Abstract
CD300a is an immunoreceptor tyrosine-based inhibitory motif (ITIM) containing molecule that belongs to the CD300 family of paired activating/inhibitory receptors. It has been shown that its ligation inhibits activation signals on cells of both myeloid and lymphoid lineages. The ligands for CD300a have not been identified. Here, we show that a CD300a-Ig fusion protein specifically binds to apoptotic cells that are evolutionary apart, such as human and insect cells, suggesting that the ligand has to be conserved. Using surface plasmon resonance, ultracentrifugation, ELISA, and reporter cell assays, we identified phosphatidylethanolamine (PE) and phosphatidylserine (PS), 2 phospholipids that translocate to the outer leaflet of the plasma membrane of dead cells, as the ligands for CD300a. Mutational and structural modeling studies identified residues that are involved in the binding of CD300a to PE and PS and that form a cavity where the hydrophilic heads of PE and PS, can penetrate. CD300a down-regulates the uptake of apoptotic cells by macrophages and its ectopic expression in CD300a-negative cell lines also decreased the engulfment of dead cells. Collectively, our results indicate that PE and PS are ligands for CD300a, and that this interaction plays an important role in regulating the removal of dead cells.
Introduction
A rising number of publications have described the diversity of paired activating and inhibitory cell surface molecules.1,2 The human CD300 family of receptors has 7 members and all of them have an extracellular immunoglobulin (Ig)V-like domain.3 The activating members of this family have a short intracellular tail and associate with immunoreceptor tyrosine-based activation motif (ITAM)–containing adaptor proteins, such as DAP12 and FcϵRIγ,3,4 whereas the inhibitory members have a long intracellular tail that carry immunoreceptor tyrosine-based inhibitory motifs (ITIM).3,5 This multi-gene family is clustered on human chromosome 17 and they are expressed on cells of both lymphoid and myeloid lineages.3
The gene encoding CD300a has undergone a very significant positive selection, suggesting an essential requirement for the host to evolve and maintain its function.6 CD300a is broadly expressed across different cell types including natural killer (NK) cells, T cells, B cells, neutrophils, plasmacytoid dendritic cells, mast cells, and eosinophils, among others.3,7–10 The cytoplasmic tail contains 3 classic and one nonclassic ITIM. Thus far, CD300a has been shown to function as an inhibitory receptor. For instance, the ligation of CD300a decreased NK cytotoxic activity,5,11 inhibited IgE-mediated degranulation of mast cells,8 B-cell receptor (BCR) and T-cell receptor (TCR)–mediated signaling,10,12 reduced FcγRIIa-triggered reactive oxygen species (ROS) production in human neutrophils,7 and suppressed the effects of eotaxin, IL-5, and granulocyte macrophage colony-stimulating factor (GM-CSF) on human eosinophils.13
A single nucleotide polymorphism (SNP) that encodes for a nonsynonymous mutation in the IgV-like domain of CD300a has been linked to psoriasis susceptibility.14 Moreover, CD300a has been proposed as a biomarker that can differentiate ulcerative colitis from Crohn disease and noninflammatory diarrhea,15 and for the detection of minimal residual disease in acute lymphoblastic leukemia.16 We have also published that B cells of HIV-infected patients express significantly lower levels of CD300a compared with healthy donors, which suggests a potential role for this immunomodulatory receptor in the B-cell dysfunction observed during HIV infection.12 Furthermore, in vivo studies with bispecific antibodies in mice have shown that ligation of CD300a is capable of reversing airway inflammation and tissue remodeling in a model of asthma,17 abrogating IgE-mediated allergic reactions18 and hampering stem cell factor (SCF) induced anaphylaxis.19 Altogether, these publications highlight, not only the clinical relevance of the CD300a receptor, but also the potential for targeting this molecule for therapeutic purposes.
Despite the intense interest in CD300a, identification of the ligand has remained elusive. In this study, using biochemical, in vitro biologic, and structural modeling analysis, we demonstrate that the aminophospholipids phosphatidylserine (PS) and especially phosphatidylethanolamine (PE) are the natural ligands for CD300a. The relevance and significance of the interaction between CD300a and PE/PS is demonstrated by the role of this receptor in modulating the engulfment of dead cells, which express these aminophospholipids in the outer leaflet of the plasma membrane.
Methods
Ig fusion proteins
The CD300a-Ig fusion protein and the previously described leukocyte-associated Ig-like receptor (LAIR)–1-Ig20 were isolated from the culture supernatants of transiently transfected HEK293 cells using protein-A-Sepharose columns (Amersham Biosciences). The purified proteins were dialyzed in PBS and further processed for conjugation with Alexa Fluor 488 using a monoclonal antibody labeling kit (Molecular Probes; Invitrogen). CD300a-Ig mutants were made using QuikChange site-directed mutagenesis kit (Stratagene). All constructs were sequenced to confirm their identities.
Flow cytometric analysis and cell binding assays
Flow cytometric experiments were performed in a FACSCalibur or LSRII (BD Biosciences) flow cytometers. Data were analyzed using FlowJo Version 9.4 software (Tree Star). For determination of CD300a expression on macrophages, cells were first blocked with 5% human serum for 15 minutes and then stained with anti-CD300a PE antibody (Beckman Coulter) and anti-CD14 PE-Cy7 (eBioscience). Isotype controls were used as negative control.
Blood samples from healthy donors were collected under an institutional review board-approved protocol at the Department of Transfusion Medicine at the National Institutes of Health (NIH). Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats by gradient centrifugation and were starved for 20 hours in PBS at 4°C. For the binding analysis of Ig fusion proteins, starved PBMCs (5 × 105) were blocked with PBS containing 5% human serum for 15 minutes at room tempera-ture (RT). Cells were washed once and incubated with binding buffer (10mM N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid [HEPES] pH 7.4, 140mM NaCl, 2.5mM CaCl2, and 2mM MgCl2) for 15 minutes at RT. The cells were washed once with binding buffer and incubated with Alexa Fluor 488–conjugated Ig fusion proteins. After 45 minutes, cells were washed twice with wash buffer (PBS containing 1% of FCS) and resuspended in PBS for flow cytometric analysis. When mentioned, cells were also stained with annexin V–APC (eBiosciences) and propidium iodide or 7-AAD (Beckman Coulter) to differentiate between dead and live cells. In some experiments the dead population was differentiated using the live/dead cell labeling kit (Molecular Probes; Invitrogen). Binding assays were also performed on dexamethasone DT40 treated cells and starved Sf-9 insect cells.
To investigate the role of divalent cations in the binding of CD300a-Ig to dead cells, starved PBMCs were pretreated with 5mM ethylenediaminetetraacetic acid (EDTA) or ethylene glyco-bis(b-aminoethyl ester)-N,N,N′,N′-tetraacetic acid (EGTA) for 15 minutes at RT, and then binding assays were performed as previously described. In certain experiments, an indirect binding assay was performed; first, cells were incubated for 45 minutes with unlabeled Ig fusion proteins. After the incubation, extensive washes were performed and a secondary detection reagent, anti–human Ig FITC was added, and cells were further incubated for 30 minutes. After that, cells were again washed and resuspended in PBS for acquisition in the flow cytometer.
In blocking experiments, starved PBMCs were blocked with 5% human serum and then were preincubated with unlabeled MFG-E8 and duramycin at 4 different concentrations of MFG-E8 (2.5-20 μg/mL) and duramycin (0.125-1μM) for 45 minutes at RT. The cells were washed twice and then stained with CD300a-Ig conjugated to Alexa Fluor 488 for 40 more minutes. Then cells were washed and resuspended in PBS before acquisition in the flow cytometer.
