level of Chk1 phosphorylation by p90 RSK increased rapidly until 20 min and reached ∼1 mol of phosphate/mol of protein (Figure 5E). These results indicate the possibility that p90 RSK governs serum-induced Chk1–Ser-280 phosphorylation likely through direct enzyme–substrate reaction.
Ser-280 phosphorylation on Chk1 by p90 RSK promotes Chk1 activation processes after UV irradiation
To elucidate the role of Chk1–Ser-280 phosphorylation, we first performed the in vitro kinase assays using immunoprecipitates of Myc-Chk1 before or after serum stimulation. As shown in Supplemental Figure S2, we observed only marginal change in the catalytic activity of Chk1 WT, although we detected Ser-280 phosphorylation on WT protein after serum stimulation. In addition, Ser-280 mutation including phosphomimic mutation did not affect the catalytic activity (Supplemental Figure S2). Thus, unlike Ser-345 phosphorylation (Zhao and Piwnica-Worms, 2001; Walker et al., 2009), Ser-280 phosphorylation has little impact on Chk1 catalytic activity.
Next we examined the relationship with the DNA damage or replication checkpoint. Compared with nontreated (asynchronous [AS]) cells, the level of Chk1–Ser-280 phosphorylation is significantly elevated in cells irradiated with UV light (Figure 6A). However, IR or hydroxyurea (HU) treatment induced only marginal change in the level of Chk1–Ser-280 phosphorylation, although Chk1 was phosphorylated at Ser-296 and Ser-345 (sites required for the checkpoint activation; see Introduction) in response to these stimuli (Figure 6A). After UV irradiation, high-level Chk1–Ser-280 phosphorylation was observed in the cells in which Chk1 was phosphorylated at Ser-345 or Ser-296 (Figure 6B). As shown in Figure 6C, Ser-345– or Ser-296xphosphorylated Chk1 was highly enriched in immunoprecipitates of Ser-280–phosphorylated Chk1 from UV-irradiated cells. These results suggested the correlation between Ser-280 and Ser-296/Ser-345 phosphorylation on Chk1 after UV irradiation.
FIGURE 6:
UV irradiation elevates Chk1–Ser-280 phosphorylation level in a p90 RSK–dependent manner. (A–C) RPE1 cells were treated with each genotoxic stimulus as follows. For UV irradiation (A–C), the culture medium was removed, and cells were irradiated in uncovered tissue culture dishes with 254-nm UV light at a dose of 20 J/m2 (FUNA-UV-LINKER; Funakoshi, Tokyo, Japan). After the growing medium was added back, cells were cultured for 30 min. For IR (A), cells were treated with 20 Gy of x-rays and then cultured for 30 min. For HU treatment (A), cells were treated as described in Materials and Methods. These treated cells were subjected to immunoblotting (A), immunocytochemistry (B), or immunoprecipitation (C). Bar, 10 μm (B). (D, E) RPE1 cells were pretreated with 10 μM U0126, 20 μM SB203580 (D), 10 μM BI-D1870, 1 μM MK-2206 (E), or an equal volume of DMSO (Control) for 30 min. After UV irradiation, cells were incubated in the growing medium containing the same chemical agent for 20 min. The phosphorylation of MK2 (D) or Akt/PKB (E) was also monitored as a control for SB203580 or MK-2206, respectively. Amounts of Chk1 phosphorylated at Ser-280, Ser-296, and Ser-345 were quantified using densitometry, normalized to the content of Chk1, and presented as fold of treatment with DMSO. Data represent mean ± SEM of three independent experiments, *p < 0.05 and **p < 0.01 vs. the treatment with DMSO (E).
Both MAPK cascade–p90 RSK and PI3-K–Akt/PKB pathways were activated in RPE1 cells after UV irradiation (Figure 6A). In addition, p38 MAPK is known to be activated after UV irradiation (Cargnello and Roux, 2011). Thus we examined the effect of U0126 (MEK1/2 inhibitor), BI-D1870 (p90 RSK inhibitor), MK-2206 (Akt/PKB inhibitor), or SB203580 (p38 inhibitor) on Chk1–Ser-280 phosphorylation after UV irradiation. As shown in Figure 6, D and E, Chk1–Ser-280 phosphorylation was attenuated by the treatment with U0126 or BI-D1870. On the other hand, the treatment with SB203580 (Figure 6D) or MK-2206 (Figure 6E) had little effect. Thus MAPK cascade–p90 RSK controls Chk1–Ser-280 phosphorylation after UV irradiation.
