Genes are involved in psychology and behaviour, and in their neuronal bases, in many species, including the geneticists' favourites: human, mouse, Drosophila and the worm Caenorhabditis elegans. The number of genes that modulate behaviour in the mouse alone is impressive: Table 1 summarizes the number of genes discovered on the basis of individual behavioural differences by examining spontaneous mutations, targeted genes and transgenic mice. A literature analysis of 23 journals revealed that, as of late August 2006, 3,923 genes are involved in mouse behaviour; when quantitative trait loci were included, the number increased to more than 4,000. This is probably an underestimate, as the analysis relied on such a small collection of journals.
Table 1. Number of reported genes and quantitative trait loci associated with behaviour in mice.
| Behaviour | Gene | Quantitative trait loci |
|---|---|---|
| Food and fluid consumption | 334 | 17 |
| Emotional reaction | 341 | 112 |
| Learning | 306 | 38 |
| Motor behaviour | 1,542 | 43 |
| Sleep and/or circadian activity | 119 | – |
| Social behaviour | 223 | 11 |
| Drug intake and related behaviour | 254 | 33 |
| Sensory and motor development | 46 | 41 |
| Seizures | 301 | – |
| Sensory behaviour | 457 | – |
| Total | 3,923 | 295 |
This number of genes gives rise to a paradox: as of late August 2006, the number of transgenic mice and mice with a targeted gene knockout identified in repositories was in the vicinity of 3,000: 2,500 were identified directly in repositories, and it was estimated that another 500 were kept in laboratories. As the mouse genome carries about 24,000 genes, a further 21,000 transgenic or knockout lines are needed to uncover the functions of all mouse genes. As most of the numbers in Table 1 were established using transgenic or gene-targeting technologies, we can therefore predict the number of genes that are involved in behavioural traits—after fully exploring all known genes—by a simple calculation: the number of genes linked to behavioural traits divided by the number of targeted and transfected genes, multiplied by the estimated number of genes in the mouse, which is 3,923/3,000 × 24,000 = 31,384. Obviously, this is more than the total number of genes in the mouse genome and is much larger than the number of genes that could be expressed in the brain, regardless of the actual proportion of brain-expressed genes. A literature analysis of the same 23 journals reveals that 3,900 additional genes have been recognized as modulating brain anatomy and function, with another 3,914 involved in sensory abilities. If we include these genes in the calculation, it would then predict that some 80,000 genes are involved in brain functioning and behaviour. The number is too high: there is no room for this horde of brain-behaviour genes in the brain.
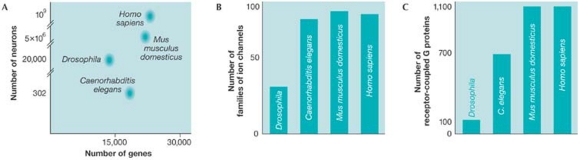
A comparison of the numerical ratios of genes to neurons in other species leads to a similar conclusion. The total number of neurons in a species is an indicator of the capability and behavioural complexity of the brain. However, the number of genes is not predictive of the number of neurons and is therefore not predictive of the complexity of behaviour (Fig 1). Drosophila has fewer genes than C. elegans, but more neurons and a larger behavioural repertoire. The mouse has fewer neurons and less behavioural complexity than Homo sapiens, but approximately the same number of genes. There is no correlation between the number of genes and the sophistication of behaviour or the number of neurons.
There is no correlation between the number of genes and the sophistication of behaviour or the number of neurons
Figure 1.
Interspecies relationships showing the ratio of the number of genes to the (A) number of neurons, (B) ion channel families and (C) categories of G proteins (adapted from Roubertoux, 2004).
This mismatch between behavioural complexity and the numbers of genes and neurons shows that the relationship between genotype and phenotype is not linear. The question then is what links DNA at the most elementary level of organization to behaviour at the highest level? An understanding of these processes should cast light on the intrinsic nature of the gene–brain–behaviour link.
Thirty years ago, Changeux & Danchin (1976) suggested that the selective stabilization of the synapse during development could explain the relationship between the high number of specialized neurons and the number of genes. Recent advances in molecular biology offer additional explanations. The concept of the multifunctional gene proposes that one gene might generate several behavioural or non-behavioural phenotypes (Roubertoux, 2004). We have found evidence for this by examining the number of phenotypes associated with experimental mutations. We reviewed 32 gene-targeting experiments with mice conducted before 1999 and compiled a list of the phenotypes reported for each of the 32 genes between 1998 and 2006. Twenty-one genes are associated with more than four behavioural phenotypes each. One example is the α-CamkII gene, which modulates differences in learning, anxiety, aggression and sensitivity to pain, and which affects long-term potentiation in the hippocampus as well as several non-behavioural phenotypes. Eleven other genes associated with more than two behavioural phenotypes each are also linked with several muscular, skeletal, immunological, digestive and respiratory phenotypes. Multifunctional genes have also been found in other species: the clk-1 gene in C. elegans affects egg-laying, pharyngeal pumping, crawling, swimming and defaecation. A single mutation in the EPM2A human gene induces seizures, dyslexia and learning defects (Ganesh et al, 2002). The multifunctionality of the gene is thus a general property throughout the genome and means that a vast phenotypic diversity can be produced from a small number of genes.
