Abstract
As part of the human gastrointestinal tract, the oral cavity represents a complex biological system and harbors diverse bacterial species. Unlike the gut microbiota which is often considered a health asset, studies of the oral commensal microbial flora have been largely limited to their implication in oral diseases such as dental caries and periodontal diseases; Little emphasis has been given to their potential beneficial roles, especially the protective effects against oral colonization by foreign/pathogenic bacteria. In this study, we used the salivary microbiota derived from healthy human subjects to investigate protective effects against the colonization and integration of Pseudomonas aeruginosa, an opportunistic bacterial pathogen, into developing and pre-formed salivary biofilms. When co-cultivated in saliva medium, P. aeruginosa persisted in the planktonic phase, but failed to integrate into salivary microbial community during biofilm formation. Furthermore, in the saliva medium supplemented with 0.05% (w/v) sucrose, the oral flora inhibited the growth of P. aeruginosa by producing lactic acid. More interestingly, while pre-formed salivary biofilms were able to prevent P. aeruginosa colonization, the same biofilms recovered from mild chlorhexidine gluconate treatment displayed a shift in microbial composition and showed a drastic reduction in protection. Our study indicates that normal oral communities with balanced microbial compositions could be important in effectively preventing the integration of foreign/pathogenic bacterial species, such as P. aeruginosa.
Keywords: bacterial interference, microbial flora, oral cavity, Pseudomonas aeruginosa, salivary biofilm
Introduction
The human oral cavity harbors over 700 different bacterial species and is one of the most complex ecosystems ever described (Paster, Boches et al. 2001; Aas, Paster et al. 2005; Paster, Olsen et al. 2006; Zaura, Keijser et al. 2009). Due to its accessibility, the oral microbial community has become one of the best studied human microbial systems (Kolenbrander and London 1993; Kolenbrander 2000; Kolenbrander, Andersen et al. 2002; Kuramitsu, He et al. 2007; Kolenbrander, Palmer et al. 2010). Extensive work has been done to 1) investigate the development and formation of multispecies oral microbial biofilms (Kolenbrander, Palmer et al. 2010), 2) reveal the antagonistic or synergistic inter-species interactions between resident bacteria within the community (Kuramitsu, He et al. 2007) and 3) evaluate the implications of interspecies interactions in oral diseases such as dental caries and periodontitis (Marsh 1994; Liljemark and Bloomquist 1996).
Accumulating evidence indicates that the indigenous microbial flora plays diverse roles in the host’s physiology. The gastrointestinal (GI)-tract associated microbiota, particularly the gut flora, has important roles in maintaining human health (Guarner and Malagelada 2003). Considered an “exteriorized organ”, the gut flora has been shown to contribute to human homeostasis with multiple functions, including harvesting energy (Sonnenburg, Xu et al. 2005), training the immune system (Cebra 1999), and protecting against epithelial cell injury (Rakoff-Nahoum, Paglino et al. 2004). More importantly, gut-associated microbial flora has also been implicated in preventing colonization of pathogenic microbes by bacterial interference (Guarner and Malagelada 2003). Unlike the gut flora, which is normally considered a health asset (O'Hara and Shanahan 2006), studies of the oral microbial flora, which is another important part of the GI-tract, has been largely limited to its implication in oral diseases such as dental caries and periodontal diseases. Although the protective roles of a healthy oral microbiotas against foreign/pathogenic bacteria have been suggested, the reports were largely descriptive of dual species antagonisms between a pathogen and a specific commensal bacterium (Uehara, Kikuchi et al. 2001; Uehara, Kikuchi et al. 2001); Demonstration of community-based interference and mechanistic studies are still lacking.
16S rRNA-based studies have revealed that despite the repeated exposure to a multitude of diverse bacterial species from different origins, the microbial compositions within oral cavities of healthy human subjects are relatively stable (Rasiah, Wong et al. 2005; Zaura, Keijser et al. 2009). While host factors have been implicated to play a significant role in shaping indigenous microbial communities (Rawls, Mahowald et al. 2006), increasing evidence also suggests that the established oral microbial community might develop invasion resistance mechanisms to protect its domain and maintain its stability. Using a mouse oral microbial community in vitro, we have recently demonstrated that an existing microbial community exerts bacterial interference effects and imposes a selective pressure on incoming foreign bacterial species, independent of host-mediated selection (“Community selection effect”); The latter may indeed play a significant role in maintaining the community stability and preventing the foreign colonization (He, Tian et al. 2010a; He, Tian et al. 2010b). In this study, we established in vitro healthy human subject-derived salivary biofilms and tested their ability to prevent the integration of P. aeruginosa, an opportunistic pathogen that can be isolated from certain oral infections (Nord, Sjöberg et al. 1972). Furthermore, the effect of shifts in biofilm microbial composition on invasive bacterial defense was investigated.
Materials and Methods
Bacterial strains and growth conditions
P. aeruginosa PAO1 (lab strain), Pa060928 (a clinical isolate from an adult cystic fibrosis (CF) patient (Kus, Tullis et al. 2004)), and salivary bacteria (S-mix) from healthy subjects were grown in salivary medium (75% (v/v) filter-sterilized saliva, 25% (v/v) brain-heart-infusion (BHI) broth). Cultures were incubated at 37°C under anaerobic condition (nitrogen 85%, carbon dioxide 5%, and hydrogen 10%). Kanamycin (150 µg/ml) was supplied in the medium to select P. aeruginosa when needed.
Saliva collection
Saliva samples were collected from 5 healthy subjects, age 25–40 under UCLA-IRB #09-08-068-02A. These individuals had no history of periodontal disease and had not used biocide-containing dentifrice for at least 6 months prior to saliva donation. Subjects had not been treated for any systemic disease nor were they taking any prescription or non-prescription medications.
Saliva collection for preparing saliva medium
Subjects were asked to refrain from any food or drink 2 hours prior to donating saliva. 10 ml of spit derived saliva was collected from each person in collection tubes. Saliva samples were pooled together and centrifuged at 14,000 g for 5 min. The supernatant was filter sterilized and used for preparing saliva medium and pre-coating 12-well plates.
Saliva collection for starting biofilms
On a separate day, subjects were asked to donate saliva; 2 ml was collected from each person as described in the previous section. Saliva samples were pooled together and centrifuged at 2,600 × g for 10 min to spin down large debris and eukaryotic cells. The supernatant containing salivary bacteria was referred to as S-mix and was used for setting up co-cultivation assays with P. aeruginosa and starting salivary biofilms.
Isolation and identification of bacterial species from salivary samples
Pooled saliva was diluted in SHI medium, an optimal medium for culturing oral bacteria that has been shown to be able to sustain the growth of highly diverse in vitro microbial communities with similar microbial profiles to the original salivary microflora (Tian, He et al. 2010). The diluted saliva was then seeded on SHI agar plates. The plates were incubated for 5 days at 37°C under anaerobic conditions. Potentially different bacterial colonies were picked from the plate based on differences in colony morphology, pigmentation, and the incubation time needed for colonies to appear. Individual colonies were grown in SHI medium at 37°C under anaerobic conditions until turbid. Bacterial cells were collected, frozen stock of each isolate was made and genomic DNA of each isolate was prepared using the MasterPure™ DNA purification kit (Epicentre)
For species identification, the universal bacterial 16S rDNA primer pair, 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5'-GGTTACCTTGTTACGACTT-3') (Martin-Laurent, Philippot et al. 2001), was used to generate an approximately 1,500-bp amplicon. Each 50 µl PCR reaction mixture contained 10 ng of genomic DNA, 200 µM of each dNTP, 4.0 mM MgCl2, 100 nM of each primer, 5 µl of 10× PCR buffer, and 2.5 U of Taq polymerase (Invitrogen). PCR conditions were as follows: 3 min at 94°C for initial denaturation and 27 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min and a final chain elongation at 72°C for 5 min. PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced at the UCLA Core DNA Sequencing Facility using 27F and 1492R primers. Obtained sequences were subjected to nucleotide BLAST searches against the NCBI (http://blast.ncbi.nlm.nih.gov/) and Human Oral Microbiome Database (http://www.homd.org/index.php). Sequences with 98% to 100% identity to those deposited in the databases were considered to be positive for taxa identification.
Co-aggregation between P. aeruginosa and oral isolates
Co-aggregation assays were performed both in saliva medium and CAB buffer which contains 150 mM NaCl, 1 mM Tris/HCl pH 8, 0.1 mM CaCl2, 0.02% NaN3, and 0.1 mM MgCl2 as previously described (Cisar, Kolenbrander et al. 1979). Bacterial cells were collected in mid-exponential phase of growth, washed and re-suspended in CAB (or saliva medium) to a final OD600nm of 1. Equal volume of different bacterial species was added to the reaction tube, vortex-mixed for 10 s and graded on a 0–4 scale after 10 min based on the degree of co-aggregation (Kaplan, Lux et al. 2009). A score of 0 was assigned for no visible co-aggregation and a score of 4 for complete sedimentation with a clear supernatant. No clumping of individual bacterial strain was observed in our experimental controls.