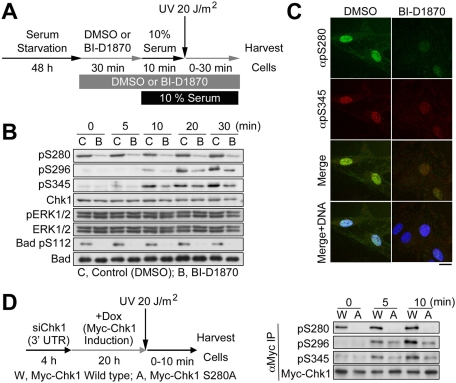
UV-induced Ser-296 and Ser-345 phosphorylations on Chk1 were moderately but significantly reduced by BI-D1870 under cultivation with the growing medium (Figure 6E). To examine the effect of Chk1–Ser-280 phosphorylation on Chk1 activation processes more clearly, we performed UV-irradiation experiments after serum stimulation (Figure 7A). At 48 h after serum starvation, BI-D1870 or DMSO (control) was added in the serum-free medium, and the cells were incubated for 30 min. Then the serum-free medium was changed to the growing medium containing the same chemical. 10 min after serum stimulation, and the cells were irradiated with UV light. As shown in Figure 7, B and C, the p90 RSK inhibitor reduced Chk1 phosphorylation not only at Ser-280, but also at Ser-296 and Ser-345. In addition, Ser-296 or Ser-345 phosphorylation after UV irradiation was reduced in RPE1 cells where Myc-Chk1 S280A replaces endogenous Chk1 compared with WT-replacing cells (Figure 7D). All these results suggested that p90 RSK modulates Chk1 activation processes through Chk1–Ser-280 phosphorylation after UV irradiation.
FIGURE 7:
P90 RSK facilitates Chk1 phosphorylation not only at Ser-280 but also at Ser-296 by Chk1 and at Ser-345 by ATR after UV irradiation. (A–C) Serum starvation, agent treatment, and serum stimulation were performed as described in Materials and Methods. At 10 min after serum addition, cells were irradiated with UV and then incubated at an indicated time (B) or 30 min (C). Experimental procedures are summarized in a schema (A). Scale bar, 10 μm (C). (D) Each Tet-On RPE1 cell line was transfected with Chk1 3′UTR siRNA according to the reverse transfection procedures (Invitrogen). At 4 h after transfection, the medium was replaced with the fresh growing medium containing Dox. At 24 h after transfection, cells were irradiated with UV light at a dose of 20 J/m2 and then incubated at an indicated time. After treatment, cells were subjected to anti-Myc immunoprecipitation (IP), followed by immunoblotting with the indicated antibody. Experimental procedures are also summarized in a schema (left).
DISCUSSION
It has long been considered that Akt/PKB directly phosphorylates Chk1 at Ser-280 for the following reasons. The minimum consensus phosphorylation motif of Akt/PKB is Arg-X-Arg-X-X-pSer/Thr (Manning and Cantley, 2007), which is completely matched with the amino acid sequence around Ser-280 on Chk1. Akt/PKB phosphorylated Ser-280 on glutathione S-transferase (GST)–Chk1 peptide (containing 39 amino acids between residues 262 and 290) in vitro (Shtivelman et al., 2002). PI3-K inhibitors (wortmannin and LY294002) attenuated insulin like growth factor-1–induced Chk1–Ser-280 phosphorylation in cells (Puc et al., 2005).
