Abstract
A recent study revealed that ES (embryonic stem) cell lines derived from the 129 murine strain carry an inactivating mutation within the caspase 11 gene (Casp4) locus [Kayagaki, Warming, Lamkanfi, Vande Walle, Louie, Dong, Newton, Qu, Liu, Heldens, Zhang, Lee, Roose-Girma and Dixit (2011) Nature 479, 117–121]. Thus, if 129 ES cells are used to target genes closely linked to caspase 11, the resulting mice might also carry the caspase 11 deficiency as a passenger mutation. In the present study, we examined the genetic loci of mice targeted for the closely linked c-IAP (cellular inhibitor of apoptosis) genes, which were generated in 129 ES cells, and found that, despite extensive backcrossing into a C57BL/6 background, c-IAP1−/− animals are also deficient in caspase 11. Consequently, data obtained from these mice should be re-evaluated in this new context.
Keywords: 129 mouse strain, caspase 1, caspase 11, inhibitor of apoptosis protein (IAP), lipopolysaccharide (LPS), sepsis
Abbreviations: c-IAP, cellular inhibitor of apoptosis; ES, embryonic stem; IAP, inhibitor of apoptosis; LPS, lipopolysaccharide; MEF, mouse embryonic fibroblast; RT, reverse transcription; TBS-T, Tris-buffered saline with 0.1% Tween 20; XIAP, X-linked inhibitor of apoptosis
INTRODUCTION
ES (embryonic stem) cell lines from the murine 129 strain are frequently used to derive genetically modified mice [1]. A recent report examining the role of caspase 1 in an endotoxin-induced murine model of sepsis revealed that the resistance to sepsis observed in caspase-1-targeted mice is largely due to a naturally occurring inactivating mutation within the caspase 11 gene (Casp4) locus that originated from the 129-derived ES cell line used in the creation of the mouse knockout [2]. This was consistent with an earlier study attributing the resistance to septic shock in the 129 strain of mice to differential expression of a gene in the region of mouse chromosome 9 where the caspase 11 gene is located [3].
Caspase 11, a mouse-specific caspase of the caspase 1 superfamily of cysteine proteases, has fundamental roles in the regulation of apoptosis and inflammation [4–6]. Caspase 11 is virtually undetectable in resting cells, but can be rapidly induced by various inflammatory or cytotoxic stimuli [4]. Once activated, caspase 11 can trigger caspase-1-dependent production of pro-inflammatory cytokines in the immune response [4], and it can also directly activate caspase 3, promoting cell death [7].
In a recent study, sequence analysis of the caspase 11 gene from several substrains of 129 mice revealed a 5 bp deletion within the gene [2]. This deletion removes the splice acceptor sequence of exon 7, resulting in a truncated transcript with exon 6 fused directly to exon 8. Absence of exon 7 from the transcript creates a frameshift mutation, resulting in a premature stop codon [2]. The predicted protein product of the shortened caspase 11 transcript, if expressed, would lack a large portion of its catalytic domain, and would be expected to be non-functional [2].
Owing to the close genetic proximity of the genes for murine caspase 1 (Casp1) and caspase 11 on chromosome 9, caspase-1-null mice, which were generated experimentally using 129 ES cells [8,9], were additionally found to bear the caspase 11 mutation, and this co-segregation was maintained despite extensive backcrossing into the caspase 11 wild-type murine strain, C57BL/6 [2]. Since deletion of caspase 11 alone has been shown to result in heightened resistance to sepsis [5], this makes it important to distinguish between the relative contributions of each protein to the inflammatory response. Indeed, the resistance to sepsis in the caspase-1-deficient caspase-11-mutated mice can be reversed by expressing exogenous caspase 11, suggesting that caspase 11 status is important for the phenotype [2].
