Abstract
A search of databases with the sequence from the 5′ untranslated region of a Hydra cDNA clone encoding a receptor protein-tyrosine kinase revealed that a number of Hydra cDNAs contain one of two different sequences at their 5′ ends. This finding suggested the possibility that mRNAs in Hydra receive leader sequences by trans-splicing. This hypothesis was confirmed by the finding that the leader sequences are transcribed as parts of small RNAs encoded by genes located in the 5S rRNA clusters of Hydra. The two spliced leader (SL) RNAs (SL-A and -B) contain splice donor dinucleotides at the predicted positions, and genes that receive SLs contain splice acceptor dinucleotides at the predicted positions. Both of the SL RNAs are bound by antibody against trimethylguanosine, suggesting that they contain a trimethylguanosine cap. The predicted secondary structures of the Hydra SL RNAs show significant differences from the structures predicted for the SLs of other organisms. Messenger RNAs have been identified that can receive either SL-A or -B, although the impact of the two different SLs on the function of the mRNA is unknown. The presence and features of SL addition in the phylum Cnidaria raise interesting questions regarding the evolution of this process.
In members of a small number of phyla, leader sequences obtained from a small nuclear RNA, the spliced leader (SL) RNA, are attached to the 5′ ends of mRNAs. To date, SL addition has been identified in three metazoan phyla (Nematoda, Platyhelminthes, and Chordata) (1–4) and in one unicellular eukaryotic phylum (Sarcomastigophora) (3, 5–7). No evidence of SL addition has been detected in any intensively studied plants, fungi, insects, echinoderms, or vertebrates. The phylogenetic distribution of SL addition (Fig. 1) is surprising and has made it difficult to discern the evolutionary history of this process. However, SL additions in unicellular eukaryotes and metazoans do share several features. Many SL RNAs have a highly conserved structure that includes three stem–loops, and all of the SL RNAs have a binding site for the Sm protein (8). In a number of uni- and multicellular species, copies of the SL RNA genes are found to be repeated within the 5S rDNA cluster (9). For all of the genes in trypanosomes, for a portion of nematode genes, and possibly in some flatworm genes, SL addition serves to separate polycistronic primary transcripts into individual mRNAs (10–12). SL addition is thus an important feature of both genome organization and gene expression in these phyla. The conservation of these features suggests the possibility that SL addition originated in unicellular eukaryotes and was retained at least into the ancestor of modern protostomes and deuterostomes but was lost after the divergence of many metazoan phyla. However, given the apparently small number and diverse phylogenetic positions of metazoan phyla that carry out SL addition, multiple independent origins for this process must also be considered. Early diverging metazoan phyla have been examined for the presence of SL addition (13), but so far no cases have been reported. Here we demonstrate the presence of SLs on mRNAs in Hydra, a member of the early-diverging metazoan phylum Cnidaria. Previous studies of Hydra genes had revealed that they undergo typical cis-splicing (14, 15), with the introns in some Hydra genes being located at sites identical to those in homologous genes in vertebrates (14). The presence and features of SL addition in Cnidaria provide additional data bearing on the puzzling evolutionary history of SL addition in metazoans.
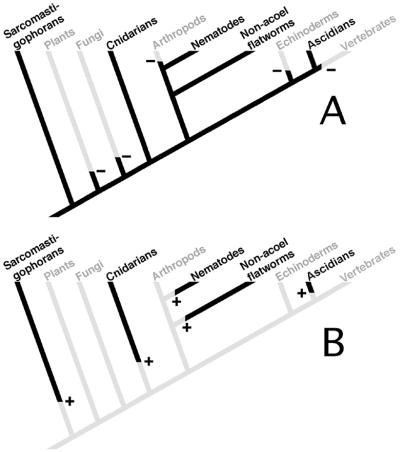
Figure 1.
Phylogenetic distribution of spliced leader addition to mRNAs. Phylogenetic relationships of only those taxa in which spliced leader addition is known to be present or likely to be absent are shown. The phylogenetic relationships between the taxa are based on multiple molecular studies (42–44). (A) A version of the tree in which spliced leader addition arose in a unicellular eukaryote and was subsequently lost from the various taxa indicated in gray. Minuses indicate points at which loss occurred. (B) A version of the tree in which spliced leader addition arose independently in the various phyla indicated in black. +, points at which origin of spliced leader addition occurred.
Materials and Methods
Database Searches.
Database searches were carried out by using the blast server at the National Center of Biotechnology Information (16).
Isolation of Hydra DNA and RNA.
Genomic DNA was isolated from the Zurich strain of Hydra vulgaris essentially as described by Davis et al. (17). Total H. vulgaris RNA was extracted with hot acidic phenol (18).
Rapid Amplification of cDNA Ends (RACE).
