Abstract
Understanding the factors responsible for variations in mutation patterns and selection efficacy along chromosomes is a prerequisite for deciphering genome sequences. Population genetics models predict a positive correlation between the efficacy of selection at a given locus and the local rate of recombination because of Hill–Robertson effects. Codon usage is considered one of the most striking examples that support this prediction at the molecular level. In a wide range of species including Caenorhabditis elegans and Drosophila melanogaster, codon usage is essentially shaped by selection acting for translational efficiency. Codon usage bias correlates positively with recombination rate in Drosophila, apparently supporting the hypothesis that selection on codon usage is improved by recombination. Here we present an exhaustive analysis of codon usage in C. elegans and D. melanogaster complete genomes. We show that in both genomes there is a positive correlation between recombination rate and the frequency of optimal codons. However, we demonstrate that in both species, this effect is due to a mutational bias toward G and C bases in regions of high recombination rate, possibly as a direct consequence of the recombination process. The correlation between codon usage bias and recombination rate in these species appears to be essentially determined by recombination-dependent mutational patterns, rather than selective effects. This result highlights that it is necessary to take into account the mutagenic effect of recombination to understand the evolutionary role and impact of recombination.
Understanding the evolutionary forces (mutation, selection) that shape genomes is a prerequisite for deciphering of their sequences. However, determining the relative contribution of mutation and selection in genome evolution is hindered by the fact that both mutation patterns and selection efficacy may vary along chromosomes. Notably, population genetics models predict that the efficacy of selection should be reduced in regions of low recombination rate because of Hill–Robertson (HR) effects (hitchhiking and background selection) (1–3). Such effects are thought to be responsible for variations in nucleotide polymorphism (4, 5), intron length (6, 7), or codon usage (8, 9) across the genome.
In a wide range of species including Caenorhabditis elegans and Drosophila melanogaster, codon usage is essentially shaped by weak selection acting for translational efficiency (10–17). In both C. elegans and D. melanogaster, synonymous codons are not used uniformly: distinct codons encoding the same amino acid occur at fairly different frequencies within genomes (14–17). We have shown that these codon usage biases are strongly correlated with gene expression level; highly expressed genes preferentially use a subset of codons called “optimal codons” (17). Moreover, it has been shown that these optimal codons correspond to the most abundant tRNAs (16, 18), which is consistent with findings from several unicellular organisms that codon usage biases are due to selection acting for translational efficiency (10–13).
Analyses of polymorphism data in bacteria and flies indicate that coefficients of selection acting on codon usage are small (19, 20). Different studies based on computer simulations have shown that in such cases of weak selection acting on multiple sites, the efficacy of selection is reduced by genetic linkage (9, 21, 22). Consistent with that model, codon usage biases are reduced in genes located in regions of low recombination rate in Drosophila (8, 9). Consequently, it is recognized that selection on codon usage is improved by recombination, and codon usage is considered one of the most striking examples that support the predictions of the HR effects at the molecular level (23–27). However, the computer simulations mentioned previously are not totally realistic because they assumed that genes evolved independently of each other (9). Furthermore, analyses of Drosophila genes have been conducted on relatively restricted data sets (8, 9). Therefore, we decided to reevaluate the relationships between codon usage and recombination by analyzing data from the complete genomes of D. melanogaster (13,877 genes) (28) and C. elegans (15,194 genes) (29).
We show that the positive correlation between codon usage bias and recombination rate previously observed in Drosophila is also found in C. elegans. However, in both nematode and fly, this effect is due to a mutational bias toward G and C bases in regions of high recombination rate, possibly as a direct consequence of the recombination process. In conclusion, the correlation between codon usage bias and recombination rate in these species appears to be essentially determined by recombination-dependent mutational patterns rather than selective effects.
Materials and Methods
Sequence Data.