Co-cultivation of salivary bacteria and P. aeruginosa (PAO1 or Pa060928) in the presence/absence of sucrose or lactic acid
To enhance the attachment of oral microbes to the wells, 12-well plates were pre-coated with saliva as previously described (Tian, He et al. 2010). Briefly, 100 µl of filtered saliva with equal amounts of PBS was added to each well of the 12-well plate to pre-coat the wells. Plates were incubated at 37 °C with their lids open for 1 hour to dry the saliva coating, followed by sterilization under UV light for 1 hour. 150 µl of fresh pooled saliva (S-mix) containing ~ 106 bacterial cells together with ~ 104 P. aeruginosa (PAO1 or Pa060928) cells, as determined by cell counting using Petroff-Hausser chamber (Hausser Scientific, PA, USA), was inoculated into pre-coated wells containing 800 µl of salivary medium supplemented with/without 0.02% and 0.05% (w/v) sucrose, or 0.02% (w/v) lactic acid. Plates were incubated at 37 °C under anaerobic conditions for 24 hours before samples (both planktonic and biofilm portion) were taken for viability counting, DNA isolation and PCR-Denaturing Gradient Gel Electrophoresis (DGGE) analysis.
Challenge of CHX (chlorhexidine gluconate)-treated salivary biofilm with P. aeruginosa PAO1
-
Establishment of salivary biofilms:
150 µl of pooled saliva was inoculated into pre-coated wells (of 12-well plates) containing 850 µl of salivary medium. Plates were incubated at 37°C under anaerobic condition overnight to allow biofilm formation.
-
CHX treatment of biofilm or/and challenge of biofilm with P. aeruginosa:
Depending on whether they would be subjected to CHX treatment/challenge with P. aeruginosa, overnight salivary biofilms were divided into three groups. Group 1 (CHX-treated only): planktonic portion was carefully removed from each well without disturbing the biofilm. 300 µl of 0.01% (w/v) CHX was added to each well, incubated for 15 or 30 seconds at room temperature before being carefully removed. Wells were immediately washed 5 times with 500 µl of PBS followed by adding 1ml of salivary medium; Group 2 (P. aeruginosa-challenged only): After removing the planktonic portion, 1 ml of salivary medium was added to the wells; Group 3 (CHX-treated and P. aeruginosa-challenged): following CHX treatment and washing with PBS 5 times, 1 ml of salivary medium was added to the wells. The 12-wells plates containing the above 3 groups were incubated at 37°C under anaerobic conditions for 24 hours to allow the CHX-treated group to recover. After 24-hour incubation, the planktonic portion of all 3 groups was removed, followed by the addition of 1 ml of salivary medium to each well in Group 1 and 1 ml of salivary medium containing ~105 P. aeruginosa cells to the wells in Group 2 and Group 3. The 12-well plates containing the above 3 groups were further incubated at 37°C under anaerobic condition for 48 hours, samples (both planktonic and biofilm portion) were taken at 24 and 48 hours and subjected to viability counts, DNA isolation and PCR-DGGE analysis.
Collection of planktonic and biofilm samples for viability counting and total DNA isolation
Samples were taken as follows: To collect the planktonic portion, the supernatant in the well was transferred to a 2 ml microfuge tube, 500 µl of PBS was then used to gently wash the biofilm to collect loosely attached cells and combined with the supernatant. To harvest the biofilm portion, 500 µl of PBS was added to the well, and a sterile spatula was used to meticulously scrape off biomass of the biofilm from the bottom of the wells. Collected cells were vortexed, 50 µl of bacterial solution was taken for each sample, subjected to serial dilution and seeded onto selective (Kanamycin 150 µg/ml) and non-selective SHI agar plates. Plates were incubated at 37°C under anaerobic condition for 5 days before colonies were counted to determine their CFU per mL. The rest of the bacterial cells collected from the planktonic and biofilm portion were spun down at 14,000 × g for 5 min, and the cell pellets were further subjected to DNA isolation.
Ethidium monoazide bromide (EMA) cross-linking
To prevent amplification of DNA from dead bacterial cells and to limit DNA-based PCR-DGGE community analysis to the viable fraction, the collected bacterial samples were treated with EMA prior to DNA extraction. EMA cross-linking was performed as described previously (Nocker and Camper 2006). Briefly, EMA (Biotium, Hayward, CA, USA) was dissolved in water to a stock concentration of 5 mg/ml and stored at −20°C in the dark. EMA was added to the culture samples to a final concentration of 100 µg/ml and samples were incubated in the dark for 5 min with occasional mixing before samples were incubated on ice and light-exposed for 1 min using a 650-W halogen light source placed about 20 cm from the samples. After photo induced cross-linking, bacterial cells were collected by centrifugation at 5,000×g for 5 min, followed by total genomic DNA isolation.
PCR-DGGE analysis
Total genomic DNA of bacterial samples was isolated using the MasterPure™ DNA purification kit (Epicentre). DNA quality and quantity were determined by a Spectronic Genesys UV spectrophotometer at 260 nm and 280 nm (Spectronic Instrument, Inc. Rochester, New York, USA)
Amplification of bacterial 16S rRNA genes by PCR was carried out as described previously by Li et al (Li, Ku et al. 2005). Briefly, the universal primer set, Bac1 (5'-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGACTACGTGCCAGCAGCC-3) and Bac2 (5'-GGACTACCAGGGTATCTAATCC-3') (Sheffield, Cox et al. 1989), was used to amply an approximately 300-bp internal fragment of the 16S rRNA gene. Each 50-µl PCR reaction contained 100 ng of purified genomic DNA, 40 pmol of each primer, 200 µM of each dNTP, 4.0 mM MgCl2, 5 µl of 10X PCR buffer, and 2.5 U of Taq DNA polymerase (Invitrogen). Cycling conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 56°C for 1 min and 72°C for 30 sec, with a final extension period of 5 min at 72°C. The resulting PCR products were evaluated by electrophoresis in 1.0 % agarose gels.
Polyacrylamide gels at an 8% concentration were prepared with a denaturing urea/formamide gradient between 40% (containing 2.8 M urea and 16 % (v/v) formamide) and 70% (containing 4.9 M urea and 28 % (v/v) formamide). Approximately 300 ng of the PCR product were applied per lane. The gels were submerged in 1 × TAE (Tris-Acetate-EDTA) buffer (40 mM Tris base, 40 mM glacial acid acetic, 1 mM EDTA) and the PCR products were separated by electrophoresis for 17 hours at 58°C using a fixed voltage of 60 V in the Bio-Rad DCode System (Bio-Rad laboratories, Inc. Hercules, CA, USA). After electrophoresis, the gels were rinsed and stained for 15 min in 1 × TAE buffer containing 0.5 µg/ml ethidium bromide, followed by 10 min of de-staining in 1 × TAE buffer. DGGE profile images were digitally recorded using the Molecular Imager Gel Documentation system (BioRad, Hercules, CA, USA). Diversity Database Software (BioRad) was used to assess the change in the relative intensity of bands corresponding to bacterial species of interest.
Identification of bacterial species from DGGE gel
The DNA bands of interest were excised from the DGGE gels and transferred to a 1.5 ml microfuge tube containing 20 µl of sterile ddH2O. Tubes were incubated at 4°C overnight before the recovered DNA samples were re-amplified with the universal primer set (Bac1 and Bac2). PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced at the UCLA Core DNA Sequencing Facility. Obtained sequences were subjected to nucleotide BLAST searches against the NCBI (http://blast.ncbi.nlm.nih.gov/) and Human Oral Microbiome (http://www.homd.org/index.php) Databases.
HPLC-ESI-MS Analysis for detecting lactic acid in spent media
Overnight cultures of salivary bacteria in saliva medium supplied with and without 0.05% (w/v) sucrose were harvested by centrifugation, spent media were collected, filtered through Millex®GP membrane (0.22 µm pore size, Millipore, Billerica, USA), and subjected to high performance liquid chromatography-mass spectrometry (HPLC/MS) analysis.
HPLC/MS was performed on a Waters 2767 HPLC separation module equipped with a XBridge BEH130 C18 column (5 µm, 4.60 × 100 mm) in tandem with a Waters 3100 mass detector (Waters, Milford, MA, USA). Separation of lactic acid was achieved using a gradient solvent system consisting of ddH2O-acetonitrile-trifluoric acid at 0.6 ml/min in the proportions 94.95:5:0.05 to 89.95:10:0.05 over 7 min, and these conditions were held for 5 min before returning to the initial conditions over 5 min, and equilibrating for 5 min. In-line MS was performed using a Waters 3100 single-quadrupole mass spectrometer with electrospray ionization (ESI) in the negative mode, specifically with a capillary voltage of −3 kV and cone voltage of −30 V. Nitrogen was used for both the cone gas (160 l/h) and desolvation gas (650 l/hr), with the source and desolvation temperatures being held at 150°C and 350°C, respectively. Total ion current (TIC) chromatograms of samples ranging between 50 and 400 m/z were displayed, and selected ion monitoring (SIM) chromatograms were used to record the abundance of the deprotonated molecule of lactic acid ([M−H]−) at m/z 89. Lactic acid in the spent medium was identified by comparing both the retention time and mass spectrum (m/z 50–110) with that of the standard lactic acid. The quantitation was based on the abundance of identified peak area in SIM chromatogram of each sample against the standard lactic acid (from 0.3 mM to 16.5 mM).