Other genes targeted using 129-derived ES cells adjacent to the caspase 11 locus may co-segregate in 129 strains. Notably, the genes encoding the c-IAP [cellular IAP (inhibitor of apoptosis)] proteins c-IAP1 and c-IAP2, Birc2 and Birc3 respectively, are in close proximity to the caspase 11 gene on mouse chromosome 9, and, as such, mice deficient in c-IAP1 and c-IAP2 also potentially carry the mutation in the caspase 11 locus. IAP proteins are a broadly expressed group of intracellular factors that function in a wide range of cellular processes, in addition to their initially described role in the suppression of programmed cell death [10]. An emerging role for the IAP proteins is in the control of the innate immune response, functioning as key enzymes in several signalling pathways [11]. Additionally, in vivo data from the published c-IAP1−/− and c-IAP2−/− mice [12] showed reduced serum levels of pro-inflammatory cytokines and chemokines, compared with controls, when treated with NOD1 and NOD2 ligands [13]. Furthermore, c-IAP1−/− and c-IAP2−/− mice were found to be protected from a lethal dose of LPS (lipopolysaccharide), suggesting a role in the response to sepsis ([14], and K. Oetjen and C.S. Duckett, unpublished work).
Since the previously reported c-IAP1- and c-IAP2-targeted mice were generated from 129 ES cells, this raised the possibility that either or both of these strains might be also deficient for caspase 11. In the present study, we have examined this question and found that, despite extensive backcrossing, c-IAP1-targeted mice, but not c-IAP2-deficient animals, have the reported caspase 11 intronic inactivating deletion. We also show that this was not due to differences in the 129 ES lines used to create the c-IAP-targeted mice, because the caspase 11 mutation was found to be penetrant in an extensive panel of 129 ES lines tested, which included the lineages used in the targeting of both the c-IAP1 and c-IAP2 genes. These data suggest first that data obtained from c-IAP1−/− mice may need further evaluation, and secondly that a selective pressure may exist for the maintenance of a wild-type caspase 11 gene during normal development.
EXPERIMENTAL
Cells
C57BL/6 and 129 ES cell lines were maintained in high glucose Dulbecco's minimal essential medium (Invitrogen) supplemented with 15% (v/v) fetal bovine serum (Harlan), 4 mM glutamine, 1 μM 2-mercaptoethanol, 1% MEM non-essential amino acids, 50 IU/ml penicillin, 50 μg/ml streptomycin and 1000 IU/ml ESGRO (Millipore) on FVB/N mouse embryonic feeder cells mitotically inactivated by irradiation. Before DNA extraction and genotyping, ES cell lines were passaged twice on gelatin-coated dishes to eliminate feeders from the cultures as described in [15].
MEF (mouse embryonic fibroblast) cells were cultured in Dulbecco's modified Eagle's medium (Mediatech) supplemented with 10% (v/v) fetal bovine serum (Mediatech) and 2 mM glutamine (Gibco) at 37°C with 5% CO2. Matched c-IAP1 wild-type and c-IAP1−/− MEFs, prepared from littermate controls, have been described previously [16].
Immunoblotting
Cell lysates from MEFs were prepared with Nonidet P40 lysis buffer (50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P40, 5 mM NaF and 1 mM Na3VO4) supplemented with protease inhibitors for 30 min on ice to ensure complete lysis. Protein quantification was determined using the Bradford assay (Bio-Rad Laboratories). Cell lysates of equal protein concentrations were prepared in lithium dodecyl sulfate sample buffer (Invitrogen) separated on denaturing NuPAGE 4–12% polyacrylamide gradient gels, and transferred on to 0.2-μm-pore-size nitrocellulose membranes (Invitrogen). Membranes were blocked in 2% (w/v) non-fat dried milk powder in TBS-T (Tris-buffered saline: 20 mM Tris/HCl and 135 mM NaCl, pH 7.6 with 0.1% Tween 20), followed by incubation overnight at 4°C with the following antibodies against the following: caspase 11 (17D9, Novus Biologicals), c-IAP1 (1E1-1-10, Enzo Life Sciences) and β-actin (AC-74, Sigma). After washing with TBS-T, membranes were incubated with secondary antibodies for 1 h at room temperature (20°C). Enhanced chemiluminescence (Pierce) was used to visualize the blots on HyBlot CL Autoradiography film (Denville Scientific).
