Summary
Although integrins are known to mediate connections between extracellular adhesion molecules and the intracellular actin cytoskeleton, the mechanisms that are responsible for coupling ligand binding to intracellular signalling, for generating diversity in signalling, and for determining the efficacy of integrin signalling in response to ligand engagement are largely unknown. By characterising the class of anti-integrin monoclonal antibodies (mAb) that stimulate integrin activation and ligand binding, we have identified integrin-ligand-mAb complexes that exhibit differential signalling properties. Specifically, addition of 12G10 mAb to cells adhering via integrin α4β1 was found to trigger disruption of the actin cytoskeleton, and prevent cell attachment and spreading, while mAb addition to cells adhering via α5β1 stimulated all of these processes. In contrast soluble ligand binding to either α4β1 or α5β1 was augmented or unaffected by 12G10. The regions of the integrin responsible for differential signalling were then mapped using chimeras. Surprisingly, a chimeric α5 integrin containing the β-propeller domain from the ligand-binding pocket of α4 exhibited the same signalling properties as the full-length α4 integrin, while exchanging or removing cytoplasmic domains had no effect. Thus the mAb 12G10 demonstrates dual functionality, inhibiting cell adhesion and spreading while augmenting soluble ligand binding, via a mechanism that is determined by the extracellular β-propeller domain of the associating α subunit. These findings therefore demonstrate a direct and variable agonistic link between the ligand-binding pocket of integrins and the cell interior that is independent of the α cytoplasmic domains. We propose that either ligand-specific transmembrane conformational changes or ligand-specific differences in the kinetics of transmembrane domain separation underlie integrin agonism.
Introduction
Integrin cell adhesion receptors are uniquely positioned at a nexus regulating the signalling flux between the outside of the cell and the cell interior. Integrins dynamically link the deformable meshwork of the extracellular matrix and the contractile actin microfilament system and thereby enable cells to direct membrane protrusions and apply contractile force to adhesive extracellular sites (1). These adhesion-dependent signals are mediated by the clustering of integrins and the congregation of signalling adaptors and enzymes into specialised morphological structures including focal complexes, focal adhesions, and fibrillar adhesions (2). In this way, the integrin-cytoskeletal junction is thought to impose temporal and spatial control on adhesion-related signalling events (3).
Integrins are non-covalently-linked αβ heterodimers. In mammals, 18 α and 8 β subunits combine to form 24 different receptors, with ligand-binding specificity being determined by the particular αβ combination. Both subunits have a conserved, modular domain structure, except that 9 α subunits contain an additional extracellular domain that is homologous to the A domain of von Willebrand factor (vWF1). This domain endows these receptors with a different mode of ligand binding and as such integrins should be classified into two subclasses depending on the presence or absence of this domain. All integrins, however, bind to their ligands in a divalent cation-dependent manner with manganese and magnesium promoting, and calcium disfavouring, ligand binding (4).
Recent NMR, electron microscopic and X-ray crystallographic studies have delivered major advances in our understanding of integrin structure (5-9). The ligand binding “head” region of αVβ3 comprises a seven bladed β-propeller module in the α-subunit and a vWF type A domain in the β subunit (βA domain, also referred to as I-like domain). This head region is attached to two legs, one formed from each subunit, that provide a link to the transmembrane and cytoplasmic domains. Extending from the α subunit β-propeller, the α subunit leg comprises three β-sandwich domains, termed thigh, calf 1 and calf 2. The β subunit domain organisation is more complex with the βA domain being inserted into an immunoglobulin hybrid domain. Therefore, although the βA domain is distal to the membrane insertion site, it is not at the N-terminus of the primary sequence. Furthermore, the hybrid domain is preceded by, and inserted into, a plexin-semaphorin-integrin (PSI) domain (9). The PSI domain is located below the hybrid domain and alongside the β subunit leg which consists of four tandem cysteine-rich EGF-like domains and a C-terminal β-sheet domain termed the β-tail domain. Both subunits are linked to relatively short (typically <60 amino acid residues) cytoplasmic tail domains via a single transmembrane pass.
The ECM components and cell surface molecules that comprise the family of integrin ligands are many and varied. Individual integrin ligands have been shown to bind to several integrins, and reciprocally, individual integrins bind to multiple ligands (10). Ligand binding to integrins requires the formation of a ligand carboxyl-divalent cation coordination complex at the metal ion-dependent adhesion site (MIDAS) in the integrin A domain (8, 11). In non-αA domain-containing integrins, residues from both the β subunit A domain and the α subunit β-propeller domain contribute to ligand binding, and ligand binding specificity has been shown to be determined by loop structures in these regions (12-14). Integrin-mediated adhesion has wide-ranging effects on cell survival, motility, differentiation and proliferation (3). Unexpectedly, therefore, the signals generated by integrins and the composition of different adhesion signalling structures initiated by integrin-ligand engagement is highly diverse (2). However, the key molecular events determining this diversity, and the mechanisms determining the variation in the signals transduced by different integrin heterodimers are largely unknown.
Distinctive cellular responses to integrin-ligand engagement have been reported on substrates recognised by the fibronectin (Fn)-binding integrin α4β1. Engagement of this integrin results in an enhanced cell migratory phenotype coupled to a reduction in cell spreading and focal adhesion formation, compared to α2β1 or α5β1. These functional properties were demonstrated to be conferred by the α4 cytoplasmic domain (15-17). These studies suggest that the α4 cytoplasmic domain modulates the association of cytoskeletal and signalling molecules with its partner β1 cytoplasmic domain differently to that of α2 or α5. Alternatively they suggest that the α4 tail binds directly to cytoplasmic factors that modulate integrin signalling. In this regard, α4 has been shown to bind directly to paxillin, with the association reducing cell spreading and promoting cell migration (18). Thus, at present, the α subunit cytoplasmic tails are believed to have a major effect on the generation of integrin signal diversity from different αβ heterodimers. Recent evidence has indicated that the extracellular βA domain also plays a role in the generation of signal diversity. The βA domains of the β1 and β3 subunits were found to display different abilities to activate the Rho family of GTPases, which are involved in cytoskeletal organisation (19, 20). Signal diversity from integrin-ligand engagement can also result from additional co-operating signalling partners. In this regard, α4β1 and α5β1 demonstrate a differential requirement for PKCα signalling during cell migration and the formation of focal adhesion structures. These processes are mediated through the co-operative signalling of the proteoglycan co-receptor syndecan-4 with α5β1, but not with α4β1(21).
To carry out its dynamic functions, integrin structure is specialised to be highly responsive and regulated (22, 23). The receptors are able to switch from an inactive (low affinity) to active (high affinity) state, and vice versa, in response to binding events taking place at both the ligand-binding pocket and the cytoplasmic domains. A large body of evidence exists to indicate that both priming of integrins, to promote ligand binding, and integrin activation, subsequent to ligand binding, involve conformational changes between and within the integrin subunits (22). A distinctive feature of integrin activation is the transition from a highly bent conformation, which represents the inactive form, to an extended, primed conformation. The bend in the molecule is located between the thigh and calf domains of αV and the EGF-like 2/3 domains of β3 (5, 7).
The studies described above raise many questions relating to integrin structure and function. For example, how is the efficacy of signalling by different integrins determined? How is ligand binding converted into a signal? What is the signalling route taken through the integrin? Are the mechanisms of priming and activation different or the same? Some of these questions have already begun to be answered. For example, engagement of integrins with their ligands results in conformational changes within the integrin βA domains leading to the swing-out of the hybrid domain in relation to the ligand-binding head region, and the propagation of integrin signalling in this manner (5, 24, 25). Furthermore, the binding of cytoplasmic factors such as talin leads to integrin priming via a route that most likely involves the separation of the cytoplasmic and transmembrane domains (6, 26). The differences between these two processes has been suggested to be the basis of the discrimination between integrin priming and activation-induced signals (27).