Cell isolation, macrophage cell culture, and siRNA knockdown
PBMCs were isolated from buffy coats by Ficoll-Hypaque gradient centrifugation. Macrophages were differentiated from monocytes obtained by either positive selection using CD14 cell isolation kit (Miltenyi) or by allowing the PBMCs to adhere on the cell culture dishes for 2 hours at 37°C. The nonadherent cells were washed thoroughly with serum-free medium and then cultured in differentiation media: Iscove modified Dulbecco media (IMDM+ Glutamax) supplemented with 10% human AB serum, 2mM Glutamine, 1% Na-pyruvate, 1% nonessential amino acid (NEAA), and 50 ng/mL of recombinant human M-CSF (R&D Systems). The media was replaced 2 to 3 times during the differentiation period. After 7 to 10 days of culture, monocytes were differentiated into macrophages.
Monocyte-derived macrophages were washed with PBS and detached from the cell culture plates by treating them with Accutase (Innovative Cell Technologies) for 30 minutes at RT. After the treatment, cells were washed and subjected to the siRNA transfection (300nM) according to the manufacturer's protocol using the Amaxa human macrophage nucleofector kit (Lonza). The nontarget and CD300a siRNA pool were obtained from ThermoScientific (Dharmacon). After 24 hours of the transfection with siRNA the cells were replaced with fresh medium and approximately 16 to 20 hours later the surface expression of CD300a was determined using flow cytometry and the phagocytosis assay was performed.
Phagocytosis assay
Plated cultured macrophages were collected after treatment with accutase for 30 minutes at RT and then replated in a 24-well plate at 2 × 105/well. They were allowed to adhere back at 37°C for at least 1 hour. DT40 cells or mouse thymocytes (from C57BL/6 mice) were incubated with 10μM dexamethasone (Sigma-Aldrich) in serum free RPMI 1640 for 4 to 6 hours to induce apoptosis. Cells were washed and cell death was confirmed by staining with annexin V and 7-AAD. Apoptotic DT40 cells or thymocytes were labeled with PKH67 and PKH26 (Sigma-Aldrich), respectively. When mentioned, the apoptotic cells were blocked with 10% human serum containing medium for 10 minutes at RT and then preincubated with Ig-fusion proteins at 15 μg/mL concentration or annexin V (5 μg/mL) for 30 to 40 minutes at RT. Labeled dexamethasone-treated thymocytes (6 × 105/well) or DT40 cells (6 × 105/well) were added to the macrophages. After 30 minutes of incubation the wells were washed using 0.05% Trypsin-EDTA (Invitrogen) and the cells were harvested using accutase. Preliminary experiments with the ImageStream System (Amnis Corporation) showed that the flow cytometry based method for the phagocytosis assay only detects engulfed cells and not adhered cells (data not shown). Collected cells were stained with PE-Cy7 conjugated anti-CD14 and analyzed by flow cytometry. The rate of phagocytosis was expressed as the percentage of CD14+ cells that are positive for PKH67 (DT40) or PKH26 (thymocytes) staining.
Mouse fibroblast L929 cells were stably transfected with either an empty vector (pcDNA3.1) or a vector encoding human CD300a. Cells were plated in a 12-well plate (5 × 105 cells/well) and incubated for 20 hours before the phagocytosis assay. Thymocytes isolated from the C57BL/6 mice were treated with 10μM dexamethasone for 5 hours at 37°C, labeled with PKH26, washed and added to the L929 cells at a ratio of 3:1. After 1 hour of incubation the cells were harvested and washed with Trypsin-EDTA and analyzed by flow cytometry.
Statistical analysis
Data were analyzed using GraphPad Prism Version 5 software and plotted as bar graphs or scatter plots. Comparisons were made with the 2-tailed t test with 99% confidence interval and the 2-way ANOVA. P < .05 was considered significant (* P < .05, **P < .01, ***P < .001).
Supplemental methods
For details on manufacturing of phospholipid liposomes, surface plasmon resonance (SPR) analysis, structural modeling, sedimentation assay, ELISA, and reporter cell assay, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
CD300a predominantly binds dead cells
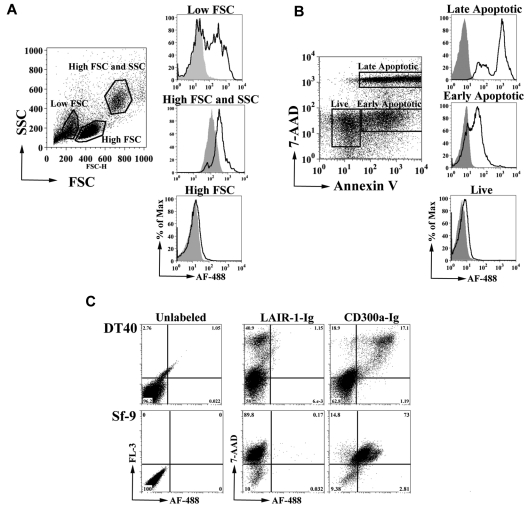
To identify the ligand of CD300a, we generated an Fc fusion protein containing the extracellular domain of CD300a (CD300a-Ig). We tested the binding of CD300a-Ig to starved PBMCs and we observed, as determined by flow cytometric analysis, that CD300a-Ig predominantly binds to cells with low forward scatter (FSC) and low side scatter (SSC), whereas the negative control fusion protein LAIR-1-Ig did not (Figure 1A). Because low FSC and SSC are features of dead cells, we resolved to further confirm that CD300a-Ig binds to cells undergoing apoptosis. Starved peripheral blood lymphocytes (PBLs) were labeled with annexin V and 7-AAD, and the binding of CD300a-Ig to live cells (annexin V− 7-AAD−), early apoptotic cells (annexin V+ 7-AAD−) and late apoptotic/necrotic cells (annexin V+ 7-AAD+) was determined. The results in Figure 1B show that CD300a-Ig predominantly binds to dead cells, with higher intensity to late apoptotic/necrotic cells, whereas live cells barely bind CD300a-Ig. Next, we determined if human CD300a binds the dead cells from other species. Our results clearly show that CD300a-Ig still is capable of binding dead cells of evolutionary distant species, such as chicken and fly (Figure 1C).
Figure 1.
CD300a-Ig binds to dead cells. (A) Human starved PBMCs from a healthy donor were labeled with Alexa Fluor 488 (AF488)–CD300a-Ig (empty histograms) or control AF488-LAIR-1-Ig (filled histograms). Three groups of cells were electronically gated based in their forward scatter (FSC) and side scatter (SSC). Data shown are representative of at least 5 experiments. (B) Starved PBLs from a healthy donor were stained with allophycocyanin (APC) annexin-V, 7-AAD, AF488-CD300a-Ig, or control AF488-LAIR-1-Ig. The binding of AF488-CD300a-Ig (empty histograms) and AF488-LAIR-1-Ig (filled histograms) was determined on live (annexin-V−7-AAD−), early apoptotic (annexin-V+7-AAD−) and late apoptotic/necrotic (annexin-V+7-AAD+) cells. Data shown are representative of at least 5 independent experiments. (C) Dexamethasone-treated DT40 chicken B cells (top panel) and serum starved Sf-9 insect cells (bottom panel) were either unlabeled or labeled with 7-AAD, AF488-CD300a-Ig or control AF488-LAIR-1-Ig and analyzed by flow cytometry. Data shown are representative of 2 independent experiments.