RESULTS
P. aeruginosa PAO1 was ineffective in integrating into oral microbial community during biofilm formation
Our previous in vitro study demonstrated that mouse oral cavity-derived microbial flora was able to prevent the integration of bacterial species from foreign origin (He, Tian et al. 2010a). In an effort to further investigate human salivary microbiota’s protective ability, the opportunistic pathogen P. aeruginosa PAO1 was initially chosen as a representative non-oral resident and co-cultivated with salivary bacteria (S-mix).
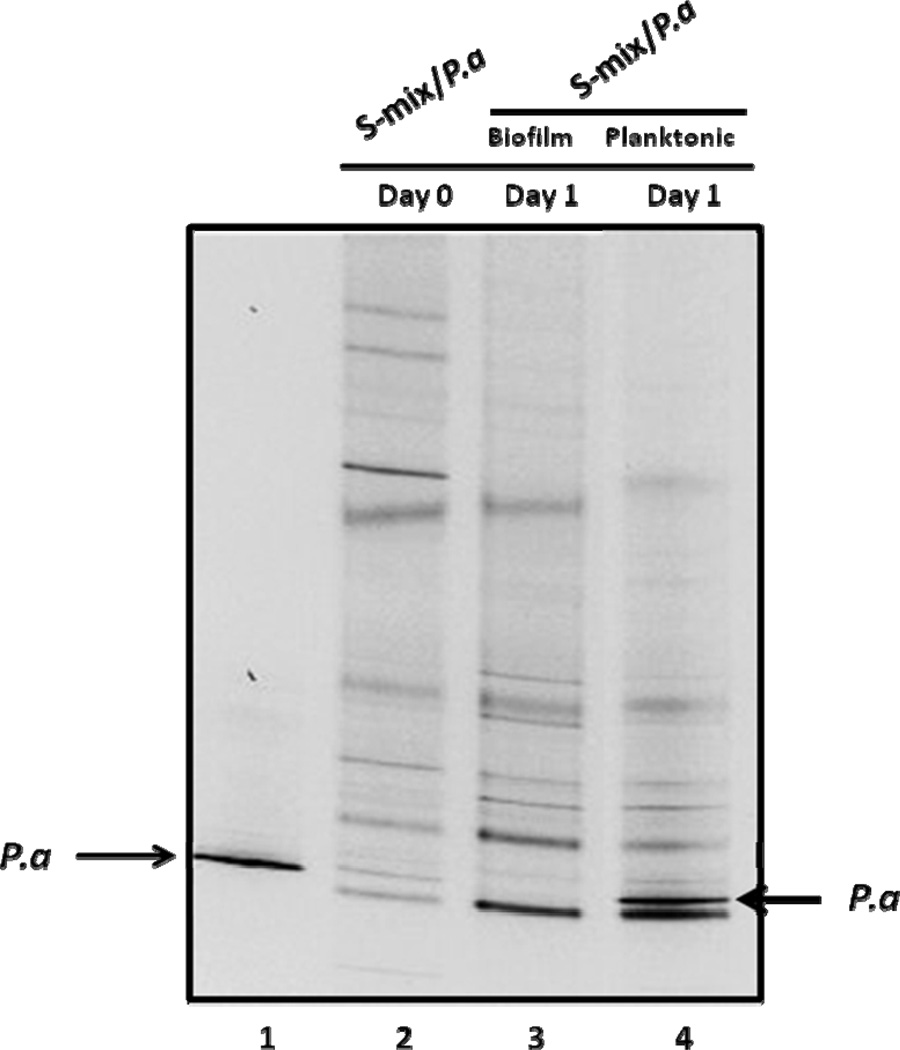
After 24-hour incubation, the co-culture resulted in the formation of biofilm attached to the bottom of the well, as well as unattached bacterial cells in the planktonic portion. PCR-DGGE analysis revealed that the banding pattern of salivary bacteria within biofilm sample was very close to that of the original saliva sample, indicating most of the salivary bacterial species were capable of integrating into surface-attached multi-species biofilm (Fig. 1, lane 2 and 3). P. aeruginosa PAO1 was capable of forming biofilm by itself under the condition used (Suppl. Fig. 1). However, when co-cultivated with S-mix, its corresponding band was not easily detectable in the biofilm portion (Fig. 1, lane 3). Separately, there was a drastic increase in the relative intensity of the same band within the planktonic portion compared to initial inoculums (Fig. 1, lanes 4). Our results suggested that although P. aeruginosa PAO1 was able to persist when co-cultivated with salivary bacteria, it was ineffective in integrating and becoming an associated member of surface-attached oral biofilm.
Figure 1.
PCR-DGGE analysis of microbial profiles obtained from biofilm and planktonic portion of the co-culture of salivary bacteria (S-mix) and P. aeruginosa (Pa) after 24 hours’ co-incubation. S-mix/Pa indicates co-culture of S-mix and Pa. The arrows indicate the DNA bands corresponding to P. aeruginosa. Two biological replicates were performed and a representative gel image is shown.
Effect of sucrose on the growth of P. aeruginosa PAO1 within the co-culture
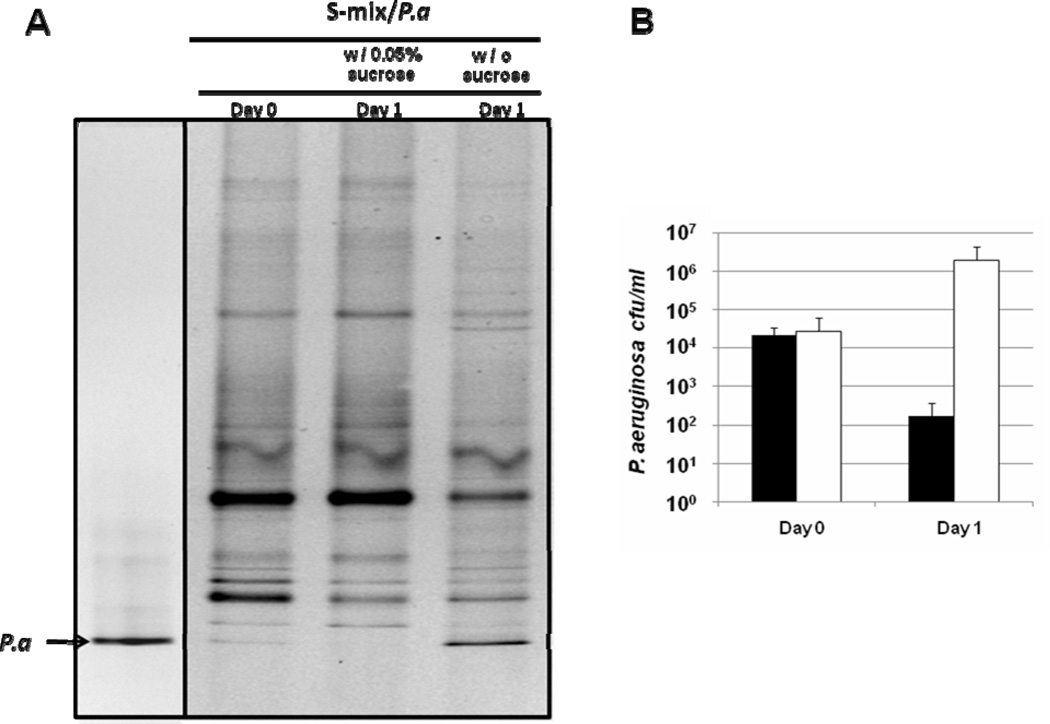
Oral bacteria are frequently exposed to different fermentable carbohydrates. Since sucrose is one of the most often consumed sugars by human, we were interested in investigating its effect on the growth of P. aeruginosa when co-cultivated with salivary bacteria (S-mix). After 24-hour incubation, the cells from the biofilm and planktonic portion of the same well were combined and subjected to PCR-DGGE analysis. Results showed that the addition of 0.05% (w/v) sucrose did not cause significant change in banding pattern of salivary bacteria within the co-cultures. However, the intensity of the band representing P. aeruginosa PAO1 was drastically reduced in the co-culture containing 0.05% sucrose (Fig. 2A), suggesting the viability of P. aeruginosa was severely affected. Viable count data showed that 24-hour incubation with S-mix in the presence of sucrose resulted in more than 100-fold reduction in the total viable count of P. aeruginosa; while in the absence of sucrose, the viable count of P. aeruginosa increased about two orders of magnitude (Fig. 2B). Since we had already shown that P. aeruginosa PAO1 was ineffective in integrating into salivary biofilms (Fig. 1), the detected P. aeruginosa most likely existed in the planktonic phase. It is worthwhile to note that the growth of P. aeruginosa was not affected when grown alone in the salivary medium supplemented with 0.05% sucrose, and the addition of sucrose resulted in a slight increase in the total viable counts of salivary bacteria compared with co-cultures without sucrose (data no shown).