We have attempted to address some of these issues by characterising the class of anti-integrin monoclonal antibodies (mAb) that stimulate integrin activation and ligand binding. Many function-modulating integrin mAbs act allosterically, displacing the conformational equilibrium of the active/inactive integrin to achieve their effects. Therefore anti-integrin mAbs are capable of acting as pseudo-agonists by stabilising integrin signalling conformations (22, 28). In this way we have identified integrin-ligand-mAb complexes that exhibit differential signalling properties, and determined a hitherto unappreciated mechanism of control of signal generation diversity that is dictated by the α subunit region of the integrin ligand-binding pocket.
Experimental Procedures
Antibodies, proteins and cell culture
Monoclonal antibodies used were: 12G10, mouse anti-human integrin β1 (29); 8E3, mouse anti-human integrin β1 (30); mAb13, rat anti-human integrin β1; mAb11 and mAb16, both rat anti-human integrin α5 (provided by K. Yamada, NIH, Bethesda, MD, USA); TS2/16, mouse anti-human integrin β1; GoH3, rat anti-human integrin α6 (provided by A. Sonnenberg, Netherlands Cancer Institute, Amsterdam, The Netherlands); K20, mouse anti-human integrin β1 (Beckman Coulter, High Wycombe, UK); 4B4, mouse anti-human integrin β1 (provided by C. Morimoto, Dana-Farber Cancer Institute, Boston, MA, USA); HUTS4, mouse anti-human integrin β1 (Chemicon, Harrow, UK); 9EG7, rat anti-mouse integrin β1 (provided by D. Westweber, University of Munster, Germany); 15/7, mouse anti-human integrin β1 (provided by T. Yednock, Elan, San Francisco, CA, USA); HP2/1 and 44H6, mouse anti-human integrin α4 (Serotec, Oxford, UK); 2B4, mouse anti-human integrin α4 (provided by J. Clements, British Biotech, Oxford, UK); P4C2, mouse anti-human integrin α4 (provided by E. Wayner, Fred Hutchinson Cancer Research Center, Seattle, WA, USA); 8F2, mouse anti-human integrin α4 (provided by C. Morimoto); and JBS5, mouse anti-human integrin α5 (Serotec). Biotinylation of mAbs was performed as previously described (31) using sulpho-LC-NHS biotin (Perbio, Chester, UK). Fab′ fragments of mAbs were prepared by ficin cleavage of purified IgG, followed by removal of Fc-containing fragments using protein A-Sepharose, according to the manufacturer’s instructions (Perbio).
Recombinant fragments of fibronectin (Fn) comprising type III repeats 12-15 plus IIICS segment (H/120), containing the recognition sequence for α4β1 and type III repeats 6-10 (FnIII(6-10)), containing the recognition sequence for α5β1, were purified and biotinylated using sulpho-LC-NHS biotin as previously described (30, 32). Purification of α4β1 from MOLT-4 cells, and α5β1 from human placenta, using mAb13, were as previously described (33, 34). Recombinant soluble human VCAM-1-Fc fusion protein, comprising Ig domains 1 and 2, was produced in COS-1 cells as previously described (35). Laminin and collagen were purchased from Sigma (Poole, Dorset, UK).
All cell lines were from the European Collection of Animal Cell Cultures (ECACC) unless otherwise stated, passaged every 3-4 days and cultured at 37°C and 5% CO2 in defined medium as follows. A375-SM human melanoma cells (provided by I. Fidler, University of Texas, USA) were cultured in Minimal Essential Medium with Earle’s salts, supplemented with 10% (v/v) fetal calf serum (FCS), minimal essential medium vitamins, non-essential amino-acids, sodium pyruvate and 1% (v/v) L-glutamine. COS-1 African green monkey kidney cells, IMR 32 human neuroblastoma cells and HT1080 human fibrosarcoma cells were cultured in Dulbecco’s minimal essential medium (DMEM) with 0.11g/l sodium pyruvate and pyridoxine supplemented with 10% (v/v) FCS, 1% (v/v) L-glutamine. Cells were detached from flasks by treatment with 1X trypsin/EDTA for 5 minutes for routine passaging. The MOLT-4 human lymphoblastic leukaemia suspension cell line, the K562 human chronic myelogenous leukaemia suspension cell line, K562-α3A cells (K562 cells stably transfected with the human α3A integrin subunit; provided by A. Sonnenberg, K562-α4 cytoplasmic deletion mutant cells (36) (provided by M. Hemler, Dana-Farber Cancer Institute, Boston, MA, USA) and the Jurkat human T cell lymphoblastic leukaemia suspension cell line (provided by P. Shore, University of Manchester, UK) were cultured in RPMI-1640 supplemented with 10% (w/v) FCS and 1% (v/v) L-glutamine. An addition of 1mg/ml G418 (Invitrogen, Paisley, UK) was made for cells transfected with the appropriate expression vector.
cDNA plasmids and construction of α4/α5 subunit chimeras
The α6A cDNA in pRC-CMV was a gift from A. Sonnenberg. Site-directed mutagenesis of α4 and α5 cDNAs was performed using the method of Kunkel (37), or using the GeneEditor site-directed mutagenesis kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. For GeneEditor, 5′ to 3′ oligonucleotides (MWG-Biotech UK Ltd, Milton Keynes, UK) containing the desired mutations were GTAGTAATTGTTGACGCTAGCTTAAGCCA CCCTGAGTCAG for α4 and ATCGTGTCCGCTAGTGCTAGCCTCACCATC TTCCCCGCC for α5. Briefly, to construct the α4/α5 chimera composed of the extracellular and transmembrane region of α4 linked to the cytoplasmic domain of the α5 subunit, a Hind III site was introduced into α4 at the equivalent position to an existing Hind III site in α5 as described (16). To construct the α4Pα5L chimera, comprising the N-terminal β-propeller domain of α4 (Y1-L440) with the α5 leg, transmembrane and cytoplasmic domains (T459-A1008), an alignment of the primary structure of α4 and α5 in the region of the β-propeller domain was performed (ClustalW, http:expasy.ch). The conserved ASL sequence (A438 in α4 and A456 in α5) was chosen to introduce an unique Nhe I restriction enzyme site into the equivalent position in the α4 and α5 cDNAs. After restriction enzyme digestion the required fragments were ligated into the mammalian expression vector pCDNA3 (Invitrogen) via Xho I / Sal I and Xba I sites to enable cell surface expression. All mutagenesis and chimera constructions were verified by DNA sequencing.
Transfection of mammalian cells
K562 cells were transfected with plasmid DNA either by electroporation, or by using GeneJammer transfection reagent (Stratagene, La Jolla, CA, USA). Electroporation was performed with subconfluent cells resuspended to 6-7×106 cells/ml (K562 cells) or 7-10×106 cells/ml (COS-1 cells). 750μl cells were aliquoted into 0.4cm electroporation cuvettes (Biorad, Hemel Hempstead, UK) and 10-20μg DNA in 50μl water added on ice for 20 minutes prior to electroporation at 230V and 950μF (K562 cells) or 250V and 950μF (COS-1 cells) with a resultant time constant of 18-22mS using a Biorad gene pulser II. Electroporated K562 cells were transferred to prewarmed medium and grown for 48hrs before addition of the selection antibiotic G418 at 1mg/ml. Alternatively, 6μl GeneJammer transfection reagent (Stratagene) was diluted in serum-free medium for 5-10 minutes and 1μg DNA added and incubated for 5-10 minutes. Sub-confluent cells were resuspended at 1-2×106 cells/ml and 1ml seeded in 35mm dishes. The DNA/GeneJammer mixture was added to the cells and incubated for 72hrs before passaging into medium containing the selection antibiotic G418 at 1mg/ml.