CD300a specifically binds PE and PS
Apoptotic cells are characterized by dynamic changes on the cell surface, including the loss of membrane phospholipid asymmetry, which is one of the mechanisms leading to phagocytic recognition of dead cells.21–23 In normal cells, PC and sphingomyelin are the main phospholipids of the outer leaflet of the plasma membrane, whereas PS and PE reside in the inner leaflet21; however, from the early stages of apoptosis, cells expose PS and PE in the outer leaflet of the plasma membrane.21,24–26 The exposure of PS is the best characterized, and one of the most important, “eat-me” signals of apoptotic cells leading to their engulfment by phagocytes.22,23
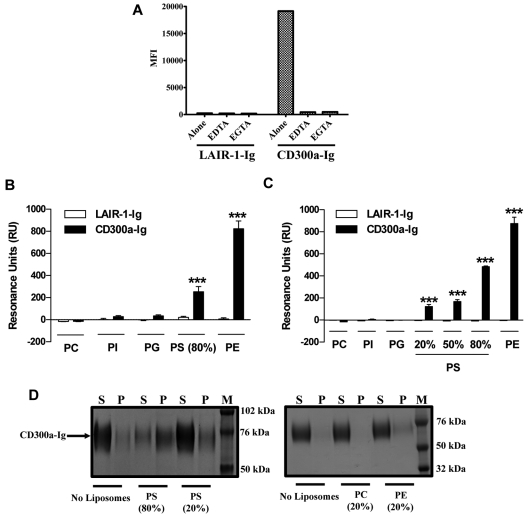
Because our results (Figure 1) suggest that the ligand of CD300a must be evolutionary conserved, we tested the possibility that CD300a binds lipids that are exposed by the outer leaflet of the plasma membrane of dead cells. First, we determined whether a metal ion is required for the binding of CD300a-Ig to dead cells, as is the case for the binding of annexin V, TIM-3, and TIM-4 to its ligand PS.27–29 Our results show that in the presence of the divalent cation chelators EDTA and EGTA the binding of CD300a-Ig is abrogated (Figure 2A), which indicates that an ion, probably Ca2+, is required for the binding of CD300a to its ligand. Next, we performed SPR experiments to directly assess the binding of CD300a to lipids. The SPR experiments were intended as an orthogonal method to search for binding partners of CD300a. We used the resonance unit (RU) values as a yes/no answer to CD300a-phospholipid interactions. The sensograms for the interaction of PE, PS, and PC-containing liposomes to CD300a-Ig at different concentrations are shown in supplemental Figure 1. We immobilized liposomes onto an L1 sensor and determined the binding of CD300a-Ig to them. The results in Figure 2B show that CD300a-Ig specifically binds to immobilized liposomes containing PS, although it does not bind liposomes containing phosphatidylglycerol (PG), phospatidylinositol (PI) purified from liver, or liposomes containing PC alone. PE is another phospholipid that is exposed on the outer leaflet of apoptotic cells.24,25 This aminophospholipid is synthesized by 2 independent pathways, and in one of them, PS, is converted to PE by a PS decarboxylase.26 Consequently, we tested the ability of CD300a-Ig to bind immobilized PE-containing liposomes. Our results show that CD300a-Ig exhibited a higher binding for immobilized PE containing liposomes than immobilized PS-containing liposomes (Figure 2B). The negative control LAIR-1-Ig did not show binding to any of the immobilized liposomes. Next, we performed SPR analysis immobilizing CD300a-Ig, and the negative control LAIR-1-Ig, to a CM5 sensor (Figure 2C). Results confirmed the specific binding of PE and PS, in a dose-dependent manner, containing liposomes to CD300a-Ig, whereas liposomes containing only PC or PC plus PG or PI purified from liver did not show any binding. In this system, PE also showed a higher level of binding to CD300a-Ig than PS.
Figure 2.
Binding and association of CD300a-Ig to PE and PS. (A) Starved PBMCs from healthy donors were left untreated (alone) or preincubated with 2 different divalent cation chelators (EDTA and EGTA). The binding of LAIR-1-Ig or CD300a-Ig to the 7-AAD positive cells was assessed and the mean fluorescence intensity (MFI) values are plotted. Data shown is representative of 3 independent experiments. (B) Liposomes of specified compositions (x-axis) were prepared and coupled to a L1 biosensor. See the supplemental Methods for specifics of liposome composition. The binding of LAIR-1-Ig (white bars) and CD300a-Ig (black bars) was analyzed by allowing the proteins to pass through the L1 sensor. The binding (resonance units or RU) for plateau values is shown. The RU are plotted after subtracting from the buffer control. Bar graph represents the average ± SEM. Results shown are from 8 independent experiments. (C) LAIR-1-Ig and CD300a-Ig were immobilized onto a CM5 sensor and a series of liposomes with different compositions (x-axis) were allowed to pass through the sensor and the binding analysis of liposomes to proteins was analyzed. See supplemental Methods for specifics of liposome composition. The binding (RU) for plateau values is shown. The RU are plotted after subtracting from the buffer control. Bar graph represents the average ± SEM. Results shown are from 2 independent experiments. (D) Liposomes of different compositions were incubated with CD300a-Ig fusion protein and allowed to associate. After 40 minutes of incubation the supernatants (S) and pellet (P) fractions were separated by ultracentrifugation and analyzed by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) analysis. The prestained protein markers (M) are also represented on the protein gel. Data shown are representative of 3 independent experiments.
We have also analyzed the association of CD300a-Ig to PE and PS liposomes using a sedimentation assay (Figure 2D). In this assay, the complexes formed by liposomes and proteins are localized in the pellet fraction, whereas the proteins that are not associated with liposomes remain in the supernatant fraction. Our results show that CD300a-Ig is in the pellet fraction when it is associated with PE and PS-containing liposomes, but absent when liposomes are made only with PC or they are not present (Figure 2D). The negative control protein LAIR-1-Ig did not appear in the pellet fraction in any condition (data not shown). Altogether, these results indicate that PS, and especially PE, are the ligands for the immunomodulatory receptor CD300a.
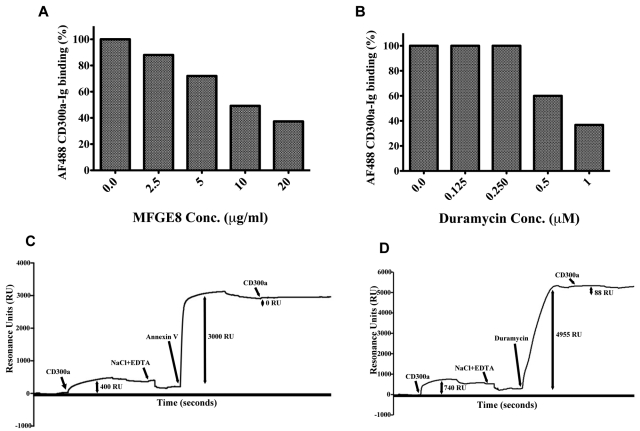
To further prove that there is a specific binding of CD300a to PE and PS, we performed blocking experiments (Figure 3). First, we show that binding of CD300a-Ig to dead cells is blocked in a dose- dependent manner in the presence of MFG-E8, a PS-binding molecule30 (Figure 3A), and in the presence of duramycin, a 19 aminoacid peptide that binds PE31 (Figure 3B). Finally, and using the same SPR technology, we showed that annexin V, another PS-binding molecule,28 blocks the binding of CD300a-Ig to immobilized PS-containing liposomes (Figure 3C) and that duramycin blocks the binding of CD300a-Ig to immobilized PE containing liposomes (Figure 3D).