However, the consensus sequence of Akt/PKB is also shared by other basophilic kinases, such as p90 RSK (Anjum and Blenis, 2008). Our in vitro analysis also reveals that Akt/PKB phosphorylates the full length of Chk1 at several sites: Ser-280 is only a minor phosphorylation site (Figure 5D). On the other hand, p90 RSK phosphorylates Chk1 predominantly at Ser-280 (Figure 5, D and E), which is consistent with the in vivo phenomena occurring after serum stimulation (Figure 2, A–D). Puc et al. (2005) reported that PI3-K–Akt/PKB pathway regulated Chk1–Ser-280 phosphorylation. However, PI3-K inhibitors (wortmannin and LY294002) also inhibited MAPK cascade under their conditions. In our experimental conditions, the inhibitors used did not show apparent cross-inhibition between MAPK cascade–p90RSK and PI3-K–Akt/PKB pathways. Our pharmacological experiments show strong dependence of Chk1–Ser-280 phosphorylation on the activity of p90 RSK but not of Akt/PKB (Figures 3C and 4C). Taking this together with the data on knockdown through siRNAs (Figure 3D) and gain of function using each kinase mutant (Figure 5, A and B), we propose that p90 RSK but not Akt/PKB is responsible for Chk1–Ser-280 phosphorylation after serum stimulation.
Our observations suggest that p90 RSK induces Chk1 translocation from cytoplasm to nucleus through Chk1–Ser-280 phosphorylation. They are in contrast with previous observations that Chk1–Ser-280 phosphorylation induced cytoplasmic sequestration of Chk1 (Puc et al., 2005). Using the system of transient overexpression of Chk1 in U2OS cells, Puc et al. (2005) reported that the nuclear-to-cytoplasmic (N/C) ratio for Chk1 WT and SA mutant was greater than for the SE mutant, regardless of DNA damage. However, using the system of inducible expression in several types of cells including U2OS cells, we found that the N/C ratio for Chk1 WT was greater than for the SA mutant but smaller than for the SE mutant (Figure 2, E–I). We consider that this contrast may be due to the difference between transient overexpression and inducible expression. We previously demonstrated that the transient transfection of exogenous Chk1 induced Chk1–Ser-345 phosphorylation even in the absence of genotoxic stimuli, whereas the inducible expression did not (Enomoto et al., 2009). Because Chk1 phosphorylation occurs predominantly at Ser-280 (but not at other known phosphorylation sites) after serum stimulation (Figure 2, A–D), the change in Chk1 localization by Ser-280 phosphorylation after serum stimulation may be more reflected by the inducible expression of Chk1 mutants (Figure 2, E–I).
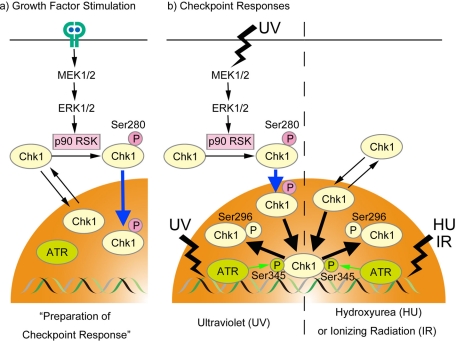
Our results point to the potential role of p90 RSK–Chk1 pathway (summarized in Figure 8). Following the stimulation of RTK with growth factor, p90 RSK is activated downstream of MAPK cascade and then phosphorylates Chk1 specifically at Ser-280. Although Chk1 constantly shuttles between cytoplasm and nucleus, Ser-280 phosphorylation promotes nuclear retention of Chk1. Because Chk1 is activated in the nucleus (Sanchez et al., 1997; Jiang et al., 2003; Matsuyama et al., 2011), such nuclear accumulation is likely to be of great use in the preparation for the DNA damage checkpoint. In support of this hypothesis, Ser-280 phosphorylation accelerates Chk1 activation processes (Chk1 phosphorylation at Ser-345 by ATR and its autophosphorylation) after UV irradiation (Figures 6 and 7).
FIGURE 8:
Model for Chk1–Ser-280 phosphorylation by p90 RSK.