To enrich the population of transfected K562 cells, cells were subjected to immunomagnetic bead selection. After 2-3 weeks of growth in medium containing selection antibiotics, cells were resuspended to 1×107 cells/ml in ice-cold RPMI 1640 supplemented with 1% (v/v) FCS (wash medium). Antibodies directed against epitopes of interest were added at 5ug/ml and incubated on ice for 30 minutes. Cells were washed with cold wash medium before addition of 2×107 goat anti-mouse IgG coated magnetic beads (Dynabeads M-450, Dynal, Bromborough, UK) and incubated on ice for 20 minutes. Bead-cell complexes were isolated using a magnetic separation device (Dynal), and unbound cells removed by washing in ice-cold wash medium. Bead-cell complexes were resuspended in growth medium plus selection antibiotic, and expression assessed by fluorescence-activated cell scanning (FACS) analysis (FACscan, Becton Dickinson, Cowley, UK). The bead-sorting procedure was repeated 2-3 times to obtain cells expressing high levels of the protein of interest. Cells were then cloned by limiting dilution. Identical results were obtained with mixed cell populations and multiple clones. Expression and folding of the α4 subunit was verified by FACS using a panel of anti-α4 and anti-α5 mAbs, or GoH3 for K562-α6 cells.
Cell attachment assays
96-well plates (Costar Corning) were coated for 60-90 minutes at room temperature with protein ligands diluted in phosphate-buffered saline (PBS, Invitrogen). After aspiration, wells were incubated for 30-60 minutes with 10mg/ml of filtered, heat-denatured bovine serum albumin (BSA). As controls, some wells were incubated with BSA only. Wells were washed with HEPES-buffered saline, HBS (150mM NaCl, 25mM HEPES pH7.4), before addition of 50μl HBS containing 2X final concentration of antibodies, cations and/or inhibitors where appropriate. Subconfluent cells were washed with HBS, resuspended to 0.2 to 1×106 cells/ml, and 50μl aliquots added to wells. Plates were incubated for 30 minutes at 37°C and 5% (v/v) CO2. Unbound or loosely bound cells were removed by aspiration and gentle washing with HBS. Wells were fixed by addition of 5% (v/v) glutaraldehyde in HBS. To assess the total number of cells added, 100%, 75%, 50%, 25% and 0% cells were added to wells and fixed by the addition of 1/10 volume of 50% (v/v) glutaraldehyde. Wells were aspirated and washed with HBS before addition of 0.1% (w/v) crystal violet in 200mM methylethanesulphonic acid (MES) pH6 for 60 minutes. Wells were then aspirated and washed with water before addition of 10% (v/v) acetic acid and the absorbance at 570nm of each well was measured with a multiscan plate reader.
Soluble ligand binding assays
Soluble VCAM-1-Fc protein was labelled with the fluorescent dye Oregon Green using a Fluoro-reporter labelling kit according to the manufacturer’s instructions (Molecular Probes, Eugene, OR, USA). K562-α4, K562-X4C0, K562-α4Pα5L or A375-SM cells were washed and resuspended in HBS containing 1% BSA and 1mM MnCl2 (HBS/Mn2+). 1×106 /ml cells were incubated with Oregon Green-VCAM-1-Fc (50μg/ml) with or without anti-integrin mAbs (20μg/ml) or EDTA (5mM) for 45 minutes at 37°C. Cells were washed in HBS/Mn2+ and fixed in 0.2% (v/v) formaldehyde in PBS. Binding of Oregon Green-VCAM-1-Fc was detected by FACS.
Cell spreading assays
Wells were coated and blocked as described for cell attachment assays. Adherent cells were detached with 0.05% (w/v) trypsin, 0.02% (w/v) EDTA and resuspended to 2×105/ml in DMEM/25mM HEPES and allowed to recover for 10 min at 37°C. For experiments examining the effects of mAbs on spreading, 50μl aliquots of the cell suspension were added to wells together with 50μl of mAbs diluted to 2x the final concentration in DMEM. Plates were incubated at 37°C and 5% (v/v) CO2 for various times ranging from 30 minutes to 2hrs. The cells were fixed with 5% (w/v) glutaraldehyde for 30 minutes and wells washed with PBS. The degree of cell spreading was assessed as a percentage of the total number of cells counted, using phase-contrast microscopy. At least 100 cells in four to six randomly chosen high-powered fields were counted per treatment point. Axiovision v4.2 software (Carl Zeiss, Hertfordshire, UK) was used to measure cell areas, and two-tailed t-tests were performed to determine statistical significance.
Immunofluoresence
To assess the effect of mAbs on pre-spread A375-SM cells, immunofluorescence was performed essentially as previously described (21). Briefly, cells were incubated with 13-mm diameter coverslips that were precoated with 10μg/ml H/120 or FnIII(6-10) and blocked with heat-denatured BSA. Cells were allowed to spread for 60 mins before the addition of mAbs (10μg/ml) for a further 30 mins. For cells spread on FnIII(6-10), the heparin-binding fragment of Fn (H/0) was added throughout to allow full development of focal adhesion structures (21). Cells were then fixed and processed to visualise vinculin (mouse anti-human hVIN-1 mAb; 1:400 dilution; Sigma) and actin (rhodamine-conjugated phalloidin; 1:1000 dilution; Sigma).
Solid phase receptor-ligand binding assay and ELISA
Assays were performed as described previously (31, 34). Briefly, wells of high binding microtitre plates (Corning Costar) were coated with 50μl aliquots of affinity-purified α4β1 or α5β1 integrin (approximately 1μg/ml in PBS) overnight. Wells were blocked for 3 hours with 200μl blocking buffer; 5% (w/v) BSA, 150mM NaCl, 0.05% (w/v) NaN3, 25mM Tris-HCl, pH 7.4. Wells were washed three times with TBS plus 1mM MnCl2 and 0.1% (w/v) BSA (TBS/Mn). To study the effects of other divalent cations, this buffer was treated with Chelex beads (Bio-Rad) prior to the addition of any cations, and then MgCl2 or CaCl2 or MnCl2 added. For solid phase assays, anti-integrin antibodies at 20μg/ml were added to wells in 50μl TBS/Mn and a range of VCAM-1-Fc, biotinylated H/120 or biotinylated FnIII(6-10) concentrations in 50μl TBS/Mn were also added to wells as indicated. For ELISA, biotinylated mAbs were added at 0.3-10μg/ml. Plates were incubated at 37°C for 2-3 hours before washing wells three times with TBS/Mn. VCAM-1-Fc binding was detected using horseradish peroxidase (HRP)-conjugated anti-human-Fc antibody (Sigma), and biotinylated Fn fragments or mAbs were detected with HRP-conjugated ExtrAvidin (Sigma). Wells were washed with TBS/Mn before addition of ABTS substrate and absorbance readings at 405nm measured using a multiscan plate reader. Each experimental condition was performed using at least three replicate wells and background attachment to BSA-blocked wells subtracted from all measurements.
Results And Discussion
12G10 selectively disrupts α4β1-mediated cell attachment, cell spreading and cytoskeletal organisation
The initial aim of these studies was to identify conditions under which individual integrins exhibited variable signalling. To this end, the effects of monoclonal antibodies, previously demonstrated to stimulate ligand binding, on the signalling properties of various integrin-ligand combinations were examined. Two integrin-ligand combinations (α4β1 binding to H/120, the IIICS region of fibronectin, or to VCAM-1, and α5β1 binding to the central cell-binding domain of fibronectin, FnIII6-10) and two activating anti-β1 mAbs (12G10 and TS2/16) were used. α4β1 and α5β1 were selected because they have been reported previously to transduce different effects on cell spreading and migration via different signals (15, 16). Specifically, α4β1 engagement promotes enhanced cell migratory activity while reducing spreading, in part due to the ability of α4 to bind paxillin (18). In addition, both receptors have a differential requirement for PKCα activation when stimulating focal adhesion formation and migration (21). 12G10 and TS2/16 were compared because they represent the two main classes of anti-integrin mAbs that enhance ligand binding and cell adhesion (4, 29, 38). These classes are distinguished by the ability of divalent cations and ligand occupancy to modulate expression of their epitopes. While both 12G10 and TS2/16 have been reported to stimulate ligand binding to purified integrins, 12G10 binding to the integrin is stimulated by ligand, Mn2+ and Mg2+, and inhibited by Ca2+, but the expression of the TS2/16 epitope is unaltered by cation and ligand occupancy (31). These findings demonstrate that both 12G10 and TS2/16 are allosteric activators of integrins, but they imply that the mAbs function in different ways, possibly by stabilising different conformations of the integrins (4, 22).