5′ RACE for the HTK32 cDNA was performed essentially according to Frohman (19). PolyA+ RNA was isolated from adult H. vulgaris polyps by using an RNeasy Midi Kit (Qiagen, Chatsworth, CA) and an Oligotex mRNA Mini Kit (Qiagen). Reverse transcription was performed by using a gene-specific primer (5′-TTGTAGCTCTTACATTAC-3′). The resulting first-strand cDNA was polydA tailed with terminal transferase (Boehringer Mannheim) before the first round of PCR. The initial amplification reaction was carried out by using a mixture of three primers as described by Frohman (19). The sequences of the primers are as follows: 5′-CAATAACTCTATATTTACC-3′; 5′-AAGGATCCGTCGACATCG-3′; 5′-AAGGATCCGTCGACATCGATAATACGACTCACTATAGGGATTTTTTTTTTTTTTTT-3′. Amplification conditions were as described by Frohman (19). The second round of amplification was for 40 cycles at a final annealing temperature of 44°C by using the following primer pair: 5′-GTATAAACACCAGCATC-3′; 5′-ATCGATAATACGACTCAC-3′. The 390-bp fragment obtained from the second amplification reaction was purified by agarose gel electrophoresis, extracted from the gel by using the Qiaex II Gel Extraction Kit (Qiagen), and cloned into the pGEM-T EZ cloning vector (Promega).
For 3′ RACE of SL-A and -B, polyA tails were added to the total RNA sample by using cloned yeast polyA polymerase (United States Biochemical). The polyadenylation reaction was carried out according to the manufacturer's protocol. First-strand cDNA was synthesized from the pool of polyadenylated RNA by using AMV reverse transcriptase (Boehringer Mannheim) and was primed from an oligonucleotide containing a dT16 sequence at its 3′ end (5′-AAGGATCCGTCGACATCGATAATACGACTCACTATAGGGATTTTTTTTTTTTTTTT-3′). Two rounds of touchdown PCR (20) were performed to obtain the 3′ ends of the SL-A and -B RNAs. For SL-A, the first round of amplification was carried out for 40 cycles at a final annealing temperature of 38°C by using the following pair of primers: 5′-GGTAGGTACCATAACAGTTTAC-3′; 5′-AAGGATCCGTCGACATCG-3′. The second round of amplification was carried out for 35 cycles at a final annealing temperature of 49°C by using the following pair of primers: 5′-TCTCTTTACGATTTTCGGG-3′; 5′-ATCGATAATACGACTCAC-3′. The 101-bp fragment obtained from the second amplification reaction was gel-purified and cloned into pGEM-T EZ (Promega). The 3′ end of SL-B was identified by the same procedure by using the first-round primers 5′-GTAGGCATCAATAAATTTTGAC-3′ and 5′-AAGGATCCGTCGACATCG-3′ to amplify at a final annealing temperature of 42°C. The second-round primers were 5′-GCAAATTTTCGAATTTCGGGG-3′ and 5′-ATCGATAATACGACTCAC-3′, and a final annealing temperature of 46°C was used. The 79-bp fragment obtained from the second round of amplification was gel-purified and cloned into pGEM-T EZ.
Amplification of 5S rRNA Gene Repeats.
5S rRNA gene repeats were amplified with primers corresponding to the conserved portions of known cnidarian 5S rRNA sequences (21–23). The sequences of the primers were as follows: 5′-GTTAAGCACCGTCAAGCCAGG-3′; 5′-CTTCCGTGATCGGACGAGAAC-3′. Amplification was carried out for 35 cycles at a final annealing temperature of 51°C. The resulting products were gel-purified and cloned into pGEM-T EZ.
Outron Cloning.
A fragment of genomic DNA containing sequence upstream of the Hydra Syk gene (24) was amplified by using the splinkerette method (25). The resulting fragment was cloned into pGEM-T EZ and sequenced.
RNA Hybridization Analysis.
For probing with SL probes, samples of total RNA (12 μg each) were separated in a formaldehyde-containing 3% NuSieve GTG agarose gel (BMA Biomedicals). Before transfer, the lane containing the RNA size markers (Ambion, Austin, TX) was separated and stained with ethidium bromide. After capillary transfer to a Genescreen II nylon membrane (DuPont), the RNA was crosslinked to the membrane by using a UV Stratalinker 2400 (Stratagene). Probes labeled with [α-32P]dATP were made by extending a primer annealed to a synthetic oligonucleotide template by using Klenow DNA polymerase (Promega) in Prime-It II dATP reaction buffer (Stratagene). For the SL-A intron probe, the template was 5′-GTAGGCATCAATAAATTTTGACGCAAATTTTCGAATTTTCGGGGTTTCGGTAGTGGGTAAA-3′, and the primer was 5′-CCCACTACCGAAACCCCGAA-3′. For the SL-B exon probe, the template was 5′-ACGGAAAAAAACACATACTGAAACTTTTTAGTCCCTGTGTAATAAG-3′, and the primer was 5′-CTTATTACACAGGGACTAAAAAG-3′. Hybridizations were carried out in an aqueous buffer [1 M NaCl, 100 mM Tris⋅HCl, pH 8.0/0.1% BSA/0.1% polyvinylpyrrolidone/0.1% Ficoll/0.05% sodium pyrophosphate/0.1% SDS/0.1 mM disodium EDTA/50 μg/ml heparin/100 μg/ml torula yeast RNA (Sigma)/500 μg/ml herring sperm DNA (Sigma)] for 18 h at room temperature. The blots were washed at room temperature twice in high salt buffer (1 M NaCl/0.1 M Tris, pH 8.0/0.1 mM disodium EDTA), then in 2× standard saline phosphate/EDTA [0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA (26)]. Hybridization was detected by exposure of the filters to x-ray film with intensifying screens at −70°C.
