Abstract
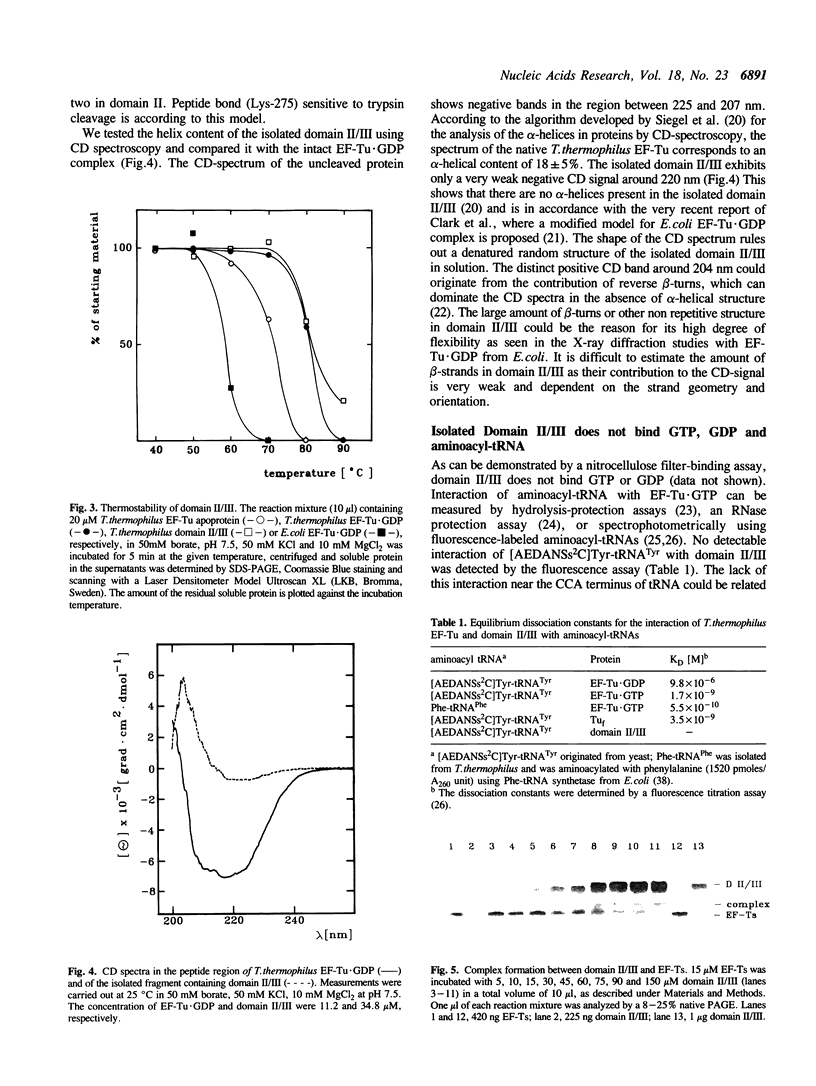
The middle and C-terminal domain (domain II/III) of elongation factor Tu from Thermus thermophilus lacking the GTP/GDP binding domain have been prepared by treating nucleotide-free protein with Staphylococcus aureus V8 protease. The isolated domain II/III of EF-Tu has a compact structure and high resistance against tryptic treatment and thermal denaturation. As demonstrated by circular dichroism spectroscopy, the isolated domain II/III does not contain any alpha-helical structure. Nucleotide exchange factor, EF-Ts, was found to interact with domain II/III, whereas the binding of aminoacyl-tRNA, GDP and GTP to this EF-Tu fragment could not be detected.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahamson J. K., Laue T. M., Miller D. L., Johnson A. E. Direct determination of the association constant between elongation factor Tu X GTP and aminoacyl-tRNA using fluorescence. Biochemistry. 1985 Jan 29;24(3):692–700. doi: 10.1021/bi00324a023. [DOI] [PubMed] [Google Scholar]
- Arai K. I., Kawakita M., Kaziro Y. Studies on polypeptide elongation factors from Escherichia coli. II. Purification of factors Tu-guanosine diphosphate, Ts, and Tu-Ts, and crystallization of Tu-guanosine diphosphate and Tu-Ts. J Biol Chem. 1972 Nov 10;247(21):7029–7037. [PubMed] [Google Scholar]
- Arai K., Ota Y., Arai N., Nakamura S., Henneke C., Oshima T., Kaziro Y. Studies on polypeptide-chain-elongation factors from an extreme thermophile, Thermus thermophilus HB8. 1. Purification and some properties of the purified factors. Eur J Biochem. 1978 Dec;92(2):509–519. doi: 10.1111/j.1432-1033.1978.tb12773.x. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Douglass J., Smith D. Conformational alteration of protein synthesis elongation factor EF-Tu by EF-Ts and by kirromycin. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3264–3267. doi: 10.1073/pnas.74.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms S., Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J Mol Biol. 1980 Apr;138(2):149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- Clark B. F., Kjeldgaard M., la Cour T. F., Thirup S., Nyborg J. Structural determination of the functional sites of E. coli elongation factor Tu. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):203–208. doi: 10.1016/0167-4781(90)90167-z. [DOI] [PubMed] [Google Scholar]
- Duffy L. K., Gerber L., Johnson A. E., Miller D. L. Identification of a histidine residue near the aminoacyl transfer ribonucleic acid binding site of elongation factor Tu. Biochemistry. 1981 Aug 4;20(16):4663–4666. doi: 10.1021/bi00519a022. [DOI] [PubMed] [Google Scholar]
- Duisterwinkel F. J., Kraal B., De Graaf J. M., Talens A., Bosch L., Swart G. W., Parmeggiani A., La Cour T. F., Nyborg J., Clark B. F. Specific alterations of the EF-Tu polypeptide chain considered in the light of its three-dimensional structure. EMBO J. 1984 Jan;3(1):113–120. doi: 10.1002/j.1460-2075.1984.tb01770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston J. F. A kinetic analysis of the interaction of elongation factor Tu with guanosine nucleotides and elongation factor Ts. J Biol Chem. 1984 Nov 10;259(21):12997–13003. [PubMed] [Google Scholar]