Multifunctionality extends the concept of pleiotropy, whereby a polypeptide has different effects in different cells. It would explain why the same allele induces one effect in a liver cell and another in a heart cell. The brain has widely diversified functions and its parts are as different from each other as they are from other organs. The same allelic form of a gene thus produces different effects in brain structure A than in brain structure B because the differences between A and B can be as great as the differences between a heart cell and a liver cell.
This was illustrated in mice with a null allele for the dopamine hydroxylase gene, which produces a dopamine deficiency throughout the body. A total lack of dopamine induces a variety of disorders, including feeding deficits, sucrose avoidance and poor interest in nest building. Normal behaviour patterns can be restored by locally injecting dopamine into the regions of the caudate putamen and nucleus accumbens. When dopamine is injected into the lateral caudate putamen, feeding is restored; when it is injected into the nucleus accumbens, only sucrose preference is restored; and when it is injected into the lateral or the central caudate putamen, nest building is restored (Szczypka et al, 2001). The product of the gene therefore has various functions depending on the brain regions in which the gene is expressed.
The product of the gene therefore has various functions depending on the brain regions in which the gene is expressed
Alternative splicing has been suggested as a mechanism behind this multifunctionality (Roubertoux, 2004; Ule et al, 2006). Through alternative splicing, the Drosophila Down syndrome cell adhesion molecule (Dscam) gene, which encodes an axon guidance receptor, can express 38,016 different messenger RNAs and thus 38,016 possible functions (Schmucker et al, 2000). This is three times the size of the Drosophila genome, which has some 13,000 genes. Other genetic events contribute to the multifunctionality of genes either alone or in interaction with alternative splicing. Interactions between the allelic forms of a gene or of several genes and genetic cascades increase the number of genotypes that subsequently foster the emergence of multiple phenotypes. Intra-locus interaction, as observed in genomic imprinting, induces different phenotypes depending on whether the imprinted allele is from the father or the mother.
The interaction between alleles of different genes, called epistasis, produces a larger set of possible genotypes; for example, two genes A and B, each with two alleles, can generate nine genotypes (AABB, AABb, AAbb, AaBB, AaBb, Aabb, aaBB, aaBb, aabb). When a single gene is either suppressed or present as multiple copies, it can affect the expression of several hundred other genes, thus giving rise to numerous phenotypes. For example, two mitochondrial genes coding for proteins show non-synonymous differences between two mice strains; the cross-transfer of mitochondrial DNA between the strains modulates cognitive performance in the resulting congenic mice (Roubertoux et al, 2003). The modulation seems to be caused not only directly by the mitochondrial alleles, but also indirectly by interactions with nuclear genes. Of the 34 behavioural measurements made, 16 showed a direct effect from mitochondrial DNA, and 15 showed interactions with nuclear genes.
The role of epistasis in generating multifunctionality could also explain the differences between humans and chimpanzees. About 1.34% of genes differ between these closely related species, and about 100 of these genes are expressed in the brain. Could these 100 genes encompass the genes of the human species, including those necessary to discover the existence of the double helix or to compose Tristan and Isolde? It is now known that these 100 genes interact with genes common to both humans and chimpanzees and that they modify the expression of 35% of the chimpanzee genes—approximately 8,400—most of which are expressed in the brain (Hacia, 2001).
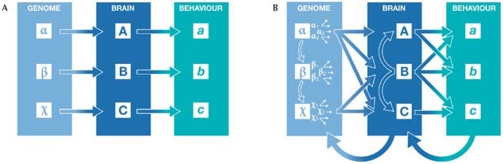
Cascade is a specific form of epistasis: the function of gene α requires the product of gene δ, and the function of both β and χ requires the product of gene α (Fig 2). Cascades frequently affect genes involved in development. The paired box 6 (PAX6/Pax6) gene is regulated by five genes and itself regulates more than 30 genes, thus indirectly contributing to several phenotypes. The same is true for the steroid sulphatase gene (Sts), which we identified as contributing to spontaneous aggression against a male of the same species (Roubertoux et al, 2005). We realized that the Sts gene was the initiator of a cascade of biological events, rather than being directly responsible for aggressive behaviour. Steroid sulphatase catalyses the hydrolysis of 3-β-hydroxysteroid sulphates. The sulphated steroids modulate type A γ-aminobutyric acid receptors (GABAA) and N-methyl-D-aspartate A receptors (NMDAA), which are known for their role in aggression (Maxson, 1996). Depending on whether these steroids are sulphated, GABAA and NMDAA contribute to a wide range of neuronal mechanisms involved in neuronal plasticity, learning, cognition and motor behaviour.