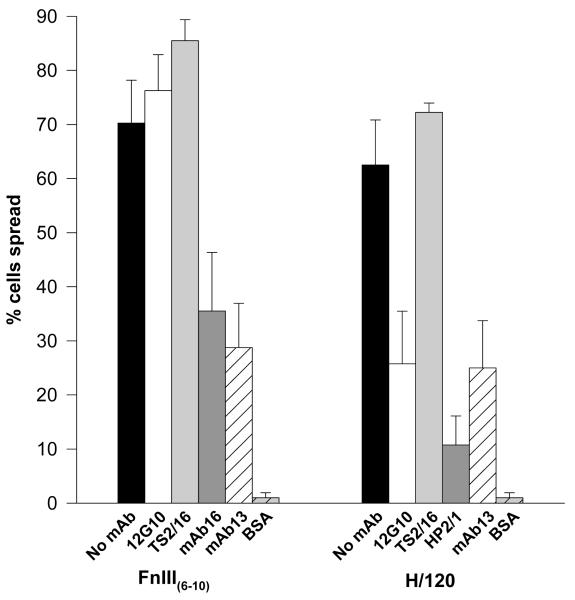
To probe potential signalling differences in cells between the activation states of α4β1 and α5β1, the effect of 12G10 and TS2/16 on α4β1- and α5β1-mediated cell spreading was studied. The ability of cells to spread on ligands has been extensively used to assess post-ligand binding events subsequent to integrin-mediated adhesion. The A375-SM cell line expresses both α4β1 and α5β1,and can be directed, via the use of specific integrin ligands, to utilise either of these receptors, facilitating cell spreading with the formation of distinct focal adhesions and actin stress fibres (21). As reported previously (32), 60-70% of A375-SM cells spread via α4β1 or α5β1, forming phase-dark, polyhedral morphologies. Surprisingly, addition of 12G10 inhibited cell spreading mediated by α4β1, but not by α5β1, resulting in phase-bright cells with a round morphology. This effect was not observed with TS2/16 (Fig. 1). A small number of cells were phase-dark on α4β1 ligands in the presence of 12G10, but the surface area occupied by these cells was much less than control cells. Similar results were observed for 12G10 and TS2/16 treatment of HT1080 and MOLT-4 cells spreading via α4β1. In other experiments, α2β1- and α6β1-mediated HT1080 cell spreading was not inhibited by 12G10 or TS2/16 (data not shown). Thus, 12G10 modulates α4β1 function differently to other integrins such as α2β1,α5β1 and α6β1 despite recognising the common β1 subunit. Subsequent experiments were designed to test the working hypothesis that the effects of 12G10 on cells occur through the induction of different mAb-induced agonistic signalling states of β1 integrins.
Figure 1. Effect of 12G10 and TS2/16 on α4β1- and α5β1-mediated A375-SM cell spreading.
A375-SM cells were allowed to spread on 10μg/ml FnIII(6-10) or H/120 for 60 minutes alone (black bars), or in the presence of 12G10 (activating anti-β1 mAb; white bars), TS2-16 (activating anti-β1 mAb; light grey bars), mAb16 or HP2/1 (anti-functional anti-α5 and α4 mAbs respectively; dark grey bars), mAb13 (anti-functional anti-β1 mAb; white hatched bars) or to BSA alone (light grey hatched bars).
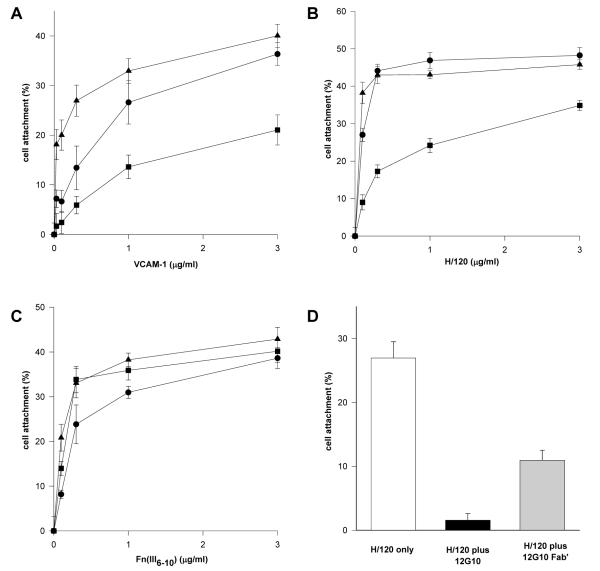
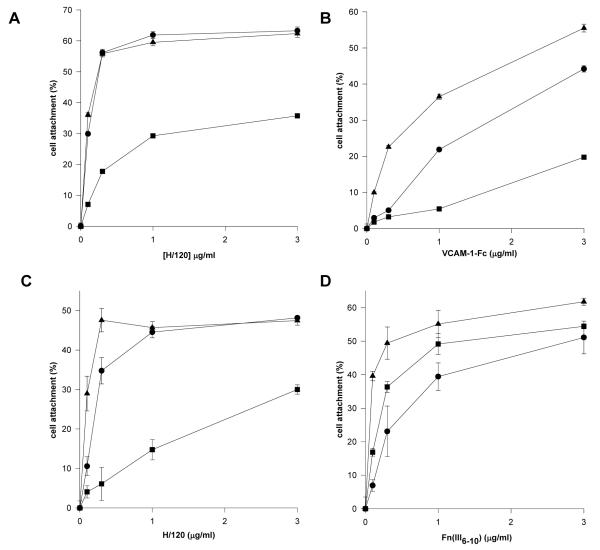
To probe potential signalling differences in cells between the activation states of α4β1 and α5β1 further, the effect of 12G10 and TS2/16 on α4β1- and α5β1-mediated cell attachment was studied. Cell attachment of α4-transfected K562 cells (K562-α4) to the α4β1 ligands VCAM-1 and H/120 was partially inhibited by addition of 12G10, whereas TS2/16 augmented cell attachment as expected, particularly at low ligand concentrations (Fig. 2A/B). By contrast, both 12G10 and TS2/16 augmented α5β1-mediated cell attachment of the same cells to FnIII(6-10) (Fig. 2C). In support of these data, five other cell lines expressing endogenous α4β1, Jurkat, MOLT-4, A375-SM, HT1080 and IMR-32, each of which displayed different basal adhesive states and expression levels of α4β1, also demonstrated reduced α4β1-mediated cell attachment in the presence of 12G10 (data not shown). These findings demonstrate that the 12G10-induced inhibition of α4β1-mediated cell attachment is not cell type-specific. Further confirmation of the specific effects of 12G10 on α4β1-mediated cell attachment was obtained in assays with the above cell lines utilising collagen and laminin as ligands. 12G10 had no inhibitory effect on α1β1-, α2β1-, α3β1- or α6β1-mediated adhesion (data not shown). In further studies, 12G10 Fab′ fragments inhibited α4β1-mediated cell attachment of K562-α4 cells (Fig. 2D). These observations are consistent with the effects of the mAbs on cell spreading. Together they suggest that 12G10 and TS2/16 stabilise different agonistic signalling conformations of α4β1, the 12G10-induced conformer of α4β1 resulting in reduced cell attachment and spreading. This was not the case for other β1 integrins such as α5β1. As 12G10 Fab′ fragments also inhibit cell adhesion it appears that the mechanism is unlikely to be due to mAb-induced steric hindrance of the ligand binding site. Furthermore, TS2/16, and other activating anti-β1 mAbs such as 15/7 and HUTS-4 (data not shown), stabilise different conformers of α4β1 that do not induce this effect. We hypothesise that the 12G10 signalling conformer of α4β1 leads to the disruption of the cytoskeleton, resulting in a less adhesive phenotype.
Figure 2. Effect of 12G10 and TS2/16 on K562-α4 cell attachment.
K562-α4 cells were allowed to attach to (A) VCAM-1-Fc, (B) H/120, or (C) FnIII(6-10) only (circles), or in the presence of 12G10 (activating anti-β1 mAb; squares), or TS2/16 (activating anti-β1 mAb; triangles). (D) K562-α4 cells were allowed to attach to 0.1 μg/ml H/120 only (white bar) or in the presence of 12G10 (black bar) or 12G10 Fab’ fragments (grey bar).
