Trafficking of glucocerebrosidase (GBA) enzyme from the endoplasmic reticulum to the lysosome requires lysosomal integral membrane protein type 2 (LIMP-2), which is a receptor for GBA. This study shows that phosphatidylinositol 4-kinase (PI4K) type IIIβ controls the exit of LIMP-2/GBA complex from the Golgi, while PI4KIIα is required for the post-Golgi trafficking of the complex via the late endosomes.
Abstract
Gaucher disease is a lysosomal storage disorder caused by a defect in the degradation of glucosylceramide catalyzed by the lysosomal enzyme β-glucocerebrosidase (GBA). GBA reaches lysosomes via association with its receptor, lysosomal integral membrane protein type 2 (LIMP-2). We found that distinct phosphatidylinositol 4-kinases (PI4Ks) play important roles at multiple steps in the trafficking pathway of the LIMP-2/GBA complex. Acute depletion of phosphatidylinositol 4-phosphate in the Golgi caused accumulation of LIMP-2 in this compartment, and PI4KIIIβ was found to be responsible for controlling the exit of LIMP-2 from the Golgi. In contrast, depletion of PI4KIIα blocked trafficking at a post-Golgi compartment, leading to accumulation of LIMP-2 in enlarged endosomal vesicles. PI4KIIα depletion also caused secretion of missorted GBA into the medium, which was attenuated by limiting LIMP-2/GBA exit from the Golgi by PI4KIIIβ inhibitors. These studies identified PI4KIIIβ and PI4KIIα as important regulators of lysosomal delivery of GBA, revealing a new element of control to sphingolipid homeostasis by phosphoinositides.
INTRODUCTION
Maintenance of cellular homeostasis in eukaryotes depends on efficient breakdown of macromolecules, such as proteins, lipids, glycoconjugates, and nucleic acids. Lysosomes are the primary sites of macromolecule degradation, with over 50 different soluble hydrolases present in their lumen (Pohl et al., 2009). Delivery of newly synthesized hydrolases from the endoplasmic reticulum (ER) and the Golgi to lysosomes is therefore critical for proper degradation. Most soluble lysosomal enzymes contain a mannose-6-phosphate (M6P) residue that is recognized in the trans-Golgi network (TGN) by cation-dependent or cation-independent M6P receptors (CD-M6PRs or CI-M6PRs, respectively; Kornfeld and Mellman, 1989). Both receptors are integral membrane proteins that release their lysosomal cargo into the lumen of sorting endosomes (SE) and late endosomes (LE) under increasingly acidic conditions. The receptors are then retrieved to the TGN by retrograde trafficking for subsequent rounds of transport (Sannerud et al., 2003). Mutations that cause loss of M6P moieties prevent lysosomal delivery of hydrolases, resulting in hydrolase secretion and compromised lysosomal function, as observed in patients with I-cell disease (Reitman et al., 1981). However, even under such conditions, certain lysosomal proteins, including sphingolipid activator proteins and β-glucocerebrosidase (GBA), retain their lysosomal distribution, indicative of the existence of M6P-independent pathways to lysosomes (Rijnboutt et al., 1991).
The lysosomal integral membrane protein type 2 (LIMP-2, encoded by SCARB2) has recently been identified as a receptor for GBA (Reczek et al., 2007). LIMP-2 is a type III transmembrane protein with a glycosylated luminal loop and a short cytosolic tail that harbors a “dileucine-based” sorting motif (DERAPLI) critical for AP-3 binding and sorting to lysosomes (Vega et al., 1991; Sandoval et al., 1994, 2000; Honing et al., 1998). LIMP-2 binds GBA in the ER, and the two proteins traverse Golgi and endocytic compartments together en route to lysosomes, at which the acidic pH facilitates dissociation of GBA from LIMP-2 (Reczek et al., 2007; Blanz et al., 2010). Lysosomal delivery of GBA is critical for regulation of sphingolipid metabolism, since GBA is the only enzyme capable of efficiently hydrolyzing lysosomal glucosylceramide (GlcCer; Peters et al., 1976; Daniels et al., 1980). Mutations in GBA in humans that abolish LIMP-2 binding result in missorting and secretion of the enzyme, leading to lysosomal accumulation of GlcCer and causing Gaucher disease, the most common metabolic storage disorder (Beutler, 1991; Reczek et al., 2007). Consistent with its role in GBA delivery, total depletion of LIMP-2 results in missorting of GBA in vitro and in vivo, leading to secretion of the enzyme. Macrophages and mouse embryonic fibroblasts (MEFs) derived from LIMP-2 knockout mice secrete as much as 50% of the total GBA, compared with 7% observed in wild-type cells (Reczek et al., 2007), and sera of LIMP-2 knockout mice showed an 11-fold increase in GBA activity, compared with GBA activity of wild-type mice (Reczek et al., 2007). While these findings unequivocally establish LIMP-2 as a GBA receptor, surprisingly little is known about the regulation of various trafficking steps occurring along LIMP-2/GBA transport from Golgi to lysosomes.
Sorting of many proteins at the Golgi depends on the availability of the regulatory lipid, phosphatidylinositol 4-phosphate (PtdIns4P; De Matteis et al., 2005). Acute depletion of PtdIns4P in Golgi membranes results in impaired CI-M6PR exit from the Golgi (Szentpetery et al., 2010). PtdIns4P synthesis in mammalian cells is carried out by four PtdIns4P kinases (PI4Ks) belonging either to type II (IIα and IIβ) or type III (IIIα and IIIβ) families. While all of these enzymes exhibit some degree of localization to the Golgi complex, synthesis of PtdIns4P in Golgi membranes is primarily mediated by PI4KIIα and PI4KIIIβ (Wang et al., 2003; Godi et al., 2004; Weixel et al., 2005). Notably, distinct PI4K isoforms have been shown to regulate specific Golgi/TGN sorting processes. For example, Golgi-localized PI4KIIα is required for Golgi recruitment of the clathrin adaptor proteins AP-1 and γ-ear–containing, ARF-binding proteins (GGAs) (Wang et al., 2003, 2007). Accordingly, depletion of PI4KIIIβ does not impair recruitment of AP-1 to the Golgi, demonstrating a preference of this sorting complex for PI4KIIα-derived PtdIns4P (Wang et al., 2003). Consequently, trafficking of AP-1 cargo molecules, such as CI-M6PR, between Golgi and LE is specifically regulated by PI4KIIα (Wang et al., 2003). Similarly, PI4KIIα associates with and regulates the AP-3 clathrin adaptor, adding a noncatalytic component to its regulation of AP-3–dependent cargo sorting (Craige et al., 2008; Salazar et al., 2009). In contrast, PI4KIIIβ plays a role in Golgi recruitment of the ceramide (Cer) transfer protein (CERT; Toth et al., 2006) via a PtdIns4P-binding pleckstrin homology (PH) domain present in CERT (Hanada et al., 2003; D'Angelo et al., 2007). In fact, Golgi recruitment of several other lipid transfer proteins possessing PH domains that bind PtdIns4P, such as the four-phosphate adaptor proteins (FAPP-1 and FAPP-2) and oxysterol-binding protein, also rely on production of PtdIns4P (Dowler et al., 2000; Godi et al., 2004; Raychaudhuri et al., 2006; Banerji et al., 2010). In these cases, the relative roles of the two enzymes, PI4KIIα and PI4KIIIβ, have not been clearly defined (Levine and Munro, 2002; Balla et al., 2005). The roles of PI4Ks, therefore, are tightly linked to the control of cellular lipid metabolism in general, with multiple connections to sphingolipid synthesis and transport.
In the present study, we describe a new level of control by which PtdIns4P indirectly affects sphingolipid metabolism. We show that PI4KIIIβ and PI4KIIα play active roles at sequential trafficking steps in the lysosomal transport of the GBA hydrolase in complex with its receptor LIMP-2. We demonstrate the requirement for the catalytic activity of PI4KIIIβ in Golgi exit of the LIMP-2/GBA complex, which is followed by PI4KIIα-mediated trafficking to lysosomes. In addition to the established function of PI4Ks in the transport of sphingolipids along the synthetic route, these results describe for the first time a role for PI4Ks in sphingolipid catabolism as regulators of lysosomal delivery of a key sphingolipid hydrolase.
RESULTS
Identification of PI4KIIIβ interaction with LIMP-2/GBA complex
The yeast homologue of PI4KIIIβ, Pik1, has previously been shown to have a role in Golgi-to-plasma membrane and Golgi-to-vacuole transport (Strahl and Thorner, 2007). Similarly, Golgi-localized mammalian PI4KIIIβ has been implicated in regulation of sorting of various cargoes to the plasma membrane (Godi et al., 1999; Weisz et al., 2000; Hausser et al., 2005), but the role of PI4KIIIβ in sorting to endocytic compartments remains less understood. Accordingly, we sought to identify new protein-binding partners of this kinase that are involved in endocytic transport. Affinity purification coupled to mass spectrometry (AP-MS) is a powerful method for identifying and characterizing interaction partners (Vasilescu and Figeys, 2006; Goudreault et al., 2009). We used FLAG-tagged human PI4KIIIβ stably expressed in HEK293 cells to perform AP-MS. FLAG-PI4KIIIβ and its interacting partners were purified, subjected to proteolytic digestion, and identified using gel-free MS. This approach readily reproduced previously detected PI4KIIIβ binding partners, such as various 14-3-3 isoforms (YWHA genes) and Rab11 (de Graaf et al., 2004; Hausser et al., 2006; Polevoy et al., 2009). In addition, several novel PI4KIIIβ interaction partners were identified (Table 1, Supplemental Table S1, and Supplemental Figure S1) including GBA and its receptor, LIMP-2, indicating that LIMP-2/GBA could potentially bind as a complex to PI4KIIIβ. Therefore we assessed the functional relevance of this interaction by focusing on the catalytic role of PI4KIIIβ at the Golgi in sorting of the subpopulation of LIMP-2/GBA traversing the Golgi, en route to lysosomes.