Figure 3.
Blocking experiments of CD300a-Ig binding to dead cells and liposomes. Starved PBMCs from healthy donors were preincubated with increasing concentrations of MFG-E8 (A) and duramycin (B). Then, the binding of Alexa Fluor 488 (AF488) CD300a-Ig to the 7-AAD positive cells was assessed and the percentage change in CD300-Ig binding as a function of different concentrations of MFG-E8 (A) and duramycin (B) is shown. Each bar graph is a representative example of 2 independent experiments. Blocking of CD300a-Ig binding to PS and PE containing liposomes by annexin V (C) and duramycin (D) is shown. The detailed procedure is described in supplemental Methods. The y-axis indicates the strength of binding by proteins to the immobilized liposomes on the L1 chip. The 1-headed arrows correspond to the reagents added as specified in the graph. The double-headed arrows indicate the binding ability of the proteins specified as RU. Note that the data points for the complete dissociation curve after the injection of NaCl plus EDTA have been removed. The data are a representative of 2 independent experiments.
Identification of residues involved in the binding of CD300a to PE and PS
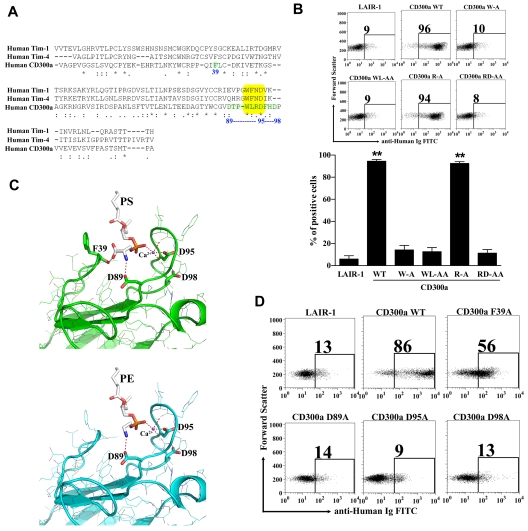
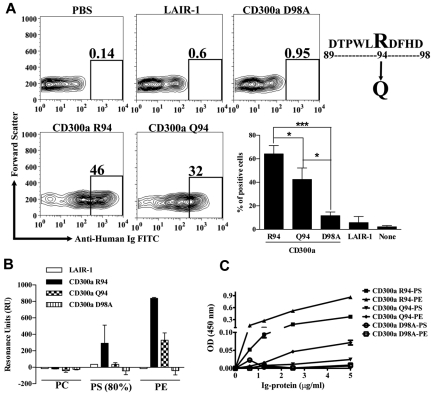
T-cell immunoglobulin and mucin (TIM)–1, TIM-3, and TIM-4 have been identified as receptors for PS.27,32,33 The crystal structures of the TIM-3 and TIM-4 proteins complexed with PS were previously described.27,29 The structures revealed a metal ion-dependent ligand-binding site in the IgV-like domain of these receptors. A narrow cavity is shaped by the CC′ loop and the hydrophobic FG loop that allows the access of the hydrophilic head of PS that coordinates with the metal ion, whereas the aromatic residues on the tip of the FG loop interacted with the fatty acid chains. The WFND (Trp Phe Asn Asp) motif, which is conserved between the TIM proteins that bind PS, is very important for binding to this aminophospholipid. The residues Asn and Asp from the WFND motif coordinate the cation in the TIM-4 structure complexed with PS, and the aromatic residues Trp and Phe at the tip of the FG loop build the binding pocket.29 Sequence comparison of the IgV-like domains of TIM-1 and TIM-4 sequences with the IgV-like domain of CD300a showed that in the latter the motif is WLRD (Trp Leu Arg Asp), which is similar to the WFND motif present in the TIM molecules (Figure 4A). Mutational analysis of the WLRD residues show that they play an important role in the binding of CD300a-Ig to dead cells (Figure 4B), echoing the importance of the WFND motif in TIM-4 for binding to PS.29
Figure 4.
Modeling of CD300a structure complexed with PE and PS and mutational studies. (A) Alignment of human CD300a and human TIM-1 and TIM-4. The extra cellular domains of the CD300a receptor and the Ig-V like domain of TIM-1 and TIM-4 were aligned using the multiple sequence alignment program “CLUSTALW.” The yellow box highlights the WFND motif of the TIM proteins and the correspondent WLRD motif in CD300a. The residues in green and numbered in blue were shown to have potential for binding to PE and PS according to the model presented in (C). (B) Binding analysis of CD300a-Ig mutant proteins to dead PBLs. Purified CD300a-Ig mutants were incubated with starved PBLs and their binding to 7-AAD+ PBLs after adding anti-human Ig FITC was determined by flow cytometry. LAIR-1-Ig was used as a negative control protein. Numbers in the dot plots are the percentage of 7-AAD+ PBLs that bind the fusion proteins. A representative experiment is shown (top panel). Bar graph represents the average ± SEM. Results shown are from 3 independent experiments (bottom panel). (C) Modeling of CD300a IgV-like domain structure complexed with PS (green, top panel) and PE (blue, bottom panel). The aminophospholipids (PS and PE) and the divalent metal ion calcium (Ca2+), in violet, are embedded into the CD300a structure. The dotted red lines represent the bonds, with and without coordination by the metal ion calcium (Ca2+), between the aminoacids in CD300a and PS or PE. (D) Binding analysis of CD300a-Ig mutant proteins to dead PBLs. The residues proposed to bind Ca2+ or directly to PE and PS according to the model described in (C) were mutated and the Ig-fusion proteins were tested for the binding to the dead PBLs. Purified CD300a-Ig mutants were incubated with starved PBLs and their binding to 7-AAD+ cells after adding anti–human Ig FITC was determined by flow cytometry. LAIR-1-Ig was used as a negative control protein. Numbers in the dot plots are the percentage of 7-AAD+ PBLs that bind the fusion proteins. Data shown is a representative of 3 independent experiments.
To further identify residues that may be involved in the binding of PE and PS, a molecular model of CD300a was generated based on the crystal structure of TIM-4 complexed with PS (Figure 4C). The overall structure of the model is similar to the crystal structure of CD300a34 (pdb id:2Q87) in the absence of ligands. The positions of the FG and CC′ loops were modeled to reflect the conformation of the corresponding loops in the TIM-4-PS complex (pdb id:3BIB), thereby creating the metal ion and phospholipid binding sites. The metal ion and a molecule of PS were placed in positions corresponding to those in the ligand-bound TIM-4 structure. Three Asp residues in the FG loop show the potential for interaction with either the metal ion or PS. The side chain of Asp 89 is positioned to form a hydrogen bond/salt bridge with the amino group of the PS molecule, and corresponds to Glu 93 of TIM-4. Asp 95 is in position to coordinate the metal ion, and Asp 98 is positioned nearby and could also be involved in an interaction with either the PS ligand or the metal ion. The position of Asp 95 in the model is similar to that of Asn 99 in TIM-4. Asp 100 in TIM-4 coordinates the metal ion and it is possible that Asp 98 in CD300a plays the same role, but this would require a change in loop conformation in the model. It does not seem probable that both Asp 95 and Asp 98 participate in metal ion binding. In the TIM-4 complex, the carboxyl group of PS forms a hydrogen bond with the backbone amide group of Lys 41 of the CC′ loop, whereas in the CD300a model the backbone carbonyl group of Phe 39 serves this function. The structure of the PE complex was modeled by removing the carboxyl group from PS. This results in loss of the hydrogen bond with Phe 39 (Lys 41 in TIM-4) of the CC′ loop.