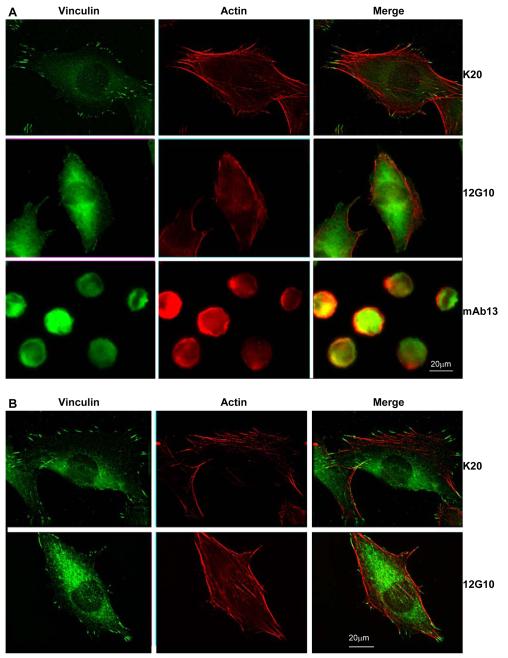
To test the hypothesis that the 12G10-stabilised conformers of α4β1 and α5β1 result in differential signalling we focused on the effects of 12G10 on the preformed cytoskeleton. Untreated cells, or cells treated with a neutral anti-β1 mAb, K20, formed prominent vinculin-containing focal adhesions and actin stress fibres when allowed to spread via α4β1 or α5β1. However, cells spread via α4β1 and then 12G10-treated, displayed a greatly reduced actin stress fibre network with a disrupted focal adhesion staining pattern (Fig. 3A). This effect preceded, and was independent of, the ability of 12G10 to reduce cell spreading, as the phenotype occurred in cells that had spread to the same extent as control cells, and was distinctly different to the effects of mAb13 which perturbs integrin-ligand binding (Fig. 3A). In addition, quantification of A375-SM cell spreading demonstrated no significant difference in the mean cell area of cells spread for 60 mins and then challenged with either 12G10 or K20 (see Fig. 3 legend). The effects of 12G10 on the cytoskeleton were not observed in cells spread via α5β1 (Fig. 3B). These data therefore support a model of differential signalling between the 12G10-stabilised conformers of α4β1 and α5β1, with the former promoting disengagement from, and retraction of, the actin cytoskeleton, leading to reduced cell attachment and spreading.
Figure 3. Effect of 12G10 on focal adhesion and actin stress fibre formation in A375-SM cells.
A375-SM cells were spread on (A) 10μg/ml H/120 or (B) FnIII(6-10) for 60 minutes before addition of the mAbs K20, 12G10 or mAb13 for 30 minutes as indicated. Cells were then fixed and processed to visualise vinculin (green) and actin (red). The mean cell area ± s.d. (arbitrary units) was 1549.5 ± 822.0 for H/120 only (n=73), 1491.3 ± 713.2 for H/120 plus 12G10 (n= 83), 1485.8 ± 758.8 for H/120 plus K20 (n=87), 1164.7 ± 590.3 for H/120 plus mAb13 (n=101), 1151.1 ± 721.5 for FnIII(6-10) only (n=87), 1128.7 ± 643.8 for FnIII(6-10) plus 12G10 (n=84), 1129.1 ± 735.8 for FnIII(6-10) plus K20 (n=99), and 687.4 ± 487.3 for FnIII(6-10) plus mAb13 (n=90). Statistical differences were not detected (p>0.1; two tailed t-test) between the means of any of the groups (for comparisons made of cells spread on the same ligand) except for H/120 only vs. H/120 plus mAb13 (p<0.01) and FnIII(6-10) only vs. FnIII(6-10) plus mAb13 (p<0.01).
Divalent cation regulation of 12G10 binding and the effects of 12G10 on ligand binding are similar for both α4β1 and α5β1
12G10 was initially characterised as an anti-β1 mAb that recognised a ligand-induced binding site (LIBS) epitope and augmented α5β1-Fn interactions (29). Subsequently, the epitope of 12G10 was found to overlap with those of other function-altering anti-β1 mAbs such as mAb13 and TS2/16. Thus, the twelve amino-acid epitope sequence (K207-K218) for these mAbs is located in the α2 helix of the β1 subunit A domain, and it was suggested that this region was involved in conformational changes that regulate integrin-ligand binding (31, 39). An interesting feature of many LIBS mAbs is the divalent cation regulation of their epitopes. Since cations also regulate ligand binding, and in some cases the pattern of effects of different cations is the same for mAb and ligand binding, it appears that some activating mAbs recognise sites that are regulated by modulators of integrin function. One possibility is that cation-responsive, activating mAbs recognise naturally occurring conformers of integrins and that they are therefore able to displace a conformational equilibrium in favour of these forms. This displacement would lead to an increase in the proportion of ligand-competent integrin in the population. Other activating mAb epitopes are unaffected by either ligand or cation binding and here the most likely mechanism of action is through inducing an activated conformation in the integrin rather than stabilising a naturally occurring conformation. The effects of 12G10 on ligand binding to α5β1 are well-characterised (29, 31). In agreement with the model described above, the binding of 12G10 to α5β1 was regulated in a cation-dependent manner that correlated with the activation state of the receptor (31). The cation-regulated component of the 12G10 epitope was subsequently mapped to residues R154 and R155 in the βA domain α1 helix, indicating that this helix is also conformationally regulated by ligand binding (24). Therefore 12G10 is believed to report the activation state of the β1 A domain by detecting the position of the α1 helix (25). Reciprocally, 12G10 is thought to augment ligand binding by stabilising the α1 and α2 helices of the β1A domain in a ligand-competent, physiologically relevant conformation (4). The binding of TS2/16 to α5β1 is not modulated by cations (31) and this mAb is therefore thought to activate the integrin by inducing a non-physiological conformer.
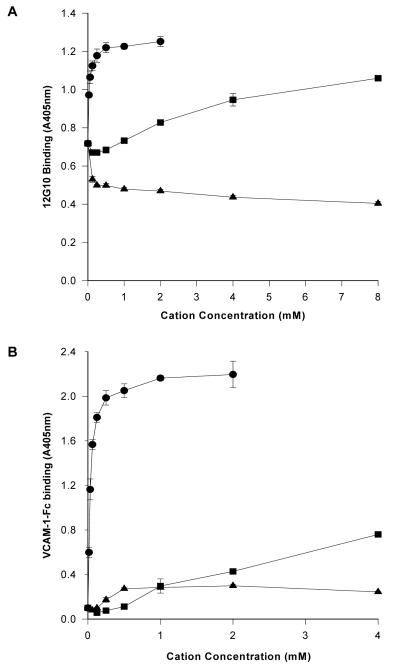
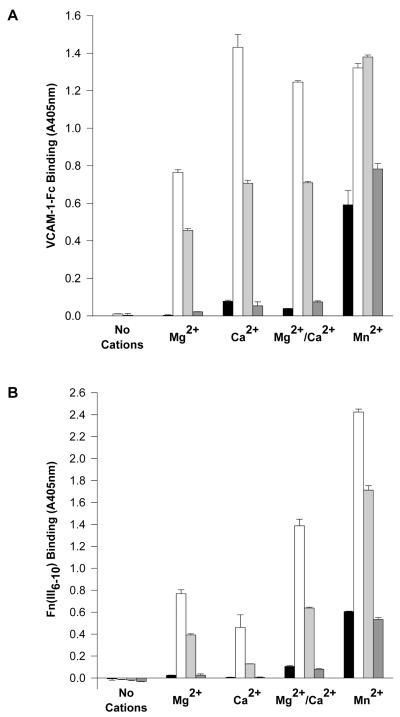
When the divalent cation-dependent binding of 12G10 to α4β1 was examined and compared to the effects of cations on ligand binding, the observed pattern mirrored that seen for α5β1 (31), i.e. Mn2+ supported high levels, Mg2+ moderate levels and Ca2+ did not support binding (Fig. 4A/B). As observed for α5β1, divalent cations did not modulate the expression of the TS2/16 epitope (data not shown). These data suggest that 12G10 binding to α4β1 correlates with the activation state of the receptor, and that the mAb recognises similar conformational changes in α4β1 and α5β1.