Our study also raises the question of why Chk1–Ser-280 phosphorylation is required for the checkpoint following UV irradiation but not IR or HU treatment in mammalian cells. We consider that more activation of Chk1 may be required for repairing UV-damaged lesions than other lesions, especially in the G1 phase. UV-induced DNA lesions are repaired mainly through the nucleotide excision repair (NER) pathway in the G1 phase (Mitchell et al., 2003). The NER process consists of sequential recruitments of proteins to a DNA damage site for damage recognition, the excision of the damaged DNA to create single-stranded DNA (ssDNA) intermediate, filling in of the ssDNA region, and ligation (Sancar et al., 2004). Such a ssDNA intermediate is known to activate the ATR-Chk1 pathway (Cimprich and Cortez, 2008; Flynn and Zou, 2011). IR-induced double-strand breaks (DSBs) are known to be repaired in a cell cycle phase–dependent manner. In G1 phase, DSBs undergo only minor nucleolytic processing and are rapidly repaired by nonhomologous end-joining. During the S or G2 phase, DSBs are resected by exonucleases to generate ssDNA and then repaired by homologous recombination. Chk1 activation in response to IR was reported to be restricted to the S and G2 phases (Jazayeri et al., 2006). In response to HU treatment, Chk1 is activated only during S phase because HU, the DNA reductase inhibitor, suppresses DNA replication through dNTP depletion. Together with a recent report that ERK1/2 plays an important role in the checkpoint following UV irradiation (Moniz and Stambolic, 2011), our data suggest that p90 RSK activation may be required for rapid activation of Chk1 (especially in the G1 phase) after UV irradiation.
Ras-MAPK and PI3-K–Akt/PKB pathways are up-regulated in a wide spectrum of human cancers (Vivanco and Sawyers, 2002; Anjum and Blenis, 2008; Engelman, 2009; Wong et al., 2010; Cargnello and Roux, 2011). The present study demonstrates the possibility that the p90 RSK–Chk1 pathway may serve as a barrier to protect genomic integrity in the case of Ras-MAPK up-regulation. Of interest, the PI3-K–Akt/PKB pathway overrides cell cycle arrest induced by the DNA-damage checkpoint (Henry et al., 2001; Shtivelman et al., 2002; Nimbalkar and Quelle, 2008; Xu et al., 2010). Detailed analyses of these pathways in DNA damage checkpoints will provide further insight into the role of these pathways in carcinogenesis.
MATERIALS AND METHODS
Cell culture
RPE1 (CRL-4000; American Type Culture Collection, Manassas, VA) cells were grown in DMEM/F12 (a 1:1 mixture of DMEM and Ham's F12 medium; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; growing medium). U2OS or HeLa cells were grown in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS (growing medium). Serum stimulation experiments were performed as follows. RPE1 cells were cultured for 48 h in the medium containing no serum (serum-free medium). U2OS or HeLa cells were cultured for 48 h in the medium containing 0.5% FBS (serum-free medium). After the serum starvation, the cells were incubated in the growing medium.
For inhibitor experiments, cells were cultured for 48 h in the serum-free medium and then pretreated with 10 μM U0126 (MEK1/2 inhibitor; Promega, Madison, WI), 10 μM LY294002 (PI3-K inhibitor; Merck-Calbiochem, Darmstadt, Germany), 10 μM BI-D1870 (p90 RSK inhibitor; Symansis, Shanghai, China), 1 μM MK-2206 (Akt/PKB inhibitor; Selleck Chemicals, Houston, TX), or an equal volume of dimethyl sulfoxide (DMSO; control) in fresh serum-free medium for 30 min. After the preincubation, 1/9 volume of FBS containing the same chemical was added in the medium (final concentration, ∼10% FBS), and then cells were incubated for an additional 5 or 10 min.
For the activation of DNA replication checkpoint, RPE1 cells were incubated in the culture medium containing 3 mM HU for 2 h. For preparation of mitotic RPE1 cells, the cells were treated with 50 ng/ml nocodazole (Sigma-Aldrich) for 4 h. Then mitotic cells were collected by mechanical shake-off.