Figure 2.
Effect of sucrose on the growth of P. aeruginosa within the co-culture. (A) Salivary bacteria (S-mix) and P. aeruginosa (Pa) was co-cultivated in the presence and absence of 0.05% (w/v) sucrose for 24 hours, DNA from total viable cells (including biofilm and planktonic portion) were isolated and subjected to PCR-DGGE analysis. (B) Total co-culture samples (including planktonic and biofilm cells) were subjected to serial dilution and plated onto selective plates. Viable counts were monitored for P. aeruginosa. Black bars represent P. aeruginosa in the co-culture supplemented with sucrose; open bars represent P. aeruginosa in the co-culture without sucrose. Two biological replicates were performed and a representative gel image is shown. Three replicates were performed for each viability count assay. Average values + SD are plotted.
Detection of lactic acid in the spent medium of salivary microbiota
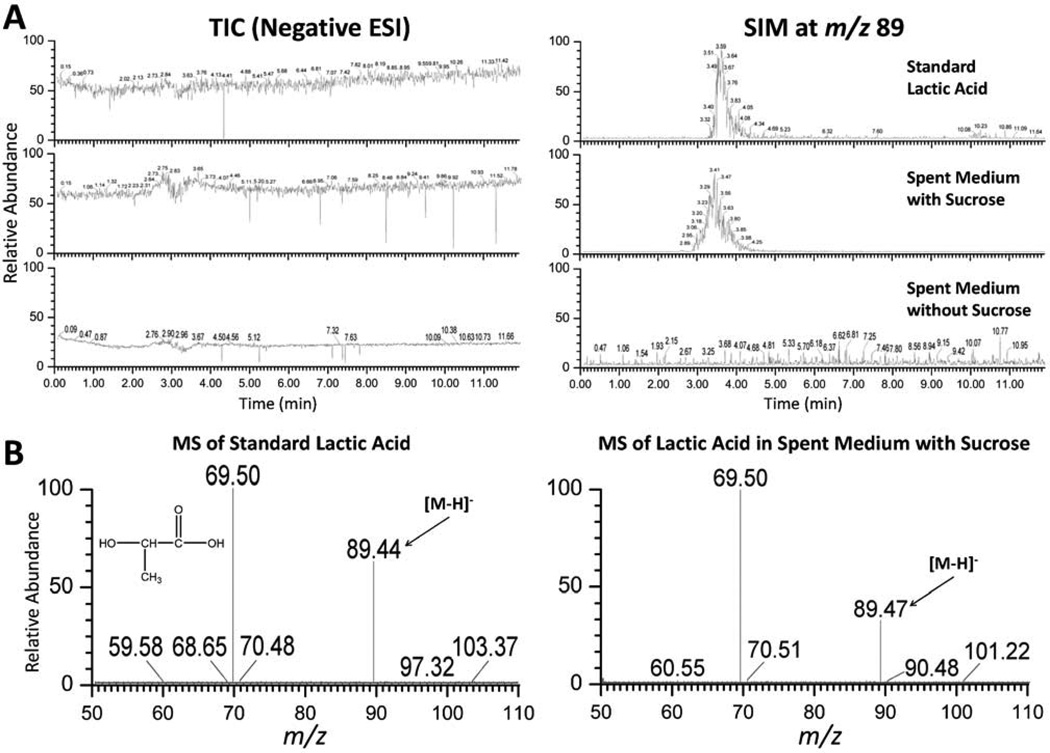
Many oral bacterial species, including Streptococcus spp. and Lactobacillus spp. are able to produce lactic acid as the major metabolic end-product of carbohydrate fermentation. Due to its antimicrobial property, particularly towards gram-negative bacteria, we reasoned that lactic acid might play a role in inhibiting the growth of P. aeruginosa cells when co-cultivated with salivary bacteria in the presence of sucrose. To test this, we performed HPLC/MS analysis on the spent medium of salivary bacteria grown in the salivary medium with and without sucrose. Lactic acid was detected in the spent medium supplemented with 0.05% sucrose by comparing its SIM chromatogram at m/z 89 (Fig. 3A) and the mass spectrum (Fig. 3B) with that of standard lactic acid (Fig. 3). After calculating the corresponding peak area against the standard, the concentration of lactic acid within spent medium (with sucrose) was determined to be 2.71±0.95 mM; sufficient to inhibit the growth of P. aeruginosa (data not show). Meanwhile, the absence of sucrose resulted in no significant amount of lactic acid detected in the spent medium (Fig. 3A). It is worthwhile to point out that the pH of the fresh saliva medium was around 7.1, while the spent saliva medium containing 0.05% sucrose had a pH of ~6.6. We also showed that saliva medium with adjusted pH of 6.6 didn’t significantly affect the growth of P. aeruginosa compared to the original saliva medium (data not shown).
Figure 3.
HPLC/MS analysis of the spent medium of oral flora grown in salivary medium supplemented with and without 0.05% (w/v) sucrose. (A) TIC (left panel) and SIM (right panel, with deprotonated molecular ion 89 m/z) chromatograms: top, a standard lactic acid sample at 3.25 mM prepared in ddH2O; middle, spent medium with 0.05% sucrose; bottom, spent medium without sucrose. (B) ESI-MS of standard lactic acid (left) and corresponding peak in the spent medium with 0.05% sucrose (right).
Inhibitory effect of lactic acid on the growth of P. aeruginosa PAO1 when co-cultivated with salivary bacteria
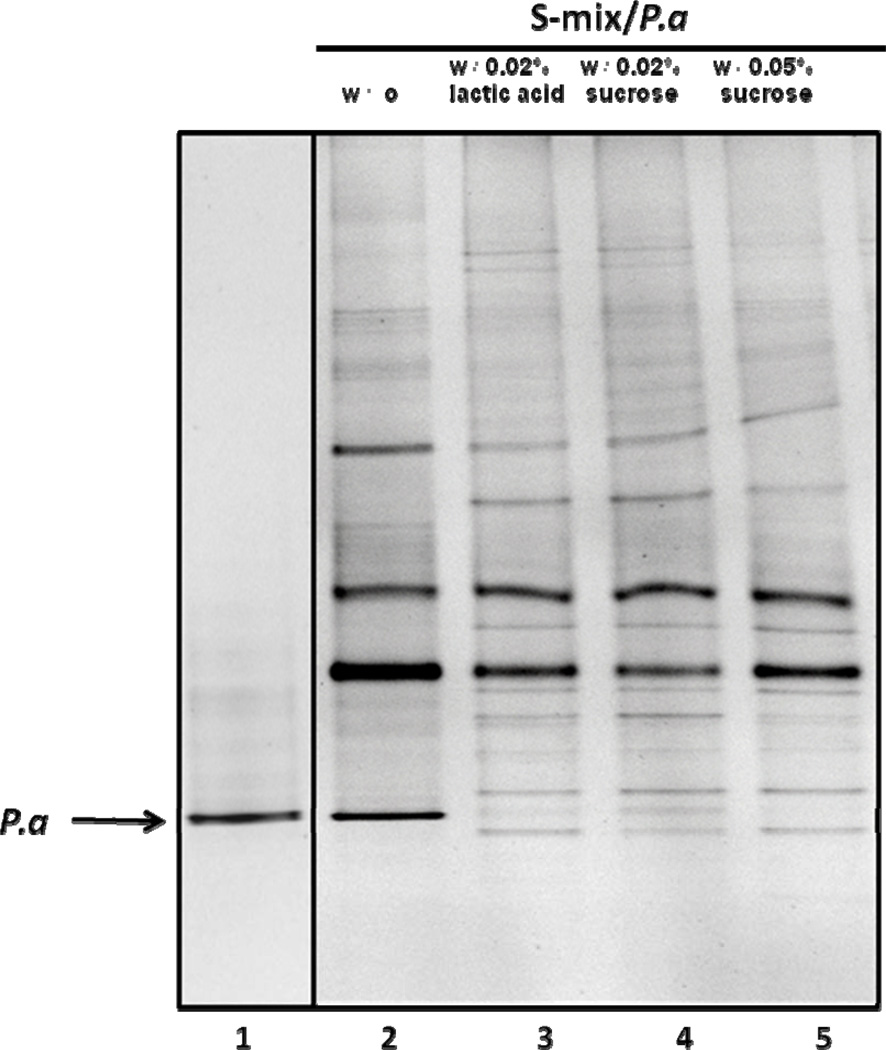
To further confirm the inhibitory effect of lactic acid towards P. aeruginosa, we added lactic acid to the P. aeruginosa PAO1/salivary bacteria co-culture to a final concentration 0.02% (2.2 mM). After 24-hour incubation, cells within the same well (including biofilm and unattached planktonic cells) were combined and subjected to PCR-DGGE analysis. The data revealed that, the sample from co-culture without addition of either sucrose or lactic acid contained the band representing P. aeruginosa PAO1 with high intensity (Fig. 4, lane 2), while samples prepared from co-cultures with the addition of sucrose or lactic acid displayed much weaker P. aeruginosa band (Fig. 4, lane 3–5).