Figure 4. Expression of the 12G10 epitope correlates with the activation state of α4β1.
Divalent cation regulation of (A) 12G10 (0.3μg/ml) binding to purified α4β1 or (B) VCAM-1-Fc (0.5μg/ml) binding to purified α4β1 in the presence of Mn2+ (circles), Mg2+ (squares) or Ca2+ (triangles).
To test if differences exist between the ligand-binding competencies of the 12G10-induced conformers of α4β1 and α5β1 that could provide alternative explanations for the above data, the effects of 12G10 on ligand binding to affinity-purified α4β1 were investigated. In solid phase assays, ligand binding to both purified α4β1 and α5β1 receptors was augmented by 12G10 (Fig 5A/B). Differences between α4β1 and α5β1, however, were observed with respect to the ability of cations to support 12G10-induced ligand binding. Most strikingly, Ca2+ supported consistently higher levels of 12G10-induced ligand binding to α4β1 compared with Mg2+, an effect not observed with α5β1. This most likely reflects subtle differences between the interaction of the 12G10-stabilised ligand binding pocket of α4β1 or α5β1 with their ligands. Other activating anti-β1 mAbs such as 15/7, 9EG7 and TS2/16 were also shown to increase α4β1-ligand binding in these assays (Fig 5A/B and data not shown). This is in accordance with previous data indicating that modulation of the activation of the β1A domain occurs in a similar manner for both α4β1 and α5β1 (40). Therefore both the 12G10-induced conformers of α4β1 and α5β1 demonstrate augmented levels of ligand binding despite displaying differential effects on cell attachment and spreading mediated by these integrins, indicating that the mechanism of action of 12G10 on cell adhesion does not involve a direct occlusion of the ligand-binding pocket of α4β1.
Figure 5. 12G10 augments β1-dependent ligand binding.
Divalent cation regulation of (A) VCAM-1-Fc binding to α4β1 or (B) FnIII(6-10) binding to α5β1. Assays were performed in the absence (black bars) or presence of 12G10 (activating anti-β1 mAb; white bars), TS2-16 (activating anti-β1 mAb; light grey bars) or K20 (non-functional anti-β1 mAb; dark grey bars). Divalent cations conditions are 1mM each as indicated. Identical results were obtained for H/120 binding to α4β1 as seen in (A) for VCAM-1-Fc.
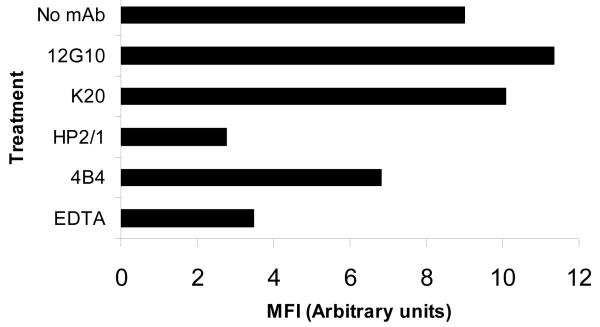
Solid phase assays allow analysis of receptor-ligand interactions in isolation, rather than in the context of cell membrane-bound proteins. To further investigate the effects of 12G10 on ligand binding, soluble ligand binding assays were employed with whole cells. Binding of soluble, fluorescently-labelled VCAM-1 to K562-α4 or A375-SM cells was inhibited by EDTA and anti-functional α4 and β1 mAbs, demonstrating the specificity of binding. In contrast to the effects of 12G10 on cell attachment, however, the mAb did not inhibit soluble VCAM-1-Fc binding to either K562-α4 or A375-SM cells (Fig. 6 and data not shown). Taken together with the solid phase assay data described above, these findings provide further evidence that the mode of action of 12G10 to selectively inhibit α4β1-mediated cell spreading and attachment is not due to steric blocking of the ligand binding interface.
Figure 6. Effect of 12G10 on soluble ligand binding to K562-α4 cells.
Binding of Oregon green-labelled VCAM-1-Fc to K562-α4 cells in suspension was detected by FACS. Cells were incubated in the absence of antibody (No mAb) or in the presence of 12G10 (activating anti-β1 mAb), K20 (non-functional anti-β1 mAb), HP2/1 (anti-functional anti-α4 mAb), 4B4 (anti-functional anti-β1 mAb) or EDTA. Mean Fluorescence Intensity (MFI) is shown for each condition (10000 events).
The ligand-binding pocket determines agonistic differences between α4β1 and α5β1
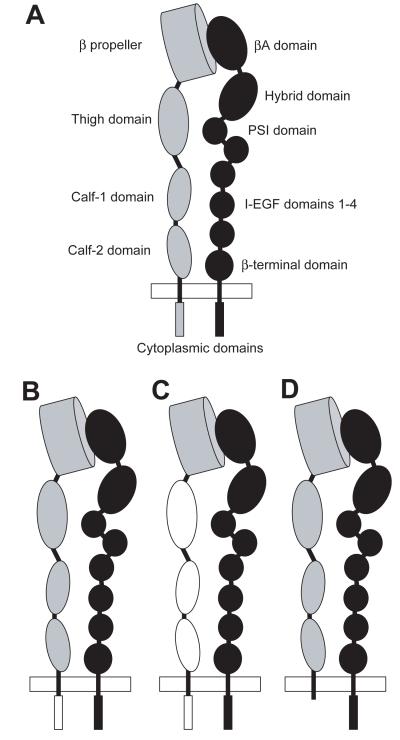
Having identified integrin-ligand-antibody complexes with different signalling properties, we next aimed to identify the regions of the integrins that were responsible for these differences. In previous studies, the cytoplasmic tails of integrins have been shown to be responsible for some functional differences between receptors. Particularly pertinent to this study is the fact that the α4 tail has the property of reducing cell spreading and increasing migration due to its interaction with the cytoplasmic signalling adaptor paxillin. To test if the specific effects of 12G10 on α4β1-mediated adhesion required the α4 tail, an integrin chimera comprising the α4 extracellular and transmembrane domains and the α5 cytoplasmic tail was generated (Fig. 7). This chimeric integrin was expressed on the surface of K562 cells and tested in cell adhesion assays. The chimera-expressing cells demonstrated α4β1-mediated cell attachment to H/120 and VCAM-1, and 12G10 inhibited this attachment in an identical manner to that seen for full-length α4 (data not shown). Furthermore, K562 X4C0 cells expressing α4 with the tail truncated after the conserved GFFKR cytoplasmic motif (36; Fig. 7), also displayed a 12G10 inhibition profile on α4β1 ligands that was identical to K562-α4 cells (Fig. 8A/B). These data demonstrate that the effects of 12G10 on α4β1 function are independent of the α tail and cannot therefore be due to modulation of the α4-paxillin interaction.
Figure 7. Schematic diagram of the domain structure of an integrin and α4/α5 chimeras.
(A) The domain structure of an integrin α (grey) / β (black) heterodimer is shown as indicated by refs 7, 8 and 9. The plasma membrane is depicted as a white rectangle. The βA domain contains the epitopes for a number of function modulating mAbs including 12G10 and TS2/16. In (B), (C) and (D) schematic representations of the α4/α5 chimeras expressed in K562 cells are shown in which the α4 subunit domains are grey and the α5 subunit domains are white. (B) Schematic of the chimera in which the α4 tail has been swapped for the α5 tail, (C) schematic of the α4Pα5L chimera which comprises the propeller domain of α4 (Y1-L440) and the α5 leg, transmembrane and cytoplasmic domains (T459-A1008), and (D) schematic of the X4C0 chimera (36) in which the α4 tail was truncated after the GFFKR motif.
Figure 8. Effect of 12G10 and TS2/16 on K562 X4C0 and K562-α4P/α5L chimera cell attachment.