To validate our structural model of CD300a complexed with PE and PS, we made mutations to analyze their effect on binding (Figure 4D). Results show that mutation of Asp 89 (D89A), that forms a hydrogen bond/salt bridge with the amino group of PE and PS, abolished the binding of CD300a-Ig to dead cells. The substitution of Asp 95, which according to our model coordinates the metal ion, for Ala (D95A) also abolished the binding of CD300a-Ig to dead cells. Mutation of Arg 94 (R94A) has no effect on the binding of CD300a-Ig to dead cells, which suggests that the loss of binding in the double-substitution of Arg 94 and Asp 95 for Ala residues (RD/AA) is exclusively because of the essential role of Asp 95 (Figure 4B). Although according to our model Asp 98 does not seem to participate in the metal ion binding, we also mutated this residue for Ala (D98A), because Asp 98 in CD300a is the equivalent of Asp 100 in TIM-4, where it coordinates the metal ion binding. Our results show that Asp 98 is also important for the binding of CD300a-Ig to dead cells (Figure 4D), to immobilized PE and PS-containing liposomes, and to purified PE and PS (see the next paragraph). These results may be explained by the fact that substitution of Asp 98 for Ala may disrupt the conformation of the FG loop. Obtaining the crystal structure of the IgV-like domain of CD300a complexed to PE and PS will clarify this issue. Finally, the mutation of Phe 39 for Ala (F39A) resulted in a decrease in the binding of CD300a-Ig to dead cells. This can be explained by the fact that Phe 39 forms a hydrogen bond with the carboxy group of PS that is absent in PE, and therefore the hydrogen bond is not required. We propose that the observed binding of CD300a-Ig F39A to dead cells is mostly because of the interaction with PE, rather than PS, expressed on the surface of apoptotic cells.
A SNP that encodes for the nonsynonymous substitution R94Q in the IgV-like domain of CD300a has been linked to psoriasis susceptibility.14 We investigated whether this clinically important Gln substitution impacts the binding of CD300a to its ligands. Our results show that CD300a-Ig R94 binds better to dead cells than CD300a-Ig Q94 (Figure 5A). SPR analysis show that CD300a-Ig Q94 barely binds to immobilized PS-containing liposomes, whereas the binding to immobilized PE-containing liposomes is reduced to one third compared with the binding of CD300a-Ig R94 (Figure 5B). The negative control LAIR-1-Ig and the mutant CD300a-Ig D98A do not bind to immobilized PE and PS-containing liposomes. Finally, in ELISA experiments, we measured the binding of several CD300a-Ig proteins to purified PE and PS immobilized on a plate (Figure 5C). Results show that CD300a-Ig R94 binds both PE and PS, the latter with lower affinity than the former. However, CD300a-Ig Q94 shows a low affinity to PE and even lower to PS, which confirms the results obtained with the SPR analysis. The mutant CD300a-Ig D98A does not bind neither to PE or PS. Altogether, these results suggest that the susceptibility to psoriasis that is linked to the SNP that encodes for Gln 94 in the IgV-like domain of CD300a is in part because of a change in the ability of the receptor to bind its ligands.
Figure 5.
Impaired binding of CD300a-Ig Q94, a SNP linked to psoriasis susceptibility. (A) Binding of CD300a-Ig Q94 to dead PBLs. Purified CD300a-Ig proteins were incubated with starved PBLs and their binding to 7-AAD+ cells after adding anti–human Ig FITC was determined by flow cytometry. LAIR1-Ig was used as a negative control protein. Numbers in the dot plots are the percentage of 7-AAD+ PBLs that bind the fusion proteins. A representative experiment is shown. Bar graph represents the average ± SEM. Results shown are from 5 independent experiments. (B) Binding of CD300a-Ig proteins to liposomes. Liposomes of specified compositions (x-axis) were prepared and coupled to a L1 biosensor. See the supplemental Methods for specifics of liposome composition. The binding of CD300a-Ig R94 (wild-type), CD300a-Ig Q94 (SNP), CD300a-Ig D98A (not binding mutant) and LAIR-1-Ig (negative control) was analyzed by allowing the proteins to pass through the L1 sensor. The binding (RU) for plateau values is shown. The RU were plotted after subtracting from the buffer control. Bar graph represents the average ± SEM. Results shown are from 2 independent experiments. (C) Binding of CD300a-Ig Q94 to purified lipids. Aminophospholipids PE and PS were coupled to 96-well plates as described in supplemental Methods. A sandwich ELISA was performed with the Ig-fusion proteins at different concentrations. The optical density at 450 nm was determined. Data shown are representative of 2 independent experiments.
CD300a inhibits the uptake of dead cells
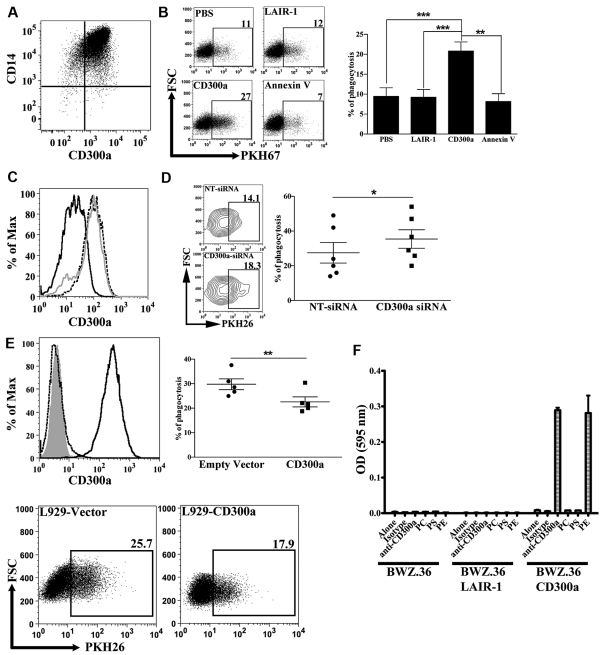
Although many cell types can remove dead cells, professional phagocytes such as macrophages, are the most efficient. This is mainly because macrophages express many receptors that are specific for ligands expressed on dead cells.22,23,35 We wanted to know whether the CD300a receptor has a role in the phagocytosis of apoptotic cells by macrophages. The human monocyte derived macrophages that we have generated are positive for CD300a cell-surface expression (Figure 6A). Accordingly, we tested the engulfment of apoptotic cells in the presence of CD300a-Ig, or the protein controls LAIR-1-Ig and annexin V. The presence of CD300a-Ig resulted in a significant increase in the number of phagocytosed dead cells by macrophages, whereas the presence of the protein control LAIR-1-Ig had no effect (Figure 6B). When annexin V was added to the assay we observed a small decrease in the number of engulfed dead cells (Figure 6B), although not statistically significant, which is in accordance with published data.36 To rule out the possibility that the Fc portion of the CD300a-Ig protein has some effect because of its binding to Fc receptors expressed by macrophages, we performed phagocytosis assays after efficiently knocking down CD300a with siRNA technology (Figure 6C). The results presented in Figure 6D show that macrophages treated with siRNA specific for CD300a exhibit a higher efficiency in the engulfment of apoptotic cells compared with macrophages that have been treated with nontarget siRNA. Altogether, these results indicate that CD300a negatively regulates the uptake of dead cells by macrophages.