TABLE 1:
List of FLAG-PI4KIIIβ interactors and average spectra detected using AP-MS.
| Hit | Averaged spectra |
|---|---|
| GBA | 60 |
| YWHAB | 40.5 |
| YWHAG | 40 |
| YWHAH | 38 |
| YWHAQ | 31.5 |
| RAB11B | 13 |
| C10orf76 | 9 |
| LIMP-2 | 5 |
| MTA2 | 3.5 |
| RAB11FIP2 | 3.5 |
| MRPL11 | 2 |
FLAG-tagged PI4KIIIβ was stably expressed in HEK293 cells and used as bait to perform biological replicates of AP-MS (n = 2). Data were analyzed with ProHits (Liu et al., 2010) and exported to an Excel file. Filtering to remove background proteins is described in Materials and Methods. Only proteins that appeared in both biological replicates are reported in the left column. The total spectral counts of the two biological replicates are averaged and presented in the right column. This is diagrammed in Figure S1. The details of each separate FLAG-PI4KIIIβ AP-MS experiment are presented in Table S1.
PtdIns4P regulates efficient LIMP-2 transport out of the Golgi
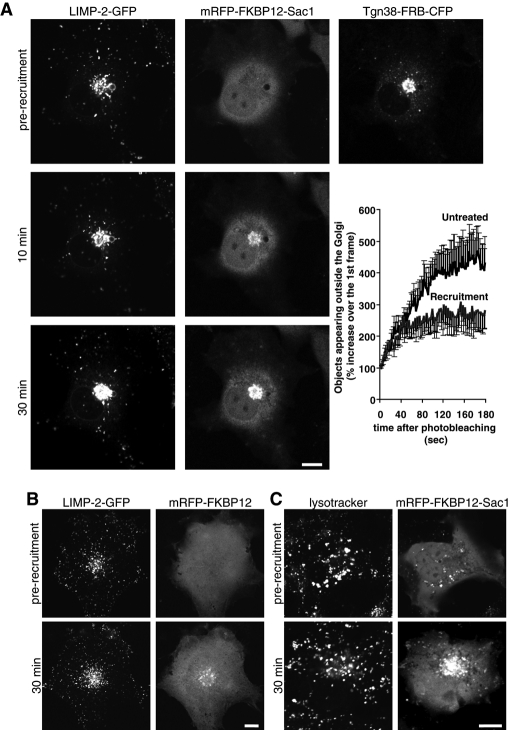
The role of PtdIns4P in regulation of LIMP-2 exit from the Golgi was assessed in live COS-7 cells using a green fluorescent protein (GFP)-tagged LIMP-2 under conditions of acute PtdIns4P depletion. Rapamycin-induced Golgi recruitment of a cytosolic version of the PtdIns4P phosphatase Sac1, lacking the ER-localization sequence, was proven to be a valuable tool in studying the role of Golgi PtdIns4P in endocytic transport (Szentpetery et al., 2010). The type I Golgi protein, Tgn38, fused to the FKBP12-rapamycin–binding (FRB) domain of mTOR was used to recruit cytosolic Sac1 phosphatase fused to the recruiter protein FKBP12. Rapamycin addition induces heterodimerization of FRB domain to the FKBP12 module, resulting in rapid depletion of PtdIns4P at the Golgi by the recruited Sac1 enzyme (Szentpetery et al., 2010).
Subcellular distribution of LIMP-2-GFP in untreated COS-7 cells was mostly confined to acidic LE/lysosomal vesicles (Figure S2A), a compartment also enriched in another abundant lysosomal resident protein, lysosomal-associated membrane protein 1 (LAMP-1). This distribution was in good agreement with published data on the distribution of the endogenous LIMP-2 protein (Vega et al., 1991; Kuronita et al., 2002). Some LIMP-2–containing vesicles also had detectable levels of internalized rhodamine-labeled epidermal growth factor (Rh-EGF), while being mostly devoid of CI-M6PR (Figure S2, B–D).
Effects of PtdIns4P elimination were studied in COS-7 cells transfected with LIMP-2-GFP, monomeric red fluorescent protein-FKBP12-Sac1 (mRFP-FKBP12-Sac1), and Tgn38-FRB-cyan fluorescent protein (Tgn38-FRB-CFP). Addition of rapamycin resulted in rapid redistribution of cytosolic Sac1 to the Golgi that was detectable after 3 min (Szentpetery et al., 2010). The resulting depletion of PtdIns4P led to Golgi accumulation of LIMP-2-GFP and a corresponding decrease in the rate of Golgi exit without noticeable effect on the peripheral lysosomal pool of the protein (Figure 1A). In control experiments in which LIMP-2-GFP and Tgn38-FRB-CFP were coexpressed together with mRFP-FKBP12 protein lacking the Sac1 phosphatase, rapamycin-induced recruitment of FKBP12 did not affect subcellular distribution of LIMP-2 (Figure 1B). Furthermore, PtdIns4P depletion at the Golgi did not cause any notable change in the distribution of acidic lysosomal vesicles marked by LysoTracker Green (Figure 1C). Taken together, these results suggested that PtdIns4P elimination at the Golgi leads to accumulation of LIMP-2 at this compartment without redistribution of peripheral lysosomes.
FIGURE 1:
The effect of PtdIns4P elimination at the Golgi on LIMP-2 distribution in COS-7 cells. Cells coexpressing either mRFP-FKBP12-Sac1 (A) or mRFP-FKBP12 (B) together with Tgn38-FRB-CFP and LIMP-2-GFP were mounted on the microscope's heated stage (35°C) and treated with 100 nM rapamycin to induce recruitment of the cytosolic-FKBP12 constructs to the Golgi. Time-lapse images of individual cells were recorded for 30 min, and representative images are shown at 0 min (prior to recruitment) and at 10 and 30 min after recruitment. Graph shows morphometric analysis of the recordings in (A) using ImageJ software. The curves represent the number of individual fluorescent vesicles appearing outside the Golgi 15 min after the addition of rapamycin and photobleaching the whole cell outside of the Golgi area. The graph illustrates the first 200 s of the recordings taken at 0.5 frames/s. Each value represents a mean ± SEM of five recordings for each sample (control and rapamycin-treated). (C) Cells expressing mRFP-FKBP12-Sac1 and Tgn38-FRB-CFP were preincubated for 10 min with 100 nM LysoTracker Green at 35°C prior to addition of rapamycin. Scale bars: 10 μm.
LIMP-2 exit from the Golgi depends on PI4KIIIβ activity
Although LIMP-2 associates with GBA in the ER (Reczek et al., 2007), steady-state distribution of LIMP-2 is primarily confined to LE/lysosomes (Vega et al., 1991; Kuronita et al., 2002). Accordingly, full-length LIMP-2 does not exhibit significant colocalization with the Golgi-associated PI4KIIIβ (Figure S3A), and shows only partial colocalization with the endosomal pool of PI4KIIα (Figure S3C). Interestingly, a LIMP-2 construct containing only the second transmembrane domain and the short, cytosolic tail with the dileucine motif exhibited a more pronounced Golgi retention, while still correctly reaching the lysosomal compartment. This truncated LIMP-2 construct showed clear colocalization with PI4KIIIβ on Golgi membranes (Figure S3B), making it a potentially useful tool in assessing the role of PI4KIIIβ in LIMP-2 Golgi sorting.
To evaluate which of the two Golgi-localized PI4Ks is important for LIMP-2 trafficking via the Golgi, we took advantage of an isoform-specific PIK inhibitor, PIK93, that potently inhibits PI4KIIIβ (IC50 = 19 nM; Knight et al., 2006; Balla et al., 2008). Since the PI4KIIIα enzyme is less affected, and the type II PI4Ks are insensitive to PIK93, this inhibitor has already been successfully used to show the importance of PI4KIIIβ in CERT-mediated transfer of Cer from the ER to the TGN (Toth et al., 2006).