Peptides and antibodies
We designed and synthesized a phosphopeptide corresponding to Chk1 phosphorylated at each site and its nonphosphorylated version of peptide as described previously (Goto and Inagaki, 2007). We immunized rats with each phosphopeptide-conjugated keyhole limpet hemocyanin and then produced each site- and phosphorylation state–specific monoclonal antibody for Ser-286, Ser-296, Ser-301, Ser-317, or Ser-345 on Chk1 (Shiromizu et al., 2006; Ikegami et al., 2008; Kasahara et al., 2010). Antibodies from commercial sources were as follows: mouse anti-Chk1 (clone name, G4) from Santa Cruz Biotechnology (Santa Cruz, CA), mouse anti–pan-Akt (40D4), anti-ERK1/2 (3A7), rabbit anti–Akt-pThr-308 (C31E5E), anti–Akt-pSer-473 (D9E), anti-Bad (D24A9), anti–Bad-pSer-112 (40A9), anti–Bad-pSer-136 (D25H8), anti–Chk1-pSer-345 (133D3), anti–ERK1/2-pThr-202/pTyr-204 (D13.14.4E), anti– MAPK-activated protein kinase-2–pThr-334, anti–p90 RSK1/RSK2/RSK3 (32D7), and anti–RSK-pThr-573 from Cell Signaling Technology (Beverly, MA), mouse anti-Chk1 (DCS-310) from Sigma-Aldrich, mouse anti-Myc (4A6) from Millipore (Bedford, MA), and anti–Chk1-pSer-280 from Epitomics (Burlingame, CA).
Immunoprecipitation and immunoblotting
We performed the immunoprecipitation as described previously (Ikegami et al., 2008). In some immunoblotting experiments, we used immunoreaction enhancer solutions (Can Get Signal; Toyobo, Osaka, Japan) for dilution of primary and secondary antibodies. Band intensities were analyzed by densitometry (ImageJ, version 1.38x, for Macintosh OS X; National Institutes of Health, Bethesda, MD).
For the detection of the in vivo phosphorylation of Chk1, we used Mn2+-Phos-tag–modified acrylamide gel (Wako Pure Chemical, Osaka, Japan) in which the phosphorylated proteins migrate more slowly than nonphosphorylated protein by the interaction of phosphate groups with Mn2+-Phos-tag (Kinoshita et al., 2006; Kinoshita-Kikuta et al., 2007). After the serum starvation, cells were treated with the growing medium serum for 0 or 10 min and then subjected to the immunoprecipitation. Each immunoprecipitate was subjected to Mn2+-Phos-tag SDS–PAGE (7.5% polyacrylamide gel including 50 μM Phos-tag acrylamide and 100 μM MnCl2) and then analyzed by immunoblotting.
siRNA transfection
Three siRNAs for human Chk1 and one nonsilencing (control) siRNA were purchased from Qiagen (Valencia, CA): Sq. 1, (AA)CTGAAGAAGCAGTCGCAGT; Sq. 2, (AA)CCAGATGCTCAGAGATTCT; 3′ untranslated region (UTR), (CT)GGTGAATATAGTGCTGCTA; and control siRNA, (AA)TTCTCCGAACGTGTCACGT. RPE1 cells were transfected with each Chk1 siRNA or control siRNA at a 10 nM concentration. Human p90 RSK1/2/3 or Akt1/2 proteins were knocked down in HeLa cells using a pool (final 5 nM concentration per each protein) of four siRNAs provided by Thermo Fisher Scientific (Waltham, MA; Dharmacon ON-TARGETplus SMARTpool: p90 RSK1, catalogue no. L-0003025-00-0005; p90 RSK2, L-003026-00-0005; p90 RSK3, L-004663-00-0005; Akt1, L-003000-00-0005; and Akt2, L-003001-00-0005). In parallel, a pool (final 15 nM concentration) of four nontargeting siRNAs was used as negative control (Dharmacon ON-TARGETplus siCONTROL, D-001810-10−05; Thermo Fisher Scientific). For all siRNA transfection experiments, we used Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's protocol.
Immunocytochemistry
Immunocytochemistry was performed as described previously (Enomoto et al., 2009), with a slight modification. For the double staining with αpS280 and αpS296 or αpS345, cells were fixed with 1.85% formaldehyde in phosphate-buffered saline (PBS) at room temperature for 10 min and then permeabilized with 0.1% Triton X-100 in PBS at room temperature for 10 min. Each fluorescence image was captured as a single optical section using a Zeiss LSM510 confocal laser-scanning microscope (Carl Zeiss, Thornwood, NY). We calculated the N/C ratio of the antibody intensity as described previously (Enomoto et al., 2009).