For identification of RNAs containing trimethylguanosine (TMG) caps, 30 μg of total RNA was immunoprecipitated by using an anti-TMG antibody/agarose conjugate [Oncogene Research Products (Cambridge, MA)]. Both the bound and unbound fractions were phenol/chloroform extracted and precipitated in 75% ethanol. Electrophoresis, transfer, hybridization, and washing were all performed as described above. A probe that recognized 5S rRNA was made with the template 5′-CCTACGACCATACCACGGTGAACACACCCGTTCTCGTCCGATCACGGAAG-3′ and the primer 5′-CTTCCGTGATCGGACGAGAAC-3′ by using the same procedure as described above for preparation of SL probes.
DNA Sequencing.
DNA sequencing was carried out by primer walking and was done by the University of California, Irvine, DNA Core Facility.
RNA Structure Modeling.
RNA secondary structures were modeled by using Version 2.3 of the mfold program (27, 28). Details of the folding conditions are described in the text.
Results
A Number of Hydra mRNAs Contain Identical Sequences at Their 5′ Ends.
A database search with the sequence of the 5′ untranslated region of the H. vulgaris receptor protein-tyrosine kinase gene HTK32 (unpublished work) revealed a conserved sequence at the 5′ end of several Hydra cDNA sequences. Subsequent comparisons among all of the Hydra cDNA sequences in the database revealed two sequences that were located at the 5′ ends of a variety of cDNAs (Fig. 2A). This finding suggested that Hydra mRNAs may undergo SL addition at their 5′ ends. We attribute the differing lengths of the sequences to premature termination of reverse transcription.
Figure 2.
(A) Sequence identities at the 5′ ends of cDNA clones from Hydra genes. Identical 5′ sequences are in lowercase; divergent downstream sequences are in uppercase. The translation start ATG codon is separated from the 5′ untranslated region sequence by a slash. The upper group of sequences contains the spliced leader A (SL-A) sequence; the lower group contains the spliced leader B (SL-B) sequence. GenBank accession nos. for the sequences are as follows: Hint, M64611; Syk, AF060949; HTK32, AF123442; HTK54, U24116; HFZ, AF209200; Csk, AF067775; Cnash, U36275; Alx, AF295531; Pax-A, U96193; enolase, U85827; PLC-βHI, AB017511; hyGK, AF031931; ECE, AF162671; hym-323, AB40074; HTK16, U00936; HZO-1, AF230482; PKC1B, Y12857; ras1, X78597; nucleoporin, U85827; annexin XII, M83736. All genes are from H. vulgaris except hyGK (Hydra oligactis), enolase (H. oligactis), Pax-A (Hydra littoralis), and PLC-βI (Hydra magnipapillata). The three different Hint sequences (labeled Hint 1–3) arise because of alternative splicing. Hint produces a long transcript with SL-A at the 5′ end and a shorter transcript that can contain either SL-A or -B (45). The single nucleotide difference (T>G) in the SL-A sequence of HTK54 may be because of an error during cDNA synthesis or the presence of multiple alleles of SL-A. A complete copy of the SL-B sequence is located internally in a H. vulgaris cDNA for cAMP-response element-binding protein (CREB) (46), where it results in the truncation of a highly conserved portion of the CREB coding sequence. We have attempted, without success, to confirm this arrangement by amplification of the corresponding region from first-strand cDNA made from H. vulgaris polyA+ RNA. We therefore believe that this clone is a hybrid produced during cDNA library construction by ligation of the 3′ end of a partial CREB cDNA to the 5′ end of a cDNA derived from an SL-B-containing mRNA. (B) Alignment of sequences from the 5′ ends of HTK32 cDNA clones containing SL-A or -B sequences. The clone containing SL-B includes four nucleotides that are not present in the SL-A-containing message. The splice acceptor dinucleotide is underlined. The SL-A-containing sequence is from a clone isolated from a cDNA library. The SL-B-containing sequence was obtained by 5′ RACE (see Materials and Methods). (C) Genomic sequence from the Hydra Syk gene (24). The splice acceptor dinucleotide is indicated by double underlining. The genomic sequence is aligned with the sequences from Syk cDNAs containing either SL-A or -B (see A). The pyrimidine-rich sequence upstream of the splice acceptor dinucleotide is shaded.