RT (reverse transcription)–PCR
c-IAP1 wild-type or c-IAP−/− MEFs were treated as indicated with LPS, and cells were then washed with PBS. Total RNA was isolated using the RNeasy minikit (Qiagen) following the manufacturer's instructions. RT with random hexamer primers and MultiScribe™ reverse transcriptase (Applied Biosystems) was performed on 1 μg of total RNA. Full-length caspase 11 cDNA was amplified using the primers 5′-ATGGCTGAAAACAAACACCCT-3′ and 5′-TCAGTTGCCAGGAAAGAGGTAG-3′. Products were resolved on 2% agarose gels and images captured on Gel Doc™ (Bio-Rad Laboratories). A 500 bp fragment of the caspase 11 sequence was amplified by PCR from genomic DNA prepared from cell pellets using the primers 5′-GTTGCCAGGCTCTGCTAAAA-3′ and 5′-TGCCCAGGAGTGGTATTATTG-3′.
RESULTS
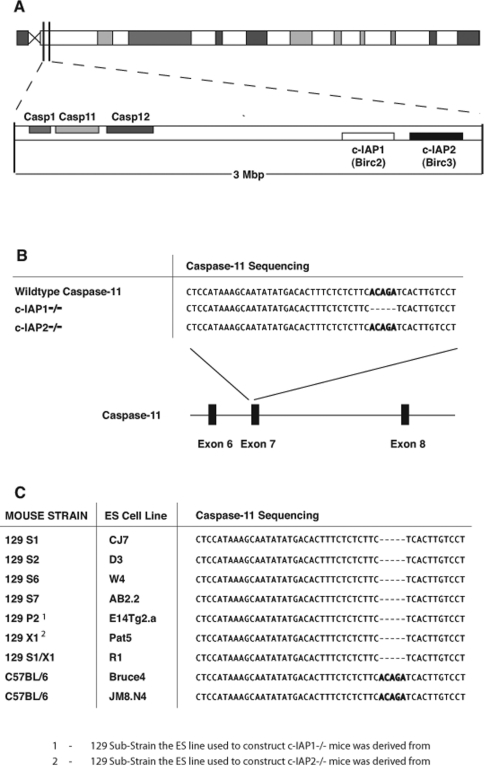
The genes encoding c-IAP1 and c-IAP2 lie in close proximity (~2.5 Mb) to that encoding caspase 11 on mouse chromosome 9 (Figure 1A). Since both c-IAP genes have been targeted previously using ES cell lines derived from 129 mice, we sought to examine the caspase 11 status of these mice. Sequence analysis from genomic DNA revealed that c-IAP1−/− mice also contain the mutant caspase 11 locus, despite the fact that these mice have been extensively backcrossed into the caspase 11 wild-type mouse stain, C57BL/6 (Figure 1B). In contrast, even with the close proximity of the c-IAP2 and caspase 11 genes, the c-IAP2−/− mice tested did not carry the caspase 11 deletion (Figure 1B).
Figure 1. Somatic mutation of caspase 11 in c-IAP1-deficient mice.
(A) Schematic of mouse chromosome 9 showing the relative positions of the caspase 1, caspase 11, caspase 12, c-IAP1 and c-IAP2 genes. Casp, caspase. (B) Alignment of the sequencing data of the exon 7 boundary from the wild-type caspase 11 locus, compared with sequencing from genomic DNA isolated from c-IAP1−/− and c-IAP2−/− mice. (C) Alignment of the sequencing data of the exon 7 boundary in the caspase 11 locus in cell lines from 129- and C57BL/6-derived ES cells.
Considerable heterogeneity exists within 129 mice used to derive many of the ES cell lines used for targeted mutagenesis, thus creating multiple individual substrains [1]. Although the c-IAP1 and c-IAP2 genes were both targeted in 129 ES cell lines, these lines were derived from different 129 substrains [12,14]. Therefore the unexpected differences between mice targeted for c-IAP1 and c-IAP2 might potentially have occurred due to differences in the ES lines used for their construction.