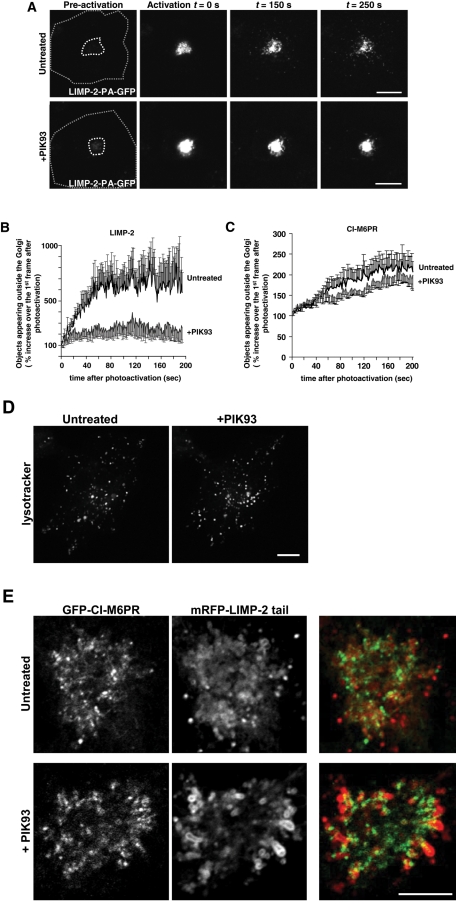
To follow the exit of LIMP-2 from the Golgi, we generated a LIMP-2 construct fused to a photoactivatable GFP. After photoactivation of LIMP-2 molecules at the Golgi, this construct enabled us to follow LIMP-2 trafficking out of the Golgi in time-lapse imaging in live cells. By quantifying the number of fluorescent vesicles appearing outside the photoactivated area, we were able to determine the rate of LIMP-2 exit from the Golgi. COS-7 cells were transfected with photoactivatable LIMP-2 (LIMP-2-PA-GFP) together with a monomeric Kusabira-Orange (mKO)-tagged Golgi enzyme galactosyltransferase (mKO-GalT) to identify the Golgi area. After photoactivation of the Golgi area, an increasing number of LIMP-2-containing vesicles were seen to emanate out of the Golgi, with a corresponding decrease in total fluorescence at the Golgi (Figure 2A, top panels). However, when cells were pretreated with PIK93 for 15 min, LIMP-2 was seen to remain at the site of photoactivation and showed significantly fewer vesicles emanating from the Golgi (Figure 2A, bottom panels). Since some LIMP-2–containing vesicles are always found in the pericentriolar area, the fluorescent vesicles still leaving the photoactivated area in PIK93-treated cells most likely represent non-Golgi vesicles present close to the Golgi at the time of photoactivation (Figure 2B). Distribution of the acidic LE/lysosomal vesicles, marked by LysoTracker (Figure 2D), did not noticeably change following the treatment with PIK93. Moreover, PIK93 treatment had only minimal effects on the rate of CI-M6PR exit from the Golgi (Figure 2C), consistent with previous reports showing CI-M6PR trafficking between Golgi and LEs was PI4KIIα-dependent (Wang et al., 2003). At the same time, sorting of LAMP-1 was also significantly impaired by PIK93 treatment (Figure S4, A and B). A LIMP-2 point mutant with significantly abrogated GBA binding (Blanz et al., 2010) was found to exhibit a similar degree of PIK93 inhibition of Golgi exit compared the wild-type protein (Figure S4C), indicating that LIMP-2 Golgi sorting did not depend on its binding to GBA.
FIGURE 2:
PI4KIIIβ inhibition affects LIMP-2 Golgi exit. (A) COS-7 cells were transfected with LIMP-2-PA-GFP together with mKO-GalT. Golgi area was photoactivated based on the GalT distribution (inner dashed line) using a 405-nm laser line. Emergence of Golgi-derived vesicles containing photoactivated LIMP-2 molecules was then followed in the GFP channel by time-lapse microscopy of control cells or cells pretreated with PIK93 (1 μM) for 20 min. Scale bars: 10 μm. (B) Morphometric analysis of the recordings in (A). Graphs show the number of individual fluorescent vesicles appearing outside the Golgi photoactivation area (dashed line in (A)) that exhibited a signal above an arbitrary threshold normalized to the value prior to photoactivation. The graph illustrates the first 200 s of the recordings taken at 1 frame/s. Each value represents a mean ± SEM of six recordings for each sample (control and PIK93-treated). (C) Morphometric analysis of COS-7 cells transfected with PA-GFP-CI-M6PR together with mKO-GalT, performed as described in (B), with each value representing a mean ± SEM of 10 recordings for each sample (control and PIK93-treated). (D) COS-7 cells were preincubated for 10 min with 100 nM LysoTracker Green and treated with 1 μM PIK93 for 30 min. Lysosomal distribution was not affected by PI4KIIIβ inhibition. (E) High-magnification image of the Golgi area of a COS-7 cell transfected with mRFP-LIMP-2-tail and GFP-CI-M6PR. LIMP-2-tail exhibits significantly increased Golgi retention in comparison with the full-length protein, but it redistributes and accumulates at peripheral Golgi membranes upon addition of PIK93, while Golgi localization of GFP-CI-M6PR remains unaltered. Scale bars: 10 μm.
To compare the relative distribution of LIMP-2 and CI-M6PR at Golgi membranes, we again used the LIMP-2 tail-mRFP, which exhibits prolonged Golgi residence. When this construct was coexpressed with CI-M6PR, the LIMP-2 tail localized to Golgi membrane domains adjacent to, but clearly distinct from, those containing CI-M6PR (Figure 2E; note that only the Golgi is shown). Remarkably, PI4KIIIβ inhibition led to LIMP-2 tail accumulation within the Golgi, at distinct peripheral membrane folds and away from CI-M6PR, the latter not showing changes in its distribution (Figure 2E). This finding demonstrated the existence of subdomains within the Golgi and the importance of PI4KIIIβ catalytic activity in proper LIMP-2, but not M6PR, sorting at the level of the Golgi.
PI4KIIα is involved in post-Golgi trafficking of LIMP-2 along the degradative pathway
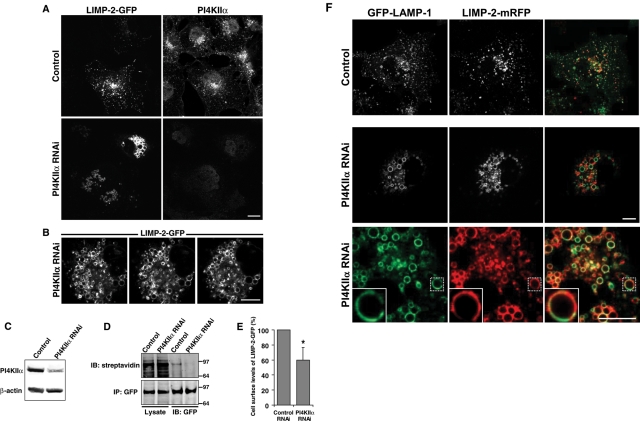
The PI4KIIIβ inhibition data showed that PI4KIIIβ plays a key role in generating the PtdIns4P pool required for proper sorting of LIMP-2 at the Golgi. However, these results do not exclude a potential role for the other Golgi PI4Ks, such as PI4KIIα, in LIMP-2 trafficking. Unfortunately, no inhibitors for type II PI4Ks have been identified to date. Therefore we studied LIMP-2 trafficking to LE/lysosomes after RNA interference (RNAi)-mediated knockdown of PI4KIIα. COS-7 cells were treated with small-interfering RNA (siRNA) targeting PI4KIIα for 2 d (Figure 3C), which was followed by transfection with the LIMP-2-GFP construct for 1 d. Cells were then studied either live or fixed and immunostained for PI4KIIα. Depletion of PI4KIIα resulted in a dramatic accumulation of LIMP-2-GFP in clusters of enlarged vesicles, without any detectable Golgi accumulation (Figures 3A and S5, A–C). Using serial optical sections, we found LIMP-2 clusters to be composed of a collection of large vesicles, many greater than 2 μm in diameter, and tightly compacted in the juxtanuclear area (Figure 3B). Furthermore, biotinylation of cell surface proteins with membrane-impermeable biotin followed by immunoprecipitation of LIMP-2-GFP revealed that the low basal levels of the surface pool of LIMP-2 were even further decreased by ∼40% following the depletion of PI4KIIα (Figure 3, D and E).
FIGURE 3:
PI4KIIα knockdown affects transport of LIMP-2 to lysosomes. Following a 2-d treatment with control siRNA oligos or oligos against PI4KIIα, COS-7 cells were transfected with LIMP-2-GFP alone (A and B) or with LIMP-2-mRFP together with LAMP-1-GFP (F). At 24 h following the transfection, cells were either fixed in 4% paraformaldehyde (A) or immediately analyzed by live-cell confocal microscopy (B and D). Fixed cells were permeabilized and incubated with a rabbit polyclonal antibody against PI4KIIα (A). Primary antibody was detected using the Alexa Fluor 568–conjugated secondary antibody. (B) Live-cell imaging was performed by mounting coverslips on the heated stage (35°C) and recording 0.5-μm-thick serial Z-sections. PI4KIIα knockdown results in accumulation of LIMP-2-GFP in enlarged endosomal structures that form perinuclear clusters spanning several micrometers, as seen in a series of consecutive optical slices. (C) The efficiency of the siRNA knockdown was shown by Western blot analysis using a rabbit anti-PI4KIIα polyclonal antibody and mouse anti-actin antibody. (D) After a 2-d treatment with siRNA (control or PI4KIIα), COS-7 cells were transfected with LIMP-2-GFP, biotinylated with membrane-impermeable biotin, and immunoprecipitated using anti-GFP beads. Total cell lysates and eluted immunoprecipitated samples were analyzed by SDS–PAGE, which was followed by Western blotting using IRDye 800–conjugated streptavidin (top) or an anti-GFP antibody (bottom). (E) Quantification of LIMP-2-GFP biotinylation in immunoprecipitated samples shown in (D) representing mean ± SEM from three experiments. Star denotes statistically significant difference, as determined by one-tailed Student's t test (p < 0.05). (F) Depletion of PI4KIIα in cells expressing LAMP-1-GFP and LIMP-2-mRFP induces accumulation of both molecules on large endocytic structures, at which the two molecules show a significant degree of segregation within the same vesicles (see insets). Scale bars: 10 μm.