Mutagenesis
The following site-directed mutagenesis was performed using KOD-Plus mutagenesis kit (Toyobo). For the construction of nonphosphorylated or phosphomimic mutation of Ser-280 on human Chk1 (Shiromizu et al., 2006), Ser-280 was changed to Ala (A) or Glu (E). For the construction of (KD Chk1 mutant, Lys-38 was changed to Met (K38M). For the construction of CA or KD mutation of human p90 RSK2 (a kind gift of N. Goshima, Biomedicinal Information Research Center, National Institute of Advanced Industrial Science and Technology, Tokyo, Japan), Tyr-707 or Lys-451 was substituted with Ala (Y707A or K451A), respectively (Poteet-Smith et al., 1999). For the construction of CA or KD mutation of human Akt1 (a kind gift of N. Goshima), Thr-308/Ser-473 or Lys-179 was replaced with Asp (T308D/S473D) or Met (K179M), respectively (Bellacosa et al., 1998).
Establishment of Tet-On cell lines
Tet-On cell lines were established with the same procedure described for Tet-On HeLa cells (Ibi et al., 2011). Enhanced green fluorescent protein (EGFP; BD Clontech, Mountain View, CA) or each kinase construct with a Myc or triple FLAG (3X) tag at the N-terminus was produced by PCR and then inserted into pENTR vectors (Invitrogen). CSII-TRE-Tight vectors expressing the foregoing proteins were constructed through the homologous recombination between the pENTR vectors and CSII-TRE-Tight-RfA (a tet-responsive lentivirus vector; Ibi et al., 2011) using Gateway technology (Invitrogen). Production and infection of recombinant retroviruses and lentiviruses were as described previously (Sasaki et al., 2009). Tet-On cells were infected with CSII-TRE-Tight lentiviruses expressing each protein.
Proteins
We generated the recombinant baculovirus encoding GST-Chk1-His KD or KD/S280A by a combination of the GATEWAY vector conversion system and Bac-to-Bac baculovirus expression system (Invitrogen). Chk1-His KD or KD/S280A was purified from Sf9 cells as described previously (Kasahara et al., 2010). GST-tagged human Cdc25C fragment (residues 195–256) was purified from Escherichia coli strain DH5α (Invitrogen) as described previously (Kasahara et al., 2010). We purchased active p90 RSK1 (catalogue no. 14-479) or Akt1 (catalogue no. 14-276) from Upstate (Millipore).
In vitro kinase assay
Chk1 phosphorylation assay was performed at 30°C in 20 μl of 25 mM Tris-Cl (pH 7.5), 0.1 mM ATP, 10 mM MgCl2, and 92.5 μg/ml Chk1-His (KD or KD/S280A) with or without 3.75 μg/ml active p90 RSK1 or 36.9 μg/ml active Akt1 (Millipore). Some experiments were performed in the presence of [γ-32P]ATP (4 μCi).
Supplementary Material
Acknowledgments
We thank N. Goshima for providing p90 RSK and Akt constructs; N. Liu and M. Kitagawa (Hamamatsu University, Hamamatsu, Japan) for helpful discussion; Y. Hayashi, K. Kobori, C. Yuhara, and E. Kawamoto for technical assistance; Y. Takada for secretarial expertise; and J. Shields for critical reading of the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and from the Ministry of Education, Science, Technology, Sports and Culture of Japan; by a Grant-in-Aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare, Japan; by the Uehara Memorial Foundation; by the Astellas Foundation for Research on Metabolic Disorders; by the Naito Foundation; by the Takeda Science Foundation; and by the Daiichi-Sankyo Foundation of Life Science.
Abbreviations used:
- ATR
ataxia telangiectasia mutated- and rad3-related kinase
- Dox
doxycycline
- ERK
extracellular signal-regulated kinase
- HU
hydroxyurea
- IR
ionizing radiation
- MAPK
mitogen-activated protein kinase
- MK2
MAPK-activated protein kinase-2
- p90 RSK
90-kDa ribosomal S6 kinase
- PH
pleckstrin homology
- PI3-K
phosphatidylinositol 3-kinase
- PIP3
phosphatidylinositol(3,4,5)phosphate
- PKB
protein kinase B
- RTK
receptor tyrosine kinase
- siRNA
small interfering RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-10-0883) on February 22, 2012.