Full-length sequences of the six C. elegans chromosomes along with gene annotations were retrieved from the Genome division of GenBank (30) (release 111, April 1999). Chromosome regions that have not yet been sequenced are represented by tracks of N corresponding to the estimated gap size. Data available in GenBank at that time (without N) totaled 94.5 Mb, corresponding to 95% of the estimated whole genome sequence (29). Full-length sequences of the euchromatic portions of the four D. melanogaster chromosomes were retrieved from the Berkeley Drosophila Genome Project database (http://fruitfly.org). These sequences correspond to the right and left arms of chromosomes X, II, and III and to 1 Mb of chromosome IV. Data available in the Berkeley Drosophila Genome Project (BDGP) database at that time (without N) totaled 120 Mb, corresponding to 90% of the estimated whole euchromatic genome sequence (28). We selected all complete protein-coding genes described in databases annotations: 15,194 genes for C. elegans and 13,877 genes for D. melanogaster.
Codon Usage.
For each gene, codon usage bias was measured by the frequency of optimal codons (Fop) (15, 17). Optimal codons are defined as those codons whose frequency increases with gene expression level (17). Fop varies between 0.36 when the bias is null to 1 when the bias is maximum.
Recombination Rate.
To analyze the rate of recombination along the C. elegans and D. melanogaster chromosomes, we used a procedure similar to the one described previously (8, 31). The C. elegans genetic map data were taken from A C. elegans Data Base (ACEDB) (release WS6, December 1998) (R. Durbin and J. Thierry-Mieg, unpublished observations; ftp://ftp.sanger.ac.uk/pub/acedb/celegans/). We selected the 225 loci that had been localized both in the genetic map and in the genomic sequence. The D. melanogaster genetic map data were taken from FlyBase (January 1998) (32). We selected the 892 loci that had been localized both in the genetic map and in the genomic sequence. The polynomial curves as a function of the genetic distance vs. the nucleotide coordinate in the genomic sequence were obtained for each chromosome (R2 ≥ 0.97 for all chromosomes). Recombination rate, as a function of nucleotide position along a chromosome, was estimated by taking the derivative of the polynomial function for each chromosome. The recombination rate in the fourth chromosome of Drosophila was considered to be null.
Genes were classified into three groups of recombination rate: low, medium, and high. The limits between these three classes were chosen so as to divide the data set into three subsets of equal size: C. elegans: low (<1.1 cM/Mb), medium (1.1–3.2 cM/Mb), high (>3.2 cM/Mb); D. melanogaster: low (<2.3 cM/Mb), medium (2.3–3.5 cM/Mb), high (>3.5 cM/Mb).
Gene Expression.
Expression levels were determined by counting the number of occurrences of each gene among expressed sequence tag (EST) sequences from different cDNA libraries, as described (17). In GenBank we selected 72,567 C. elegans ESTs from whole animal cDNA libraries at two developmental states (adult and embryo) and 86,121 ESTs from D. melanogaster (adult ovary and head and embryo). Genes (coding regions) were then compared with the species-specific EST data set with the use of blastn2 (33). blastn2 alignments showing at least 95% identity over 100 nt or more were counted as a sequence match.
Genes were classified into four groups of expression level: very low (no EST detected), low, medium, and high. The limits between these three latter classes were chosen so as to obtain subsets of equal size: C. elegans: very low (0 EST detected, n = 9, 392), low (1–5 ESTs detected, n = 2,009), medium (6–16 ESTs detected, n = 2,025), high (>17 ESTs detected, n = 1, 768); D. melanogaster: very low (no EST detected, n = 5,288), low (1–5 ESTs detected, n = 2,721), medium (6–11 ESTs detected, n = 3,453), high (>12 ESTs detected, n = 2,415).
All of the data are available at http://pbil.univ-lyon1.fr/datasets/Marais2001/data.html.
Results
Codon Usage Bias and Recombination.