PI4KIIα knockdown has already been reported to cause enlarged LAMP-1–positive endosomal structures in fixed cells (Craige et al., 2008), similar to LAMP-1 accumulation on large vesicles described in this study using live-cell microscopy. When LIMP-2-mRFP and LAMP-1-GFP were coexpressed following PI4KIIα knockdown, a fraction of both proteins localized to the same vesicular clusters, although they exhibited a significant degree of segregation (Figure 3F). In addition, LIMP-2 and LAMP-1 appeared segregated to different subdomains within the same individual vesicle in many of the largest vesicles (Figure 3F, insets). The heterogeneity of the enlarged endosomes was evident, as some of the largest vesicles in clusters were quite acidic, while most of the smaller showed no LysoTracker fluorescence, suggesting a larger defect in lysosomal function (Figure S5A). Interestingly, transfection of GFP-CI-M6PR and LIMP-2-mRFP in PI4KIIα siRNA-treated cells resulted in partial but detectable accumulation of CI-M6PR and LIMP-2 on common large vesicles (Figure S5B). Taken together, these data indicated that PI4KIIα is required for proper transport of LIMP-2 between TGN and LEs en route to lysosomes and that this defect was not confined to the LIMP-2 protein.
Proper morphology of LIMP-2–enriched lysosomes requires AP-3 binding
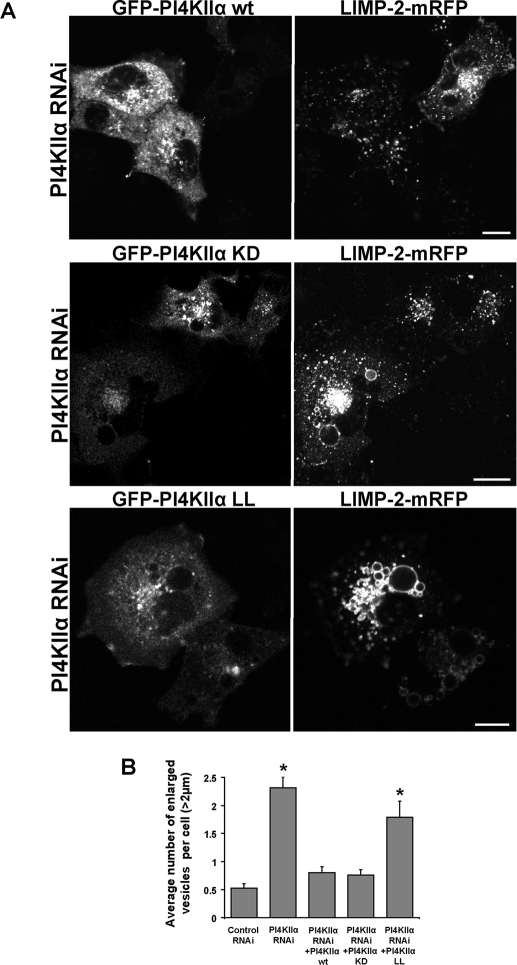
In addition to its PI4K activity, the endosomal pool of PI4KIIα was shown to have a key role in the assembly of sorting complexes via interaction with the AP-3 adaptor protein and with the biogenesis of lysosome-related organelle complex 1 (BLOC-1; Salazar et al., 2009). A dileucine motif within PI4KIIα was found essential for AP-3 binding, with the kinase activity further stabilizing this interaction. Interestingly, a kinase-dead mutant still exhibited 50% of its AP-3–binding affinity, while a mutation in the dileucine motif (PI4KIIα LL mutant) resulted in only 10% of the wild-type binding (Craige et al., 2008; Salazar et al., 2009). We therefore assessed the ability of PI4KIIα wild-type, kinase-dead (KD), and AP-3 binding–deficient LL mutants to rescue the altered morphology of LIMP-2–enriched vesicles in cells knocked down for PI4KIIα. Cells were treated for 2 d with siRNA oligos designed against PI4KIIα; this was followed by 1-d cotransfection with LIMP-2-mRFP, together with wild-type GFP-PI4KIIα, GFP-PI4KIIα KD, or GFP-PI4KIIα LL. In spite of the PI4KIIα siRNA treatment, the kinase was efficiently reexpressed in knockdown cells transfected with PI4KIIα in a construct using a cytomegalovirus-driven promoter (as also observed by Craige et al., 2008; Salazar et al., 2009). Also, expression of the GFP-tagged enzymes did not change the efficiency of the PI4KIIα knockdown, as determined by Western blotting (unpublished data). Knockdown of PI4KIIα resulted in LIMP-2 accumulation in large endocytic structures. These were largely reverted in cells expressing either GFP-PI4KIIα or GFP-PI4KIIα KD (Figure 4A). However, expression of GFP-PI4KIIα LL was not sufficient to elicit significant rescue of the morphological phenotype as confirmed by counting large vesicles with a diameter larger than 2 μm (Figure 4B). Taken together, these findings suggest that proper AP-3 binding is more important than kinase activity for recovery of lysosomal transport of LIMP-2.
FIGURE 4:
Role of the PI4KIIα AP-3–binding and catalytic activity in LIMP-2 trafficking. (A) COS-7 cells were transfected with control siRNA oligos or oligos designed against PI4KIIα for 2 d; this was followed by cotransfection of LIMP-2-mRFP with either GFP-PI4KIIα or catalytically inactive PI4KIIα mutant GFP-PI4KIIα KD. Cells were fixed 1 d later and analyzed by confocal microscopy. Cells expressing only LIMP-2 under PI4KIIα knockdown conditions show accumulation of LIMP-2 in bright perinuclear clusters of enlarged vesicles. Lysosomal distribution is recovered in knockdown cells coexpressing either GFP-PI4KIIα, GFP-PI4KIIα KD, or GFP-PI4KIIα LL together with LIMP-2. (B) The bar diagram shows the summary of the effect of PI4KIIα depletion on LIMP-2-mRFP accumulation to large vesicles. The vesicles larger than 2 μm in diameter were counted per each cell in control siRNA-treated (n = 116) and PI4KIIα siRNA-treated (n = 117) cells expressing LIMP-2-mRFP, in comparison with cells treated for PI4KIIα knockdown and transfected with GFP-PI4KIIα (n = 80), GFP-PI4KIIα KD (n = 92), or GFP-PI4KIIα LL (n = 69) together with LIMP-2-mRFP (means ± SEM). Stars denote significance of differences relative to the values of the control siRNA-treated sample based on one-way analysis of variance (ANOVA; p < 0.001).
Unique roles of PI4KIIα and PI4KIIIβ in LIMP-2–mediated lysosomal transport of GBA
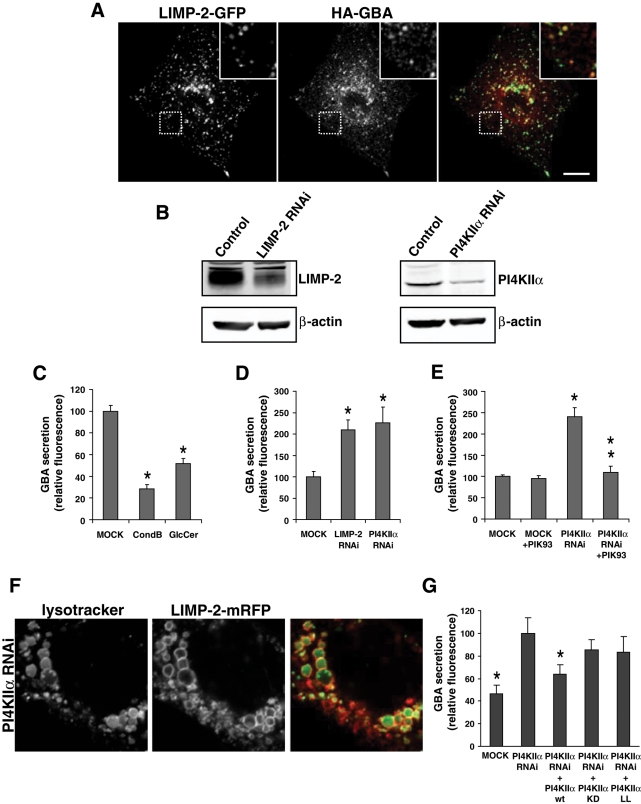
Proper localization of LIMP-2 is essential for lysosomal delivery of GBA and its role in sphingolipid metabolism (Reczek et al., 2007). Nevertheless, subcellular distribution of GBA does not fully mirror that of LIMP-2. Initial immunofluorescence studies on endogenous GBA detected the protein in lysosomes, as well as in ER and Golgi (Willemsen et al., 1987). Similarly, when hemagglutinin (HA)-tagged GBA was coexpressed with LIMP-2 in our studies, HA-GBA mostly exhibited ER and Golgi distribution (Figure S6), and only a small fraction of GBA was found to colocalize with LIMP-2 in lysosomes (Figure 5A).
FIGURE 5:
Regulation of GBA transport by PI4Ks. (A) Following cotransfection of HA-GBA with LIMP-2-GFP for 24 h, COS-7 cells were fixed, permeabilized, and incubated with rabbit polyclonal antibody against the HA epitope. HA-GBA was found to colocalize with LIMP-2-GFP in LE/lysosomes, although a significant fraction of the enzyme also localizes to the ER and Golgi under the overexpression conditions. Scale bar: 10 μm. (B) COS-7 cells were transfected with either control siRNA duplexes or duplexes directed against LIMP-2 or PI4KIIα. After 2 d, cells were lysed and the efficiency of depletion was determined by Western blot analysis using a goat anti–LIMP-2, rabbit anti-PI4KIIα, or mouse anti-actin antibody. (C) Cells transfected with HA-GBA for 1 d were washed prior to addition of fresh media. After 6 h, the culture medium was collected and activity of secreted GBA on its fluorogenic substrate PFB-FDGlu was measured at pH 5.0, as described in Materials and Methods. GBA activity in untreated medium was compared with that of the medium incubated with GBA inhibitor CondB or with GlcCer. Bar graphs represent mean values ± SEM. (D and E) Cells were treated for 2 d with control or PI4KIIα siRNA oligos prior to transfection with HA-GBA. On the following day, cells were incubated for 6 h, either in fresh medium (D) or fresh medium containing 100 nM PIK93 (E). Activity of secreted GBA in collected medium was measured in three independent samples from three separate experiments and represented as mean fluorescence values ± SEM. (F) After a 2-d treatment with siRNA oligos (control or PI4KIIα), COS-7 cells were transfected with LIMP-2-GFP for 24 h; this was followed by preincubation with 100 nM LysoTracker Green for 10 min. Scale bars: 10 μm. (G) Following a 2-d treatment with siRNA oligos, cells were transfected with HA-GBA together with HA-PI4KIIα wild-type, KD, or LL mutant for 24 h. Cells were then washed and incubated for 6 h in fresh medium and assayed for the activity of secreted GBA in the collected medium. Shown are the mean fluorescence values ± SEM from eight independent experiments. Rescue efficiency of individual PI4KIIα mutants was assessed in comparison with the secretion recorded for the PI4KIIα knockdown sample (taken as 100%). Stars denote statistically significant differences relative to the PI4KIIα knockdown sample, as determined by one-way ANOVA (p < 0.05).