The putative spliced leader sequences are found on mRNAs encoding a variety of proteins, including receptor and nonreceptor protein-tyrosine kinases, transcription factors, metabolic enzymes, and structural proteins. SL sequences may not be added to all mRNAs. Of the 50 Hydra cDNA sequences in the database for which 5′ untranslated region sequence is available, ≈30% contain a SL (N.A.S., unpublished observation). Given that some of these sequences may be incomplete, 30% represents a minimum for the fraction of mRNAs sampled in this manner that are trans-spliced.
In two of the three cases where cDNAs containing SL-A and cDNAs containing SL-B have been reported for a given gene, the junction of the SL-A sequence and the SL-B sequence with the remainder of the cDNA sequence is located at the identical position. The one exception is HTK32, where clones containing different SL sequences show SL-B to be attached at a site four nucleotides upstream of the SL-A attachment site (Fig. 2B). In HTK32 clones containing SL-B, the AG splice acceptor dinucleotide used for splicing to SL-A is revealed, indicating that spliced leader attachment in Hydra occurs at a typical splice acceptor site and that HTK32 undergoes alternative trans-splicing. To further confirm that leader sequences are spliced onto the ends of mRNAs in Hydra, we amplified the sequences upstream of the gene encoding the Hydra Syk protein-tyrosine kinase (24), which has been found to yield both SL-A- and -B-containing mRNAs. As predicted, the genomic sequence diverged from the cDNA sequences at the point where the spliced leader sequences begin (Fig. 2C), and a splice acceptor dinucleotide is located at the predicted position. The splice acceptor dinucleotide is immediately preceded by a pyrimidine-rich sequence (shaded in Fig. 2C), a feature also found immediately upstream of the splice acceptor dinucleotides of introns in cnidarians (R.E.S., unpublished observation).
SL-A and -B Are Encoded by Genes Located in the 5S rRNA Gene Clusters.
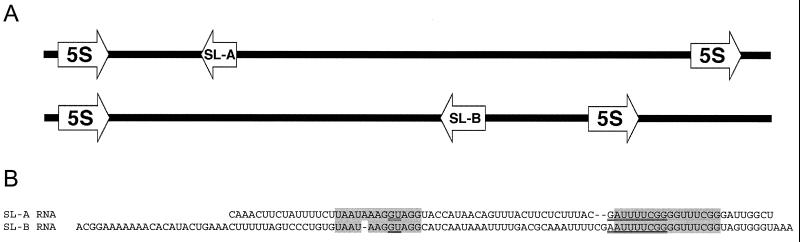
If Hydra mRNAs undergo spliced leader addition, we would expect to find genes encoding small RNAs that contain the putative SL sequences followed by a splice donor dinucleotide. In a number of species that have been shown to use spliced leader RNAs, the genes encoding the SL RNAs are present in the spacer between the repeated 5S rRNA genes (9). We thus amplified the 5S rDNA repeats from H. vulgaris and examined the spacer region for the presence of sequences corresponding to the two leader sequences. Amplification generated two products, a major one of ≈1.3 kb and a minor one of ≈1.6 kb, indicating that H. vulgaris contains two types of 5S gene repeats. The 1.6-kb repeat contained the SL-A gene, and the 1.3-kb repeat contained the SL-B gene (Fig. 3A). Both SL genes had the same transcriptional orientation relative to the flanking 5S genes. Sequencing of the SL genes revealed a candidate GT splice donor dinucleotide at the predicted position in each of the genes (Fig. 3B).
Figure 3.
SL-A and -B RNA sequences and gene arrangement. (A) Arrangement of the SL-A and -B genes in the 5S rRNA clusters. The 5′ and 3′ ends of the 5S gene were identified by comparison to available cnidarian 5S rRNA sequences (21–23). (B) The SL-A and -B RNA sequences were aligned manually. Conserved sequences surrounding the splice donor site and the predicted Sm-binding site are shaded. The predicted Sm-binding sequence and the splice donor dinucleotide are doubly underlined.
We have assumed the 5′ ends of the SL-A and -B genes to be at or very near the position defined by the longest of each type of SL sequence found in the cDNA clones. To map the 3′ ends of the SL genes, we used a modification of the RACE procedure (19) in which a preparation of total RNA from Hydra was tailed at the 3′ end with polyA polymerase. Synthesis of cDNA from this tailed RNA preparation was primed with an oligo-dT containing RACE primer. SL-A- and -B-specific cDNA products were then amplified by using nested primers corresponding to the predicted intron portions of the SL genes described above. Multiple clones from the resulting PCR products were sequenced. For SL-A RNA, the 3′ end could be identified unambiguously. However for SL-B, the DNA sequence contains a sequence of three A residues at the position where the polyA tail was attached. Thus one or more of the three As shown at the 3′ end of SL-B RNA in Fig. 3B may be derived from the polyA tail.