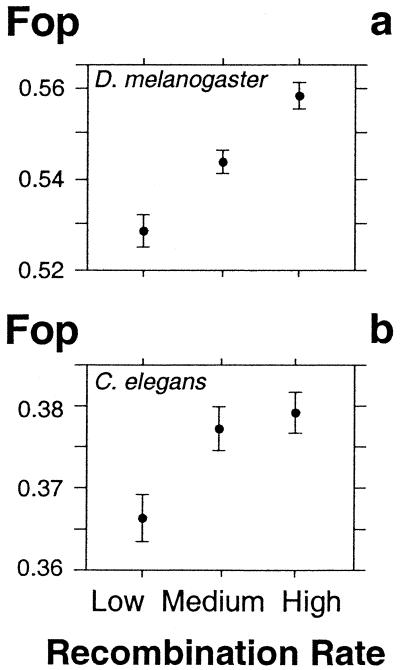
Because recombination is predicted to increase the efficacy of selection (1–3), one should expect a positive correlation between the frequency of optimal codons and the rate of recombination (23–27). To test this prediction, we conducted an exhaustive analysis of the relationships between codon usage and recombination rate in the complete genomes of nematode (29) (15,194 genes) and Drosophila (28) (13,877 genes). Recombination rates were estimated with the use of the genes that have been localized both in the genetic map and in the genomic sequence, as described previously (8, 31). For each chromosome, a polynomial regression curve of the genetic distance as a function of the nucleotide coordinate in the genomic sequence was generated, from which recombination rates were derived (see Materials and Methods). Fig. 1 shows that both in C. elegans and D. melanogaster there is a positive correlation between the Fop and the local recombination rate (Spearman's nonparametric correlation coefficient ρ C. elegans, ρ = 0.09; D. melanogaster, ρ = 0.13). These correlations are statistically significant (P < 0.0001), although they are relatively weak. We have previously shown that in both nematode and Drosophila, codon usage bias is strongly correlated not only with expression level but also with protein length (17). However, the relationship between codon usage bias and recombination rate is independent of these two factors (data not shown).
Figure 1.
Variation of the frequency of optimal codons (Fop) according to the recombination rate in D. melanogaster (a) and C. elegans (b). The recombination rate (in centimorgans per megabase, cM/Mb) along each chromosome was estimated as described in Materials and Methods. Error bars indicate the 95% confidence interval.
Such a relationship between codon usage and recombination has already been found in Drosophila and was interpreted as evidence of HR effects, i.e., that the efficacy of selection was improved by recombination (8, 9) (referred to as the HR model in the rest of the text). However, this correlation might also be explained by a variation in mutation patterns along the genome according to the recombination rate [referred to as the mutation bias variation model (MBV) in the rest of the text]. It should be noted that if such variations of mutation patterns do occur, they should affect all base positions within the gene, not only coding or synonymous sites, but also noncoding DNA. Thus, a prediction of the MBV model is that the base composition of noncoding DNA should also vary with recombination rate.
Noncoding DNA G + C Content and Recombination.
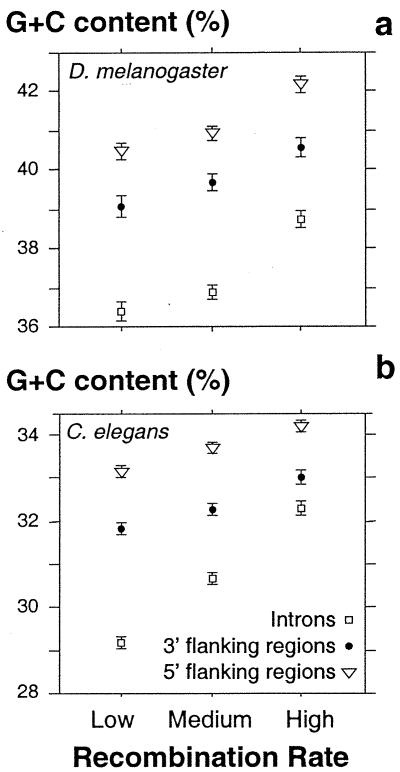
C. elegans has 21 optimal codons, of which 16 end in G or C bases (15, 17), and D. melanogaster has 22 optimal codons, of which 21 end in G or C bases (14, 17). The positive correlation between Fop and recombination rate thus implies that the G + C content of synonymous sites increases with recombination. In Fig. 2 we show that the G + C content of noncoding DNA (flanking regions and introns of genes) is also positively correlated with the recombination rate in both nematode (introns, ρ = 0.28; 5′ flanking regions, ρ = 0.09; 3′ flanking regions, ρ = 0.12; with P < 0.0001) and drosophila (introns, ρ = 0.20; 5′ flanking regions, ρ = 0.15; 3′ flanking regions, ρ = 0.12 with P < 0.0001). This observation, in agreement with the MBV model, suggests that there might be a mutational bias toward G and C bases in regions of high recombination rate. However, this observation is not sufficient to totally exclude the HR model because the G + C content in some noncoding regions might also be under selection (e.g., regulatory elements).