REFERENCES
- Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Bellacosa A, Chan TO, Ahmed NN, Datta K, Malstrom S, Stokoe D, McCormick F, Feng J, Tsichlis P. Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene. 1998;17:313–325. doi: 10.1038/sj.onc.1201947. [DOI] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature Rev. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature Rev. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Enomoto M, Goto H, Tomono Y, Kasahara K, Tsujimura K, Kiyono T, Inagaki M. Novel positive feedback loop between Cdk1 and Chk1 in the nucleus during G2/M transition. J Biol Chem. 2009;284:34223–34230. doi: 10.1074/jbc.C109.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–140. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Inagaki M. Production of a site- and phosphorylation state-specific antibody. Nature Protoc. 2007;2:2574–2581. doi: 10.1038/nprot.2007.374. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Henry MK, Lynch JT, Eapen AK, Quelle FW. DNA damage-induced cell-cycle arrest of hematopoietic cells is overridden by activation of the PI-3 kinase/Akt signaling pathway. Blood. 2001;98:834–841. doi: 10.1182/blood.v98.3.834. [DOI] [PubMed] [Google Scholar]
- Ibi M, et al. Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. J Cell Sci. 2011;124:857–864. doi: 10.1242/jcs.075705. [DOI] [PubMed] [Google Scholar]
- Ikegami Y, Goto H, Kiyono T, Enomoto M, Kasahara K, Tomono Y, Tozawa K, Morita A, Kohri K, Inagaki M. Chk1 phosphorylation at Ser286 and Ser301 occurs with both stalled DNA replication and damage checkpoint stimulation. Biochem Biophys Res Commun. 2008;377:1227–1231. doi: 10.1016/j.bbrc.2008.10.119. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Jiang K, Pereira E, Maxfield M, Russell B, Goudelock DM, Sanchez Y. Regulation of Chk1 includes chromatin association and 14–3-3 binding following phosphorylation on Ser-345. J Biol Chem. 2003;278:25207–25217. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- Kasahara K, Goto H, Enomoto M, Tomono Y, Kiyono T, Inagaki M. 14-3-3gamma mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage. EMBO J. 2010;29:2802–2812. doi: 10.1038/emboj.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King FW, Skeen J, Hay N, Shtivelman E. Inhibition of Chk1 by activated PKB/Akt. Cell Cycle. 2004;3:634–637. [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Kinoshita-Kikuta E, Aoki Y, Kinoshita E, Koike T. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol Cell Proteomics. 2007;6:356–366. doi: 10.1074/mcp.T600044-MCP200. [DOI] [PubMed] [Google Scholar]
- Lewis TS, Shapiro PS, Ahn NG. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- Lindsley CW. The Akt/PKB family of protein kinases: a review of small molecule inhibitors and progress towards target validation: a 2009 update. Curr Top Med Chem. 2010;10:458–477. doi: 10.2174/156802610790980602. [DOI] [PubMed] [Google Scholar]
- Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi S, et al. Inhibition of phosphorylation of BAD and Raf-1 by Akt sensitizes human ovarian cancer cells to paclitaxel. J Biol Chem. 2002;277:33490–33500. doi: 10.1074/jbc.M204042200. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama M, Goto H, Kasahara K, Kawakami Y, Nakanishi M, Kiyono T, Goshima N, Inagaki M. Nuclear Chk1 prevents premature mitotic entry. J Cell Sci. 2011;124:2113–2119. doi: 10.1242/jcs.086488. [DOI] [PubMed] [Google Scholar]
