Stem cell activities have to be terminated during flower development. A nucleostemin-like 1 gene, NSN1, is described in Arabidopsis. NSN1 is a midsize nucleolar GTPase required for proper termination of stem cell activities during flower development.
Abstract
Mammalian nucleostemin (NS) is preferentially expressed in stem cells and acts to promote cell cycle progression. In plants, stem cell activities have to be terminated during flower development, and this process requires the activation of AGAMOUS (AG) gene expression. Here, a nucleostemin-like 1 gene, NSN1, is shown to be required for flower development in Arabidopsis. The NSN1 mRNA was found in the inflorescence meristem and floral primordia, and its protein was localized to the nucleoli. Both heterozygous and homozygous plants developed defective flowers on inflorescences that were eventually terminated by the formation of carpelloid flowers. Overexpression of NSN1 resulted in loss of apical dominance and formation of defective flowers. Expression of the AG gene was found to be up-regulated in nsn1. The carpelloid flower defect of nsn1 was suppressed by the ag mutation in the nsn1 ag double mutant, whereas double mutants of nsn1 apetala2 (ap2) displayed enhanced defective floral phenotypes. These results suggest that in the delicately balanced regulatory network, NSN1 acts to repress AG and plays an additive role with AP2 in floral organ specification. As a midsize nucleolar GTPase, NSN1 represents a new class of regulatory proteins required for flower development in Arabidopsis.
INTRODUCTION
Mammalian nucleostemin (NS) was first discovered in a subtractive screen between embryonic stem cells and differentiated tissue cultures (Tsai and McKay, 2002). It is expressed preferentially in embryonic and adult stem cells, as well as in several cancer cell lines (Tsai and McKay, 2002; Fan et al., 2006; Han et al., 2005). Depletion or overexpression of NS in cultured stem cells impairs cell proliferation, suggesting that it may regulate cell cycle progression (Tsai and McKay, 2002). Targeted deletion of NS in a mutant mouse line results in developmental arrest at the implantation stage, suggesting that NS is required for early embryogenesis (Beekman et al., 2006; Zhu et al., 2006).
Animal NS is a midsize (538 amino acids) GTPase localized to the nucleoli and may shuttle rapidly between the nucleolus and nucleoplasm (Tsai and McKay, 2002, 2005). GTP binding is required for its nucleolar localization, as deletion of the GTP-binding domain of NS results in accumulation of the protein in the nucleoplasm (Tsai and McKay, 2005). One of the major functions of the nucleoli is rRNA transcription and ribosome assembly (Politz et al., 2005). NS is found in the rRNA-free sites within the nucleolar granular component, suggesting that NS serves a function other than ribosome assembly (Politz et al., 2005; Romanova et al., 2009). The unique ability of NS in the regulation of cell cycle progression and stem cell proliferation has been linked both directly and indirectly to p53, MDM2, ribosomal protein RSL1D1, and telomeric repeat–binding factor TRF1 (Beekman et al., 2006; Zhu et al., 2006; Ma and Pederson, 2007; Dai et al., 2008; Jafarnejad et al., 2008). Mammalian NS has also been shown to play a critical role in pre-rRNA processing. Purified NS exists in a large complex that contains three nucleolar proteins involved in pre-rRNA processing (Romanova et al., 2009). Retention of these proteins in the nucleoli is dependent on the presence of NS. The processing of pre-rRNA into 28S rRNA is delayed by the knockdown of NS and promoted by the overexpression of NS. This biochemical evidence demonstrates a role of NS in ribosome biogenesis and is consistent with data obtained in yeast and nematode (Du et al., 2006; Kudron and Reinke, 2008). Thus, at the molecular level, NS appears to have multiple biochemical roles.
Pattern formation during flower development has been well studied in the last two decades. Most of the homeotic genes identified so far encode DNA-binding transcription factors. AGAMOUS (AG), encoding a MADS domain–containing transcription factor, commences to express at the central part of stage 3 flower primordia and is accumulated in whorls 3 and 4 in older floral primordia (Yanofsky et al., 1990; Bowman et al., 1991). AG functions to specify the stamen and carpel identities and controls floral meristem determinacy (Mizukami and Ma, 1997; Lohmann et al., 2001; Sun et al., 2009). In ag-1, stamens and carpels are replaced by a new flower that only has sepals and petal (Bowman et al., 1989). When AG is overexpressed, inflorescence meristem becomes determinate and produces terminal flowers (Mizukami and Ma, 1992, 1997). HUA1, HUA2, HUA1 ENHANCER 1 (HEN1), and HUA2 ENHANCER2 (HEN2) are components of the AG pathway. Among them, HUA2 and HEN1 play important roles in specifying stamen and carpel identity (Chen and Meyerowitz, 1999; Chen et al., 2002). APELATA2 (AP2), encoding a transcription factor that does not contain a MADS box, acts to specify the identity of whorl 1 and 2 floral organs (Jofuku et al., 1994). In ap2 mutants, sepals are transformed into carpelloid structures (Bowman et al., 1991). AP2 and AG reciprocally repress gene expression of each other (Bowman et al., 1991; Drews et al., 1991).
Regulation of AG transcription requires sequences located in its 3-kb second intron, which serves as a binding site for positive and negative regulators (Sieburth and Meyerowitz, 1997; Busch et al., 1999). As positive regulators, WUSCHEL (WUS) and LEAFY (LFY) bind to this regulatory intron sequence and activate AG gene expression (Lenhard et al., 2001; Lohmann et al., 2001). Negative regulators, including AP2, LEUNIG (LUG), SEU (SEUSS), BELLRINGER (BLR), and PERIANTHIA (PAN), are also known to regulate AG via this intron sequence (Liu and Meyerowitz, 1995; Deyholos and Sieburth, 2000; Franks et al., 2002; Bao et al., 2004; Sridhar et al., 2004; Das et al., 2009).
We were interested in understanding a potential role of NS homologues in plants. In this study, we identified a nucleostemin-like 1 (NSN1) gene in Arabidopsis and characterized a knockout mutant of this gene (nsn1-1). As a nucleolar GTPase, NSN1 represents a new class of proteins that play a critical role in controlling floral identity and maintenance of inflorescence meristem. At the genetic level, NSN1 acts as an AG repressor and is required for the maintenance of inflorescence meristem identity and flower development in Arabidopsis.