Since GBA trafficking defects usually manifest in increased secretion, we next investigated the effects of PI4K inhibitors or knockdown on GBA secretion by measuring the enzymatic activity of GBA in the culture medium. Normally, most GBA is delivered to lysosomes and only a small (although detectable) fraction enters the constitutive secretory pathway (Reczek et al., 2007). Following a 1-d expression of HA-GBA in COS-7 cells, fresh medium was added for 6 h and enzymatic activity of GBA was measured from the collected medium using the fluorogenic GBA substrate 5-(pentafluorobenzoylamino) fluorescein di-β-d-glucopyranoside (PFB-FDGlu) at pH 5.0 (Lorincz et al., 1997; Steet et al., 2006). The specificity of this fluorometric assay was tested in several ways. First, COS-7 cells transfected with HA-GBA exhibited a dramatic increase in signal intensity compared with untransfected cells. Second, when GBA activity of the medium was assayed in the presence of the GBA inhibitor conduritol B epoxide (CBE, 500 μM), an almost 80% decrease in enzymatic activity was observed (Figure 5C). Third, addition of 1 mM GlcCer to collected media led to a competitive inhibition of GBA, resulting in an almost 50% decrease in the conversion of the fluorogenic substrate (Figure 5C).
In agreement with previous reports (Reczek et al., 2007), cells treated with siRNA oligos against LIMP-2 for 2 d prior to HA-GBA transfection showed a 2.1-fold increase in GBA activity in the medium, compared with cells treated with control siRNA oligos (Figure 5, B and D). Treatment with siRNA oligos against PI4KIIα also led to an average 2.2-fold increase in GBA secretion, compared with control cells (Figure 5D). Notably, inhibition of PI4KIIIβ by PIK93 in PI4KIIα siRNAi-treated cells abolished the increased leakage of GBA, reducing it back to the basal levels (Figure 5, E and F). Increased GBA secretion after PI4KIIα knockdown was efficiently rescued by expression of the wild-type kinase (Figure 5G). However, neither PI4KIIα KD nor the AP-3 binding–deficient LL mutants showed significant rescue following the PI4KIIα siRNAi treatment (Figure 5G).
DISCUSSION
An increasing body of evidence suggests a tight connection between inositol phospholipids and sphingolipid metabolism. In Saccharomyces cerevisiae, one of the PI4Ks, Stt4p, is linked to the regulation of sphingolipid biosynthesis (Tabuchi et al., 2006), as is Sac1p (Brice et al., 2009; Breslow et al., 2010), the phosphatase that regulates PtdIns4P levels both in the ER and the plasma membrane (Foti et al., 2001; Stefan et al., 2011). In mammalian cells, several lipid transport proteins contain PtdIns4P-binding PH domains, including the ceramide transfer protein CERT (Hanada et al., 2003) and the GlcCer transfer protein FAPP-2 (D'Angelo et al., 2007). PtdIns4P production is therefore critical to the regulation of sphingolipid biosynthetic pathways, ensuring that Cer and GlcCer reach the appropriate Golgi compartment for synthesis of sphingomyelin and complex glycosphingolipids, respectively. Remarkably, the PI4K enzyme that produces PtdIns4P differs for the two processes: PI4KIIIβ is primarily responsible for CERT (Toth et al., 2006), whereas PI4KIIα is more important for FAPP-2 function (D'Angelo et al., 2007). The present study extends our understanding of the regulation by these PI4Ks to the catabolism of sphingolipids by revealing a concerted role of the two kinases in the trafficking and delivery of one of the key enzymes of sphingolipid breakdown to the lysosome.
Our analysis has clearly shown that PtdIns4P, especially the pool made by PI4KIIIβ, plays an important role in exit of LIMP-2 from the Golgi. This conclusion was reached by several observations using a variety of approaches. First, either acute elimination of PtdIns4P from the Golgi by a recruited Sac1 phosphatase or inhibition of PI4KIIIβ with a selective inhibitor, PIK93, resulted in the retention of LIMP-2-GFP in the Golgi. Second, PIK93 treatment prevented exit of a LIMP-2-PA-GFP construct from the Golgi compartment. Third, PIK93 also inhibited the missorting of the GBA enzyme into the medium under conditions that prevented GBA reaching the lysosomes. An important observation of this series of experiments was the selective effect of PI4KIIIβ inhibition by PIK93 on LIMP-2-GFP but not on CI-M6PR, another receptor that is responsible for lysosomal delivery of several hydrolases. This finding is in agreement with earlier reports that CI-M6PR, a bona fide AP-1 cargo, requires PtdIns4P synthesized by PI4KIIα for proper trafficking along the degradative pathway (Wang et al., 2003) and that total elimination of PtdIns4P also prevented exit of this receptor from the Golgi (Szentpetery et al., 2010). This critical observation was further underlined by a clear segregation within the Golgi of the CI-M6PR and an mRFP-tagged LIMP-2 tail after PIK93 treatment. Together these data suggest the existence of Golgi membrane subdomains, in which PtdIns4P made by distinct PI4Ks controls the exit of select cargo molecules.
Owing to the lack of inhibitors for type II PI4Ks, the role of this enzyme was studied by RNAi-mediated knockdown, which alters trafficking pathways on a different timescale. Nevertheless, PI4KIIα knockdown cells displayed a very characteristic phenotype, namely accumulation of LIMP-2-GFP molecules in large, closely aggregated vesicles that are likely prelysosomal sorting intermediates. This effect was reversed either by a catalytically active or an inactive PI4KIIα, but not by the AP-3–binding LL mutant. Interestingly, both kinase-dead and AP-3 binding–deficient mutants failed to significantly rescue the GBA secretion defect induced by PI4KIIα knockdown, indicating the apparent rescue of the morphological defect by this variant does not reflect full recovery of the segregation between endocytic/lysosomal compartments and correct acidification. Since PI4KIIα KD still exhibits ∼50% of AP-3 binding (Craige et al., 2008), this explains the partial ability of PI4KIIα KD to alleviate the morphological defect. A similar observation was made recently by Craige et al., who showed enlarged prelysosomal vesicles containing LAMP-1 after PI4KIIα knockdown. However, unlike LIMP-2, this LAMP-1 phenotype was partially reversed only by a catalytically active enzyme, but not by an inactive kinase. These authors also showed that association of the PI4KIIα via a dileucine motif with AP-3 was important for supporting the role of the kinase in maturation of LE (Craige et al., 2008). The connection between AP-3 and LIMP-2 has already been revealed in several studies showing selective binding of the dileucine-based sorting motif of LIMP-2 to AP-3 (Honing et al., 1998; Le Borgne et al., 1998; Janvier et al., 2003).
An intriguing observation in the course of our studies was revealed by a comparison of the distribution of LIMP-2 and LAMP-1 in cells depleted in PI4KIIα. Remarkably, while the enlarged vesicles entrapped both of these proteins, in most of these enlarged vesicles the two molecules were clearly segregated into distinct subdomains (see Figure 3). This observation suggested a lateral separation of these molecules and different processing during their normal trafficking through these compartments. Indeed, unlike LIMP-2, most LAMP-1 is first transported to the plasma membrane and endocytosed prior to reaching lysosomes (Janvier and Bonifacino, 2005). Also, during maturation of LE, LAMP-1 preferentially clusters within cholesterol-rich domains of the growing limiting membrane (Du et al., 2011). Depletion of PI4KIIα, therefore, attenuates plasma membrane-to-lysosome trafficking of LAMP-1, as well as Golgi-to-lysosome LIMP-2 transport, resulting in accumulation of the two molecules at an enlarged common compartment. Importantly, LIMP-2 accumulation in the partially acidic enlarged endosomes leads to a dramatic increase in GBA secretion into the medium under PI4KIIα knockdown. Consistent with the role of PI4KIIIβ in LIMP-2 exit from the Golgi, inhibition of PI4KIIIβ in the presence of PI4KIIα RNAi prevented this increased GBA secretion.