- McGowan CH, Russell P. The DNA damage response: sensing and signaling. Curr Opin Cell Biol. 2004;16:629–633. doi: 10.1016/j.ceb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Meuillet EJ, et al. Molecular pharmacology and antitumor activity of PHT-427, a novel Akt/phosphatidylinositide-dependent protein kinase 1 pleckstrin homology domain inhibitor. Mol Cancer Ther. 2010;9:706–717. doi: 10.1158/1535-7163.MCT-09-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Hoeijmakers JH, Niedernhofer LJ. Divide and conquer: nucleotide excision repair battles cancer and ageing. Curr Opin Cell Biol. 2003;15:232–240. doi: 10.1016/s0955-0674(03)00018-8. [DOI] [PubMed] [Google Scholar]
- Moniz LS, Stambolic V. Nek10 mediates G2/M cell cycle arrest and MEK autoactivation in response to UV irradiation. Mol Cell Biol. 2011;31:30–42. doi: 10.1128/MCB.00648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis. 2006;21:3–9. doi: 10.1093/mutage/gei063. [DOI] [PubMed] [Google Scholar]
- Nimbalkar D, Quelle FW. Phosphoinositide 3-kinase signaling overrides a G2 phase arrest checkpoint and promotes aberrant cell cycling and death of hematopoietic cells after DNA damage. Cell Cycle. 2008;7:2877–2885. doi: 10.4161/cc.7.18.6675. [DOI] [PubMed] [Google Scholar]
- Nishimoto S, Nishida E. MAPK signalling: ERK5 versus ERK1/2. EMBO Rep. 2006;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteet-Smith CE, Smith JA, Lannigan DA, Freed TA, Sturgill TW. Generation of constitutively active p90 ribosomal S6 kinase in vivo. Implications for the mitogen-activated protein kinase-activated protein kinase family. J Biol Chem. 1999;274:22135–22138. doi: 10.1074/jbc.274.32.22135. [DOI] [PubMed] [Google Scholar]
- Puc J, et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Sapkota GP, et al. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J. 2007;401:29–38. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki R, Narisawa-Saito M, Yugawa T, Fujita M, Tashiro H, Katabuchi H, Kiyono T. Oncogenic transformation of human ovarian surface epithelial cells with defined cellular oncogenes. Carcinogenesis. 2009;30:423–431. doi: 10.1093/carcin/bgp007. [DOI] [PubMed] [Google Scholar]
- Shiromizu T, Goto H, Tomono Y, Bartek J, Totsukawa G, Inoko A, Nakanishi M, Matsumura F, Inagaki M. Regulation of mitotic function of Chk1 through phosphorylation at novel sites by cyclin-dependent kinase 1 (Cdk1) Genes Cells. 2006;11:477–485. doi: 10.1111/j.1365-2443.2006.00955.x. [DOI] [PubMed] [Google Scholar]
- Shtivelman E, Sussman J, Stokoe D. A role for PI 3-kinase and PKB activity in the G2/M phase of the cell cycle. Curr Biol. 2002;12:919–924. doi: 10.1016/s0960-9822(02)00843-6. [DOI] [PubMed] [Google Scholar]
- Tan Y, Ruan H, Demeter MR, Comb MJ. p90(RSK) blocks bad-mediated cell death via a protein kinase C-dependent pathway. J Biol Chem. 1999;274:34859–34867. doi: 10.1074/jbc.274.49.34859. [DOI] [PubMed] [Google Scholar]
- Tonic I, Yu WN, Park Y, Chen CC, Hay N. Akt activation emulates Chk1 inhibition and Bcl2 overexpression and abrogates G2 cell cycle checkpoint by inhibiting BRCA1 foci. J Biol Chem. 2010;285:23790–23798. doi: 10.1074/jbc.M110.104372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NL, Adams DG, Vaillancourt RR, Heimark RL. Signal transduction from N-cadherin increases Bcl-2. Regulation of the phosphatidylinositol 3-kinase/Akt pathway by homophilic adhesion and actin cytoskeletal organization. J Biol Chem. 2002;277:32905–32914. doi: 10.1074/jbc.M200300200. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Walker M, Black EJ, Oehler V, Gillespie DA, Scott MT. Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene. 2009;28:2314–2323. doi: 10.1038/onc.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Hegarat N, Black EJ, Scott MT, Hochegger H, Gillespie DA. Akt/PKB suppresses DNA damage processing and checkpoint activation in late G2. J Cell Biol. 2010;190:297–305. doi: 10.1083/jcb.201003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.