Figure 6.
Inhibitory role of CD300a on the engulfment of dead cells. (A) Expression of CD300a on macrophages. Human monocyte derived macrophages were stained with anti-CD14 PE-Cy7 and anti-CD300a PE and analyzed by flow cytometry. The quadrants in the dot plot were determined according to the binding of isotype negative control antibodies (not shown). (B) Effect of CD300a-Ig on the phagocytic function of macrophages. PKH67 labeled dexamethasone treated DT40 cells were incubated with purified proteins, that is, CD300a-Ig, LAIR-1-Ig, and annexin V, and then they were subjected to a phagocytosis assay as described in “Methods.” Macrophages, electronically gated by the expression of CD14, that have ingested DT40 cells are positive for PKH67. A representative experiment is shown. Numbers in the dot plots represent the percentage of macrophages that have engulfed dead DT40 cells. Bar graph represents the average ± SEM. Results shown are from 5 independent experiments. (C) Histogram representation of CD300a expression on macrophages after siRNA knockdown. The broken line histograms shown CD300a expression on nontransfected cells; the gray line corresponds to the CD300a expression on cells transfected with nontarget siRNA (NT-siRNA); and the black line histogram shows CD300a expression after transfection with siRNA specific for CD300a (CD300a-siRNA). Data shown is a representative of 6 independent donors. (D) Knockdown of CD300a increases the phagocytic activity of macrophages. PKH26 labeled dexamethasone treated thymocytes were subjected to a phagocytosis assay with macrophages transfected with NT-siRNA or with CD300a-siRNA. Macrophages, electronically gated by the FSC and SSC parameters that have ingested thymocytes are positive for PKH26. A representative experiment is shown (left panel). Numbers in the contour plots represent the percentage of macrophages that have engulfed dead thymocytes. The graph represents the percentage of macrophages that have engulfed (% of phagocytosis) dead cells (right panel). Each pair of symbols corresponds to a different donor. (E) L929 cells were stably transfected with empty or CD300a vector. The histograms (top left panel) show the staining with anti-CD300a Ab of empty vector transfected cells (dotted black line) and CD300a transfected cells (black line). The gray histogram represents unstained cells. L929 cells were mixed with PKH26 labeled dexamethasone treated thymocytes for 60 minutes, treated with trypsin/EDTA to detach the cells, and analyzed by flow cytometry. L929 cells, electronically gated by the FSC and SSC parameters that have ingested dead thymocytes, are positive for PKH26. A representative experiment is shown (bottom panel). Numbers in the dot plots represents the percentage of L929 cells that have engulfed dead thymocytes. The graph represents the percentage of L929 cells that have engulfed (% of phagocytosis) dead cells (top right panel). Each pair of symbols corresponds to an independent experiment. (F) Functional recognition of PE by CD300a. Levels of β-galactosidase activity generated by culture of BWZ.36, BWZ.36 LAIR-1/CD3ζ (BWZ.36 LAIR-1), and BWZ.36 CD300a/CD3ζ (BWZ.36 CD300a) cells on uncoated plates or plates coated with isotype-control antibody, anti-CD300a antibody, PC, PS, or PE. The lipids (PC, PS, and PE) were air-dried and the isotype controls and anti-CD300a antibodies were immobilized on a 24-well plate. Data shown is representative of 2 independent experiments.
To completely rule out any role played by the Fc receptors, we transfected the mouse fibroblast L929 cells with a plasmid encoding human CD300a and analyzed the ability of the transfected cells to phagocytose apoptotic thymocytes. The results show that CD300a-expressing cells phagocytosed between 18%-35% less apoptotic cells compared with control transfected L929 cells (Figure 6E).
Finally, to further study the functional interaction between CD300a and its ligands, we used the BWZ.36 T cell mouse hybridoma reporter cell line transfected with a chimeric receptor consisting of the human CD300a extracellular portion and human CD3ζ transmembrane and intracytoplasmic portions (BWZ.36 CD300a cells). In Figure 6F, we show that CD300a is able to functionally interact with PE, but not with PS. These results suggest that the recognition of PE on the cell surface of dead cells by CD300a delivers a signal that modulates the uptake of apoptotic cells.
Discussion
In this report we identify PS, and predominantly PE, as the ligands for the human ITIM-containing receptor CD300a. The role of PS exposure by the outer leaflet of the plasma membrane and many of its ligands has been extensively studied and several receptors recognizing PS that deliver “eat-me” signals have been identified.22,23 Here we show for the first time that PE, an aminophospholipid that is also exposed on the outer leaflet of the plasma membrane of dead cells,24,25 has an important role in the processes leading to the uptake of apoptotic cells through its recognition by CD300a.
Our results show that CD300a binds late apoptotic/necrotic cells that are characterized by a permeable plasma membrane to a greater extent than early apoptotic cells (Figure 1B). This may be explained by the higher levels of PE and PS exposed on the outer leaflet of the plasma membrane and also by the possibility that the CD300-Ig fusion protein may even be entering to the late necrotic/apoptotic cells. We have studied the interaction of CD300a with PE and PS using liposomes containing different combinations of phospholipids. We show that PC:PS and PC:PE-containing liposomes bind to CD300a, and that this binding is specific to PE and PS, because liposomes that do not contain PE or PS do not bind to CD300a. Our attempts to make usable pure PE, and PS, liposomes have not been successful. This was not surprising because it is well known that liposomes containing only PE (or PS) aggregate in the presence of calcium (see Figure 2A for the importance of calcium for binding CD300a-Ig to dead cells). However, when vesicles are prepared using PE or PS in combination with PC, stable bilayers are generated, thereby overcoming the problem of aggregation in the presence of calcium.37 We have also studied the interaction of CD300a with single pure phospholipids and we show a dose-dependent binding of CD300a-Ig to pure PE and PS immobilized on a plate. Furthermore, we have addressed the functional recognition of single phospholipids immobilized on a plate by reporter cells expressing a CD300a-CD3ζ chimeric receptor (Figure 6F). The results from the functional recognition differ somewhat from what would have been expected from the binding of CD300a-Ig to liposomes and single pure lipids, which suggestis that the particular steric environment of the extracellular part of CD300a affects its binding. Finally, our structural models in Figure 4C support the binding of CD300a to PE and PS. Altogether, our results demonstrate that CD300a binds PE and PS.
Very recently it has been published that mouse CD300-Ig molecules are able to bind overlapping but distinctive patterns of lipids immobilized on a plate.38 Interestingly, mouse CD300a, also known as MAIR-I and LMIR1,39,40 was shown not to bind any tested lipid, including PE and PS.38 The discrepancy between mouse and human CD300a binding to phospholipids may be explained by the lack of the WLRD motif and the Phe 39 in the mouse CD300a. Our structural model and mutational studies have demonstrated that those residues are involved in the binding of CD300a to PE and PS (Figure 4).