Figure 4.
Effect of lactic acid on the growth of P. aeruginosa within co-culture. Salivary bacteria (S-mix) and P. aeruginosa (Pa) was co-cultivated in the absence (lane 2), or the presence of 0.02% lactic acid (lane 3), 0.02% (lane 4) or 0.05% (lane 5) sucrose for 24 hours, DNA from total viable cells (including planktonic and biofilm) were isolated and subjected to PCR-DGGE analysis. Arrow indicates the band of P. aeruginosa. Two biological replicates were performed and a representative gel image is shown.
Pa060928, a clinical P. aeruginosa isolate was ineffective in integrating into salivary biofilm and suffered viability loss when co-cultivated with salivary bacteria in the presence of sucrose
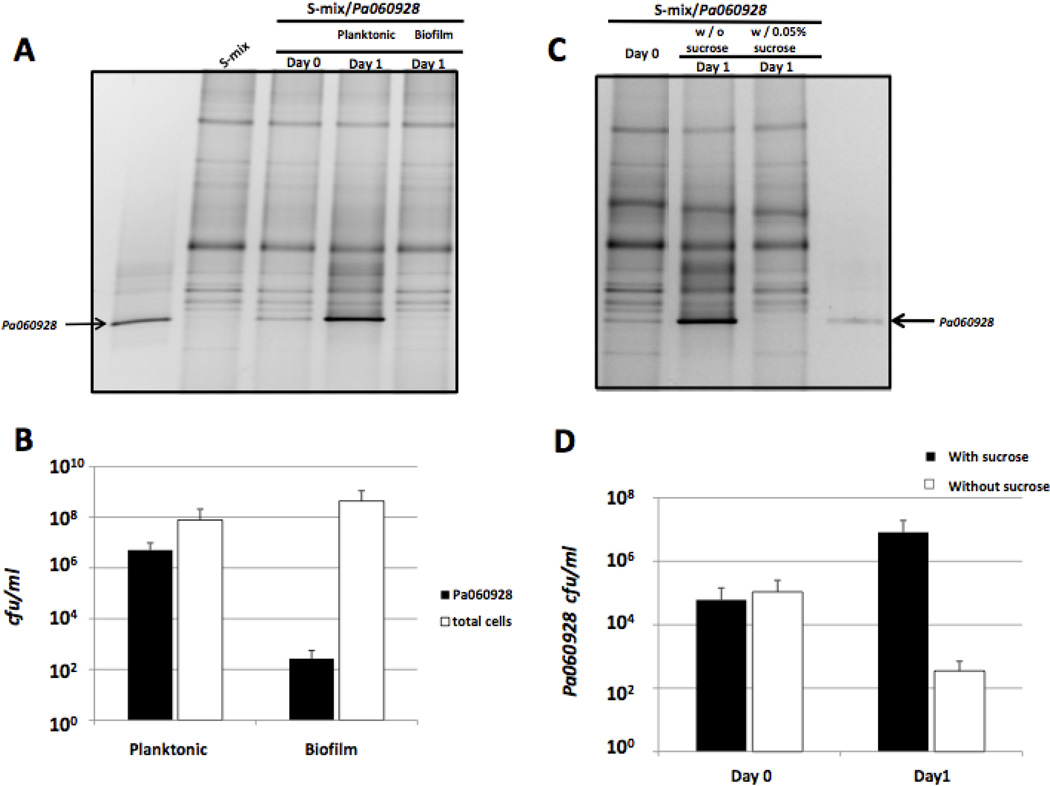
To further demonstrate that salivary bacterial community was effective in preventing the integration of pathogenic species, a clinical P. aeruginosa isolate (Pa060928) from an adult cystic fibrosis patient, was co-cultivated with salivary flora. PCR-DGGE analysis revealed that the band representing Pa060928 became the most dominant one within the planktonic portion, while it was not easily detectable in the biofilm portion (Fig 5A). This is corroborated by the viability data, which showed that after 24-hour co-cultivation, the majority of Pa060928 cells stayed in the planktonic phase, while those found within biofilm only accounted for less than 0.01% of the total Pa cells (Fig 5B). When sucrose was added to the co-culture medium, Pa060928 suffered drastic viability loss as revealed both by DGGE analysis and viable count (Fig 5C and 5D), a phenomenon similar to what had been observed when PAO1 strain was tested.
Figure 5.
Co-cultivation of Pa060928 with salivary bacteria (S-mix) in the absence and presence of sucrose. (A) PCR-DGGE analysis of microbial profiles obtained from biofilm and planktonic portion of the co-culture after 24-hour co-incubation. S-mix/Pa060928 indicates co-culture of S-mix and Pa060928. The arrows indicate the DNA bands corresponding to Pa060928. Three biological replicates were performed and a representative gel image is shown. (B) Viability count of Pa060928 and total bacteria within planktonic and biofilm portions after 24-hour incubation in saliva medium. Three replicates were performed for each viability count assay. Average values + SD are plotted. (C) PCR-DGGE analysis of total bacterial profile (including planktonic and biofilm portion) after 24-hour co-cultivation in the absence and presence of 0.05% sucrose. Three biological replicates were performed and a representative gel image is shown. (D) Viability count of Pa060928 within the co-culture (combining planktonic and biofilm portion) after 24-hour incubation in the absence and presence of sucrose. Black bars represent Pa060928 in the co-culture without sucrose; open bars represent Pa060928 in the co-culture supplemented with 0.05% sucrose. Three replicates were performed for each viability count assay. Average values + SD are plotted.
Co-aggregation between P. aeruginosa and oral isolates
In an effort to test the co-aggregation ability of P. aeruginosa with oral bacteria, we isolated major bacterial species from saliva samples of healthy subjects, and their ability to adhere to P. aeruginosa (both the lab strain PAO1 and the clinical isolate Pa060928) was determined by a co-aggregation assay. Results showed that all the tested salivary bacterial isolates, including Streptococcus spp, Lactobacillus spp., Actinomyces spp. and Prevotella spp. demonstrated substantial levels (with co-aggregation scores of 2 to 4) of interspecies co-aggregation with F. nucleatum, while no detectable co-aggregation was observed between P. aeruginosa (both PAO1 and Pa060928 strains) and oral isolates, including F. nucleatum (Table 1). Similar results were observed when co-aggregation assay was performed in saliva medium (Suppl. Table 1).
Table1.
Co-aggregation between P. aeruginosa and oral isolatesa in co-aggregation buffer
| Strains | F. nucleatum (oral isolate) | P. aeruginosa PAO1 | Pa060928 |
|---|---|---|---|
| Gram positive oral isolate | |||
| Streptococcus mitis | 4 | 0 | 0 |
| Streptococcus salivarius | 4 | 0 | 0 |
| Streptococcus australis | 3 | 0 | 0 |
| Streptococcus parasanguinis | 4 | 0 | 0 |
| Lactobacillus casei | 2 | 0 | 0 |
| Actinomyces naeslundii | 3 | 0 | 0 |
| Gram negative oral isolate | |||
| Prevotella denticola | 2 | 0 | 0 |
| F. nucleatum | 0 | 0 | 0 |
| Non-oral isolate | |||
| E. coli | 0 | 0 | 0 |
The method for assigning co-aggregation scores is described in Materials and Methods section.