RESULTS
A midsize GTPase with homology to animal nucleostemins
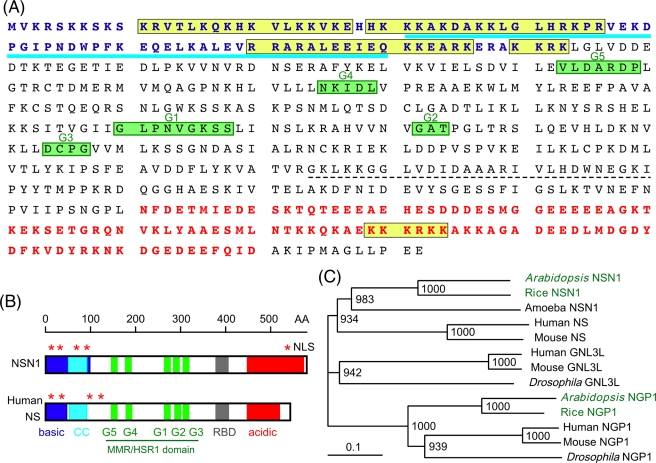
In a search for GTP-binding proteins that may play a role in cell division control, we cloned the Arabidopsis NSN1 (At3g07050) cDNA. The deduced NSN1 polypeptide, consisting of 582 amino acid residues with calculated molecular mass of 65.8 kDa (Figure 1A), contains multiple functional motifs, including five potential nuclear localization signals, a basic domain, a coiled-coil domain, a GTP-binding region, an RNA-binding domain, and an acidic domain (Figure 1B). The conserved GTP-binding motifs are arranged in a circularly permutated order (G5-G4-G1-G2-G3; Daigle et al., 2002). NSN1 shares 30 and 31% amino acid sequence identity with human NS and GNL3L, respectively, but is less homologous to human NGP1 (25% identity; Supplemental Figure S1). In Arabidopsis, the most closely related protein, At1g52980, has only 25% sequence identity with NSN1, suggesting that NSN1 is a unique gene. At1g52980, referred to as AtNGP1, represents an uncharacterized paralogue of NSN1 in Arabidopsis. Both NSN1 and NGP1 proteins belong to a new GTPase family with a signature MMR/HSR1 domain (Daigle et al., 2002). This GTPase family is conserved in organisms ranging from Archaea to vertebrates (Reynaud et al., 2005; Meng et al., 2007) and can be grouped into at least four subfamilies (Figure 1C). The human and mouse genomes contain three members for each genome—NS, GNL3L, and NGP1—whereas Arabidopsis and rice have two members—NSN1 and NGP1—per genome. Dictyostelium, on the other hand, has only one NSN1 member.
FIGURE 1:
Functional motifs and phylogenetic tree of Arabidopsis nsn1. (A) Amino acid sequence of NSN1 (At3g07050). Blue, basic domain (basic); blue underline, coiled-coil domain (CC); yellow boxes, nuclear localization signals (NLS); green boxes, GTP-binding motifs (G1–G5); dashed underline, RNA-binding domain (RBD); red, acidic domain (acidic). (B) Comparison of Arabidopsis NSN1 with human NS. The GTP-binding motifs (G1–G5) in NSN1 are arranged in a circularly permuted manner known as the MMR/HSR1 domain. Five NLS motifs are indicated by asterisks. Shaded areas correspond to the structural domains indicated in A. (C) Phylogenetic tree of NSN1 homologues. Human and mouse contains three NS-related genes (NS, GNL3L, and NGP1), whereas Arabidopsis and rice have two (NSN1 and NGP1). The amoeba Dictyostelium has only one NSN1 gene. Peptide sequences used for phylogenetic analysis include At3g07050 (AtNSN1), At1g52980 (AtNGP1), Os01g0375000 (rice NSN1), Os03g0352400 (rice NGP1), AAV74413 (human NS), AAH11720 (human GNL3L), Q13823 (human NGP1), AAO19472 (mouse NS), NP_932778 (mouse GNL3L), NP_663527 (mouse NGP1), Q8MT06 (Drosophila NGL3L), EAL25205 (Drosophila NGP1), and Q54KS4 (amoeba NSN1). Bootstrap values from 1000 trials are displayed on nodes.
Loss-of-function mutants
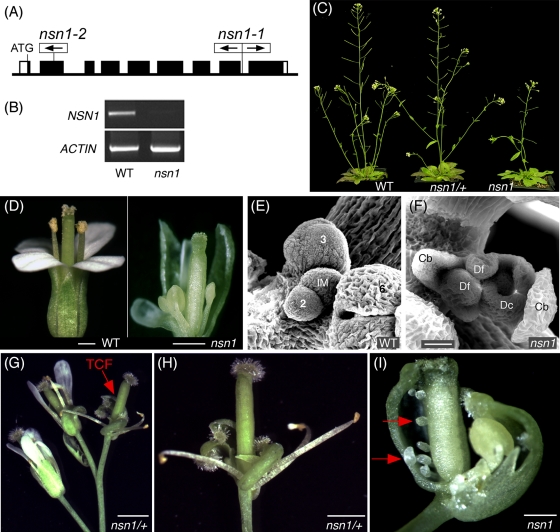
To investigate the function of NSN1, we characterized two Arabidopsis mutants nsn1-1 and nsn1-2, the former having T-DNA inserted in the eighth intron and the latter containing a transposon in the second exon (Figure 2A). The two mutant lines had indistinguishable growth and reproduction phenotypes (Supplemental Figure S2), suggesting that the defects observed are caused by mutations of the same locus, At3g07050. Further molecular characterization was conducted in nsn1-1 unless indicated otherwise. T-DNA insertion in the NSN1 gene was confirmed by genomic PCR (Supplemental Figure S3), and the knockout of NSN1 gene expression in homozygous lines was verified by reverse transcriptase (RT)-PCR (Figure 1B). This analysis suggests that nsn1-1 is a null mutation. The mutant exhibited pleiotropic defects in growth and development, with the most striking phenotypes in the formation of defective flowers and siliques (see later discussion).
FIGURE 2:
Termination of inflorescence meristem and homeotic transformation of floral organs in nsn1. (A) nsn1-1 (SALK_029201) contained two T-DNA inserts in the eighth intron of the NSN1 gene, and nsn1-2 (RAFL11-2045-1) had one transposon in the second exon. Framed arrows indicate the direction of T-DNA or transposon. (B) RT-PCR showing the lack of detectable NSN1 mRNA in homozygous nsn1-1 plants. ACTIN mRNA served as a control. Nsn1-LP and Nsn1-RP were used as primers (for their positions see Supplemental Figure S3A) for RT-PCR, which generated a cDNA fragment of 1050 base pairs. (C) Phenotypes of 30-d-old WT, nsn1-1/+, and nsn1 plants. Heterozygous nsn1/+ plants showed no obvious growth phenotype but had significant defects in reproduction. Homozygous nsn1 plants were very small and yielded only a few seeds per plant. (D) A flower at position 25 from the lower part of inflorescence of a 40-d-old nsn1-1 plant had a reduced number of petals and shortened stamen filaments as compared with a WT flower at the same position of inflorescence. (E, F) Scanning electronic images of the inflorescence meristem (IM) of 40-d-old WT (E) and nsn1 (F) plants. Note that the inflorescence meristem (F) was terminated by the formation of a terminal carpelloid flower (TCF). Cb, carpelloid bract; Dc, defective carpel; Df, defective floral meristem developed inside a TCF. Stages of the WT floral primordia (E) are indicated. (G–I) Inflorescences of nsn1/+ and nsn1 were terminated by the formation of a TCF (red arrow in G), which is shown in a higher magnification (H). A typical TCF of nsn1 would contain homeotically transformed carpelloid bracts, severely defective stamens, and no petals (I). Ectopically developed ovules are indicated by red arrows (I). Shown are the terminal carpelloid flowers at position 35 of nsn1-1/+ (H) and position 36 of nsn1-1 (I). Scale bars, 25 μm (E, F), 2 mm (G), 1 mm (D, H, I).