Impaired sorting of cargoes to lysosomes after PI4KIIα knockdown has already been observed by Minogue and colleagues, who reported that endocytosed EGF receptors were not efficiently degraded in cells depleted in PI4KIIα (Minogue et al., 2006). An important question, however, is whether the lysosomal sorting defects are caused by altered sphingolipid metabolism, since inhibition of sphingolipid synthesis also impacts trafficking of other enzymes to lysosome-related organelles. For example, in melanocytes derived from GlcCer synthase knockout mice, delivery of tyrosinase to melanosomes is impaired, resulting in Golgi accumulation of the enzyme and loss of pigmentation (Sprong et al., 2001). M6PR-mediated lysosomal transport in these cells, however, remains unaltered, consistent with a specific role for glycosphingolipid regulation of M6PR-independent trafficking pathways, including LIMP-2-mediated GBA transport. Similarly, loss of GlcCer synthase led to altered sorting of LAMP-1 to lysosomes, inducing a switch to an alternate, intracellular trafficking route, rather than LAMP-1 traversing via the plasma membrane (Groux-Degroote et al., 2008). Since PI4KIIα regulates GlcCer transport and formation of complex glycosphingolipids (D'Angelo et al., 2007), and even appears to play a role in the enhanced sphingomyelin synthesis induced by 25-oxysterols (Banerji et al., 2010), it remains to be determined whether the blockade of LIMP-2/GBA delivery to lysosomes is a direct effect in the maturation and fusion of these compartments due to lack of the kinase or an indirect consequence of altered sphingolipid synthesis.
The question then arises how PtdIns4P can regulate the trafficking of LIMP-2 and, even more importantly, how the same lipid made by two different enzymes can regulate the transport of the molecule through different compartments, namely the Golgi and LE. There is an increasing body of evidence that phosphoinositides act in concert with small GTP-binding proteins and clathrin adaptors to regulate cargo selection (Brown et al., 2001; Christoforidis et al., 1999; Wang et al., 2003, 2007). Therefore it is conceivable that PtdIns4P made at the Golgi by PI4KIIIβ regulates the formation of a protein complex that drives the exit of LIMP-2 from the Golgi, while PI4KIIα at the TGN is important at a subsequent stage, helping to assemble a signaling domain that interacts with LIMP-2 and directs it to the LE/lysosome. The exact proteins involved in these different stages of transport must be identified in future studies, but it is likely that AP-3 is an important component at the sorting step regulated by PI4KIIα (Craige et al., 2008; Salazar et al., 2009).
It is important to note that neither of the PI4Ks showed a particularly prominent colocalization with LIMP-2 (or GBA) in COS-7 cells. The differential steady-state localization of PI4KIIIβ at the Golgi and LIMP-2 primarily in lysosomes suggests an interaction that may be confined to a transition state only affecting a small fraction of the respective proteins at any given time. This may explain our inability to find these proteins in complex using immunoprecipitation coupled with Western blotting. Nonetheless, the more sensitive proteomic analysis was able to reveal the association of a small fraction of the LIMP-2 and GBA proteins with PI4KIIIβ that remains below the detection limit of Western analysis following immunoprecipitation. It will be important to determine in future studies whether PI4KIIIβ directly interacts with LIMP-2/GBA or via other protein intermediates.
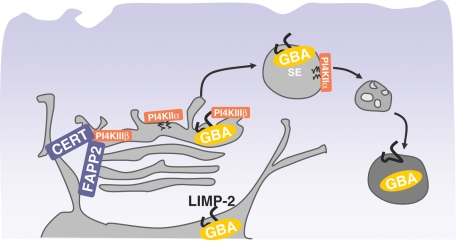
In summary, the present studies demonstrate a sequential role of two distinct PI4Ks in the lysosomal delivery of the GBA enzyme via its receptor, LIMP-2 (Figure 6). PI4KIIIβ has a major role in exit of LIMP-2 from the Golgi, whereas PI4KIIα has a role supporting trafficking steps between the LE and lysosomes. These results are the first to implicate PtdIns4P and PI4Ks in the regulation of the catabolism of complex sphingolipids, adding new details to the biology of the GBA enzyme, a protein known for its prominent role in Gaucher disease. The central role of PI4Ks in integrating the synthetic and degradative pathways of sphingolipids in mammalian cells suggests an evolutionarily conserved connection between phosphoinositides and sphingolipids from yeast to humans. The role of these lipid kinases in the controlling lysosomal function may explain the late onset spinocerebellar degeneration observed in PI4KIIα gene-trap mice (Simons et al., 2009) and raises the possibility that impaired PI4K functions may contribute to other neurodegenerative disorders.
FIGURE 6:
Model of PI4K regulation of LIMP-2-mediated GBA transport. Binding of GBA to LIMP-2 in the ER lumen results in trafficking of this complex to the Golgi. Efficient exit out of this compartment then depends on availability of PtdIns4P and the catalytic activity of PI4KIIIβ, a kinase that also regulates the recruitment of CERT and FAPP-2. Once the GBA/LIMP-2 complex leaves the Golgi, PI4KIIα regulates its efficient transport from SE/LE to lysosomes, such that depletion of PI4KIIα leads to missorting of GBA away from lysosomes, resulting in GBA secretion.
MATERIALS AND METHODS
DNA constructs
Bovine PI4KIIIβ (a kind gift from Lewis C. Cantley, Harvard Medical School, Boston, MA) was cloned between the EcoRV and XhoI sites of the FLAG-pcDNA3.1 plasmid (Gingras et al., 2005). GFP-PI4KIIα and its kinase-dead version have been described previously (Balla et al., 2002). PI4KIIα dileucine mutants containing L60P and L61P mutation, as previously described (Craige et al., 2008), were generated with the following primer pair: forward 5′-GCGGCAGCCAGCTGCTGATCGGGCCCGGGGCGCG-3′ and reverse 5′-CGGGCCCGATCAGCAGCTGGCTGCCGCTCGCGGTC-3′, using site-directed mutagenesis (QuickChange; Stratagene, La|Jolla, CA). Human LIMP-2 (accession number BC021892) was obtained as a full-length EST clone (3872778; Open Biosystems, Huntsville, AL). LIMP-2 was then amplified from this template using the primer pair: forward 5′-CCCAAGCTTATGGGCCGATGCTGCTTCTACAC-3′, and reverse 5′-GGCGAATTCGTTGGGTTCGAATGAGGGGTGCTCTTTCATCC-3′, and subcloned into the pEGFP-N1 or photoactivatable pEGFP-N1 plasmids (generously provided by Jennifer Lippincott-Schwartz, National Institute of Child Health and Human Development, National Institutes of Health [NICHD, NIH]) using the HindIII/EcoRI restriction sites. LIMP-2-GFP L160P was generated by QuickChange site-directed mutagenesis (Stratagene) using the following primer pair: forward 5′-GATCATCGAGGCCATGCCGAAAGCCTATCAGC-3′ and reverse 5′- GCTGATAGGCTTTCGGCATGGCCTCGATGATC-3′. We also generated an mRFP-LIMP-2-tail construct by amplifying LIMP-2 starting at residue 413, containing only the second transmembrane domain with the cytosolic tail using the primer pair: forward 5′-GGCGAATTCGAGTGTTCACATTGATAAAGAGACGGCGAG-3′, and reverse 5′-CGGGGTACCTTAGGTTCGAATGAGGGGTGCTC-3′. This fragment (LIMP-2-tail) was then subcloned, using the EcoRI/KpnI restriction sites, into a previously described construct (Varnai et al., 2007) containing a STIM1 signal sequence (MDVCVRLALWLLWGLLLHQGQS) followed by mRFP in pEGFP-C1 plasmid backbone and a linker (KLGAGAGAGAILNS) placed between the C-terminus of mRFP and the LIMP-2 tail sequence. Human HA-GBA was amplified from a full-length EST (clone 2899915; Open Biosystems) using the following primers: forward 5′-GGCGAATTCATGGAGTTTTCAAGTCCTTCCAGAG-3′ and reverse 5′-TCCCCCCGGGACTGGCGACGCCACAGGTAGGTG-3′, and subcloned, using the EcoRI/XmaI sites, into an pEGFP-N1 backbone vector containing an HA epitope sequence in the place of the GFP sequence between the XmaI and NotI restriction sites. Human GOLPH3 (accession number NM_022130.3) was obtained as a full-length EST clone (SC112810; Open Biosystems), amplified using forward primer: 5′-AAAATCTCGAGCGACCTCGCTGACCCAGCGCAG-3′ and reverse primer: 5′-ATATGAATTCGAGAATGGTTCACCCCGAGCAGAG-3′, and cloned between the XhoI and EcoRI sites of the pEGFP-C1 vector, as described previously (Dippold et al., 2009). Human mRFP-FKBP12-Sac1, Tgn38-FRB-CFP, mRFP-FKBP12 only, GFP-CI-M6PR, and PA-GFP-CI-M6PR have been described previously (Szentpetery et al., 2010). LAMP-1-GFP was kindly provided by Jose A. Martina (National Heart, Lung, and Blood Institute, NIH, Bethesda, MD). mKO-GalT was a gift from Jennifer Lippincott-Schwartz (NICHD, NIH, Bethesda, MD).
Materials and antibodies
Rapamycin was purchased from Calbiochem, and conduritol β-epoxide (CBE) was purchased from Sigma-Aldrich (St. Louis, MO), while PIK93 synthesis was performed as previously described (Knight et al., 2006). PFB-FDGlu, LysoTracker Green, and Rh-EGF were all obtained from Invitrogen (Carlsbad, CA). Rabbit polyclonal antibody against PI4KIIα was a kind gift from Pietro De Camilli (Yale School of Medicine, New Haven, CT), and goat antibody against human LIMP-2 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor 568 donkey anti-rabbit secondary antibody was obtained from Invitrogen. β-d-GlcCer-C16 (860539P) was purchased from Avanti (Alabaster, AL), and streptavidin IRDye 800 was purchased from Rockland Biosciences (Gilbertsville, PA).