Phagocytosis of dead cells is of fundamental importance to an array of biologic responses, including tissue homeostasis, remodeling, development, and resolution of inflammation, that are centered in the immune system. It is a very complex and dynamic process that is regulated by a multitude of positive and negative signals.22,23,41,42 It is well established that the recognition of PS on apoptotic cells within tissues acts as an “eat-me” signal and drives their engulfment. To date, multiple distinct PS-binding proteins have been identified. Direct binding receptors include members of the TIM family (TIM-1, TIM-3, and TIM-4),27,29,32,33 BAI1,43 and Stabilin-2.44 In addition, soluble PS-binding molecules have been identified, and these include MFG-E8,30 Gas6,45 protein S,46 and annexin V.23,28 The latter is commonly used to detect PS on the outer leaflet of the plasma membrane on apoptotic cells. In addition, thrombospondins, CD36 and CD68 were suggested to be capable of binding PS.23 Very recently we have published that CD300f, another member of the CD300 family of receptors, recognizes PS exposed on the outer leaflet of the plasma membrane and can modulate phagocytosis.47 In this paper, we demonstrate that CD300a binds PE and PS and inhibits the uptake of apoptotic cells by macrophages and L929 cells (Figure 6). Our results have strong potential to alter the existing “eat me” signal paradigm in that during the death process, when cells expose PE they may provide a “do not eat me yet” signal. This concept fits nicely with the fact that essentially all immunologic responses are the result of tipping the balance between inhibition and activation. The amount of PE exposure is probably a dynamic process that depends on many factors and would be a very interesting area of research to pursue. Many others have described the expression of PE on the outer leaflet of the plasma membrane of apoptotic cells.24–25
Our data, along with those from recent publications,38,47,48 which show that CD300 molecules bind lipids and that they are involved in autoimmune processes, are portraying a picture where the CD300 family of proteins is emerging as a sensing network that would be receptive to signals from complex physiologic environments, such as those that occur during the engulfment of dead cells. This complexity is further highlighted by differences in the intracellular signaling cascades (ITIMs vs ITAMs), in the heterodimerization potential between different family members and in their diverse pattern of cellular expression.3,4
Given that CD300a is expressed on cells from both myeloid and lymphoid lineages,3 and that PS (and probably PE) is not only exposed on apoptotic cells, but also in activated B cells49 and T cells,47 and in monocytes,50 future studies addressing the role of this receptor in modulating key immune functions, such as cell mediated cytotoxicity and cytokine production, cell differentiation, inflammation, and phagocytosis during health and disease are also warranted.
Supplementary Material
Acknowledgments
The authors thank Jasmin Herz for her dedication in providing the mouse thymii whenever needed. They also thank John Mariano for his help in isolating monocytes and for critically reading the manuscript. They also express thanks to the National Institutes of Health Clinical Center Department of Transfusion Medicine for the blood samples.
This work was funded by the intramural programs of the Food and Drug Administration and the National Institute of Allergy and Infectious Diseases.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: V.R.S., J.F.A., E.C., and S.-C.C. performed the experiments; V.R.S., J.F.A., E.C., and F.B. analyzed results; V.R.S. and F.B. designed the research; and V.R.S., J.E.C., and F.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Francisco Borrego, Laboratory of Molecular and Developmental Immunology, Division of Monoclonal Antibodies, Office of Biotechnology Products, Center for Drug Evaluation and Research, Food and Drug Administration, Bdg 29B, Rm 3NN18, 29 Lincoln Drive, HFD-123, Bethesda, MD, 20892; e-mail: francisco.borrego@fda.hhs.gov.
References
- 1.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290(5489):84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 2.Stanietsky N, Mandelboim O. Paired NK cell receptors controlling NK cytotoxicity. FEBS Lett. 2010;584(24):4895–4900. doi: 10.1016/j.febslet.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 3.Clark GJ, Ju X, Tate C, Hart DN. The CD300 family of molecules are evolutionarily significant regulators of leukocyte functions. Trends Immunol. 2009;30(5):209–217. doi: 10.1016/j.it.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Barriocanal A, Comas-Casellas E, Schwartz S, Jr., Martin M, Sayos J. CD300 heterocomplexes, a new and family-restricted mechanism for myeloid cell signaling regulation. J Biol Chem. 2010;285(53):41781–41794. doi: 10.1074/jbc.M110.140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lankry D, Simic H, Klieger Y, Levi-Schaffer F, Jonjic S, Mandelboim O. Expression and function of CD300 in NK cells. J Immunol. 2010;185(5):2877–2886. doi: 10.4049/jimmunol.0903347. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen R, Bustamante C, Clark AG, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3(6):e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez Y, Tang X, Coligan JE, Borrego F. The CD300a (IRp60) inhibitory receptor is rapidly up-regulated on human neutrophils in response to inflammatory stimuli and modulates CD32a (FcgammaRIIa) mediated signaling. Mol Immunol. 2008;45(1):253–258. doi: 10.1016/j.molimm.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachelet I, Munitz A, Moretta A, Moretta L, Levi-Schaffer F. The inhibitory receptor IRp60 (CD300a) is expressed and functional on human mast cells. J Immunol. 2005;175(12):7989–7995. doi: 10.4049/jimmunol.175.12.7989. [DOI] [PubMed] [Google Scholar]
- 9.Narayanan S, Silva R, Peruzzi G, et al. Human Th1 cells that express CD300a are polyfunctional and after stimulation up-regulate the T-box transcription factor eomesodermin. PLoS One. 2010;5(5):e10636. doi: 10.1371/journal.pone.0010636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simhadri VR, Mariano JL, Zhou Q, Debell KE, Borrego F. Differential expression of CD300a/c on human TH1 and TH17 cells. BMC Immunol. 2011;12:62. doi: 10.1186/1471-2172-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantoni C, Bottino C, Augugliaro R, et al. Molecular and functional characterization of IRp60, a member of the immunoglobulin superfamily that functions as an inhibitory receptor in human NK cells. Eur J Immunol. 1999;29(10):3148–3159. doi: 10.1002/(SICI)1521-4141(199910)29:10<3148::AID-IMMU3148>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 12.Silva R, Moir S, Kardava L, et al. CD300a is expressed on human B cells, modulates BCR-mediated signaling, and its expression is down-regulated in HIV infection. Blood. 2011;117(22):5870–5880. doi: 10.1182/blood-2010-09-310318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munitz A, Bachelet I, Eliashar R, Moretta A, Moretta L, Levi-Schaffer F. The inhibitory receptor IRp60 (CD300a) suppresses the effects of IL-5, GM-CSF, and eotaxin on human peripheral blood eosinophils. Blood. 2006;107(5):1996–2003. doi: 10.1182/blood-2005-07-2926. [DOI] [PubMed] [Google Scholar]
- 14.Speckman RA, Wright Daw JA, Helms C, et al. Novel immunoglobulin superfamily gene cluster, mapping to a region of human chromosome 17q25, linked to psoriasis susceptibility. Hum Genet. 2003;112(1):34–41. doi: 10.1007/s00439-002-0851-y. [DOI] [PubMed] [Google Scholar]