K562-X4C0 cells were allowed to the α4β1 ligands (A) H/120 or (B) VCAM-1-Fc. Assays were performed in the absence (circles) or presence of 12G10 (activating anti-β1 mAb; squares), or TS2-16 (activating anti-β1 mAb; triangles). Identical experiments were performed with K562-α4Pα5L cells on (C) H/120 or the α5β1 ligand (D) FnIII(6-10).
To define further the region of the α4 subunit responsible for the effects of 12G10 on α4β1 function, a second α4/α5 chimera, comprising the N-terminal β-propeller domain of α4 (Y1-L440) with the α5 leg, transmembrane and cytoplasmic domains (T459-A1008), was generated (Fig. 7) and again expressed in K562 cells (termed K562-α4Pα5L). Cells expressing this chimera attached to α4β1 ligands in a manner that was identical to that observed with wild type α4β1 (data not shown). This finding confirms previous observations for integrins such as α5β1 and αVβ3, where residues in the β-propeller domain have been shown to contribute to the ligand binding pocket and determine ligand-binding specificity. The K562-α4Pα5L-expressing cells also demonstrated reduced cell attachment to α4β1 ligands in the presence of 12G10 but not TS2/16 (Fig. 8C and data not shown), as seen for wild type α4β1. Again this contrasted with the effect of 12G10 on these cells to attach to the α5β1 ligand FnIII(6-10) (Fig. 8D). Therefore the α4-propeller domain of the α4 subunit is sufficient to confer the ability of 12G10 to inhibit cell attachment on the α5 subunit. Furthermore, the effects of 12G10 and TS2/16 on soluble VCAM-1-Fc binding to K562-X4C0 and K562-α4Pα5L cells (performed as in Fig. 6) were identical to that observed for K562-α4 cells, i.e. 12G10 and TS2/16 did not inhibit soluble ligand binding (data not shown). These results suggest that the βA domain conformation, at least in the region of the conformationally important 12G10 epitope, can be modulated by the β-propeller domain of the associating α subunit. These data demonstrate that the different 12G10-induced agonistic states of α4β1 and α5β1 are determined by the extracellular ligand binding pocket of α4β1, i.e. the N-terminal β-propeller domain of the α4-subunit in conjunction with the β1A-domain.
A series of previous studies have suggested that the cytoplasmic tails of integrins, or associating signalling molecules, are responsible for the differential signalling observed between integrins sharing a common β-subunit. There is some limited evidence, however, that the integrin extracellular domain makes a contribution (19, 20). Specifically it was found that extracellular domains, in particular the βA-domain, differentially regulated Rho-family GTPase signalling from β1 and β3 integrins. The results described here support the concept of extracellular domain control of integrin signalling, but fundamentally differ to that of published data in that they describe a process by which the extracellular domain of the α-subunit influences the function of the associating β-subunit, thereby suggesting another possible mechanism by which integrin heterodimers can initiate specific signals and generate signal diversity from a common β subunit.
Our findings raise the issue of how different conformations of the ligand-binding pocket of integrins are transduced through the molecule to the cytoplasm. Initially, shape shifting in the βA-domain needs to be propagated to the underlying hybrid/PSI domains. A recent crystal structure analysis of αIIbβ3 complexed with different peptidomimetic ligands (9) suggests how this might take place. One possibility is that different α subunits might influence the degree of hybrid domain swing-out induced by ligand binding. Alternatively, the kinetics of leg separation might be different for different α-β combinations. The net result of either mechanism would be to alter the proportion of integrin molecules in an activated conformation. The question remains as to how the link between α4β1 and the cytoskeleton is severed. Is this mediated by modulation of a direct β1-cytoskeletal link or via indirect signals to the cytoskeleton from the β subunit? Treatment of cells with 12G10 perturbed the formation of stress fibres and resulted in the disassembly of existing stress fibres and FAs. As these effects are reminiscent of the effects of the Rho-family of GTPases on cell morphology (41), we speculated that 12G10 might modulate the activity of this family of proteins. However, constitutively active or dominant negative Rho-family GTPases did not accentuate or rescue the effects of 12G10 observed in these assays, and the levels of GTP-bound Rho GTPases was also not affected by 12G10 (data not shown). It therefore seems unlikely that the mechanism of 12G10 action is a direct one on Rho GTPase signalling.
Recently cAMP-dependent protein kinase (PKA) was found to be required for the 12G10-induced cell-cell and cell-substrate adhesion of HT1080 cells (42). This study implied a role for PKA in β1 integrin activation dependent signalling. We found that inhibition of PKA with MPKI peptide did not modulate the effects of 12G10 on α4β1- or α5β1-mediated cell attachment (data not shown). We can therefore discount PKA as a downstream target of the 12G10 signalling effects we observe. Inhibition of other signalling molecules such as MEK, PI-3K and tyrosine kinase also had little or no effect on the 12G10 modulation of α4β1. Tyrosine phosphorylation (Y-P) plays a central role in integrin signalling and therefore its role in this phenomenon was assessed. 12G10 treatment of A375-SM cell spreading was found to reduce the overall Y-P pattern for α4β1 but not α5β1 spread cells (data not shown). This Y-P reduction, however, paralleled the reduction in A375-SM cell spreading, and was therefore likely to be downstream of the effects on cell spreading. This close correlation of Y-P with the 12G10 effect suggests that 12G10 is modulating a direct link to cytoskeleton.
An alternative signalling mechanism for α4β1 could be via a signalling co-receptor, analogous to the way that syndecan-4 co-operates with α5β1 to activate PKCα (21, 43). Previously, mutations in the extracellular β-propeller domain of the α4 subunit displayed defects in α4β1-dependent static cell adhesion and adhesion under shear flow, but not to soluble ligand binding, similar to the data reported here (44). These β-propeller mutations were found to be defective in their interactions with CD81, a transmembrane-4 superfamily member protein that associates laterally with α4β1 (45), and modulates co-operative signalling events with α4β1 (46, 47). The possibility that modulation of α4β1-CD81 association by 12G10 could be responsible for the effects of the mAb on α4β1, was discounted however as the α4Pα5L integrin chimera did not associate with CD81, unlike wild type α4β1, as determined by co-immunoprecipitation experiments (data not shown).
In summary, the mAb 12G10 has been shown to stimulate ligand binding to both α4β1 and α5β1, reporting and stabilising conformational changes within these integrins that correlate with their activation states. The 12G10-induced conformers, which likely reflect physiologically relevant agonistic states, differentially modulate α4β1 and α5β1 adhesive functions apparently via a severing of the connection with the cytoskeleton. Other activating mAbs, such as TS2/16, the epitope of which overlaps with that of 12G10, do not selectively modulate α4β1 and α5β1 function, illustrating the specific and novel nature of the 12G10-induced effects. Furthermore, these data indicate that the extracellular β-propeller domain of the α-subunit dictates the agonistic differences between α4β1 and α5β1. The 12G10-induced inhibition of cell adhesion is controlled independently of the α4 thigh, stalk and tail domains suggesting that the conformation of the β1-subunit can be regulated by the associating α-subunit. As the effects of 12G10 on α4β1 are independent of the non-ligand-binding domains of the α4-subunit, these data indicate that signals can be propagated through the β-subunit to adjust cytoskeletal attachments independently from the α-subunit. Thus, these findings demonstrate a direct and variable agonistic link between the ligand-binding pocket of integrins and the cell interior that is independent of the α cytoplasmic domains, and suggest that regulated signalling via changes in integrin extracellular domain conformation is more important than previously suspected.
Acknowledgements
We thank Martin Hemler (Dana Farber Cancer Inst., Boston, MA. USA) for provision of K562-X4C0 cells used in this study. These studies were supported by Wellcome Trust grant 045225 (to MJH).
Footnotes
The abbreviations used are: mAb, monoclonal antibody; VCAM-1, vascular cell adhesion molecule;; vWF1, von Willebrand factor; PSI, plexin-semaphorin-integrin; MIDAS, metal ion-dependent adhesion site; Fn, fibronectin; H/120, recombinant fragment of Fn comprising type III repeats 12-15 plus IIICS segment; FnIII(6-10), recombinant fragment of Fn comprising type III repeats 6-10; HBS, HEPES-buffered saline.