The predicted SL-A and -B RNA sequences differ significantly both in length and sequence (Fig. 3B). Alignment of the sequences shows regions of conservation only around the splice donor site and the predicted Sm-binding site. To confirm that the SL sequences are transcribed as small RNAs, gel-fractionated total Hydra RNA was probed for SL-A and -B sequences. The SL-A probe detected a single RNA of the expected size (Fig. 4A, lane 1). The SL-B probe detected an RNA of the expected size (Fig. 4A, lane 2) as well as a second smaller RNA. The identity of the second RNA is unknown.
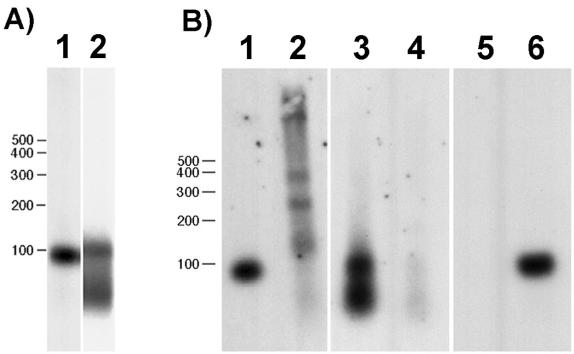
Figure 4.
(A) Northern blot of Hydra total RNA hybridized with probes to the intron region of SL-A (lane 1) and the exon region of SL-B (lane 2). Both probes hybridize to RNA of the predicted sizes as described in the text. (B) TMG-containing RNA was isolated from an aliquot of total RNA by using an anti-TMG antibody–agarose conjugate. Bound (lanes 1, 3, and 5) and unbound (lanes 2, 4, and 6) RNAs were hybridized with the SL-A (lanes 1 and 2) and -B (lanes 3 and 4) probes used in A and with a probe for 5S rRNA (lanes 5 and 6).
Hydra SL RNAs Contain TMG Caps.
SL RNAs in other metazoans contain a TMG cap. To determine whether Hydra SL RNAs contain a TMG cap, we tested whether the SL RNAs could be bound by an antibody against TMG. Both SL-A and -B RNAs were recovered in the bound fraction (Fig. 4B, lanes 1–4). The specificity of the binding was confirmed by showing that 5S RNA, which lacks a TMG cap, remains in the unbound fraction (Fig. 4B, lanes 5 and 6).
The Predicted Structures of the Hydra SL RNAs.
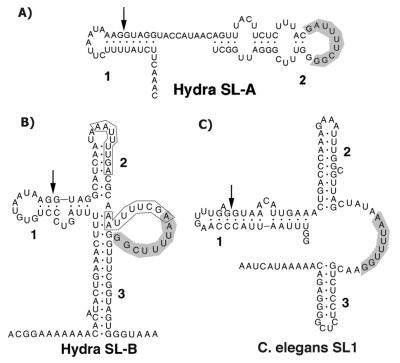
SL RNAs identified in other systems are predicted to adopt a conserved structure that consists of a stem–loop containing the splice donor dinucleotide, followed by two additional stem–loops that flank a single-stranded region containing a binding site for the Sm protein. The exceptions to this structure are the SL of the flatworm Schistosoma mansoni, in which the second stem–loop is absent (29), and the SL of the ascidian Ciona intestinalis, in which both the second and third stem–loops are absent (1). We have attempted to model the secondary structures of the Hydra SL RNAs by using Version 2.3 of mfold (27, 28). Two folding constraints were applied. First, five nucleotides including the splice donor dinucleotide (GGUAG) were required to be base paired. Second, the 3′-most predicted Sm-binding sequence was required to be single-stranded. Both of these constraints are conserved features of the structures predicted in previous SL RNA models. Folding was carried out at 18°C, the temperature at which Hydra is typically cultured. All other parameters were used at their default settings. Using these conditions, mfold yielded a single structural prediction for SL-B, which is shown in Fig. 5B. This same model was obtained at temperatures up to 37°C. Either requiring the middle predicted Sm-binding site to be single-stranded or requiring both it and the 3′-most predicted Sm-binding site to be single-stranded did not change the structure significantly. We did not produce a model in which the 5′-most candidate Sm-binding site was forced to be single-stranded, because this site is located significantly 5′ of the location of the Sm-binding sites of all other SL RNAs. By using the same constraints, except for a temperature of 25°C, the SL1 RNA of Caenorhabditis elegans was folded as a control. Two structures were obtained for SL1 RNA. Both contained the expected three stem–loops but differed in the details of the folding of the first stem–loop. The structure that most closely resembles those published previously is shown in Fig. 5C. This structure contains features in the first stem–loop that have been confirmed by NMR spectroscopy (30). The Hydra SL-B model differs significantly from models of other SL RNAs. Of particular interest is the long stem produced by base pairing between the 5′ and 3′ portions of the molecule. Such a structure has not been reported for other SL RNAs. This stem is a robust feature of the model. It is obtained in completely unconstrained foldings and in foldings obtained when the mfold percent suboptimality is increased from the default value of 5% to a value of 20%. It thus seems likely that this stem is a feature of the RNA in vivo. Interestingly, however, the SL-B structure does contain three stems. The significant difference is that the third stem is formed by base pairing with sequences from the 5′ end of the RNA, whereas in other species it forms from the folding back of 3′ sequences. The predicted structure for SL-A (Fig. 5A) differs significantly from that of SL-B and lacks the structural features predicted for most SL RNAs. Surprisingly, the putative Sm-binding site is located on the loop of the single stem–loop in the SL-A model.