CHX-induced shift in microbial profile within salivary biofilms resulted in reduced defense against P. aeruginosa integration
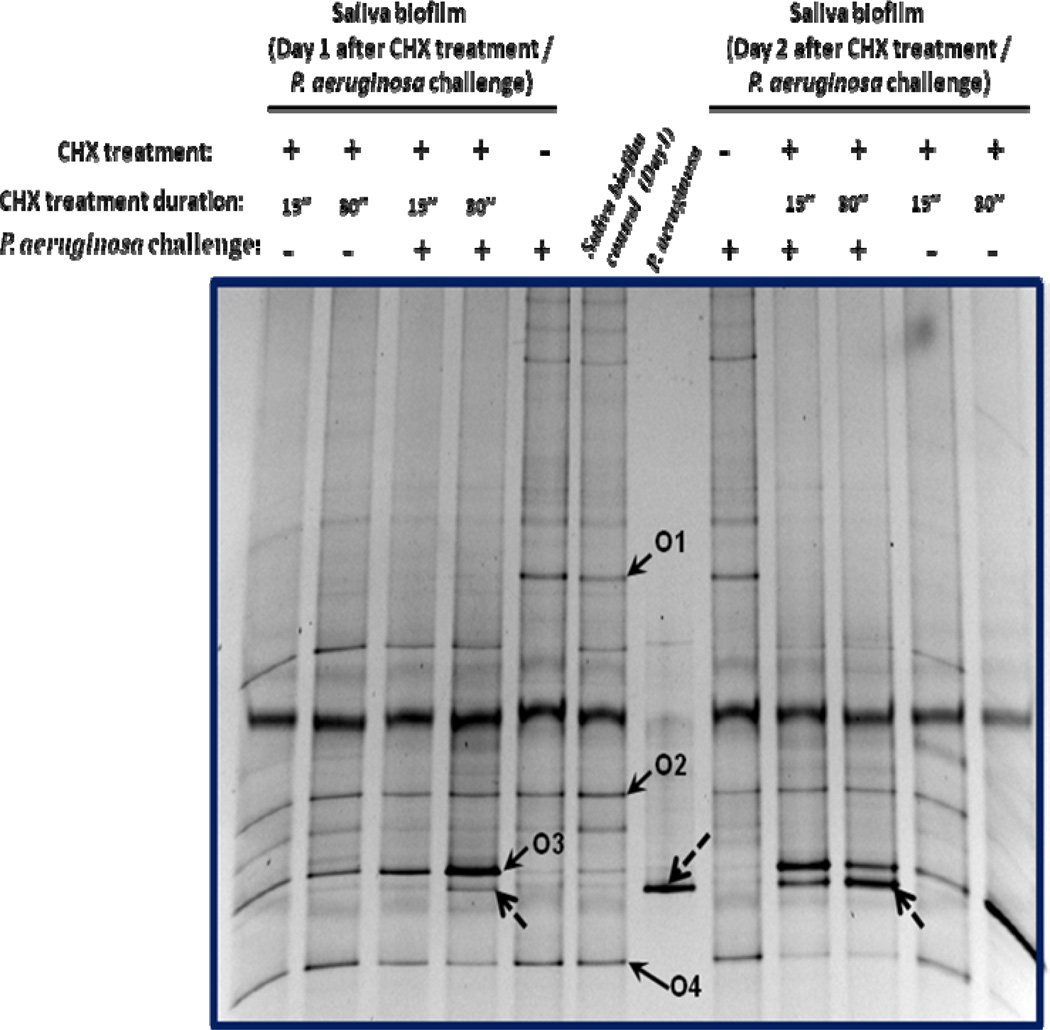
In an effort to test the protective capability of pre-formed salivary biofilm against integration by foreign bacteria, we challenged 24 hour-old saliva biofilms with P. aeruginosa PAO1 and monitored the microbial profiles using PCR-DGGE analysis. Results showed that even after 48-hour co-incubation, there was no detectable P. aeruginosa band within the salivary biofilm (Fig. 6: samples with CHX treatment (−) and P. aeruginosa challenge (+)), suggesting the pre-formed biofilms were effective in preventing P. aeruginosa integration.
Figure 6.
PCR-DGGE analysis of CHX-treatment effect on biofilm’s capability to prevent P. aeruginosa PAO1 colonization. Pre-formed (24 hour-old) saliva biofilm was subjected to single-dose CHX treatment for 15 or 30 sec, followed by 24 hours re-growth. The recovered biofilm was challenged with P. aeruginosa and followed by extended incubation for 48 hours. The microbial profiles of the biofilms were monitored by PCR-DGGE analysis. CHX treatment or P. aeruginosa challenge is indicated by (+), while (−) represents samples without CHX treatment or P. aeruginosa challenge. Bacterial species corresponding to the bands of interest were identified a s : O 1—Streptococcus spp. ; O 2—Porphyromonas spp. ; O 3—Neisseria spp. ; O 4—Peptostreptococcus stomatis. Doted arrow indicates P. aeruginosa PAO1. Two biological replicates were performed and a representative gel image is shown.
We further investigated the effect of shifts in microbial composition within the same biofilms on their defense capability. By mildly treating pre-formed salivary biofilms with diluted (0.01%) chlorhexidine gluconate (CHX) followed by 24-hour regrowth, we generated a community with a drastic shift in the microbial profile and reduction (less than 20%) in the biodiversity (Fig. 6—sample with CHX treatment (+)), while still maintaining similar amounts of viable cells compared with non-treated biofilm (data not shown). The treatment resulted in reduced population of certain oral microbes, e.g. Streptococcus spp. (Fig. 6, O1), and clonal expansion of bacteria species that were less susceptible to CHX, such as Neisseria spp (Fig. 6, O3) as indicated by the increased band intensity at 24 and 48 hours. Meanwhile, the abundance of certain oral residents, including Porphyromonas spp. (Fig 6, O2) and Peptostreptococcus spp. (Fig 6, O4) remained relatively stable. When challenged with P. aeruginosa cells, CHX treated biofilms suffered drastically reduced protective ability in preventing P. aeruginosa from colonizing the biofilm, as indicated by the increased intensity of the band representing P. aeruginosa within the biofilm samples taken 24 and 48 hours after CHX treatment/P. aeruginosa challenge (Fig. 6—samples with CHX treatment (+) and P. aeruginosa challenge (+)). For CHX treated samples, longer treatment time (30 seconds) and longer incubation time (48 hours) resulted in a more intense signal of the P. aeruginosa band recovered from biofilm samples (Fig. 6)
Discussion
The interaction between the human indigenous microflora and exogenously acquired pathogens has been the subject of continuous investigation for the past few decades (Sanders 1969; Brook 1999; Reid, Howard et al. 2001). It has been speculated that indigenous flora may enhance the host’s ability to resist infection, and the notion that it may be manipulated to the host’s advantage makes the topic an interesting one (Reid, Howard et al. 2001; Falagas, Rafailidis et al. 2008). Unlike the indigenous microbial flora associated with other parts of the human body, such as the intestine, the upper respiratory track and the female genital tract, whose beneficial and protective roles have been well studied (Larsen 1993; Brook 2005; O'Hara and Shanahan 2006), the demonstration and investigation of oral microbial community-based bacterial interference in preventing pathogenic/foreign colonization is still lacking.
Despite its repeated exposure to various bacteria from the nose, the respiratory and intestinal tract, as well as contaminated water and food sources, the microbial composition within oral cavities of healthy subjects is relatively stable (Rasiah, Wong et al. 2005; Zaura, Keijser et al. 2009); However, patients with certain oral disease conditions have been shown to carry altered oral microbial communities and are prone to the colonization by bacteria of foreign origin (Leung, Jin et al. 1998; Almståhl, Wikström et al. 2008). These intriguing phenomena suggest that the normal commensal oral microbial communities might play beneficial roles and exert protective functions against pathogenic/foreign colonization.
Using an in vitro model of mice GI-tract associated microflora, we demonstrated that the oral microbiota was able to prevent the integration of bacterial species originating from the gut (He, Tian et al. 2010a). In this study, we further investigate the protective role of the human oral microbiota by establishing saliva-derived biofilms and testing their ability to prevent the integration by two P. aeruginosa strains: the PAO1 lab strain and Pa060928, a clinical isolate from a cystic fibrosis patient. Saliva medium was used for cultivation to mimic the nutritional condition within oral cavity. We demonstrated that although both P. aeruginosa strains were able to persist during co-cultivation with oral flora in saliva medium, they mainly existed in the planktonic phase and were ineffective in integrating into surface-attached oral microbial communities during biofilm formation (Fig. 1 and Fig. 5).
Development into structured oral biofilms requires co-aggregation between different bacterial species and attachment to the extracellular matrix (Lamont and Jenkinson 2000; Kolenbrander, Palmer et al. 2010). Co-aggregation among indigenous oral bacterial species is a fairly common phenomenon. It has been demonstrated that while oral bacterial species don’t co-aggregate well with non-oral bacteria, such as intestinal species (Ledder, Timperley et al. 2008), all of the roughly 1,000 oral bacterial strains examined so far have at least one co-aggregation partner. Indeed, coaggregation plays a pivotal role in the formation of structured multispecies oral biofilms (Kolenbrander, Palmer et al. 2010). As non-oral commensal bacteria, neither the P. aeruginosa lab strain PAO1 nor the clinical isolate Pa060928 were able to co-aggregate with any of the tested oral bacterial species isolated from healthy subjects in this study (Table 1), including the “bridging” organism—F. nucleatum, which has been suggested to play critical roles in facilitating the development of oral community due to its ability to co-aggregate with a variety of oral bacteria (Kolenbrander, Palmer et al. 2010). Our results suggested that the inability of P. aeruginosa to adhere to oral bacterial species might contribute to its ineffectiveness in integrating into the developing salivary biofilm. As an opportunistic pathogen, P. aeruginosa can be frequently isolated from certain oral infection sites (Nord, Sjöberg et al. 1972) and is one of the most common pathogens identified within the oral cavity and sputum of patients with cystic fibrosis (Komiyama, Tynan et al. 1985) and ventilator-associated pneumonia (Bonten, Bergmans et al. 1999). Interestingly, in the case of cystic fibrosis patients, P. aeruginosa isolates can effectively co-aggregate with certain oral isolates obtained from the same patients (Komiyama, Habbick et al. 1987), further indicating that co-aggregation might play an important role in determining if P. aeruginosa can integrate into developing oral microbial communities. The CF clinical isolate Pa060928 used in this study was unable to coaggregate with any of the oral isolates from healthy subjects tested (Table 1). One possible explanation could be that in CF patients, the changes in oral ecological conditions could result in the colonization of specific oral bacterial strains with distinct outer membrane characteristics, which could allow them to interact and co-aggregate with certain P. aeruginosa strains.