To explore the possibility the caspase 11 mutation might only occur in a subset of 129 strains, caspase 11 was sequenced in an extensive panel of 129-derived ES lines extending across different lineages. The relevant region of the caspase 11 gene was sequenced in multiple 129-derived ES cell lines, including the 129 P2 and 129 X1 lines which were used to target the c-IAP1 and c-IAP2 genes respectively [12,14]. All 129-derived ES cell lines tested harboured the mutation in the caspase 11 locus, suggesting that this is a true reflection of the 129 strain, rather than a genetic peculiarity of a lineage of substrains (Figure 1C). This suggests that the c-IAP2, but not the c-IAP1 locus, has separated from the mutant caspase 11 locus in the course of backcrossing. This mutation is specific to the 129 strain, because two ES cell lines derived from C57BL/6 mice, Bruce4 and JM8.N4, were found to be wild-type for the caspase 11 locus (Figure 1C).
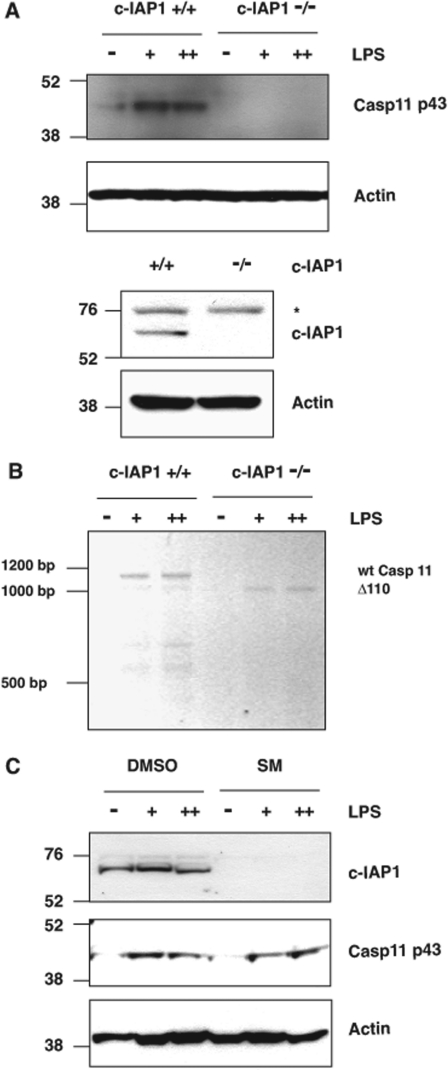
Caspase 11 cannot be readily detected in resting cells, but is induced by inflammatory stimuli such as LPS [4]. Caspase 11 induction in wild-type MEFs was compared with matched c-IAP1−/− littermates, in which c-IAP1 deficiency was confirmed by immunoblot analysis (Figure 2A). When MEFs were exposed to bacterial LPS, wild-type cells demonstrated a robust induction of pro-caspase 11, as predicted, but c-IAP1−/− MEFs lacked detectable caspase 11 (Figure 2A). To determine whether this was due to the splicing defect predicted by the 5 bp deletion described previously [2], rather than a c-IAP1-dependent defect in the signalling pathway driving caspase 11 expression, RT–PCR was performed on RNA harvested from wild-type or c-IAP1−/− MEFs, following LPS stimulation. Primers designed to amplify full-length caspase 11 mRNA recovered only a truncated form of caspase 11 cDNA in c-IAP1−/− MEFs (Δ110) exposed to LPS, as compared with the wild-type controls (Figure 2B), consistent with c-IAP1-deficient MEFs harbouring the somatic mutation in their caspase 11-coding sequence (Figure 1C).
Figure 2. c-IAP1−/− MEFs are unable to induce caspase 11 in response to LPS.