Figure 5.
Predicted secondary structures of SL-A RNA (A), SL-B RNA (B), and the C. elegans SL1 RNA (C). The structures were generated by using Version 2.3 of the mfold RNA secondary structure modeling program (27, 28). The parameters and constraints used for folding are detailed in the text. The stem–loop structures are numbered for reference. Arrows in Stem 1 indicate the 3′ end of the exon sequence in each RNA. Putative Sm protein-binding sequences are shaded. Two other possible Sm-binding sequences discussed in the text are boxed in B. The diagrams were produced by inputting the structural data generated by mfold into RnaViz (47).
Discussion
The evolutionary history of spliced leader addition to mRNAs has been difficult to discern because of lack of knowledge of how widespread the phenomenon is and to what degree the features of the process are conserved. With the identification of spliced leader addition in a cnidarian, we now know that members of four metazoan phyla and one phylum of unicellular eukaryotes carry out this reaction. In all cases, the spliced leader is derived by trans-splicing of an exon contained in a small RNA molecule, and the exon is followed by a canonical splice donor dinucleotide. At least some of the genes encoding spliced leader RNA in a given species in each of these phyla are present in tandem arrays (summarized in ref. 9). However, in C. elegans, where several classes of SL RNA genes are present, some of the SL genes are present in tandem arrays and some are present as single copies (31). In a number of cases, but not all, the tandemly repeated SL RNA genes are interspersed with other repeated genes, most commonly the 5S rRNA genes (9). It has been argued that this is the result of a propensity for 5S rRNA genes to insert into preexisting SL RNA gene clusters (9). However, given the presence of 5S rRNA gene clusters in organisms lacking SL genes, it seems more likely that SL genes have inserted into preexisting 5S clusters.
Except for the Sm-binding sequence and the splice donor dinucleotide, cross-phylum sequence conservation among SL RNAs is absent. Within phyla, sequence conservation is variable. The exon portion of the major SL RNA in nematodes, SL1, is identical in sequence in all members of the phylum that have been examined (4). In flatworms, however, SL exon sequences vary considerably across genera (13). The sequence conservation in the nematode SL1 exon is thought to be because of the presence of transcription promoter elements within the SL exon (32, 33). The promoters of flatworm SL RNAs have not yet been mapped, so it is at present unclear why sequence variation is tolerated in the exons of their SL RNA genes. The SL-B exon is identical in sequence among four different species of Hydra. Studies of additional cnidarians will be required to determine the extent of trans-splicing and the degree of conservation of SL sequences in this phylum.
Most of the SL RNAs described previously can be folded into a conserved secondary structure that contains three stem–loops and a single-stranded binding site for the Sm protein (8). Exceptions to this structure include the SL RNA from the flatworm Schistosoma mansoni, in which the second stem–loop is absent (29), and the Ciona SL RNA, which lacks the second and third stem–loops (1). Our modeling of the secondary structure of the Hydra SL-B RNA have yielded the surprising finding that sequences from the 5′ and 3′ ends of the molecule apparently base pair with each other to form a long stem. Although this is quite different from other SL RNA structures, it does conserve a structural element, the presence of a stem 3′ to the Sm-binding sequence. It will be interesting to determine whether this unusual way of forming the third stem is actually used by the RNA.
Perhaps the most unusual feature of SL addition in Hydra is the alternative usage of two very different leader sequences. SL-A and -B show very little similarity in sequence and are quite different in length. Although this level of dissimilarity has not been shown in other species containing multiple spliced leaders, this finding is not necessarily unexpected. Mutagenesis studies have shown there is considerable flexibility in the sequence and structural requirements for splicing of the C. elegans SL-1 RNA (34, 35). We now have multiple examples of genes whose mRNAs can accept either SL-A or -B. Whether other genes accept only one or the other SL is unclear, because we have not yet searched specifically for examples of each SL on particular RNAs.