Termination of inflorescence meristem and homeotic floral organ transformation
We characterized both heterozygous (nsn1/+) and homozygous nsn1 plants because they showed different degrees of defects in seedling growth and reproductive fertility. Both heterozygous and homozygous plants had highly reduced self-fertility, and the latter phenotype was much more severe (Figure 2C and Supplemental Figure S2C). Heterozygous nsn1/+ plants were comparable to the wild type in vegetative growth but developed defective flowers and produced short siliques. The main and branch inflorescence meristems in nsn1/+ were also characteristically transformed into determinate apexes by developing a terminal carpelloid flower on each apex. A typical terminal carpelloid flower of nsn1/+ exhibited a complete absence of petals in whorl 2 and contained a reduced number of deformed stamens in whorl 3 (Figure 2, G and H). Both heterozygous and homozygous nsn1 plants produced carpelloid flowers that were characterized by the homeotic transformation of sepals in whorl 1 into carpelloid sepals (Figure 2, H and I). Carpelloid flowers were also produced at various flower positions of the upper inflorescences of nsn1-1 plants (Supplemental Figure S5). The pistil of nsn1/+ carpelloid flowers morphologically resembled that of the control flowers, except that the two carpels often became dehiscent and contained aberrant ovule-like and stamen-like structures inside the locule. Homozygous nsn1 plants exhibited much more severe flower defects. The flowers lacked floral organs of whorls 2 and 3 and instead developed stigma-like, style-like, and ovule-like structures (Figure 2I). Homozygous nsn1 plants also had severe developmental defects in shoot apex (Figure 2F). The transformation of an inflorescence meristem to a terminal floral primordium took place in 40- to 45-d-old nsn1 plants. In wild-type (WT) plants, the inflorescence meristems stop to develop floral organs when they are ∼50 d old (Figure 2, F and G). This supports that NSN1 is required for the maintenance of inflorescence meristem. Scanning electronic microscopy revealed that the inflorescence meristem dome of nsn1 was smaller than that of WT plants at young stages (20 d old; Supplemental Figure S6). Taken together, these data suggest that the loss of function of NSN1 results in an increase in the chance of changing earlier cell fate of inflorescence meristem and floral meristem from indeterminate to determinate.
Complementation of nsn1
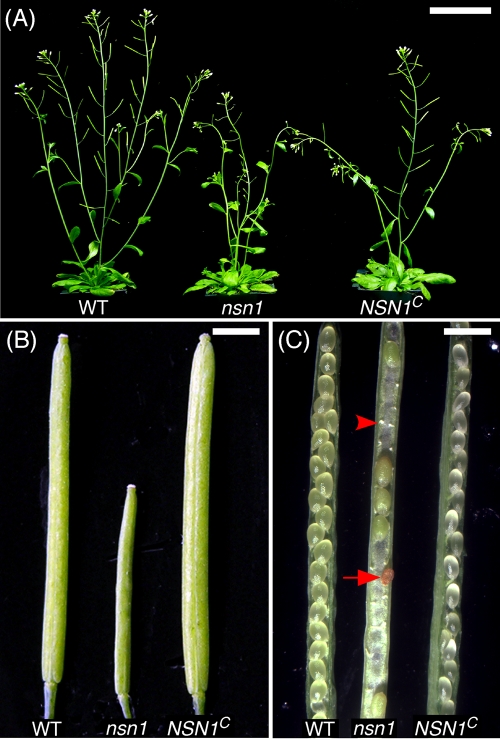
To confirm that the phenotypes of nsn1 were caused by a mutation in the NSN1 locus, we transformed the heterozygous nsn1-1/+ plants with JAtY64G16, a TAC clone containing At3g07050 and 18 other putative genes. After genome typing, T2 plants carrying JAtY64G16 in the homozygous nsn1 genomic background were maintained. Their T3 progeny (NSN1C) were scored phenotypically for complementation. Reproductive phenotypes of NSN1C were significantly different from those of the heterozygote nsn1 plants (see later discussion) and were fully restored to those of the WT control (Figure 3, A–C). Flower morphology and silique length of NSN1C were indistinguishable from the WT. In opened siliques of heterozygote nsn1/+, there were frequent unfertilized ovules, arrested embryos, and aborted seeds, which were not observed in NSN1C siliques (Figure 3C). On average, an nsn1 silique contained only 6.4 ± 3.8 seeds (n = 10 siliques), whereas WT and NSN1C produced 60.6 ± 1.7 and 64.4 ± 7.9 seeds per silique, respectively. This complementation test demonstrated that 1 of the 19 genes in the JAtY64G16 TAC clone is responsible for the observed floral and fertility phenotypes. When the complementation result was combined with the observation that two independent mutant alleles of nsn1 (i.e., nsn1-1 and nsn1-2; see Supplemental Figure S2) exhibited identical growth and reproduction phenotypes, we concluded that the defective phenotypes of nsn1 plants are caused by the mutation at the NSN1 locus (At3g07050).
FIGURE 3:
Complementation of nsn1-1 using a genomic DNA fragment containing At3g07050. (A) The 30-d-old adult plants of the wild-type (WT), nsn1-1, and the homozygous mutant nsn1-1 containing At3g07050 (NSN1C). The length of siliques was longer in NSN1C than in nsn1 and was comparable to that of the wild type. (B) Siliques were thicker in NSN1C plants than in nsn1-1 and were comparable to that of the wild type. (C) Unfertilized ovules (arrowhead) and aborted embryos (arrow) were present in nsn1-1 siliques but absent in NSN1C and wild type plants, suggesting full complementation in NSN1C. Scale bars, 5 cm (A), 6 cm (B, C).
Localization in the nucleoli
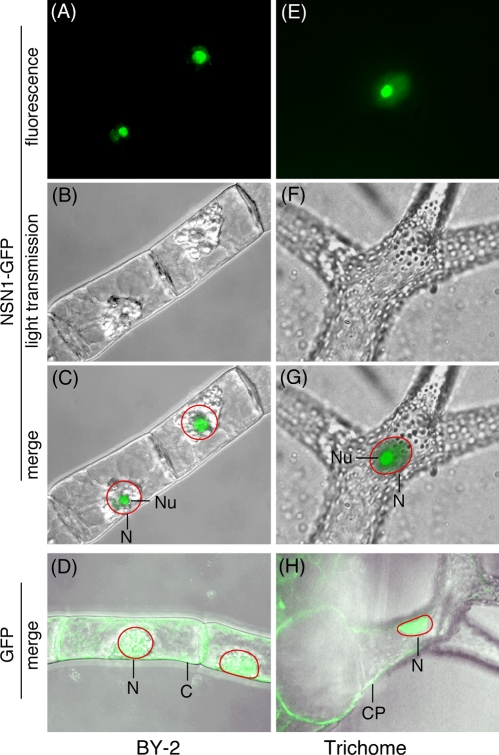
We expressed a green fluorescent protein (GFP)–tagged NSN1 in tobacco BY-2 cells and in transgenic Arabidopsis plants. NSN1 was localized to the nucleoli in BY-2 cells (Figure 4C). In transgenic plants, GFP alone was distributed uniformly in the nuclei and cytoplasm (Figure 4H). However, GFP-tagged NSN1 was concentrated in the nucleoli with a lower but detectable level of presence in the nucleoplasm (Figure 4G). It has been shown that the N-terminal lysine-rich domain (KRD) of the animal NS and GNL3L serves as the nucleolar localization signal, whereas the GTP-binding domain is required for efficient retention of the protein in the nucleoli (Meng et al., 2007). It is not known whether the N-terminal KRD of NSN1 is responsible for its nucleolar targeting.