Figure 2.
Relationship between G + C content (%) in noncoding DNA and the recombination rate. Only genes with flanking regions and intron sequences larger than 200 nucleotides were selected. C. elegans: n = 10,486 genes; D. melanogaster: n = 7,337 genes. The recombination rate is as in Fig. 1. Error bars indicate the 95% confidence interval.
Test of HR and MBV Models in C. elegans.
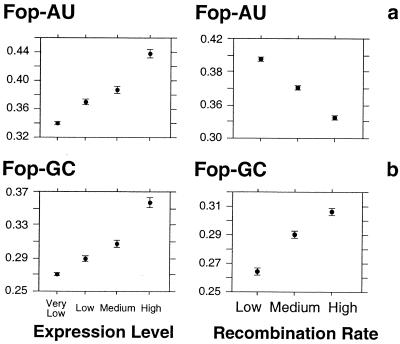
The presence in C. elegans of five optimal codons ending in A or U allowed us to directly test the HR model. If recombination improves selection on codon usage, as was previously proposed (8, 9), then the frequency of optimal codons should increase with recombination, whatever their ending base. In contrast, if the positive correlation between synonymous codon usage and recombination is due to a mutational bias toward G and C bases, then the frequency of GC-ending optimal codons (Fop-GC) should increase with recombination, whereas the frequency of AU-ending optimal codons (Fop-AU) should decrease. Fig. 3 shows that both Fop-AU and Fop-GC are positively correlated with gene expression level, consistent with the fact that the selective pressure for using optimal codons increases with gene expression level. However, this figure shows that whereas Fop-GC is positively correlated with recombination rate (ρ = 0.3 with P < 0.0001), Fop-AU decreases with increasing recombination rate (ρ = −0.35 with P < 0.0001). The fact that the overall frequency of optimal codons is positively correlated with recombination (Fig. 1) simply reflects the fact that there are only five AU-ending codons among the 21 optimal codons.
Figure 3.
Variation in the frequency of optimal codons ending in A, U or G, C according to the recombination rate and the gene expression level in C. elegans. (a) Frequency of the optimal codons ending in A or U (Fop-AU). (b) Frequency of the optimal codons ending in G or C (Fop-GC). C. elegans has 21 optimal codons, of which 16 end in G or C and 5 end in A or U (15, 17). Fop-AU (and, respectively, Fop-GC) was calculated by dividing the number of occurrences of optimal codons ending in A or U (G or C) by the number of occurrences of codons encoding amino acids that have optimal codons ending in A or U (G or C). The recombination rate is as in Fig. 1. The expression level was estimated with EST data, as described in Materials and Methods. Error bars indicate the 95% confidence interval.
Test of HR and MBV Models in D. melanogaster.
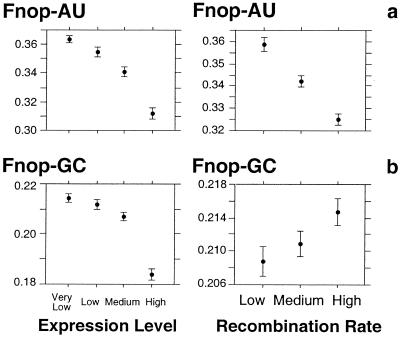
In Drosophila, Fop-AU measurements are not possible for statistical reasons because there is only one optimal codon ending in A or U bases in this species (14, 17). We chose another strategy to directly test the HR model: we used the nonoptimal codons ending in G or C. If recombination improves selection on codon usage, then the frequency of nonoptimal codons should decrease with recombination, whatever their ending base. In contrast, if the positive correlation between synonymous codon usage and recombination is due to a mutational bias toward G and C bases, then the frequency of AU-ending nonoptimal codons (Fnop-AU) should decrease with recombination, whereas the frequency of GC-ending nonoptimal codons (Fnop-GC) should increase. Fig. 4 shows that both Fnop-AU and Fnop-GC are negatively correlated with gene expression level, consistent with the fact that the selective pressure for avoiding nonoptimal codons increases with gene expression level. However, this figure shows that whereas Fnop-AU is negatively correlated with recombination rate (ρ = −0.15 with P < 0.0001), Fnop-GC increases with increasing recombination rate (ρ = 0.06 with P < 0.0001). Therefore, in both nematode and Drosophila, the positive correlation between the frequency of optimal codons and recombination rate is not due to improved selection but to a mutational bias toward G and C bases in regions of high recombination rate.