Another interesting finding of this study was that in the presence of sucrose, saliva-derived microbial flora was not only able to prevent P. aeruginosa from integrating into biofilms, but it could also inhibit its growth within the planktonic portion of the co-culture by producing lactic acid (Fig. 2, 3 and 5). As an antimicrobial agent, lactic acid is able to inhibit the growth of many bacteria, particularly gram-negative species of the families Enterobacteriaceae and Pseudomonadaceae (Ray and Sandine 1992; Alakomi, Skytta et al. 2000). One aspect of its antibacterial action is to lower the pH, which by itself might have certain inhibitory effects (Pasricha, Bhalla et al. 1979). However, this is unlikely to be the key factor in our study since the high buffering capacity of salivary medium used in this study didn’t result in significant drop in pH, even when lactic acid could be detected at millimolar levels in the spent medium (data not shown). Another important antimicrobial property of lactic acid is its ability, in the undissociated form, to permeabilize gram-negative bacteria by disrupting the outer membrane; it is also capable of penetrating the cytoplasmic membrane, resulting in reduced intra-cellular pH and disruption of the transmembrane proton motive force (Ray and Sandine 1992). Lactic acid produced by probiotic Lactobacillus spp. has been shown to play an important role in fending off pathogenic Salmonella strains in the intestinal tract (Fayol-Messaoudi, Berger et al. 2005). In the oral cavity, gram-positive microbes are the major species detected in saliva and supra-gingival plaque where the first encounter between exogenous gram-negative pathogens, such as P. aeruginosa, and oral microbes takes place. The fact that many oral bacterial species, including Streptococcus spp. and Lactobacillus spp., are able to produce lactic acid as the major metabolic end-product of carbohydrate fermentation (Dashper and Reynolds 1996; Ljungh and Wadström 2006) strongly suggests that it could be one of the defense mechanisms used by the oral flora to inhibit the growth and prevent the colonization of exogenous gram-negative pathogens, such as P. aeruginosa, when the fermentable sugars are available.
From the community perspective, the production of lactic acid could be a double-edged sword: while it inhibits the pathogens, it might also have negative effects on certain residents within the community. How the residential bacteria cope with this situation is not clear and is currently under investigation. However, a recent report by Jakubovics et al showed that the co-aggregation with Actinomyces naeslundii protected another oral resident, Streptococcus gordonii from oxidative damage due to catalase production by A. naeslundii (Jakubovics, Gill et al. 2008). This suggests that co-aggregation between two oral bacterial species might help some species better tolerate the environmental stress experienced within multispecies communities.
Our results demonstrated that pre-formed salivary biofilms were effective in preventing P. aeruginosa colonization (Fig. 6). For a bacterium to successfully colonize the pre-existing biofilm microbial community, it needs to overcome the invasion resistance developed by the existing flora. The invasion resistance often includes: 1) depletion of attachment sites, 2) production of antibiotic substance, or 3) the establishment of a restrictive physiological microenvironment, such as altered pH (Bernet, Brassart et al. 1994; Liévin, Peiffer et al. 2000; Kreth, Merritt et al. 2005). Although the underlying mechanism is unclear and currently under investigation, recent studies by Kreth et al demonstrated that, depending on the sequence of inoculation, preformed Streptococcus sanguinis biofilms can prevent the integration of late coming Streptococcus mutans strains by producing the antimicrobial agent hydrogen peroxide (Kreth, Merritt et al. 2005). In the context of this study, it is possible that the presence of certain species in the established oral biofilm could exert inhibitory effects against P. aeruginosa. Our result is consistent with the report by Li et al, where they showed that the presence of existing oral biofilms can greatly reduce S. mutans colonization (Li, Guo et al. 2010).
The most intriguing finding of our study was that a shift in salivary microbial composition that resulted from mild CHX treatment led to a drastic reduction in the biofilm’s protection against P. aeruguinosa colonization (Fig. 6). CHX is an cationic bis-biguanide biocide with broad-spectrum antibacterial activity, which can induce concentration-dependent growth inhibition (Hugo and Longworth 1966) and has been shown to be able to induce a bacterial profile change in existing microbial floras (McBain, Bartolo et al. 2003). In our study, a low concentration of CHX was used to generate a microbial community with a shifted population profile.
The observed reduction in the biofilm’s protective capability was not due to the reduction in the total microbe population within the biofilm because 24-hour re-growth allowed the bacterial counts of the microbial community to recover to pre-treatment levels (data not shown). After treatment, previously minor species, such as Neisseria spp., became dominant while Streptococcus spp. suffered severe reduction in its population within the biofilm. The change in microbial composition could potentially affect community dynamics, including its invasion resistance and its response to the presence of bacteria of foreign origin (Ley, Peterson et al. 2006).
Furthermore, the reduced defense ability could also result from the decrease in biodiversity within the biofilm. In other ecosystems, it has been observed that species-rich communities are more resistant to invasion by exotic species than species-poor communities (Elton 1958). This is also in agreement with our previous studies showing the maximum inhibitory effect exerted by an in vitro mouse oral microbial community toward a gut bacterium was achieved when the whole community was involved, suggesting a community-based antagonistic action (He, Tian et al. 2010a). Our data corroborated the documented reports, which show that microbial composition within oral cavities of healthy subjects are relatively stable (Rasiah, Wong et al. 2005; Zaura, Keijser et al. 2009) while patients with altered oral microbial communities can be more susceptible to colonization by bacteria of foreign origins (Leung, Jin et al. 1998; Almståhl, Wikström et al. 2008).
The human oral cavity has evolved complex and sophisticated mechanisms to fend off bacterial pathogens, including physical barriers, protective immunity conferred by the mucosal lining (Walker 2004), and defense components within saliva (Tabak 2006). Our study supports the notion that bacterial interference exerted by oral commensal flora could also play a significant role in protecting against the colonization by foreign/pathogenic bacteria. In this regard, a normal and balanced oral commensal microbiota can greatly contribute to ecologic stability.
Acknowledgements
We would like to thank Dr. Ian McHardy for his critical review of this manuscript. This study is supported by NIH grants (DE20102 and GM095373) and funds from C3 Jian Inc.