FIGURE 4:
Nucleolar localization of NSN1. (A–D) Tobacco BY-2 cells expressing GFP-tagged NSN1 (A–C) or GFP control (D). (E–H) Arabidopsis trichomes expressing GFP-tagged NSN1 (E–G) or GFP control (H). Green fluorescence images of cells (A, E) were superimposed with the light transmission views (B, F), generating merged images (C, G). For GFP controls, only the merged images (D, H) are shown. CP, cytoplasm; N, nucleus; Nu, nucleolus. Red circles in C, D, G, and H indicate nuclear boundaries.
Expression in the inflorescence and floral meristems
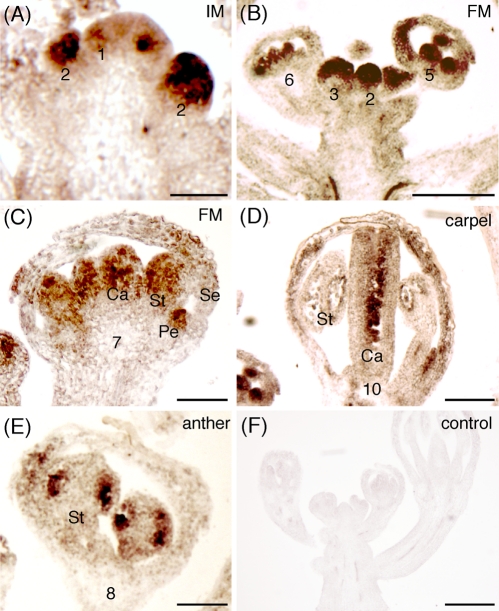
We measured the expression of NSN1 mRNA in different tissues of Arabidopsis plants by RT-PCR and found that its expression was highly concentrated in apical meristem and developing flowers and siliques, although its mRNA could be detected at relatively low levels in most tissues tested (Supplemental Figure S3D). This expression pattern was consistent with data deposited in the online database GENEVISTIGATOR (www.genevestigator.com), which shows that high expression levels of NSN1 are detected in inflorescence, flower, seed, and shoot apex (Supplemental Figure S4). We performed in situ RNA hybridization to examine the cellular expression pattern of NSN1 in inflorescence meristems and developing flowers. The expression of NSN1 was found to be very low in the inflorescence meristem dome as compared with that in the floral primordia (Figure 5A). Its expression was detectable in floral primordia as early as at stage 1 of floral primordia (Figure 5A). The expression was highly elevated at stages 2 to 3 (Figure 5B). NSN1 mRNA became concentrated in the primordia of stamens and carpels at stages 5 to 6 (Figure 5C). At the later stages of floral development, NSN1 was expressed primarily in developing microspores in the stamens and ovules in the carpels (Figure 5, E and F). This expression pattern of NSN1 is consistent with its putative role in the regulation of floral development.
FIGURE 5:
In situ hybridization of NSN1 mRNA in the inflorescence meristem (IM) and floral meristem (FM). (A–E) Sections of Arabidopsis IM (A), FM (B, C), carpels (D), and anther (E) probed with the antisense NSN1 mRNA labeled with digoxigenin. The floral developmental stages are designated by numbers. Note that NSN1 was expressed at high levels in the stage 2 floral primordia, in the stamens and carpels at stage 6, in pollen grains and ovules at stage 10, and in all four whorls at stage 7. Ca, carpel; Pe, petal; Se, sepal; St, stamen. (F) The control section reacted with the sense NSN1 mRNA. Scale bars, 25 μm (A, C–E), 100 μm (B, F).
Ectopic AG expression in nsn1 mutants
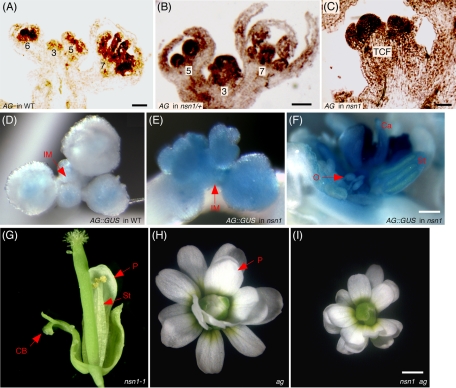
The nsn1 carpelloid flowers were anatomically similar to those developed on the bellringer (blr) mutant that is defective in a homeobox-containing transcription factor. The BLR protein binds to the promoter of the floral homeotic gene AG, repressing its expression (Bao et al., 2004). We reasoned that NSN1 might also exert its effects on floral organ identity through regulation of AG gene expression in a yet-to-be-identified manner since NSN1 is not a transcription factor but a nucleolar GTP-binding protein. We examined the expression pattern of AG in the nsn1/+ mutant. In wild type, AG is not expressed in the inflorescence meristem at all, nor is it in the floral meristem prior to stage 3 (Sieburth et al., 1995; Lenhard et al., 2001; Lohmann et al., 2001). In nsn1/+, AG mRNA could be detected in floral primordia of as early as stage 2, as well as in the whole terminal carpelloid floral primordium (Figure 6C). In flowers of stages 3 to 4 of nsn1/+, AG was expressed not only in the central parts (whorls 3 to 4) of a floral meristem, but also in whorl 1 sepal primordia (Figure 6B), suggesting that AG expression may be repressed by NSN1.
FIGURE 6:
NSN1 represses AG expression in inflorescence meristem and floral meristem. (A–C) In situ RNA hybridization of AG mRNA expression in the floral primordia of wild-type (A) and nsn1/+ plants (B), as well as in the terminal carpelloid floral primordium of homozygous nsn1 plant (C). (D–F) GUS staining of plants harboring KB9 in the control (D) and nsn1 genetic background (E, F). GUS activity was elevated in the inflorescence meristem (IM) and terminal carpelloid flower of nsn1 (F) as compared with the staining in the control plant (D). Ca, carpel-like structure; O, ectopic ovule; St, stamen-like structure. (G–I) A flower of the nsn1 ag double mutant (I) was morphologically identical to that of ag (H) but distinct from that of nsn1-1 (G). The reduced flower size of the double mutant was a result of the difference in ecotype (ag in Landsberg and nsn1 in Columbia-0). Note that the terminal carpelloid flower phenotype of nsn1 was suppressed in the double mutant. CB, carpelloid bract; P, petal. Scale bars, 50 μm (A–F), 1 mm (G–I).