(A) c-IAP1+/+ or c-IAP1−/− MEFs were exposed to 0.1 mg/ml LPS (+) or 1 mg/ml LPS (++) for 6 h before lysis. Whole-cell lysates were prepared and Western blot analysis was conducted for the levels of caspase 11, c-IAP1 or β-actin. Molecular masses are indicated in kDa. The asterisk (*) indicates a non-specific protein recognized by the anti-c-IAP1 antibody. (B) c-IAP1+/+ or c-IAP1−/− MEFs were treated as in (A). RNA was extracted and subjected to RT–PCR analysis for full-length caspase 11. Sizes are indicated in bp. wt, wild-type. (C) Wild-type MEFs were pre-treated with the Smac (second mitochondrial-derived activator of caspase) mimetic AEG40730 (50 nM) for 30 min (SM), before being exposed to LPS for 6 h. Whole-cell lysates were prepared and Western blot analysis was conducted for the levels of caspase 11, c-IAP1 or β-actin. Molecular masses are indicated in kDa. Casp, caspase.
To exclude further the possibility that c-IAP1 participates in the LPS-dependent induction of caspase 11, MEFs were exposed to AEG40730, an IAP antagonist that rapidly depletes cells of c-IAP1, c-IAP2 and XIAP (X-linked inhibitor of apoptosis) [17,18]. As expected, c-IAP1 protein levels were abrogated in wild-type MEFs treated with the IAP antagonist, as confirmed by Western blot analysis (Figure 2C). Importantly, LPS-induced expression of caspase 11 was unaltered in MEFs chemically depleted of c-IAP1, suggesting that, in these cells, c-IAP1 is dispensable for the induction of pro-caspase 11 (Figure 2C).
DISCUSSION
The findings described above suggest that the mice created by targeting c-IAP1 in a 129-derived ES cell line [12] contain an added layer of complexity if used to investigate roles of c-IAP1 in innate immunity and programmed cell death, because of a concealed defect in the caspase 11 gene (Figure 1B). It should be noted that, in that original study, the primary phenotype observed in c-IAP1−/− cells was a marked stabilization of c-IAP2, a result confirmed subsequently by Darding et al. ([19] and references therein), indicating that this effect is independent of caspase 11 status. However, a role for c-IAP1 in the regulation of the innate immunity is not ruled out. Indeed, in Drosophila, an IAP orthologue termed DIAP2 (Drosphila IAP2) is critical for activation of innate immune responses mediated by IMD (immune deficiency), the Drosophila orthologue of mammalian RIPK1 (receptor-interacting serine/threonine protein kinase 1) [20–24]. Furthermore, the syntenic nature of the c-IAP, caspase 1 and caspase 11 genes may represent a functional gene cluster. This synteny is also conserved in humans with caspases 4 and 5, the human orthologues of murine caspase 11, closely linked to the c-IAP genes.
Intriguingly, the caspase 11 defect was not detected in the c-IAP2−/− mice, despite the close proximity of the genes encoding caspase 11, c-IAP1 and c-IAP2 on mouse chromosome 9. This was not due to differences in the cell lines used in their construction, since ES cells derived from both 129 P2 and the 129 X1 mice, which were used to target c-IAP1 and c-IAP2 genes respectively, contain the mutation in the caspase 11 gene (Figure 1C). This suggests that in c-IAP2−/− mice, the caspase 11 and c-IAP2 alleles have recombined over successive crosses into the C57BL/6 background. It is intriguing that all of the c-IAP2−/− mice are invariably homozygous for wild-type caspase 11, suggesting that the crossover event must have occurred at least twice within the population to become the predominant genotype in the c-IAP2−/− colony. These observations suggest that there may be a selective advantage for expression of the wild-type caspase 11 gene during murine development. Since the c-IAP1 locus is closer to that of caspase 11 (Figure 1A), the likelihood of this crossover event may be more rare, or the selective pressure for wild-type caspase 11 may only exist in the context of the c-IAP2 deletion.