Figure 4.
Variation of the frequency of nonoptimal codons ending in A, U or G, C according to recombination rate and gene expression level in D. melanogaster. (a) Frequency of the nonoptimal codons ending in A or U (Fnop-AU). (b) Frequency of the nonoptimal codons ending in G or C (Fnop-GC). D. melanogaster has 37 nonoptimal codons, of which 29 end in A or U and 8 end in G or C (14, 17). Fnop-GC (and, respectively, Fnop-AU) was calculated by dividing the number of occurrences of nonoptimal codons ending in G or C (A or U) by the number of occurrences of codons encoding amino acids that have nonoptimal codons ending in G or C (A or U). The recombination rate is as in Fig. 1. The expression level is as in Fig. 3. Error bars indicate the 95% confidence interval.
Impact of the Method for Estimating the Recombination Rate.
In their previous study, Kliman and Hey (8) tested the MBV model by looking at the G + C content of introns. In their data set (n = 142; i.e., about 100 times less than in the present study) they did not detect any significant variation of intron G + C content with recombination, in contradiction to our observations (see their table 2 and our Fig. 2). In addition to possible errors due to limited sampling, this discrepancy might also be due to the fact that we did not use the same method to estimate the local recombination rate. Here we considered the recombination rate as a continuous variable, which might be inaccurate for some regions of the genome. Notably, because of the limiting number of points, the polynomial curves overestimate the recombination rate in telomeric regions of Drosophila chromosomes. To determine whether such a problem might have biased our results, we examined the relationship between recombination rate and codon usage bias by comparing regions of known reduced recombination with the remainder of the genome of Drosophila, as described in the previous work by Kliman and Hey (8). These regions of low recombination correspond to the entire fourth chromosome (which does not recombine) and regions adjacent to centromeres and telomeres in the other chromosomes (sections 1, 20, 21, 40–41, 60–61, 80–81, 100, 101–102) (8). We noticed that telomeric regions have peculiar characteristics: a nearly null recombination rate and a high G + C content. Therefore, we analyzed separately the 751 telomeric genes from the 123 other genes located in regions of known reduced recombination. Table 1 shows that Fop covaries with intron G + C content but not with recombination rate: in nontelomeric regions of reduced recombination rate, both Fop and intron G + C content are lower than in regions of high recombination rate, whereas genes located in telomeric regions have a very low recombination rate but a relatively high Fop and a high intron G + C content. This relationship was overlooked in previous works because nontelomeric genes with low recombination rates were pooled with telomeric genes, although the two kinds of regions have evidently distinct evolutionary dynamics. Note that the peculiar characteristics of telomeric genes (representing 5% of our data set) do not affect the results presented in the previous sections: if these telomeric genes are removed from the data set, or if their estimated recombination rate is set to zero, the correlations between recombination rate and Fop, Fnop-GC, Fnop-AU, or base composition in noncoding regions remain unchanged (data not shown). The reasons for the overall high G + C content of telomeric regions are not understood. But clearly, the covariation of Fop and intron G + C content according to chromosome location observed in D. melanogaster is totally consistent with the model of variation of mutation bias affecting codon usage and cannot be explained simply by HR effects.
Table 1.
Frequency of optimal codons (Fop) and intron G+C content in regions of high or low recombination rate in Drosophila
| Recombination rate | Number of genes | Fop | Intron G+C content | |
|---|---|---|---|---|
| Telomeric regions | Low | 751 | 0.56 ± 0.10 | 40 ± 6% |
| Centromeric regions and chromosome IV | Low | 123 | 0.33 ± 0.12 | 32 ± 5% |
| Other regions | High | 13,226 | 0.55 ± 0.10 | 37 ± 6% |
Discussion
By analyzing nematode and fly complete genomes, we show that DNA G + C content is positively correlated with recombination rate, both in noncoding regions and in synonymous positions of codons. The relationship between codon usage and recombination is not due to HR effects (selectionist model), inasmuch as both optimal and nonoptimal codons are affected. Therefore, this correlation must reflect a variation of mutational patterns with recombination rate (neutralist model).