References
- 1.Hynes RO. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Geiger B, Bershadsky A, Pankov R, Yamada KM. Nat. Revs. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 3.Miranti CK, Brugge JS. Nat. Cell Biol. 2002;4:83–90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- 4.Humphries MJ. Biochem. Soc. Trans. 2000;28:311–339. [PubMed] [Google Scholar]
- 5.Takagi J, Petre BM, Waltz T, Springer TA. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 6.Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas TA, Plow EF, Qin J. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 7.Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong JP, Stehle T, Zhang R, Joachimiak, Frech M, Goodman SL, Arnaout MA. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 9.Xiao T, Takagi J, Coller BS, Wang J-H, Springer TA. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Flier A, Sonnenberg A. Cell Tissue Res. 2001;305:285–298. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- 11.Emsley J, Knight CJ, Farndale RW, Barnes MJ, Liddington RC. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 12.Takagi J, Kamata T, Meredith J, Puzon-McLaughlin W, Takada Y. J. Biol. Chem. 1997;272:19794–197800. doi: 10.1074/jbc.272.32.19794. [DOI] [PubMed] [Google Scholar]
- 13.Humphries JD, Askari JA, Zhang X-P, Takada Y, Humphries MJ, Mould AP. J. Biol. Chem. 2000;275:20337–20345. doi: 10.1074/jbc.M000568200. [DOI] [PubMed] [Google Scholar]
- 14.Mould AP, Askari JA, Humphries MJ. J. Biol. Chem. 2000;275:20324–20336. doi: 10.1074/jbc.M000572200. [DOI] [PubMed] [Google Scholar]
- 15.Chan BM, Kassner PD, Schiro JA, Byers HR, Kupper TS, Hemler ME. Cell. 1992;68:1051–1060. doi: 10.1016/0092-8674(92)90077-p. [DOI] [PubMed] [Google Scholar]
- 16.Kassner PD, Hemler ME. J. Exp. Med. 1993;178:649–660. doi: 10.1084/jem.178.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassner PD, Alon R, Springer TA, Hemler ME. Mol. Cell Biol. 1995;6:661–674. doi: 10.1091/mbc.6.6.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Thomas SM, Woodside DG, Rose DM, Kiosses WB, Pfaff M, Ginsberg MH. Nature. 1999;402:676–681. doi: 10.1038/45264. [DOI] [PubMed] [Google Scholar]
- 19.Miao H, Li S, Hu Y-L, Yuan S, Zhao Y, Chen BPC, Puzon-McLaughlin W, Tarui T, Shyy JY-J, Takada Y, Usami S, Chien S. J. Cell Sci. 2002;115:2199–2206. doi: 10.1242/jcs.115.10.2199. [DOI] [PubMed] [Google Scholar]
- 20.Danen EHJ, Sonneveld P, Brakebusch C, Fassler R, Sonnenberg A. J. Cell Biol. 2002;259:1071–1086. doi: 10.1083/jcb.200205014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostafavi-Pour Z, Askari J, Parkinson SJ, Parker P, Ng TTC, Humphries MJ. J. Cell Biol. 2003;161:155–167. doi: 10.1083/jcb.200210176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphries MJ, McEwan PA, Barton SJ, Buckley PA, Bella J, Mould AP. Trends Biochem. Sci. 2003;28:313–320. doi: 10.1016/s0968-0004(03)00112-9. [DOI] [PubMed] [Google Scholar]
- 23.Travis MA, Humphries JD, Humphries MJ. Trends Pharmacol. Sci. 2003;24:192–197. doi: 10.1016/S0165-6147(03)00069-5. [DOI] [PubMed] [Google Scholar]
- 24.Mould AP, Askari JA, Barton SJ, Kline AD, McEwan PA, Craig SE, Humphries MJ. J. Biol. Chem. 2002;277:19800–19805. doi: 10.1074/jbc.M201571200. [DOI] [PubMed] [Google Scholar]
- 25.Mould AP, Barton SJ, Askari JA, McEwan PA, Buckley PA, Craig SE, Humphries MJ. J. Biol. Chem. 2003;278:17028–17035. [Google Scholar]
- 26.Tadokoro S, Shattil SJ, Eto K, Tai V, Linddington RC, de Pereda JM, Ginsberg MH, Calderwood DA. Science. 2003;302:103–6. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 27.Luo B-H, Springer TA, Takagi J. PLoS Biol. 2004;2:776–786. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphries MJ. Trends Pharmacol. Sci. 2000;21:29–32. doi: 10.1016/s0165-6147(99)01410-8. [DOI] [PubMed] [Google Scholar]
- 29.Mould AP, Garratt AN, Askari JA, Akiyama SK, Humphries MJ. FEBS Lett. 1995;363:118–122. doi: 10.1016/0014-5793(95)00301-o. [DOI] [PubMed] [Google Scholar]
- 30.Mould AP, Askari JA, Aota S, Yamada KM, Irie A, Takada Y, Mardon HJ, Humphries MJ. J. Biol. Chem. 1997;272:17283–17292. doi: 10.1074/jbc.272.28.17283. [DOI] [PubMed] [Google Scholar]
- 31.Mould AP, Garratt AN, Puzon-McLaughlin W, Takada Y, Humphries MJ. Biochem. J. 1998;331:821–828. doi: 10.1042/bj3310821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mould AP, Askari JA, Craig SE, Garratt AN, Clements J, Humphries MJ. J. Biol. Chem. 1994;269:27224–27230. [PubMed] [Google Scholar]
- 33.Newham P, Craig SE, Clark K, Mould AP, Humphries MJ. J. Immunol. 1998;160:4508–4517. [PubMed] [Google Scholar]
- 34.Mould AP, Akiyama SK, Humphries MJ. J. Biol. Chem. B. 1995:26270–26277. doi: 10.1074/jbc.270.44.26270. [DOI] [PubMed] [Google Scholar]
- 35.Newham P, Craig SE, Seddon GN, Schofield NR, Rees A, Edwards RM, Jones EY, Humphries MJ. J. Biol. Chem. 1997;272:19429–19440. doi: 10.1074/jbc.272.31.19429. [DOI] [PubMed] [Google Scholar]
- 36.Kassner PD, Kawaguchi S, Hemler ME. J. Biol. Chem. 1994;269:19859–19867. [PubMed] [Google Scholar]
- 37.Kunkel TA. Proc. Natl. Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchida J, Ueki S, Saito Y, Takagi J. FEBS Letts. 1997;416:212–216. doi: 10.1016/s0014-5793(97)01206-4. [DOI] [PubMed] [Google Scholar]
- 39.Takada Y, Puzon W. J. Biol. Chem. 1993;268:17597–17601. [PubMed] [Google Scholar]
- 40.Barton SJ, Travis MA, Askari JA, Buckley PA, Craig SE, Humphries MJ, Mould AP. Biochem. J. 2004;380:401–407. doi: 10.1042/BJ20031973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nobes CD, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 42.Whittard JD, Akiyama SK. J. Cell Sci. 2001;114:3265–3272. doi: 10.1242/jcs.114.18.3265. [DOI] [PubMed] [Google Scholar]
- 43.Saoncella S, Echtermeyer F, Denhez F, Nowlen JK, Mosher DF, Robinson SD, Hynes RO, Goetinck PF. Proc. Natl. Acad. Sci. USA. 1999;96:2805–2810. doi: 10.1073/pnas.96.6.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pujades C, Alon R, Yauch RL, Masumoto A, Burkly LC, Chen C, Springer TA, Lobb RR, Hemler ME. Mol. Biol. Cell. 1997;8:2647–2657. doi: 10.1091/mbc.8.12.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mannion BA, Berditchevski F, Kraeft S-K, Chen LB, Hemler ME. J. Immunol. 1996;157:2039–2047. [PubMed] [Google Scholar]
- 46.Berditchevski F, Odintosova E. J. Cell Biol. 1999;146:477–492. doi: 10.1083/jcb.146.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]