- Ehresmann B., Imbault P., Weil J. H. Spectrophotometric determination of protein concentration in cell extracts containing tRNA's and rRNA's. Anal Biochem. 1973 Aug;54(2):454–463. doi: 10.1016/0003-2697(73)90374-6. [DOI] [PubMed] [Google Scholar]
- Gilman A. G. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Hingorani V. N., Ho Y. K. A structural model for the alpha-subunit of transducin. Implications of its role as a molecular switch in the visual signal transduction mechanism. FEBS Lett. 1987 Aug 10;220(1):15–22. doi: 10.1016/0014-5793(87)80867-0. [DOI] [PubMed] [Google Scholar]
- Jonák J., Petersen T. E., Clark B. F., Rychlík I. N-Tosyl-L-phenylalanylchloromethane reacts with cysteine 81 in the molecule of elongation factor Tu from Escherichia coli. FEBS Lett. 1982 Dec 27;150(2):485–488. doi: 10.1016/0014-5793(82)80795-3. [DOI] [PubMed] [Google Scholar]
- Jurnak F. Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science. 1985 Oct 4;230(4721):32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- Kushiro M., Shimizu M., Tomita K. Molecular cloning and sequence determination of the tuf gene coding for the elongation factor Tu of Thermus thermophilus HB8. Eur J Biochem. 1987 Dec 30;170(1-2):93–98. doi: 10.1111/j.1432-1033.1987.tb13671.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Louie A., Ribeiro N. S., Reid B. R., Jurnak F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J Biol Chem. 1984 Apr 25;259(8):5010–5016. [PubMed] [Google Scholar]
- Metz-Boutigue M. H., Reinbolt J., Ebel J. P., Ehresmann C., Ehresmann B. Crosslinking of elongation factor Tu to tRNA(Phe) by trans-diamminedichloroplatinum (II). Characterization of two crosslinking sites on EF-Tu. FEBS Lett. 1989 Mar 13;245(1-2):194–200. doi: 10.1016/0014-5793(89)80220-0. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Ohta S., Arai K., Arai N., Oshima T., Kaziro Y. Studies on polypeptide-chain-elongation factors from an extreme thermophile, Thermus thermophilus HB8. 3. Molecular properties. Eur J Biochem. 1978 Dec;92(2):533–543. doi: 10.1111/j.1432-1033.1978.tb12775.x. [DOI] [PubMed] [Google Scholar]
- Ott G., Faulhammer H. G., Sprinzl M. Interaction of elongation factor Tu from Escherichia coli with aminoacyl-tRNA carrying a fluorescent reporter group on the 3' terminus. Eur J Biochem. 1989 Sep 15;184(2):345–352. doi: 10.1111/j.1432-1033.1989.tb15025.x. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Kabsch W., Krengel U., Holmes K. C., John J., Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989 Sep 21;341(6239):209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A., Swart G. W., Mortensen K. K., Jensen M., Clark B. F., Dente L., Cortese R. Properties of a genetically engineered G domain of elongation factor Tu. Proc Natl Acad Sci U S A. 1987 May;84(10):3141–3145. doi: 10.1073/pnas.84.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M. E., Brockmöller J., Jonák J., Sprinzl M. Identification of the N-tosyl-L-phenylalanyl chloromethylketone modification site in Thermus thermophilus elongation factor Tu. FEBS Lett. 1989 Nov 6;257(2):219–222. doi: 10.1016/0014-5793(89)81538-8. [DOI] [PubMed] [Google Scholar]