Recombination and Mutational Pattern.
The positive correlation between recombination rate and G + C content seems to be a general trend in eukaryotes because it has also been observed in humans (23, 34) and yeast (35, 36). Different models can account for that correlation: (i) G + C-rich sequence motifs might enhance the recombination rate, (ii) recombination might induce mutations toward G and C, or (iii) both parameters might be linked to a third unknown factor. Several lines of evidence support the second model. Notably, it has been shown in yeast that the recombination process per se is mutagenic because of errors in double-strand DNA break repair (37). In mammals recombination also appears to be mutagenic (38). The recombination rate in pseudoautosomal regions of sex chromosomes is very high because they are the site of an obligatory pairing and recombination during male meiosis. This high recombination rate in pseudoautosomal regions compared with X-unique sequences is associated with a huge acceleration (estimated to be 170-fold) in the rate of substitution during mouse species evolution. Interestingly, this high rate of sequence divergence in pseudoautosomal regions is accompanied by an increase in G + C content (38). This observation might be due to the fact that, in mammals, the repair of mismatches that arise during recombination is biased toward G + C-richness (39).
HR Effects and Codon Usage.
Although the cause for the relationship between mutational patterns and the recombination process remains putative, we show that in nematode and in Drosophila the positive correlation between synonymous codon usage bias and recombination is due to a mutational bias toward G and C in regions of high recombination. We do not exclude the possibility that recombination might weakly improve the efficacy selection on codon usage. However, this selective effect, if any, is swamped by the mutagenic effect of recombination. Is it possible to detect a positive effect of recombination on selection efficacy once the correlation with GC content has been accounted for? Introns are probably the best possible indicator of local mutational patterns. Therefore, we computed the expected Fop-AU (C. elegans) and Fnop-GC (D. melanogaster) according to intron G + C content. For this purpose, we computed a regression between Fop-AU (Fnop-GC) and intron G + C content, and we analyzed the residuals of this correlation. As expected, the correlation between these residuals and the recombination rate is decreased slightly compared with the original correlation between Fop-AU (Fnop-GC) and the recombination rate (C. elegans, ρ = −0.3 with P < 0.0001; D. melanogaster, ρ = 0.05 with P < 0.0001). However, the slope remains negative for Fop-AU and positive for Fnop-GC, whereas the contrary would have been expected according to the HR model. Hence, we failed to find any evidence that recombination improves selection on codon usage.
Evolutionary Role of Recombination.
The conclusion of our work is that synonymous codon usage does not support the natural selection–genetic linkage interference models. We do not exclude the possibility, however, that such HR effects do exist, but their impact on codon usage is masked by variations in mutation pressures associated with recombination rates. We have also previously shown that the distribution of transposable elements in the C. elegans genome does not fit with the predictions of HR models (40). These models, however, are apparently supported by some other genomic features. For instance, it has been shown that variation of nucleotide polymorphism across the genome is positively correlated with the recombination rate in Drosophila (4, 5). HR effects are also thought to be responsible for variations in intron length (6, 7). We suggest that these relationships have to be reevaluated by taking account of the variations of mutation patterns linked to recombination along chromosomes. Our results remind us that recombination per se might be a source of mutation and that the mutational biases linked to recombination play an important role in shaping codon usage and genomes in general. It is essential to take account of the mutagenic effect of recombination to understand the evolutionary role and impact of recombination.
Acknowledgments
We thank Nicolas Galtier, Manolo Gouy, Vincent Daubin, Vincent Vanoosthuyse, Didier Casane, and all of the team of the Diplôme d'Etudes Approfondies “Biodiversité: Génétique, Histoire, et Mécanismes de l'Évolution” for comments and discussion. This work was supported by the Ministère de la Recherche and the Centre National de la Recherche Scientifique.