References
- Aas JA, Paster BJ, et al. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005;43(11):5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakomi H-L, Skytta E, et al. Lactic Acid Permeabilizes Gram-Negative Bacteria by Disrupting the Outer Membrane. Appl. Environ. Microbiol. 2000;66(5):2001–2005. doi: 10.1128/aem.66.5.2001-2005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almståhl A, Wikström M, et al. Microflora in oral ecosystems in subjects with radiation-induced hyposalivation. Oral Diseases. 2008;14(6):541–549. doi: 10.1111/j.1601-0825.2007.01416.x. [DOI] [PubMed] [Google Scholar]
- Bernet FM, Brassart D, et al. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut. 1994;35:483–489. doi: 10.1136/gut.35.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonten MJM, Bergmans DJJ, et al. Characteristics of polyclonal endemicity of Pseudomonas aeruginosa colonization in intensive care units. Implications for infection control. Am. J. Respir. Crit. Care Med. 1999;160(4):1212–1219. doi: 10.1164/ajrccm.160.4.9809031. [DOI] [PubMed] [Google Scholar]
- Brook I. Bacterial Interference. Critical Reviews in Microbiology. 1999;25(3):155–172. doi: 10.1080/10408419991299211. [DOI] [PubMed] [Google Scholar]
- Brook I. The role of bacterial interference in otitis, sinusitis and tonsillitis. Otolaryngology - Head and Neck Surgery. 2005;133(1):139–146. doi: 10.1016/j.otohns.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- Cisar JO, Kolenbrander PE, et al. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun. 1979;24:742–752. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper SG, Reynolds EC. Lactic acid excretion by Streptococcus mutans. Microbiology. 1996;142(1):33–39. doi: 10.1099/13500872-142-1-33. [DOI] [PubMed] [Google Scholar]
- Elton CS. The ecology of invasions by plants and animals. London: Chapman and Hall; 1958. [Google Scholar]
- Falagas ME, Rafailidis PI, et al. Bacterial interference for the prevention and treatment of infections. International Journal of Antimicrobial Agents. 2008;31(6):518–522. doi: 10.1016/j.ijantimicag.2008.01.024. [DOI] [PubMed] [Google Scholar]
- Fayol-Messaoudi D, Berger CN, et al. pH-, Lactic Acid-, and Non-Lactic Acid-Dependent Activities of Probiotic Lactobacilli against Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2005;71(10):6008–6013. doi: 10.1128/AEM.71.10.6008-6013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F, Malagelada J-R. Gut flora in health and disease. The Lancet. 2003;361(9356):512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- He X, Tian Y, et al. In Vitro Communities Derived from Oral and Gut Microbial Floras Inhibit the Growth of Bacteria of Foreign Origins. Microbial Ecology. 2010a;60(3):665–676. doi: 10.1007/s00248-010-9711-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Tian Y, et al. Oral-Derived Bacterial Flora Defends Its Domain by Recognizing and Killing Intruders—A Molecular Analysis Using <i>Escherichia coli</i> as a Model Intestinal Bacterium. Microbial Ecology. 2010b;60(3):655–664. doi: 10.1007/s00248-010-9708-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo WB, Longworth AR. The effect of chlorhexidine on the electrophoretic mobility, cytoplasmic constituents, dehydrogenase activity and cell walls of Escherichia coli and Staphylococcus aureus. J Pharm Pharmaco. 1966;18:569–578. doi: 10.1111/j.2042-7158.1966.tb07935.x. [DOI] [PubMed] [Google Scholar]
- Jakubovics NS, Gill SR, et al. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiology Ecology. 2008;66(3):637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CW, Lux R, et al. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Molecular Microbiology. 2009;71(1):35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, et al. Communication among Oral Bacteria. Microbiol. Mol. Biol. Rev. 2002;66(3):486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 1993;175(11):3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, et al. Oral multispecies biofilm development and the key role of cell–cell distance. Nat Rev Micro. 2010;8(7):471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- Komiyama K, Habbick BF, et al. Interbacterial adhesion between Pseudomonas aeruginosa and indigenous oral bacteial isolated from patients with cystic fibrosis. Can J Microbiol. 1987;33:27–32. doi: 10.1139/m87-005. [DOI] [PubMed] [Google Scholar]
- Komiyama K, Tynan JJ, et al. Pseudomonas aeruginosa in the oral cavity and sputum of patients with cystic fibrosis. Oral Surgery, Oral Medicine, Oral Pathology. 1985;59(6):590–594. doi: 10.1016/0030-4220(85)90187-2. [DOI] [PubMed] [Google Scholar]
- Kreth J, Merritt J, et al. Competition and Coexistence between Streptococcus mutans and Streptococcus sanguinis in the Dental Biofilm. J. Bacteriol. 2005;187(21):7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, et al. Interspecies Interactions within Oral Microbial Communities. Microbiol. Mol. Biol. Rev. 2007;71(4):653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kus JV, Tullis E, et al. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) and non-CF patients. Microbiol. 2004;150:1315–1326. doi: 10.1099/mic.0.26822-0. [DOI] [PubMed] [Google Scholar]
- Lamont R, Jenkinson H. Adhesion as an ecological determinant in the oral cavity. In: Kuramitsu KK, Ellen RP, editors. Oral bacterial ecology: the molecular basis. Norfolk: Horizon Scientific Press; 2000. pp. 131–168. [Google Scholar]
- Larsen B. Vaginal flora in health and disease. Clin Obstet Gynecol. 1993;36:107–121. doi: 10.1097/00003081-199303000-00016. [DOI] [PubMed] [Google Scholar]
- Ledder RG, Timperley AS, et al. Coaggregation between and among human intestinal and oral bacteria. FEMS Microbiology Ecology. 2008;66(3):630–636. doi: 10.1111/j.1574-6941.2008.00525.x. [DOI] [PubMed] [Google Scholar]
- Leung WK, Jin LJ, et al. Subgingival microbiota of shallow periodontal pockets in individuals after head and neck irradiation. Oral Microbiol Immunol. 1998;(13):1–10. doi: 10.1111/j.1399-302x.1998.tb00743.x. [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, et al. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Li L, Guo L, et al. Targeted antimicrobial therapy against Streptococcus mutans establishes protective non-cariogenic oral biofilms and reduces subsequent infection. Int J Oral Sci. 2010;2:66–73. doi: 10.4248/IJOS10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ku, J CYX, et al. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res. 2005;84:559–564. doi: 10.1177/154405910508400614. [DOI] [PubMed] [Google Scholar]
- Liévin V, Peiffer I, et al. Gastrointestinal infection: Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut. 2000;47:646–652. doi: 10.1136/gut.47.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljemark WF, Bloomquist C. Human Oral Microbial Ecology and Dental Caries and Periodontal Diseases. Critical Reviews in Oral Biology & Medicine. 1996;7:180–198. doi: 10.1177/10454411960070020601. [DOI] [PubMed] [Google Scholar]
- Ljungh A, Wadström T. Lactic acid bacteria as probiotics. Curr Issues Intest Microbiol. 2006;7:73–89. [PubMed] [Google Scholar]
- Marsh PD. Microbial Ecology of Dental Plaque and its Significance in Health and Disease. Advance in Dental Research. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- Martin-Laurent F, Philippot L, et al. DNA Extraction from Soils: Old Bias for New Microbial Diversity Analysis Methods. Appl. Environ. Microbiol. 2001;67(5):2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain AJ, Bartolo RG, et al. Effects of a Chlorhexidine Gluconate-Containing Mouthwash on the Vitality and Antimicrobial Susceptibility of In Vitro Oral Bacterial Ecosystems. Appl. Environ. Microbiol. 2003;69(8):4770–4776. doi: 10.1128/AEM.69.8.4770-4776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocker A, Camper AK. Selective Removal of DNA from Dead Cells of Mixed Bacterial Communities by Use of Ethidium Monoazide. Appl. Environ. Microbiol. 2006;72(3):1997–2004. doi: 10.1128/AEM.72.3.1997-2004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord C-E, Sjöberg L, et al. Pseudomonas Aeruginosa in Oral Infections. Acta Odontologica Scandinavica. 1972;30(3):371–381. doi: 10.3109/00016357209004604. [DOI] [PubMed] [Google Scholar]
- O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasricha A, Bhalla P, et al. Evaluation of lactic acid as an antibacterial agent. Indian Journal of dermatology, Venereology and Leprology. 1979;45:149–161. [PubMed] [Google Scholar]
- Paster BJ, Boches SK, et al. Bacterial Diversity in Human Subgingival Plaque. J. Bacteriol. 2001;183(12):3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, et al. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000. 2006;42(1):80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, et al. Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rasiah IA, Wong L, et al. Variation in bacterial DGGE patterns from human saliva: over time, between individuals and in corresponding dental plaque microcosms. Archives of Oral Biology. 2005;50(9):779–787. doi: 10.1016/j.archoralbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, et al. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-free Recipients Reveal Host Habitat Selection. Cell. 2006;127(2):423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B, Sandine WE. Acetic, propionic, and lactic acids of starter culture bacteria as biopreservatives. In: Ray B, Daeschel M, editors. Food preservatives of microbial origin. Boca Raton, Fla: CRC Press; 1992. pp. 103–136. [Google Scholar]
- Reid G, Howard J, et al. Can bacterial interference prevent infection? Trends in Microbiology. 2001;9(9):424–428. doi: 10.1016/s0966-842x(01)02132-1. [DOI] [PubMed] [Google Scholar]
- Sanders E. Bacterial Interference: I. Its Occurrence among the Respiratory Tract Flora and Characterization of Inhibition of Group a Streptococci by Viridans Streptococci. The Journal of Infectious Diseases. 1969;120(6):698–707. doi: 10.1093/infdis/120.6.698. [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Cox DR, et al. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, et al. Glycan Foraging in Vivo by an Intestine-Adapted Bacterial Symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Tabak LA. In defense of the oral cavity: the protective role of saliva secretions. Pediatric Dentistry. 2006;28:110–117. [PubMed] [Google Scholar]
- Tian Y, He X, et al. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Molecular Oral Microbiology. 2010;25(5):357–367. doi: 10.1111/j.2041-1014.2010.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara Y, Kikuchi K, et al. H2O2 Produced by Viridans Group Streptococci May Contribute to Inhibition of Methicillin-Resistant Staphylococcus aureus Colonization of Oral Cavities in Newborns. Clinical Infectious Diseases. 2001;32(10):1408–1413. doi: 10.1086/320179. [DOI] [PubMed] [Google Scholar]
- Uehara Y, Kikuchi K, et al. Inhibition of Methicillin-Resistant Staphylococcus aureus Colonization of Oral Cavities in Newborns by Viridans Group Streptococci. Clinical Infectious Diseases. 2001;32(10):1399–1407. doi: 10.1086/320147. [DOI] [PubMed] [Google Scholar]
- Walker DM. Oral mucosal immunology: an overview. Annals Academy of Medicine. 2004;33(No.4) suppl:27–30. [PubMed] [Google Scholar]
- Zaura E, Keijser B, et al. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiology. 2009;9(1):259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]