- Peter M. E., Schirmer N. K., Reiser C. O., Sprinzl M. Mapping the effector region in Thermus thermophilus elongation factor Tu. Biochemistry. 1990 Mar 20;29(11):2876–2884. doi: 10.1021/bi00463a033. [DOI] [PubMed] [Google Scholar]
- Peter M. E., Wittmann-Liebold B., Sprinzl M. Affinity labeling of the GDP/GTP binding site in Thermus thermophilus elongation factor Tu. Biochemistry. 1988 Dec 27;27(26):9132–9139. doi: 10.1021/bi00426a010. [DOI] [PubMed] [Google Scholar]
- Pingoud A., Urbanke C. Aminoacyl transfer ribonucleic acid binding site of the bacterial elongation factor Tu. Biochemistry. 1980 May 13;19(10):2108–2112. doi: 10.1021/bi00551a017. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Seidler L., Peter M., Meissner F., Sprinzl M. Sequence and identification of the nucleotide binding site for the elongation factor Tu from Thermus thermophilus HB8. Nucleic Acids Res. 1987 Nov 25;15(22):9263–9277. doi: 10.1093/nar/15.22.9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J. B., Steinmetz W. E., Long G. L. A computer-assisted model for estimating protein secondary structure from circular dichroic spectra: comparison of animal lactate dehydrogenases. Anal Biochem. 1980 May 1;104(1):160–167. doi: 10.1016/0003-2697(80)90292-4. [DOI] [PubMed] [Google Scholar]
- Thompson R. C., Karim A. M. The accuracy of protein biosynthesis is limited by its speed: high fidelity selection by ribosomes of aminoacyl-tRNA ternary complexes containing GTP[gamma S]. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4922–4926. doi: 10.1073/pnas.79.16.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Noort J. M., Kraal B., Bosch L. A second tRNA binding site on elongation factor Tu is induced while the factor is bound to the ribosome. Proc Natl Acad Sci U S A. 1985 May;82(10):3212–3216. doi: 10.1073/pnas.82.10.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Sprinzl M. Enzymic binding of aminoacyl-tRNA to Escherichia coli ribosomes using modified tRNA species and tRNA fragments. Methods Enzymol. 1979;60:615–628. doi: 10.1016/s0076-6879(79)60058-7. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A., Guariguata R., Leberman R. Bacterial elongation factor Ts: isolation and reactivity with elongation factor Tu. J Bacteriol. 1983 Mar;153(3):1266–1271. doi: 10.1128/jb.153.3.1266-1271.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos A. M., Tong L., Milburn M. V., Matias P. M., Jancarik J., Noguchi S., Nishimura S., Miura K., Ohtsuka E., Kim S. H. Three-dimensional structure of an oncogene protein: catalytic domain of human c-H-ras p21. Science. 1988 Feb 19;239(4842):888–893. doi: 10.1126/science.2448879. [DOI] [PubMed] [Google Scholar]