- 15.Burakoff R, Chao S, Perencevich M, et al. Blood-based biomarkers can differentiate ulcerative colitis from crohn's disease and noninflammatory diarrhea. Inflamm Bowel Dis. 2011;17(8):1719–1725. doi: 10.1002/ibd.21574. [DOI] [PubMed] [Google Scholar]
- 16.Coustan-Smith E, Song G, Clark C, et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2011;117(23):6267–6276. doi: 10.1182/blood-2010-12-324004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munitz A, Bachelet I, Levi-Schaffer F. Reversal of airway inflammation and remodeling in asthma by a bispecific antibody fragment linking CCR3 to CD300a. J Allergy Clin Immunol. 2006;118(5):1082–1089. doi: 10.1016/j.jaci.2006.07.041. [DOI] [PubMed] [Google Scholar]
- 18.Bachelet I, Munitz A, Levi-Schaffer F. Abrogation of allergic reactions by a bispecific antibody fragment linking IgE to CD300a. J Allergy Clin Immunol. 2006;117(6):1314–1320. doi: 10.1016/j.jaci.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Bachelet I, Munitz A, Berent-Maoz B, Mankuta D, Levi-Schaffer F. Suppression of normal and malignant kit signaling by a bispecific antibody linking kit with CD300a. J Immunol. 2008;180(9):6064–6069. doi: 10.4049/jimmunol.180.9.6064. [DOI] [PubMed] [Google Scholar]
- 20.Tang X, Narayanan S, Peruzzi G, et al. A single residue, arginine 65, is critical for the functional interaction of leukocyte-associated inhibitory receptor-1 with collagens. J Immunol. 2009;182(9):5446–5452. doi: 10.4049/jimmunol.0804052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: A matter of life and death. Annu Rev Physiol. 2003;65:701–734. doi: 10.1146/annurev.physiol.65.092101.142459. [DOI] [PubMed] [Google Scholar]
- 22.Bratton DL, Henson PM. Apoptotic cell recognition: will the real phosphatidylserine receptor(s) please stand up? Curr Biol. 2008;18(2):R76–79. doi: 10.1016/j.cub.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Ravichandran KS. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J Exp Med. 2010;207(9):1807–1817. doi: 10.1084/jem.20101157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emoto K, Toyama-Sorimachi N, Karasuyama H, Inoue K, Umeda M. Exposure of phosphatidylethanolamine on the surface of apoptotic cells. Exp Cell Res. 1997;232(2):430–434. doi: 10.1006/excr.1997.3521. [DOI] [PubMed] [Google Scholar]
- 25.Maulik N, Kagan VE, Tyurin VA, Das DK. Redistribution of phosphatidylethanolamine and phosphatidylserine precedes reperfusion-induced apoptosis. Am J Physiol. 1998;274(1 Pt 2):H242–248. doi: 10.1152/ajpheart.1998.274.1.H242. [DOI] [PubMed] [Google Scholar]
- 26.Vance JE. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J Lipid Res. 2008;49(7):1377–1387. doi: 10.1194/jlr.R700020-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.DeKruyff RH, Bu X, Ballesteros A, et al. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J Immunol. 2010;184(4):1918–1930. doi: 10.4049/jimmunol.0903059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meers P, Mealy T. Calcium-dependent annexin V binding to phospholipids: stoichiometry, specificity, and the role of negative charge. Biochemistry. 1993;32(43):11711–11721. doi: 10.1021/bi00094a030. [DOI] [PubMed] [Google Scholar]
- 29.Santiago C, Ballesteros A, Martinez-Munoz L, et al. Structures of T cell immunoglobulin mucin protein 4 show a metal-Ion-dependent ligand binding site where phosphatidylserine binds. Immunity. 2007;27(6):941–951. doi: 10.1016/j.immuni.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417(6885):182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- 31.Zhao M, Li Z, Bugenhagen S. 99mTc-labeled duramycin as a novel phosphatidylethanolamine-binding molecular probe. J Nucl Med. 2008;49(8):1345–1352. doi: 10.2967/jnumed.107.048603. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi N, Karisola P, Pena-Cruz V, et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007;27(6):927–940. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450(7168):435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 34.Dimasi N, Roessle M, Moran O, Candiano G, Svergun DI, Biassoni R. Molecular analysis and solution structure from small-angle X-ray scattering of the human natural killer inhibitory receptor IRp60 (CD300a). Int J Biol Macromol. 2007;40(3):193–200. doi: 10.1016/j.ijbiomac.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140(5):619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Krahling S, Callahan MK, Williamson P, Schlegel RA. Exposure of phosphatidylserine is a general feature in the phagocytosis of apoptotic lymphocytes by macrophages. Cell Death Differ. 1999;6(2):183–189. doi: 10.1038/sj.cdd.4400473. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh S, Bell R. Liposomes. Applications in protein-lipid interaction studies. Methods Mol Biol. 2002;199:49–60. doi: 10.1385/1-59259-175-2:49. [DOI] [PubMed] [Google Scholar]
- 38.Cannon JP, O'Driscoll M, Litman GW. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics. 2012;64(1):39–47. doi: 10.1007/s00251-011-0562-4. [DOI] [PubMed] [Google Scholar]
- 39.Kumagai H, Oki T, Tamitsu K, et al. Identification and characterization of a new pair of immunoglobulin-like receptors LMIR1 and 2 derived from murine bone marrow-derived mast cells. Biochem Biophys Res Commun. 2003;307(3):719–729. doi: 10.1016/s0006-291x(03)01245-2. [DOI] [PubMed] [Google Scholar]
- 40.Yotsumoto K, Okoshi Y, Shibuya K, et al. Paired activating and inhibitory immunoglobulin-like receptors, MAIR-I and MAIR-II, regulate mast cell and macrophage activation. J Exp Med. 2003;198(2):223–233. doi: 10.1084/jem.20021825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green DR, Ferguson T, Zitvogel L, Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol. 2009;9(5):353–363. doi: 10.1038/nri2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steevels TA, Meyaard L. Immune inhibitory receptors: essential regulators of phagocyte function. Eur J Immunol. 2011;41(3):575–587. doi: 10.1002/eji.201041179. [DOI] [PubMed] [Google Scholar]
- 43.Park D, Tosello-Trampont AC, Elliott MR, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450(7168):430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 44.Park SY, Jung MY, Kim HJ, et al. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. 2008;15(1):192–201. doi: 10.1038/sj.cdd.4402242. [DOI] [PubMed] [Google Scholar]
- 45.Lemke G, Rothlin CV. Immunobiology of the TAM receptors. Nat Rev Immunol. 2008;8(5):327–336. doi: 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson HA, Maylock CA, Williams JA, Paweletz CP, Shu H, Shacter E. Serum-derived protein S binds to phosphatidylserine and stimulates the phagocytosis of apoptotic cells. Nat Immunol. 2003;4(1):87–91. doi: 10.1038/ni871. [DOI] [PubMed] [Google Scholar]
- 47.Choi SC, Simhadri VR, Tian L, et al. Cutting edge: mouse CD300f (CMRF-35-like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. J Immunol. 2011;187(7):3483–3487. doi: 10.4049/jimmunol.1101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xi H, Katschke KJ, Jr., Helmy KY, et al. Negative regulation of autoimmune demyelination by the inhibitory receptor CLM-1. J Exp Med. 2010;207(1):7–16. doi: 10.1084/jem.20091508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dillon SR, Mancini M, Rosen A, Schlissel MS. Annexin V binds to viable B cells and colocalizes with a marker of lipid rafts upon B cell receptor activation. J Immunol. 2000;164(3):1322–1332. doi: 10.4049/jimmunol.164.3.1322. [DOI] [PubMed] [Google Scholar]
- 50.Appelt U, Sheriff A, Gaipl US, Kalden JR, Voll RE, Herrmann M. Viable, apoptotic and necrotic monocytes expose phosphatidylserine: cooperative binding of the ligand Annexin V to dying but not viable cells and implications for PS-dependent clearance. Cell Death Differ. 2005;12(2):194–196. doi: 10.1038/sj.cdd.4401527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.