Cell culture, transfection procedures, and affinity purification
COS-7 cells grown on 25-mm glass coverslips were transfected with 0.5 μg of plasmid DNA with the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. For siRNA studies, the double-stranded siRNA duplexes corresponding to nucleotides 656–676 of human LIMP-2 (NM_005506) were used. PI4KIIα siRNA duplexes were used as described previously (Balla et al., 2005), while control siRNA oligos (AllStars RNAi control) were obtained from Qiagen (Valencia, CA). COS-7 cells (105 cells in 2 ml) were plated in 35-mm culture dishes 1 d prior to transfection with 20 μl of 20 μM siRNA using Oligofectamine (Invitrogen). After 6 h, the medium was replaced with DMEM containing 10% fetal bovine serum (FBS). Transfection with the siRNAs was repeated 1 d later, and live cells were studied by confocal microscopy on day 4. The efficacy of siRNA treatment on the PI4K expression levels was determined from parallel dishes by Western blot analysis. Live cells were studied by confocal microscopy on day 4.
Transfection of HEK 293 cells and AP-MS were performed essentially as previously described (Goudreault et al., 2009), using pools of HEK 293 cells stably expressing FLAG-PI4KIIIβ. Samples were analyzed on a Thermo-Finnigan LTQ, as described in the next section.
MS analysis
Acquired RAW files were converted to mgf format and searched with the Mascot search engine (Matrix Sciences, London, UK) against the human RefSeq database (release 37) with a precursor ion mass tolerance of 3.0 Da and a fragment ion mass tolerance of 0.6 Da. Methionine oxidation was allowed as a variable modification, and trypsin specificity (with two missed cleavage sites allowed) was selected. The data were analyzed in the Analyst module of ProHits (Liu et al., 2010).
Data were exported into Excel files and manually curated. Biological duplicates of FLAG-PI4KIIIβ were analyzed. To generate lists of high-confidence specific interactors, proteins detected in any of the FLAG only samples (n = 8) were subtracted from the final list. In addition, proteins were checked against an internal Samuel Lunenfeld Research Institute database of human FLAG interactors that contains >700 independent AP-MS experiments; those detected with >30% occurrence in the database were removed. (Note that many of the PI4KIIIβ interactors, including GBA and LIMP-2, were detected in the database with <2% occurrence. Only 14-3-3 proteins, MRPL11 and MTA2 were detected in the database with >2% occurrence). Proteins that appeared in both PI4KIIIβ AP-MS runs are listed in Tables 1 and S1 and Figure S1 (n = 2). Proteins with a Mascot score of <80 were removed. Only those proteins that were detected with spectral counts >2 on both biological replicates are reported.
Confocal microscopy analysis
COS-7 cells were washed in modified Krebs-Ringer buffer (120 mM NaCl, 4.7 mM KCl, 1.2 mM CaCl2, 0.7 mM MgSO4, 10 mM glucose, 10 mM Na-HEPES, pH 7.4), and coverslips were then mounted onto live-cell microscopy chambers. All live-cell experiments were performed on a heated stage with the Krebs-Ringer medium temperature (and the objective) kept at 35°C using an inverted Zeiss LSM-510 scanning-laser confocal microscope (Carl Zeiss, Thornwood, NY) and an objective heater (Bioptechs, Butler, PA).
For immunostaining, COS-7 cells were grown on coverslips and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.4), for 10 min at room temperature. After three washes with PBS, fixed cells were incubated for 1 h with primary antibody (rabbit anti-PI4KIIα) using a staining solution (0.5% bovine serum albumin with 0.2% saponin). After three washes, cells were incubated in the same solution with appropriate secondary antibody (1:1000) for 1 h, washed with PBS, and mounted on glass slides.
Photoactivation and photobleaching of Golgi-localized LIMP-2-PA-GFP, PA-GFP-CI-M6PR, and LAMP-1-PA-GFP were performed immediately upon placing the cells at the 35°C microscope stage. The emergence of the photoactivated LIMP-2 and PA-GFP-CI-M6PR out of the Golgi area was assessed using the particle analysis function (ImageJ). The sequential time-lapse pictures were thresholded (using identical settings for all images within the series) and the individual fluorescent vesicles appearing outside the photoactivation area were counted and plotted against time.
Cell surface biotinylation
Confluent COS-7 cells were washed three times with ice-cold PBS (pH 8.0), and incubated on ice for 45 min in PBS (pH 8.0) containing 1 mg/ml of Hook-Sulfo-NHS-LC-Biotin (Genotec, St. Louis, MO), a membrane-impermeable biotin reagent. Cells were then washed three times in 100 mM glycine in PBS, and cell lysates were prepared in ice-cold lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.25% deoxycholate, 1% Nonidet P-40, 1 mM Na3VO4, 1 mM dithiothreitol, 10 μg/ml aprotinin, 10 μg/ml leupeptin). Lysates were incubated for 1 h with anti-GFP magnetic beads (GFP-TRAP; ChromoTek, Martinsried, Germany), and washed five times with lysis buffer. Bound proteins were then eluted from the beads by being heated in SDS–PAGE sample buffer at 55°C for 30 min; proteins were then resolved by SDS–PAGE and transferred to nitrocellulose membrane.
GBA secretion
Secretion of GBA into the culture medium was assessed by measuring enzymatic activity of GBA in collected medium at pH 5.0 following published protocols (Steet et al., 2006). Briefly, COS-7 cells were seeded on 100-mm dishes at a density of 4 × 105 cells/dish and treated with appropriate siRNA oligos for 48 h, as described under Transfection procedures. On day 3, cells were transfected with HA-GBA, together with HA-PI4KIIα wild-type, KD, or LL mutant for 24 h. The GBA secretion assay was performed on day 4. Cells were washed 3 times with PBS and incubated for 6 h in DMEM containing 10% FBS with or without 100 nM PIK93. Medium was then collected and centrifuged at 4000 rpm for 5 min. In control experiments, the collected supernatant was incubated for 15 min with either 500 μM CondB or GlcCer, or the supernatant was immediately adjusted to pH 5.0 prior to addition of PFB-FDGlu (at the final concentration of 375 ng/μl). The mixture was transferred to 96-well black microplates and assayed for PFB-F fluorescence in a Mithras LB940 plate reader (Berthold, Bad Wildbad, Germany) using the 485-nm excitation filter and emission at 530 nm. Recorded GBA activity was averaged for each sample and normalized to the fluorescence values of the medium collected from untransfected cells.
Supplementary Material
Acknowledgments
Confocal imaging was performed at the Microscopy and Imaging Core of the NICHD, NIH, with the kind assistance of Vincent Schram and James T. Russell. We thank Jason Burgess for reading the manuscript. This research was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver NICHD, NIH. Work in the Gingras and Brill laboratories was supported by the Canadian Institutes of Health Research (CIHR MOP-84314 to A.-C.G. and CIHR MOP-102546 to J.A.B.). A.-C.G. holds the Lea Reichmann Chair in Cancer Proteomics and a Canada Research Chair in Functional Proteomics. M.J.K. is supported by CIHR through a Banting and Best Canada Graduate Scholarship.
Abbreviations used:
- ANOVA
analysis of variance
- AP-MS
affinity purification coupled to mass spectrometry
- CBE
conduritol B epoxide
- CD-M6PR
cation-dependent M6P receptor
- Cer
ceramide
- CERT
ceramide transfer protein
- CFP
cyan fluorescent protein
- CI-M6PR
cation-independent M6P receptor
- ER
endoplasmic reticulum
- FAPP
four-phosphate adaptor proteins
- FBS
fetal bovine serum
- FRB
FKBP12-rapamycin–binding
- GBA
β-glucocerebrosidase
- GFP
green fluorescent protein
- GlcCer
glucosylceramide
- HA
hemagglutinin
- LAMP-1
lysosomal-associated membrane protein 1
- LE
late endosomes
- LIMP-2
lysosomal integral membrane protein type 2
- LIMP-2-PA-GFP
LIMP-2 tagged with photoactivable GFP
- LL
dileucine
- M6P
mannose-6-phosphate
- M6PR
mannose-6-phosphate receptor
- MEF
mouse embryonic fibroblasts
- mKO
monomeric Kusabira-Orange
- mKO-GalT
mKO-tagged Golgi enzyme galactosyltransferase
- mRFP
monomeric red fluorescent protein
- PBS
phosphate-buffered saline
- PFB-FDGlu
5-(pentafluorobenzoylamino) fluorescein di-β-d-glucopyranoside
- PH
pleckstrin homology
- PtdIns4P
phosphatidylinositol 4-phosphate
- Rh-EGF
rhodamine-labeled epidermal growth factor
- RNAi
RNA interference
- SE
sorting endosomes
- siRNA
small interfering RNA
- TGN
trans-Golgi network
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0553) on February 15, 2012.