A final feature related to SL addition for which we currently lack information in Hydra is whether any of the genes in this species are organized into operons. In trypanosomes, all of the genes are organized into operons, and SL addition separates the resulting polycistronic mRNAs into individual species (5, 36). It is not known whether any of the genes in euglenoids, the other unicellular eukaryotes that are known to carry out spliced leader addition (6, 7), are organized into operons. In C. elegans, about 25% of the genes are organized into operons (37). SL1 is spliced onto the 5′ end of the resulting polycistronic mRNAs (38), and SL2 is trans-spliced onto the downstream mRNAs (39). Recent evidence suggests that operons may also be present in flatworms (10), and that spliced leader addition may play a role in separating polycistronic transcripts there, too. There is no information on the presence of operons in Ciona. Thus, to date there are no reports of organisms that use SL addition but lack operons. This suggests the intriguing possibility that SL addition is present or may persist only in organisms at least some of whose genes are organized into operons. It has been proposed that spliced leaders in C. elegans originated as molecular parasites (40). In such a scenario, spliced leaders would allow the formation of operons, which would then depend on trans-splicing to resolve the downstream mRNAs.
The distribution of spliced leader addition can be explained in multiple ways. SL addition could have arisen once in a unicellular eukaryotic ancestor to metazoans and have been lost from many metazoan lineages. Alternatively, SL addition could have arisen multiple times. Its history could also have involved a combination of losses and gains. From existing information about the presence or absence of SL addition, its distribution cannot be explained by fewer than five independent gain or loss events (Fig. 1). Thus, the available phylogenetic distribution data do not allow one to decide between gain or loss models. Either case would involve drastic changes in the manner in which genes are organized and expressed.
Clues to the evolutionary history of spliced leader addition may lie in the history of the eIF4e proteins, the proteins that bind mRNA caps. The presence of TMG caps on mRNAs as the result of an organism acquiring SL addition requires that the organism also evolve eIF4e proteins that can bind to TMG caps. C. elegans contains five genes encoding eIF4e isoforms (41). Three of these isoforms are closely related and bind TMG caps in addition to the monomethyl guanosine (MMG) caps, which are present on non-SL-containing mRNAs. A phylogenetic comparison of eIF4e proteins from Hydra with those from C. elegans should allow us to determine whether TMG cap-binding eIF4e isoforms have originated once in metazoans (consistent with multiple loss of SL addition during metazoan evolution) or multiple times (consistent with multiple independent gain of SL addition). Parsimony analysis of the eIF4e sequences available in public databases (R.E.S., unpublished observations) supports the hypothesis that the TMG-binding isoforms of C. elegans were derived from a MMG-binding form at some point within the nematode lineage. This result thus provides preliminary support for the multiple independent gain model of SL addition.
Acknowledgments
This paper is dedicated to the memory of Robert E. Steele, Sr. We thank Diane Bridge for her many helpful comments on the manuscript and Tim Nilsen, Michael Zuker, Klemens Hertel, and Chris Greer and members of his lab for their interest and technical advice. Support for this work was provided by National Science Foundation Grant IBN-9808828 (to R.E.S.). N.A.S. was supported by National Institutes of Health Training Grant 5T32CA09054–23.
Abbreviations
- SL
spliced leader
- RACE
rapid amplification of cDNA ends
- TMG
trimethylguanosine
Footnotes
References
- 1.Vandenberghe A E, Meedel T H, Hastings K E M. Genes Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis R E. Parasitol Today. 1996;12:33–40. doi: 10.1016/0169-4758(96)80643-0. [DOI] [PubMed] [Google Scholar]
- 3.Nilsen T W. Infect Agents Dis. 1992;1:212–218. [PubMed] [Google Scholar]
- 4.Nilsen T W. Annu Rev Microbiol. 1993;47:413–440. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 5.Agabian N. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 6.Ebel C, Frantz C, Paulus F, Imbault P. Curr Genet. 1999;35:542–550. doi: 10.1007/s002940050451. [DOI] [PubMed] [Google Scholar]
- 7.Tessier L H, Keller M, Chan R L, Fournier R, Weil J H, Imbault P. EMBO J. 1991;10:2621–2625. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruzik J P, Van Doren K, Hirsh D, Steitz J A. Nature (London) 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- 9.Drouin G, de Sa M M. Mol Biol Evol. 1995;12:481–493. doi: 10.1093/oxfordjournals.molbev.a040223. [DOI] [PubMed] [Google Scholar]