Although the c-IAP2-deficient mice tested in the present study were found to be wild-type for caspase 11, the point at which the c-IAP2−/− and mutant caspase 11 loci separated is unknown. Clearly, in vivo studies to elucidate the role of c-IAP1 in the innate immune response, or indeed to study cell death, should ideally be performed in a background that is wild-type for caspase 11. Fortunately, this model currently exists. An elegant murine model has been described recently that facilitates independent or simultaneous inactivation of the linked c-IAP1 and c-IAP2 genes [25]. Importantly, these mice were generated using the C57BL/6 ES cell line Bruce4, which we have demonstrated does not carry the caspase 11 mutation (Figure 1C).
The present study has focused on the linkage between c-IAP1, c-IAP2 and the caspase 11 mutation carried from the constructed 129 ES cells. However, it is important to note that other genetic knockouts derived from the 129 ES cells may be inadvertently carrying the concealed mutation within the caspase 11 locus. Since the gene for caspase 12 (Casp12) is in close proximity to that of caspase 11 on mouse chromosome 9 (Figure 1A), it might be helpful to test mice experimentally targeted for caspase 12 constructed using a 129-derived ES cell line for the caspase 11 mutation [26]. Caspase 12 has subsequently been targeted in a C57BL/6-derived ES cell line [27], and this strain may be more appropriate for studying caspase 12 function as the wild-type status of caspase 11 has been confirmed [27].
AUTHOR CONTRIBUTION
Niall Kenneth, J. Michael Younger and Colin Duckett designed the research. Niall Kenneth and J. Michael Younger performed the research. Elizabeth Hughes and Thomas Saunders prepared and provided ES cell lines. Danielle Marcotte and Philip Barker prepared and provided genomic DNA from IAP-deficient mice. All authors analysed the data. Niall Kenneth and Colin Duckett wrote the paper.
ACKNOWLEDGEMENTS
We thank the University of Michigan DNA Sequencing Core for their assistance in sequencing the caspase 11 locus, and M. Saleh (McGill University), for providing detailed information of the caspase-12-targeted mice and a critical reading and discussion of the paper before submission. We are particularly grateful to members of the Duckett laboratory for a critical reading of the paper before submission.
FUNDING
This work was supported in part by the National Institutes of Health [grant number R01 CA142809] and the American Asthma Foundation (to C.S.D.), and a Training Grant Award [grant number T32 HL007517] (to J.M.Y.).
References
- 1.Simpson E. M., Linder C. C., Sargent E. E., Davisson M. T., Mobraaten L. E., Sharp J. J. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat. Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 2.Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 3.Yang I. V., Rutledge H. R., Yang J., Warg L. A., Sevilla S. D., Schwartz D. A. A locus on chromosome 9 is associated with differential response of 129S1/SvImJ and FVB/NJ strains of mice to systemic LPS. Mamm. Genome. 2011;22:518–529. doi: 10.1007/s00335-011-9340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang S., Miura M., Jung Y., Zhu H., Gagliardini V., Shi L., Greenberg A. H., Yuan J. Identification and characterization of Ich-3, a member of the interleukin-1β converting enzyme (ICE)/Ced-3 family and an upstream regulator of ICE. J. Biol. Chem. 1996;271:20580–20587. doi: 10.1074/jbc.271.34.20580. [DOI] [PubMed] [Google Scholar]
- 5.Wang S., Miura M., Jung Y. K., Zhu H., Li E., Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 6.Kang S. J., Wang S., Hara H., Peterson E. P., Namura S., Amin-Hanjani S., Huang Z., Srinivasan A., Tomaselli K. J., Thornberry N. A., et al. Dual role of caspase-11 in mediating activation of caspase-1 and caspase-3 under pathological conditions. J. Cell Biol. 2000;149:613–622. doi: 10.1083/jcb.149.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang S. J., Wang S., Kuida K., Yuan J. Distinct downstream pathways of caspase-11 in regulating apoptosis and cytokine maturation during septic shock response. Cell Death Differ. 2002;9:1115–1125. doi: 10.1038/sj.cdd.4401087. [DOI] [PubMed] [Google Scholar]