To confirm the AG ectopic expression pattern observed by in situ hybridization, we introduced the KB9 construct (AG::GUS) into nsn1 plants. This construct contained the regulatory elements of AG, including the 3-kb second intron, and would allow for observing AG gene expression by monitoring the activity of the β-glucuronidase (GUS) reporter (Busch et al., 1999; Lohmann et al., 2001). In the control plant harboring KB9, the GUS activity was detected only in the post–stage 3 floral meristems but not in the inflorescence meristem (Figure 6D), which is consistent with the in situ hybridization result. In nsn1 plants expressing the same construct, GUS staining was drastically enhanced across the regions of inflorescence meristem and floral meristem and floral primordia (Figure 6E). In nsn1 carpelloid flowers, GUS was expressed in all carpelloid structures, including the ovule-like ones developed on the homeotically transformed carpelloid bracts (Figure 6F). These data suggest that the termination of the inflorescence meristem and the homeotic transformation of floral organs in nsn1 are caused by AG ectopic expression and indicate a pivotal role of NSN1 as a negative regulator for AG expression.
Suppression of the terminal carpelloid flower defects of NSN1 by AG mutation
To examine the genetic relationship between NSN1 and AG, we generated nsn1-1 ag double mutation plants by crossing nsn1/+ (Columbia-0) into ag/+ (Landsberg). Seven F1 plants were confirmed to contain the nsn1 mutation using genomic PCR (unpublished data). These plants also exhibited the characteristic ag/+ silique phenotype with three carpels in one silique. From the segregated F2 individuals, nsn1-1 ag double mutant was identified on the basis of genomic PCR and flower phenotype of ag mutant. The double-mutant plants phenocopied ag mutation and produced flowers with multiple layers of sepals and petals with no staminoid and carpelloid tissues (Figure 6I). This was in contrast to the carpelloid flowers of nsn1-1, which typically contained carpelloid bracts at whorl 1 and a reduced number of petals and stamens (Figure 6G). The size of nsn1-1 ag double-mutant flowers was smaller than those of ag (Figure 6H), which was likely to have resulted from the difference in the ecotype background. This result suggests that the terminal carpelloid flower phenotype of nsn1 depends on the activity of AG and may be caused by expanded ectopic AG expression in the nsn1 floral meristem. These findings support the hypothesis that NSN1 may act as an upstream regulator of AG.
Genetic enhancement of ap2 phenotypes by nsn1
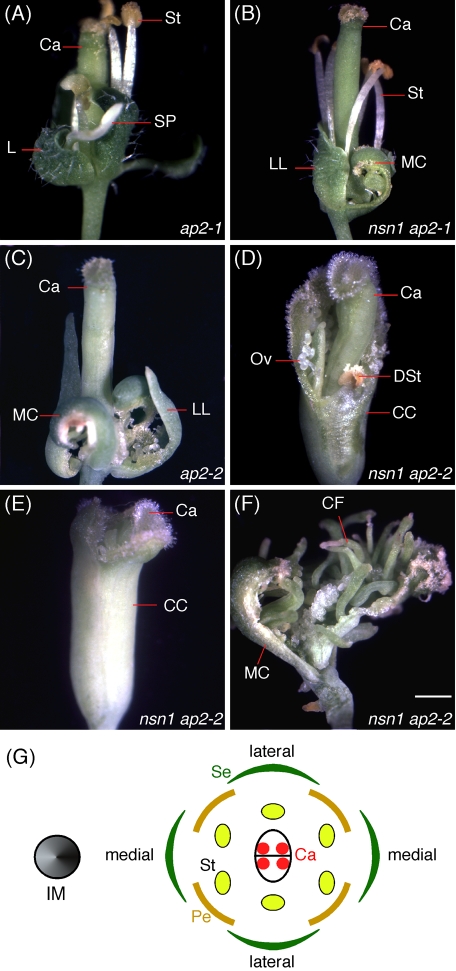
To analyze whether the ectopic AG expression during floral development in nsn1 was dependent upon APETALA2 (AP2), we introduced two ap2 loss-of-function mutations (ap2-1 and ap2-2) into the nsn1-1 mutant. In flowers of ap2-1, a weak ap2 allele, organs of whorls 1 and 2 were converted into leafy bracts (4.0 ± 0) and staminoid petals (4.0 ± 0), respectively, whereas the stamens (6.0 ± 0) and carpels (2.0 ± 0) remained normal in whorls 3 and 4 (Figure 7A). Flowers of nsn1-1 ap2-1 double mutants were more severely defective than those of either of the single mutant (Supplemental Table S1). The number of whorl 1 organs was reduced from 4.0 ± 0 to 3.4 ± 0.9, and they included leafy bracts in lateral positions and carpelloid bracts in medial positions (Figure 7B and Supplemental Table S1). In addition, whorl 2 organs disappeared completely. This phenotype of the double mutant was more severe than that of ap2-1 single mutant and very close to that of ap2-2, a strong mutant allele. In ap2-2, the four whorl 1 organs were converted into two medial carpel and two lateral leafy bracts. There were no organs in whorls 2 and 3, whereas whorl 4 contained normal carpels (Figure 7C). In nsn1-1 ap2-2 double mutant, the whorl 1 organs were converted into a carpelloid cylinder that partially or completely surrounded a normal whorl 4 gynoecium (Figure 7, D and E, and Supplemental Table S1). Stigmatic tissue was developed on the edge of the carpelloid cylinder, and ovules and degenerated stamens were present on the interior wall of the cylinder (Figure 7D). Some double-mutant flowers became an open gynoecium consisting of two medial carpel and numerous carpelloid filaments with stigmatic tissue on their tips (Figure 7F). Thus the homeotic transformation was enhanced in nsn1 ap2-2 double mutant as compared with that of either single mutant. The additive effect of AP2 and NSN1 on flower formation suggests that the two genes have distinct roles in floral development, possibly via independent repression of the AG gene.
FIGURE 7:
Analysis of nsn1-1 ap2 double mutants. (A–C) Flowers of the ap2-1 weak allele (A) and ap2-2 severe allele (C). The ap2-1 flower contained leafy bracts (L) in whorl 1 and staminoid petals (SP) in whorl 2, stamens (St) in whorl 3, and carpels (Ca) in whorl 4. The ap2-2 flower had medial carpels (MC) and lateral leafy bracts (LL) in whorl 1, and no floral organs appear in whorls 2 and 3. B, Flower of nsn1-1 ap2-1 double mutant contained medial carpels (MC) and lateral leafy bracts (LL) in whorl 1. No floral organ appears in whorl 2. (D, E) In flowers of nsn1-1 ap2-2 double mutant, the whorl 1 organs were converted into a carpelloid cylinder (CC) that encapsulated normal whorl 4 carpels (Ca) partially (D) or completely (E). Ovules (O) and degenerated stamens (DSt) were present on the interior wall of the cylinder. (F) A terminal flower of nsn1-1 ap2-2 contained an open gynoecium consisting of two medial carpel (MC) and numerous carpelloid filaments (CF) with stigmatic tissue on their tips. Scale bars, 1 mm (A–F). (G) Diagram of a wild-type flower with respect to the inflorescence meristem (IM). A medial position is on the same axis as IM, whereas a lateral position is on the sides of the axis. A WT flower consists of four green sepals (Se) in whorl 1, four petals (Pe) in whorl 2, six stamens (St) in whorl 3, and two fused carpels (Ca) in whorl 4. Red circles inside the carpels represent ovules.