Abbreviations
- Fop
frequency of optimal codons
- Fop-AU
frequency of optimal codons ending in A or U
- Fop-GC
frequency of optimal codons ending in G or C
- Fnop-AU
frequency of nonoptimal codons ending in A or U
- Fnop-GC
frequency of nonoptimal codons ending in G or C
- EST
expressed sequence tag
- MBV
mutation bias variation
- HR
Hill–Robertson
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hill W G, Robertson A. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- 2.Maynard-Smith J, Haigh J. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 3.Charlesworth B, Morgan M T, Charlesworth D. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begun D J, Aquadro C F. Nature (London) 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 5.Munte A, Aguade M, Segarra C. Genetics. 1997;147:165–175. doi: 10.1093/genetics/147.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho A B, Clark A G. Nature (London) 1999;401:344. doi: 10.1038/43827. [DOI] [PubMed] [Google Scholar]
- 7.Hurst L D, Brunton C F, Smith N G. Trends Genet. 1999;15:437–439. doi: 10.1016/s0168-9525(99)01832-6. [DOI] [PubMed] [Google Scholar]
- 8.Kliman R M, Hey J. Mol Biol Evol. 1993;10:1239–1258. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- 9.Comeron J M, Kreitman M, Aguade M. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Nucleic Acids Res. 1981;9:43–74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikemura T. J Mol Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- 12.Bulmer M. Nature (London) 1987;325:728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- 13.Sorensen M A, Kurland C G, Pedersen S. J Mol Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 14.Shields D C, Sharp P M, Higgins D G, Wright F. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 15.Stenico M, Lloyd A T, Sharp P M. Nucleic Acids Res. 1994;22:2437–2446. doi: 10.1093/nar/22.13.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriyama E N, Powell J R. J Mol Evol. 1997;45:514–523. doi: 10.1007/pl00006256. [DOI] [PubMed] [Google Scholar]
- 17.Duret L, Mouchiroud D. Proc Natl Acad Sci USA. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duret L. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- 19.Hartl D L, Moriyama E N, Sawyer S A. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akashi H. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McVean G A, Charlesworth B. Genetics. 2000;155:929–944. doi: 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W H. J Mol Evol. 1987;24:337–345. doi: 10.1007/BF02134132. [DOI] [PubMed] [Google Scholar]
- 23.Charlesworth B. Curr Biol. 1994;4:182–184. doi: 10.1016/s0960-9822(94)00045-x. [DOI] [PubMed] [Google Scholar]
- 24.Sharp P M, Averof M, Lloyd A T, Matassi G, Peden J F. Philos Trans R Soc London B. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- 25.Akashi H, Kliman R M, Eyrewalker A. Genetica (The Hague) 1998;103:49–60. [PubMed] [Google Scholar]
- 26.Kreitman M, Comeron J M. Curr Opin Genet Dev. 1999;9:637–641. doi: 10.1016/s0959-437x(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 27.Hey J. Tree. 1999;14:35–38. doi: 10.1016/s0169-5347(98)01497-9. [DOI] [PubMed] [Google Scholar]
- 28.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 29.C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 30.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F, Rapp B A, Wheeler D L. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnes T M, Kohara Y, Coulson A, Hekimi S. Genetics. 1995;141:159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flybase Consortium. Nucleic Acids Res. 1998;26:85–88. doi: 10.1093/nar/26.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eyre-Walker A. Proc R Soc Lond B. 1993;252:237–243. doi: 10.1098/rspb.1993.0071. [DOI] [PubMed] [Google Scholar]
- 35.Baudat F, Nicolas A. Proc Natl Acad Sci USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerton J L, Derisi J, Shroff R, Lichten M, Brown P O, Petes T D. Proc Natl Acad Sci USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strathern J N, Shafer B K, McGill C B. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry J, Ashworth A. Curr Biol. 1999;9:987–989. doi: 10.1016/s0960-9822(99)80430-8. [DOI] [PubMed] [Google Scholar]
- 39.Brown T C, Jiricny J. Cell. 1988;54:705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- 40.Duret L, Marais G, Biemont C. Genetics. 2000;156:1661–1669. doi: 10.1093/genetics/156.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]