- 8.Kuida K., Lippke J. A., Ku G., Harding M. W., Livingston D. J., Su M. S., Flavell R. A. Altered cytokine export and apoptosis in mice deficient in interleukin-1β converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 9.Li P., Allen H., Banerjee S., Franklin S., Herzog L., Johnston C., McDowell J., Paskind M., Rodman L., Salfeld J., et al. Mice deficient in IL-1β-converting enzyme are defective in production of mature IL-1β and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 10.Srinivasula S. M., Ashwell J. D. IAPs: what's in a name? Mol. Cell. 2008;30:123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez J., Meier P. To fight or die: inhibitor of apoptosis proteins at the crossroad of innate immunity and death. Curr. Opin. Cell Biol. 2010;22:872–881. doi: 10.1016/j.ceb.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 12.Conze D. B., Albert L., Ferrick D. A., Goeddel D. V., Yeh W. C., Mak T., Ashwell J. D. Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo. Mol. Cell. Biol. 2005;25:3348–3356. doi: 10.1128/MCB.25.8.3348-3356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertrand M. J., Doiron K., Labbe K., Korneluk R. G., Barker P. A., Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–901. doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Conte D., Holcik M., Lefebvre C. A., Lacasse E., Picketts D. J., Wright K. E., Korneluk R. G. Inhibitor of apoptosis protein cIAP2 is essential for lipopolysaccharide-induced macrophage survival. Mol. Cell. Biol. 2006;26:699–708. doi: 10.1128/MCB.26.2.699-708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes E. D., Qu Y. Y., Genik S. J., Lyons R. H., Pacheco C. D., Lieberman A. P., Samuelson L. C., Nasonkin I. O., Camper S. A., Van Keuren M. L., Saunders T. L. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm. Genome. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 16.Rumble J. M., Bertrand M. J., Csomos R. A., Wright C. W., Albert L., Mak T. W., Barker P. A., Duckett C. S. Apoptotic sensitivity of murine IAP-deficient cells. Biochem. J. 2008;415:21–25. doi: 10.1042/BJ20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galbán S., Hwang C., Rumble J. M., Oetjen K. A., Wright C. W., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Duckett C. S. Cytoprotective effects of IAPs revealed by a small molecule antagonist. Biochem. J. 2009;417:765–771. doi: 10.1042/BJ20081677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Darding M., Feltham R., Tenev T., Bianchi K., Benetatos C., Silke J., Meier P. Molecular determinants of Smac mimetic induced degradation of cIAP1 and cIAP2. Cell Death Differ. 2011;18:1376–1386. doi: 10.1038/cdd.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gesellchen V., Kuttenkeuler D., Steckel M., Pelte N., Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005;6:979–984. doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huh J. R., Foe I., Muro I., Chen C. H., Seol J. H., Yoo S. J., Guo M., Park J. M., Hay B. A. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to Gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J. Biol. Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- 22.Kleino A., Valanne S., Ulvila J., Kallio J., Myllymaki H., Enwald H., Stoven S., Poidevin M., Ueda R., Hultmark D., et al. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leulier F., Lhocine N., Lemaitre B., Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist Gram-negative bacterial infection. Mol. Cell. Biol. 2006;26:7821–7831. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valanne S., Kleino A., Myllymaki H., Vuoristo J., Ramet M. Iap2 is required for a sustained response in the Drosophila Imd pathway. Dev. Comp. Immunol. 2007;31:991–1001. doi: 10.1016/j.dci.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Gardam S., Turner V. M., Anderton H., Limaye S., Basten A., Koentgen F., Vaux D. L., Silke J., Brink R. Deletion of cIAP1 and cIAP2 in murine B lymphocytes constitutively activates cell survival pathways and inactivates the germinal center response. Blood. 2011;117:4041–4051. doi: 10.1182/blood-2010-10-312793. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B. A., Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 27.Saleh M., Mathison J. C., Wolinski M. K., Bensinger S. J., Fitzgerald P., Droin N., Ulevitch R. J., Green D. R., Nicholson D. W. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature. 2006;440:1064–1068. doi: 10.1038/nature04656. [DOI] [PubMed] [Google Scholar]