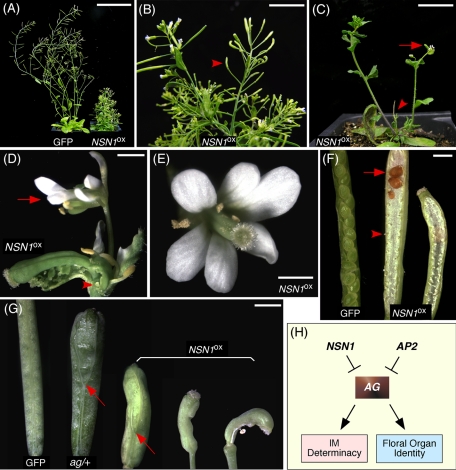
Deleterious growth and reproductive defects caused by NSN1 overexpression
After several transformation attempts, we obtained a total of six transgenic plants expressing 35SPRO:NSN1-GFP in WT genome background (NSN1OX) from a population of ∼30,000 T0 seeds. In contrast, transformation with 35SPRO:GFP (the control) generated 28 transgenic lines from the same size of T0 seed population under the same growth conditions. It is very likely that overexpression of NSN1 may have deleterious effects on plant development. In situ RNA hybridization data showed that the NSN1 mRNA transcript was increased drastically in the SAM, flower primordia, and other tissues such as cortex and pith (Supplemental Figure S3H). Plants of NSN1OX were severely dwarf and bushy and lost apical dominance (Figure 8, A–C), suggesting that NSN1 may have a role in meristem development. All branches eventually became determinate inflorescences by the development of homeotically transformed terminal flowers (Figure 8D). As compared with the terminal flowers of nsn1-knockout mutants (Figure 2, H and I), NSN1OX flowers contained sepals and petals (Figure 8, D and E) and had curled carpels (Figure 8D). Siliques of NSN1OX were short and curled and contained few seeds (Figure 8, F and G). Embryos were arrested at different stages of seed development (Figure 8F). We also compared the phenotypes between NSN1OX and ag mutants. Homozygous ag plants are known to be sterile, whereas heterozygous ag/+ flowers have an increased number of carpels (Figure 8G). Similarly, the gynoecium of NSN1OX flowers often consisted of three carpels (Figure 8G). This observation is intriguing because it further supports the notion that NSN1 may function to repress AG activity during floral organ development (Figure 8H).
FIGURE 8:
Growth and reproductive phenotypes of NSN1OX plants. (A–D) Adult plants of NSN1OX were dwarf and bushy (A) and produced mostly short siliques (arrowhead, B). They lost apical dominance in the main shoot (arrowhead, C) and developed a terminal flower (arrow, C) consisting of deformed sepals, petals, and stamens (arrow, D), as well as unfused carpels with ectopic ovules (arrowhead, D). (E–G) NSN1OX plants produced flowers with an increased number of petals (E) and short and curled siliques (F) containing unfertilized ovules (arrowhead, G) and aborted embryos (arrow, G). Siliques developed from a gynoecium of three fused carpels were often seen in NSN1OX as in ag/+ mutant (arrows, F). Scale bars, 70 mm (A), 10 mm (B), 20 mm (C), 2 mm (D–G). (H) A proposed role of NSN1 as a general negative regulator of AG in the control of inflorescence meristem (IM) determinacy and floral organ identity in Arabidopsis. It may exert its role in a pathway independent of AP2.
DISCUSSION
There are three NS-related genes—NS, GNL3L, and NGP1—in the human and mouse genomes and only two—AtNGP1 and AtNSN1—in Arabidopsis (Figure 1C). There have been limited studies on the biological functions of NGP1 and GNL3L, whereas the research on mammalian NS has recently attracted significant attention, partly because it is expressed specifically in adult and embryonic stem cells and in many tumors and tumor-derived cell lines (Tsai and McKay, 2002; Han et al., 2005; Fan et al., 2006). In this study, we present evidence showing that Arabidopsis NSN1 is required for the maintenance of normal apical meristems and flower formation. It may act as a general negative regulator of AG gene expression, and this function is independent of AP2 (Figure 8H).
NSN1 may act in a gene dosage–dependent manner and is required for flower development
Floral primordia develop from the peripheral zone of the floral meristems in plants. During floral primordia initiation, cells in the peripheral zone continue to divide and differentiate into the primordia of specialized floral organs (Lenhard et al., 2001). Our in situ hybridization data (Figure 5 and Supplemental Figure S3) showed that NSN1 mRNA is expressed in the central and peripheral zones but not in the rib zone that contributes cells to stem growth. During the development of floral primordia, NSN1 mRNA was detectable as early as in the stage 1 primordia and was highly expressed at stages 2 to 3. After stage 3, NSN1 expression was confined to the primordia of sepals, stamens, petals, and gyneocium.
The fact that heterozygous nsn1/+ plants exhibited drastically defective flower phenotypes (Figure 2) implies that NSN1 may act in a gene dosage–sensitive manner. Wild-type plants of Arabidopsis produce indeterminate inflorescences and only occasionally form terminal flowers under stress conditions. However, heterozygous nsn1/+ plants produced determinate inflorescences. One to several terminal carpelloid flowers would always be found in the terminal part of inflorescences in nsn1/+ plants (Figure 2). Homozygous nsn1 plants exhibited severe defects in both vegetative growth and flower formation. The hypothesis of gene dosage effect of NSN1 is also supported by evidence obtained from the overexpression of NSN1 in a wild-type background. Transgenic plants carrying 35SPRO:NSN1-GFP were difficult to obtain. When they survived, they displayed an array of defects in shoot growth and flower development (Figure 8). Thus NSN1 may act as a key regulator of flower development in Arabidopsis, and its function appears to depend on the gene dosage.
NSN1 controls flower organ identity by repressing AG gene expression
In the floral meristem, the meristematic cells sequentially produce sepal, petal, stamen, and carpel primordia. In contrast to the indeterminate shoot meristem, the floral meristem terminates at the end of flower development. The stem cells differentiate completely after the carpel primordia are formed. Several genes, including AP1, LFY, and AG, are known to play important roles during floral meristem initiation. In particular, the MADS-box transcription factor AG has a central role in the eventual termination of stem cell activity after the initiation of stamens and carpels (Yanofsky et al., 1990; Mizukami and Ma, 1997; Lohmann et al., 2001; Deyhle et al., 2007). AG is not expressed in the inflorescence meristem; its expression is highly enhanced after stage 3 of floral development (Lohmann et al., 2001). The relationship of AG and WUS is complicated because the activation of AG during floral meristem initiation requires WUS. It is a suicidal loop: WUS participates in AG activation in the center of the floral meristem and, in turn, the activated AG represses WUS expression, leading to the termination of WUS and stem cell activities.