- 10.Davis R E, Hodgson S. Mol Biochem Parasitol. 1997;89:25–39. doi: 10.1016/s0166-6851(97)00097-2. [DOI] [PubMed] [Google Scholar]
- 11.Blumenthal T. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 12.Blumenthal T, Thomas J. Trends Genet. 1988;4:305–308. doi: 10.1016/0168-9525(88)90107-2. [DOI] [PubMed] [Google Scholar]
- 13.Davis R E. Mol Biochem Parasitol. 1997;87:29–48. doi: 10.1016/s0166-6851(97)00040-6. [DOI] [PubMed] [Google Scholar]
- 14.Bosch T C G, Unger T F, Fisher D A, Steele R E. Mol Cell Biol. 1989;9:4141–4151. doi: 10.1128/mcb.9.10.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher D A, Bode H R. Gene. 1989;84:55–64. doi: 10.1016/0378-1119(89)90139-x. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Davis R W, Thomas M, Cameron J, John T P S, Scherer S, Padgett R A. Methods Enzymol. 1980;65:404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, JA, S, Struhl K. Current Protocols in Molecular Biology. Vol. 2. New York: Wiley; 1993. [Google Scholar]
- 19.Frohman M A. In: PCR Primer: A Laboratory Manual. Dieffenbach C W, Dveksler G S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 381–409. [Google Scholar]
- 20.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendriks L, De Baere R, Vandenberghe A, De Wachter R. Nucleic Acids Res. 1987;15:2773. doi: 10.1093/nar/15.6.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori H, Ohama T, Kumazaki T, Osawa S. Nucleic Acids Res. 1982;10:7405–7408. doi: 10.1093/nar/10.22.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker W F, Doolittle W F. Nucleic Acids Res. 1983;11:5159–5164. doi: 10.1093/nar/11.15.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steele R E, Stover N A, Sakaguchi M. Gene. 1999;239:91–97. doi: 10.1016/s0378-1119(99)00373-x. [DOI] [PubMed] [Google Scholar]
- 25.Devon R S, Porteous D J, Brookes A J. Nucleic Acids Res. 1995;23:1644–1645. doi: 10.1093/nar/23.9.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Mathews D H, Sabina J, Zuker M, Turner D H. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 28.Zuker M, Mathews D H, Turner D H. In: RNA Biochemistry and Biotechnology. Barciszewski J, Clark B F C, editors. Dordrecht, The Netherlands: Kluwer; 1999. pp. 11–43. [Google Scholar]
- 29.Rajkovic A, Davis R E, Simonsen J N, Rottman F M. Proc Natl Acad Sci USA. 1990;87:8879–8883. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Lapham J, Crothers D M. Proc Natl Acad Sci USA. 1996;93:44–48. doi: 10.1073/pnas.93.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross L H, Freedman J H, Rubin C S. J Biol Chem. 1995;270:22066–22075. doi: 10.1074/jbc.270.37.22066. [DOI] [PubMed] [Google Scholar]
- 32.Hannon G J, Maroney P A, Ayers D G, Shambaugh J D, Nilsen T W. EMBO J. 1990;9:1915–1921. doi: 10.1002/j.1460-2075.1990.tb08318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie H, Hirsh D. Proc Natl Acad Sci USA. 1998;95:4235–4240. doi: 10.1073/pnas.95.8.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson K C, Rothman J H. Mol Cell Biol. 1999;19:1892–1900. doi: 10.1128/mcb.19.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maroney P A, Hannon G J, Shambaugh J D, Nilsen T W. EMBO J. 1991;10:3869–3875. doi: 10.1002/j.1460-2075.1991.tb04956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanhamme L, Pays E. Microbiol Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blumenthal T. BioEssays. 1998;20:480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 38.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 40.Lawrence J. Curr Opin Genet Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- 41.Keiper B D, Lamphear B J, Deshpande A M, Jankowska-Anyszka M, Aamodt E J, Blumenthal T, Rhoads R E. J Biol Chem. 2000;275:10590–10596. doi: 10.1074/jbc.275.14.10590. [DOI] [PubMed] [Google Scholar]
- 42.Aguinaldo A M, Turbeville J M, Linford L S, Rivera M C, Garey J R, Raff R A, Lake J A. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 43.Baldauf S L, Doolittle W F. Proc Natl Acad Sci USA. 1997;94:12007–12012. doi: 10.1073/pnas.94.22.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wainright P O, Hinkle G, Sogin M L, Stickel S K. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 45.Kroiher M, Reidling J C, Steele R E. Gene. 2000;241:317–324. doi: 10.1016/s0378-1119(99)00474-6. [DOI] [PubMed] [Google Scholar]
- 46.Galliot B, Welschof M, Schuckert O, Hoffmeister S, Schaller H C. Development (Cambridge, UK) 1995;121:1205–1216. doi: 10.1242/dev.121.4.1205. [DOI] [PubMed] [Google Scholar]
- 47.De Rijk P, De Wachter R. Nucleic Acids Res. 1997;25:4679–4684. doi: 10.1093/nar/25.22.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]