REFERENCES
- Balla A, Kim YJ, Varnai P, Szentpetery Z, Knight Z, Shokat KM, Balla T. Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIα. Mol Biol Cell. 2008;19:711–721. doi: 10.1091/mbc.E07-07-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem. 2002;277:20041–20050. doi: 10.1074/jbc.M111807200. [DOI] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-IIIα: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Ngo M, Lane CF, Robinson CA, Minogue S, Ridgway ND. Oxysterol binding protein-dependent activation of sphingomyelin synthesis in the Golgi apparatus requires phosphatidylinositol 4-kinase IIα. Mol Biol Cell. 2010;21:4141–4150. doi: 10.1091/mbc.E10-05-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E. Gaucher's disease. N Engl J Med. 1991;325:1354–1360. doi: 10.1056/NEJM199111073251906. [DOI] [PubMed] [Google Scholar]
- Blanz J, Groth J, Zachos C, Wehling C, Saftig P, Schwake M. Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand β-glucocerebrosidase. Hum Mol Genet. 2010;19:563–572. doi: 10.1093/hmg/ddp523. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice SE, Alford CW, Cowart LA. Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem. 2009;284:7588–7596. doi: 10.1074/jbc.M808325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Bio. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Craige B, Salazar G, Faundez V. Phosphatidylinositol-4-kinase type IIα contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol Biol Cell. 2008;19:1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G, et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- Daniels LB, Glew RH, Radin NS, Vunnam RR. A revised fluorometric assay for Gaucher's disease using conduritol-β-epoxide with liver as the source of β-glucosidase. Clin Chim Acta. 1980;106:155–163. doi: 10.1016/0009-8981(80)90168-0. [DOI] [PubMed] [Google Scholar]
- de Graaf P, et al. Phosphatidylinositol 4-kinase beta is critical for functional association of Rab11 with the Golgi complex. Mol Biol Cell. 2004;15:2038–2047. doi: 10.1091/mbc.E03-12-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Di Campli A, Godi A. The role of the phosphoinositides at the Golgi complex. Biochim Biophys Acta. 2005;1744:396–405. doi: 10.1016/j.bbamcr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Dippold H, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Kumar J, Ferguson C, Schulz TA, Ong YS, Hong W, Prinz WA, Parton RG, Brown AJ, Yang H. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192:121–135. doi: 10.1083/jcb.201004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinase-β and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Goudreault M, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groux-Degroote S, van Dijk SM, Wolthoorn J, Neumann S, Theos AC, De Maziere AM, Klumperman J, van Meer G, Sprong H. Glycolipid-dependent sorting of melanosomal from lysosomal membrane proteins by lumenal determinants. Traffic. 2008;9:951–963. doi: 10.1111/j.1600-0854.2008.00740.x. [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Hausser A, Link G, Hoene M, Russo C, Selchow O, Pfizenmaier K. Phospho-specific binding of 14-3-3 proteins to phosphatidylinositol 4-kinase III beta protects from dephosphorylation and stabilizes lipid kinase activity. J Cell Sci. 2006;119:3613–3621. doi: 10.1242/jcs.03104. [DOI] [PubMed] [Google Scholar]
- Hausser A, Storz P, Martens S, Link G, Toker A, Pfizenmaier K. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIβ at the Golgi complex. Nat Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honing S, Sandoval IV, von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J. 1998;17:1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier K, Bonifacino JS. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol Biol Cell. 2005;16:4231–4242. doi: 10.1091/mbc.E05-03-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier K, Kato Y, Boehm M, Rose JR, Martina JA, Kim BY, Venkatesan S, Bonifacino JS. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 γ-σ1 and AP-3 δ-σ3 hemicomplexes. J Cell Biol. 2003;163:1281–1290. doi: 10.1083/jcb.200307157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, et al. A pharmacological map of the PI3-K family defines a role for p110α in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- Kuronita T, Eskelinen EL, Fujita H, Saftig P, Himeno M, Tanaka Y. A role for the lysosomal membrane protein LGP85 in the biogenesis and maintenance of endosomal and lysosomal morphology. J Cell Sci. 2002;115:4117–4131. doi: 10.1242/jcs.00075. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Alconada A, Bauer U, Hoflack B. The mammalian AP-3 adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J Biol Chem. 1998;273:29451–29461. doi: 10.1074/jbc.273.45.29451. [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Liu G, et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol. 2010;28:1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz M, Herzenberg LA, Diwu Z, Barranger JA, Kerr WG. Detection and isolation of gene-corrected cells in Gaucher disease via a fluorescence-activated cell sorter assay for lysosomal glucocerebrosidase activity. Blood. 1997;89:3412–3420. [PubMed] [Google Scholar]
- Minogue S, Waugh MG, De Matteis MA, Stephens DJ, Berditchevski F, Hsuan JJ. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J Cell Sci. 2006;119:571–581. doi: 10.1242/jcs.02752. [DOI] [PubMed] [Google Scholar]
- Peters SP, Coyle P, Glew RH. Differentiation of beta-glucocerebrosidase from beta-glucosidase in human tissues using sodium taurocholate. Arch Biochem Biophys. 1976;175:569–582. doi: 10.1016/0003-9861(76)90547-6. [DOI] [PubMed] [Google Scholar]
- Pohl S, Marschner K, Storch S, Braulke T. Glycosylation- and phosphorylation-dependent intracellular transport of lysosomal hydrolases. Biol Chem. 2009;390:521–527. doi: 10.1515/BC.2009.076. [DOI] [PubMed] [Google Scholar]
- Polevoy G, Wei HC, Wong R, Szentpetery Z, Kim YJ, Goldbach P, Steinbach SK, Balla T, Brill JA. Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J Cell Biol. 2009;187:847–858. doi: 10.1083/jcb.200908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Im YJ, Hurley JH, Prinz WA. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol. 2006;173:107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Schwake M, Schroder J, Hughes H, Blanz J, Jin X, Brondyk W, Van Patten S, Edmunds T, Saftig P. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of β-glucocerebrosidase. Cell. 2007;131:770–783. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Reitman ML, Varki A, Kornfeld S. Fibroblasts from patients with I-cell disease and pseudo-Hurler polydystrophy are deficient in uridine 5′-diphosphate-N-acetylglucosamine: glycoprotein N-acetylglucosaminylphosphotransferase activity. J Clin Invest. 1981;67:1574–1579. doi: 10.1172/JCI110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnboutt S, Aerts HM, Geuze HJ, Tager JM, Strous GJ. Mannose 6-phosphate-independent membrane association of cathepsin D, glucocerebrosidase, and sphingolipid-activating protein in HepG2 cells. J Biol Chem. 1991;266:4862–4868. [PubMed] [Google Scholar]
- Salazar G, Zlatic S, Craige B, Peden AA, Pohl J, Faundez V. Hermansky-Pudlak syndrome protein complexes associate with phosphatidylinositol 4-kinase type IIα in neuronal and non-neuronal cells. J Biol Chem. 2009;284:1790–1802. doi: 10.1074/jbc.M805991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval IV, Arredondo JJ, Alcalde J, Gonzalez Noriega A, Vandekerckhove J, Jimenez MA, Rico M. The residues Leu(Ile)475-Ile(Leu, Val, Ala)476, contained in the extended carboxyl cytoplasmic tail, are critical for targeting of the resident lysosomal membrane protein LIMP II to lysosomes. J Biol Chem. 1994;269:6622–6631. [PubMed] [Google Scholar]
- Sandoval IV, Martinez-Arca S, Valdueza J, Palacios S, Holman GD. Distinct reading of different structural determinants modulates the dileucine-mediated transport steps of the lysosomal membrane protein LIMPII and the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2000;275:39874–39885. doi: 10.1074/jbc.M006261200. [DOI] [PubMed] [Google Scholar]
- Sannerud R, Saraste J, Goud B. Retrograde traffic in the biosynthetic-secretory route: pathways and machinery. Curr Opin Cell Biol. 2003;15:438–445. doi: 10.1016/s0955-0674(03)00077-2. [DOI] [PubMed] [Google Scholar]
- Simons JP, et al. Loss of phosphatidylinositol 4-kinase 2α activity causes late onset degeneration of spinal cord axons. Proc Natl Acad Sci USA. 2009;106:11535–11539. doi: 10.1073/pnas.0903011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprong H, Degroote S, Claessens T, van Drunen J, Oorschot V, Westerink BH, Hirabayashi Y, Klumperman J, van der Sluijs P, van Meer G. Glycosphingolipids are required for sorting melanosomal proteins in the Golgi complex. J Cell Biol. 2001;155:369–380. doi: 10.1083/jcb.200106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steet RA, Chung S, Wustman B, Powe A, Do H, Kornfeld SA. The iminosugar isofagomine increases the activity of N370S mutant acid β-glucosidase in Gaucher fibroblasts by several mechanisms. Proc Natl Acad Sci USA. 2006;103:13813–13818. doi: 10.1073/pnas.0605928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z, Varnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci USA. 2010;107:8225–8230. doi: 10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth B, Balla A, Ma H, Knight ZA, Shokat KM, Balla T. Phosphatidylinositol 4-kinase IIIβ regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J Biol Chem. 2006;281:36369–36377. doi: 10.1074/jbc.M604935200. [DOI] [PubMed] [Google Scholar]
- Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- Vasilescu J, Figeys D. Mapping protein-protein interactions by mass spectrometry. Curr Opin Biotechnol. 2006;17:394–399. doi: 10.1016/j.copbio.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Vega MA, Segui-Real B, Garcia JA, Cales C, Rodriguez F, Vanderkerckhove J, Sandoval IV. Cloning, sequencing, and expression of a cDNA encoding rat LIMP II, a novel 74-kDa lysosomal membrane protein related to the surface adhesion protein CD36. J Biol Chem. 1991;266:16818–16824. [PubMed] [Google Scholar]
- Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- Weisz OA, Gibson GA, Leung SM, Roder J, Jeromin A. Overexpression of frequenin, a modulator of phosphatidylinositol 4-kinase, inhibits biosynthetic delivery of an apical protein in polarized Madin-Darby canine kidney cells. J Biol Chem. 2000;275:24341–24347. doi: 10.1074/jbc.M000671200. [DOI] [PubMed] [Google Scholar]
- Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem. 2005;280:10501–10508. doi: 10.1074/jbc.M414304200. [DOI] [PubMed] [Google Scholar]
- Willemsen R, van Dongen JM, Ginns EI, Sips HJ, Schram AW, Tager JM, Barranger JA, Reuser AJ. Ultrastructural localization of glucocerebrosidase in cultured Gaucher's disease fibroblasts by immunocytochemistry. J Neurol. 1987;234:44–51. doi: 10.1007/BF00314009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.