In ag mutants, WUS expression persists, the stem cell activity in the floral meristem is not turned off on time, and new whorls of organs are produced indefinitely (Yanofsky et al., 1990). In addition, stamens are replaced by petals and carpels are substituted by a reiteration of sepals–petals–petals in ag plants. AG expression triggers stamen and carpel development by temporally and spatially activating an array of genes involved in floral organ formation (Mizukami and Ma, 1997). Elevated AG expression in blr mutants can result in the formation of terminal carpelloid flowers and other striking floral phenotypes, including homeotic transformations from sepals to carpels (Bao et al., 2004). These phenotypes coincide with the floral defects observed in nsn1 (Figure 4). We reasoned that NSN1 might act like BLR as a corepressor of AG and tested this hypothesis by assaying the GUS reporter activity in nsn1 plants harboring the KB9 construct (AG::GUS). As compared with the reporter activity in the wild-type background, the GUS activity in nsn1 was highly enhanced (Figure 6, D–F). This elevated AG expression level was even prominent in the terminal carpelloid flowers developed in nsn1 plants (Figure 6F). The conclusion drawn from the AG::GUS reporter experiments is consistent with data obtained using AG mRNA in situ hybridization (Figure 6, A–C). Taken together, these data support the hypothesis that NSN1 acts as a negative regulator of AG expression.
A possible role of NSN1
Proper regulation of AG transcription requires the presence of the cis-DNA sequences located in its 3-kb second intron, which serves as the binding site for both positive and negative regulators (Busch et al., 1999; Deyholos and Sieburth, 2000; Lohmann et al., 2001). AP2 is a major negative regulator of AG. In fact, AG and AP2 reciprocally repress one another (Bowman et al., 1991; Mizukami and Ma, 1992). In addition to AP2, several other genes, including LUG, LEUNIG_HOMOLOG (LUH), SEU, and BLR, have been identified as negative regulators of AG gene expression (Drews et al., 1991; Liu and Meyerowitz, 1995; Franks et al., 2002; Bao et al., 2004; Sridhar et al., 2004; Sitaraman et al., 2008; Das et al., 2009). AP2, BLR, and PAN have been shown to bind directly to the second intron of AG (Deyholos and Sieburth, 2000; Bao et al., 2004; Das et al., 2009). In this article, we present evidence showing that NSN1 is a new and major regulator of AG. It is possible that NSN1 may regulate AG expression via direct or indirect control of the protein complex bound to the second intro of AG. Because our data also suggest that NSN1 functions in an AP2-independent manner (Figure 7), the regulation of AG by NSN1 may occur through regulators rather than AP2 (Figure 8H). The fact that NSN1 is not a putative transcription factor but instead is a nucleolar GTP-binding protein implies the existence of a new mechanism underlying the regulation of AG gene expression during flower development. Further biochemical and genetic investigations are needed in order to understand the molecular basis of the regulation of AG by NSN1.
MATERIALS AND METHODS
Arabidopsis lines and plant growth
The nsn1-1 T-DNA insertion line (SALK_029201) and seeds of ap2-2 were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). The transposon insertion allele of nsn1-2 (RAFL11-2045-1) was received from RIKEN (www.brc.riken.go.jp). Seeds of ag/+ harboring KB9 were gifts from M. Yanofsky and D. Weigel (Busch et al., 1999; Lohmann et al., 2001). Plants were grown in soil under long-day conditions (16 h light/8 h darkness) at 21°C.
Genetic analysis of double mutants
To introduce KB9 into the nsn1-1 mutant, pollen grains from the KB9 plant were crossed to the heterozygote nsn1/+ stigma. F2 plants were analyzed for their genetic background by genomic-PCR, and heterozygote nsn1/+ plants carrying KB9 were used for GUS staining. After GUS staining, the plant specimens were treated in 70% ethanol overnight. Images were taken under a dissection microscope (Leica, Wetzlar, Germany).
Pollen grains from ap2-2 were used to fertilize heterozygote nsn1/+ stigma. F1 plants were allowed for self-pollination. F2 plants were analyzed for their genetic backgrounds by genomic PCR using primers for NSN1 and the dCAPs marker developed for ap2-2. F2 plants of nsn1-1 ap2-2 double mutants were analyzed for their flower phenotypes. For generation of nsn1-1 ag double mutant, ag/+ pollen grains were used to fertilize heterozygote nsn1/+ stigma. F1 generation plants were allowed for self-pollination. F2 plants were analyzed by PCR to identify homozygote nsn1-1 ag double mutant. Flowers of nsn1-1 ag plants were photographed.
RNA isolation and RT-PCR
Total RNA was isolated from plants using RNeasy Plant Mini Kit (Qiagen, Valencia, CA). The first-strand cDNA was synthesized from 2 μg of total RNA with SuperScript First-Strand Synthesis system for RT-PCR (Invitrogen, Carlsbad, CA). A 1-μl amount of the synthesized cDNA was used for PCR amplification. Three pairs of primers were used to identity nsn1-1 as a null mutant. One pair of primers NSN1-F and N857R1 located on the first and second exons, respectively (see Supplemental Figure S3A), were used to amplify a PCR fragment with an expected size of 133 base pairs. Nsn1-LP and Nsn1-RP located on the both sides of the T-DNA insertion were used for verification of the T-DNA. The third pair of primers, Nsn1-F and Nsn1-R, was used to determine whether the full-length NSN1 mRNA (1760 base pairs) was present. The ACTIN mRNA used as an internal control was amplified using ACTIN primers ACT-F and ACT-R.
Peptide motif and phylogenetic analysis
Motif analysis of NSN1 was performed using ExPASy tools (www.expasy.ch/tools). For phylogenetic analysis, peptide sequences were aligned using ClustalX 2.0, and the tree was generated using Bootstrap neighboring joining program with a bootstrap value of 1000 trials. The phylogenetic tree was displayed in Phylip format on TreeView X.
Plant transformation
The coding region of NSN1 cDNA was amplified by RT-PCR using primers NSN1-F and NSN1-R and into pCR2.1 (Invitrogen). The insert was excised with XhoI and SacI and was subcloned into pMON-GFP. The entire cassette containing CaMV 35SPRO:NSN1-GFP:NOS3′ was excised by digestion with NotI and subcloned into binary vector pMON18342 (Hong et al., 2001), generating pMON-NSN1-GFP. The plasmid was introduced into Agrobacterium tumefaciens strain ABI by electroporation. Transformation of cultured tobacco BY-2 cells was performed as described previously (Hong et al., 2001). Wild-type plants of Arabidopsis thaliana ecotype Columbia were transformed by the floral dip method. Transgenic BY-2 cells were selected on 1/2 MS agar plates containing 200 mg/l kanamycin and 500 mg/l carbencillin, and transgenic plants were selected on 1/2 MS agar plates containing 50 mg/l kanamycin. GFP images of the transgenic cells were taken with a Zeiss confocal laser scanning microscope (Carl Zeiss, Jena, Germany).
Complementation of nsn1-1 mutation
A TAC clone (JatY64G16) containing an insert of a 77.6-kb genomic fragment in pYLTAC17 vector, obtained from the John Innes Center (Norwich, United Kingdom), was used for complementation of the nsn1-1 mutation. The plasmid was transferred into Agrobacterium tumefaciens strain ABI cells by electroporation. This clone contains 19 genes, including At3g07050 (NSN1). Heterozygote nsn1-1/+ plants were transformed by the Agrobacterium-mediated floral dip method. Transgenic plants were selected with phosphinothricin (BASTA). T2 plants that carried the transgene on the homozygote nsn1 background were maintained. T3 plants were tested for complementation.
In situ RNA hybridization
Digoxigenin-labeled riboprobes complementary to parts of NSN1, AG, CLAVATA 3 (CLV3), and WUS genes were used to detect their mRNAs in inflorescence and floral meristems as described (Goodrich et al., 1997). The NSN1 cDNA fragment corresponding to the C-terminal half of its coding region was amplified by PCR using primers NSN1-LP and NSN1-RP and cloned into pCRII (Invitrogen) to yield pNSN. The plasmid was linearized with ApaI and transcribed with SP6 RNA polymerase (Roche, Indianapolis, IN) to produce antisense RNA probe. For the control with the NSN1 sense RNA probe, pNSN1 was linearized with BamH1 and transcribed with T7 RNA polymerase (Roche). For AGAMOUS probe, cDNA fragment was amplified by AG primers AG-F and AG-R as described previously (Yanofsky et al., 1990).
Histology and GUS staining
Plant tissues were fixed in FAA solution (3.7% formaldehyde, 5% acetic acid, and 50% ethanol) at room temperature for 16 h. The fixed tissues were dehydrated in a series of ethanol (30, 50, 70, 85, 90, and 100%), cleared with xylene, and embedded in Paraplast (Sigma-Aldrich, St. Louis, MO) as described previously (Causier et al., 2005). Serial 8-μm-thick sections were transferred to polylysine-coated slides (Sigma-Aldrich) and stained with 0.01% toludine blue for 5–10 min at room temperature after dewaxing. GUS staining was performed as described (Schoof et al., 2000).
Scanning electronic microscopy
Plant tissues were fixed overnight in 50 mM phosphate butter containing 2% glutaraldehyde and 2% paraformaldehyde. Fixed tissues were rinsed three times with 50 mM phosphate buffer and kept in osmium tetroxide (1%) overnight at 4°C. The tissues were rinsed three times with 50 mM phosphate buffer and dehydrated in a 30, 50, 70, 95, and 100% alcohol gradient. After critical point drying, the tissues were coated with gold and observed under scanning electronic microscopy.
Supplementary Material
Acknowledgments
We thank A. Caplan for critical comments on the manuscript and the Arabidopsis Biological Resource Center, RIKEN, and M. Yanofsky, T. Laux, and D. Weigel for providing Arabidopsis seeds. We also thank Haiqing Sheng, Shizhen Gu, and Christina Mitchell for technical assistance during the course of this study. This work was supported by National Science Foundation grants (MCB 0548525 and IOB 0543923 to Z.H.) and an IDeA Network of Biomedical Research Excellence Scholars Program (National Institutes of Health/National Center for Research Resources P20RR016454) Fellowship (to D.K.G.).
Abbreviations used:
- AG
AGAMOUS
- AP
APETALA
- CLV
CLAVATA
- NS
nucleostemin
- NSN1
nucleostemin-like 1
- TCF
terminal carpelloid flower
- WUS
WUSCHEL
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0797) on February 22, 2012.
REFERENCES
- Bao X, Franks RG, Levin JZ, Liu Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell. 2004;16:1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman C, Nichane M, De Clercq S, Maetens M, Floss T, Wurst W, Bellefroid E, Marine JC. Evolutionarily conserved role of nucleostemin: controlling proliferation of stem progenitor cells during early vertebrate development. Mol Cell Biol. 2006;26:9291–9301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285:585–587. doi: 10.1126/science.285.5427.585. [DOI] [PubMed] [Google Scholar]
- Causier B, Castillo R, Zhou J, Ingram R, Xue Y, Schwarz-Sommer Z, Davies B. Evolution in action: following function in duplicated floral homeotic genes. Curr Biol. 2005;15:1508–1512. doi: 10.1016/j.cub.2005.07.063. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Meyerowitz EM. HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol Cell. 1999;3:349–360. doi: 10.1016/s1097-2765(00)80462-1. [DOI] [PubMed] [Google Scholar]
- Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle DM, Rossi L, Berghuis AM, Aravind L, Koonin EV, Brown ED. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry. 2002;41:11109–11117. doi: 10.1021/bi020355q. [DOI] [PubMed] [Google Scholar]
- Das P, Ito T, Wellmer F, Vernoux T, Dedieu A, Traas J, Meyerowitz EM. Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development. 2009;136:1605–1611. doi: 10.1242/dev.035436. [DOI] [PubMed] [Google Scholar]
- Deyhle F, Sarkar AK, Tucker J, Laux T. WUSCHEL regulates cell differentiation during anther development. Dev Biol. 2007;302:154–159. doi: 10.1016/j.ydbio.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Deyholos MK, Sieburth LE. Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell. 2000;12:1799–1810. doi: 10.1105/tpc.12.10.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Du X, Rao MR, Chen XQ, Wu W, Mahalingam S, Balasundaram D. The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA. Mol Biol Cell. 2006;17:460–474. doi: 10.1091/mbc.E05-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Liu Z, Zhao S, Lou F, Nilsson S, Ekman P, Xu D, Fang X. Nucleostemin mRNA is expressed in both normal and malignant renal tissues. Br J Cancer. 2006;94:1658–1662. doi: 10.1038/sj.bjc.6603145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development. 2002;129:253–263. doi: 10.1242/dev.129.1.253. [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- Han C, Zhang X, Xu W, Wang W, Qian H, Chen Y. Cloning of the nucleostemin gene and its function in transforming human embryonic bone marrow mesenchymal stem cells into F6 tumor cells. Int J Mol Med. 2005;16:205–213. [PubMed] [Google Scholar]
- Hong Z, Delauney AJ, Verma DP. A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell. 2001;13:755–768. doi: 10.1105/tpc.13.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarnejad SM, Mowla SJ, Matin MM. Knocking-down the expression of nucleostemin significantly decreases rate of proliferation of rat bone marrow stromal stem cells in an apparently p53-independent manner. Cell Prolif. 2008;41:28–35. doi: 10.1111/j.1365-2184.2007.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudron MM, Reinke V. C. elegans nucleostemin is required for larval growth and germline stem cell division. PLoS Genet. 2008;4:e1000181. doi: 10.1371/journal.pgen.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development. 1995;121:975–991. doi: 10.1242/dev.121.4.975. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell. 2007;18:2630–2635. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Zhu Q, Tsai RY. Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol Cell Biol. 2007;27:8670–8682. doi: 10.1128/MCB.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Polena I, Trask I, Bazett-Jones DP, Pederson T. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell. 2005;16:3401–3410. doi: 10.1091/mbc.E05-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud EG, Andrade MA, Bonneau F, Ly TB, Knop M, Scheffzek K, Pepperkok R. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 2005;3:21. doi: 10.1186/1741-7007-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L, Grand A, Zhang L, Rayner S, Katoku-Kikyo N, Kellner S, Kikyo N. Critical role of nucleostemin in pre-rRNA processing. J Biol Chem. 2009;284:4968–4977. doi: 10.1074/jbc.M804594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowitz EM. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman J, Bui M, Liu Z. LEUNIG HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol. 2008;147:672–681. doi: 10.1104/pp.108.115923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA. 2004;101:11494–11499. doi: 10.1073/pnas.0403055101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Xu Y, Ng KH, Ito T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol. 2006;26:9279–9290. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.