Figure 2.
The relationship between genes, the brain and behaviour. (A) An obsolete view assuming isomorphism between the three levels. (B) A more recent view based on nonlinear relationships. Each gene α, β and χ has several allelic forms that can undergo alternative splicing as shown by the small arrows; genetic cascades are the vertical arrows. The resulting brain modules, A, B and C—consisting of one neuron or a set of neurons—interact as indicated by the vertical arrows. Each module, either alone or in combination with other brain modules, can produce differences in one or more behaviour patterns. The modification in behaviour regulates brain functioning and modulates gene expression, as illustrated by the lower curved arrows.
Regardless of how genes interact, it is difficult to identify the one gene that triggers a chain of biochemical and cellular events that eventually lead to a phenotype. It is therefore absurd to ascribe a certain phenotype to a single gene in cases in which the phenotype does not display the main effect of the gene in question. Instead, a genetic network, not a single gene, produces the phenotype in almost all brain-related phenotypes. We should thus consider the genome as a site where genes interact in constant dialogue and exchange, and not as a static alignment of functional units.
It is […] absurd to ascribe a certain phenotype to a single gene in cases where the phenotype does not display the main effect of the gene in question
The concept of genotype–environment interactions is often used to explain phenotypic variation—and even more often misused. It is the trump card for zealous environmentalists and a tool for unrepentant advocates of heredity, and provides a safe haven for the virtuous defendants of ecumenism. Genotype–environment interactions cannot operate in one direction only, in which the environment affects the gene. Instead, the effect of a gene depends on the environment in which the organism develops and lives, but the same environment does not have the same effects on different genes. Studies investigating the combined use of ovary transplantation plus adoption in mice showed that certain genes tend to accelerate development, but the effect was modulated by a complex equilibrium between components in the maternal milk and the quality of care, such as the frequency of licking and lactating episodes, and nesting characteristics.
This does not necessarily mean that one environment is always better than another. A rich milk composition and frequent care can have beneficial consequences for certain genotypes, but will have null or negative effects on others (Carlier et al, 1999). In fact, most of the genes involved in behavioural differences are susceptibility genes: the environment increases the effect of the gene and the diversity of environments might increase the diversity of phenotypes produced by that gene. Overexpression or underexpression of genes in different environmental conditions is the most probable mechanism of genotype–environment interactions. Modifications induced in the brain during learning indicate how the brain is able to integrate new rules and develop new strategies by triggering changes in neuronal connectivity and long-term changes in gene expression.
The onset of a certain phenotype requires proteins, which are the final stage in a series of transitional steps from transcription to translation. Each step is an integrative process, whereby the generation of information at one level relies on information provided by previous elementary steps. The price for this generation of information is weaker causality of the link from DNA to protein in a given cell, which has several immediate consequences.
First, most of the genes involved in integrative functions are susceptibility genes. The allelic form associated with the integrated phenotype does not determine the phenotype, but defines a lesser or greater probability for expressing the phenotype. There are some exceptions—a person carrying one allele of a specific gene with more than 40 CAG repeats will develop Huntington disease; the number of repeats does not, however, predict the age of onset, the severity of the disorder or the rate of neurodegeneration.
Second, most phenotypes require several alleles acting cumulatively or by interaction. The probability of receiving from the parents all alleles necessary to develop a given phenotype is lower than the probability of receiving alleles in the case of monogenic traits. Genetics does not necessarily mean heredity.
Third, if causality were linear, it should be possible to deduce the phenotype from the genotype and to infer the genotype from the phenotype. This is not the case for most phenotypes. A mouse with an NZB/BlNJ Sts allele, for example, has a high probability of attacking males from the same species (Roubertoux et al, 2005). In turn, allelic variants for more than 50 genes can be inferred from the fact that a male mouse attacks another male mouse (Maxson, 1996). In general, the knowledge of a person's genetic make-up might be used to predict the probability of a certain phenotype, but in most cases it is impossible to make a posteriori calculations of the genetic variants on the basis of the phenotype. In humans, for example, it would not be possible to infer the mutation associated with a given intellectual disability phenotype if more than 1,370 loci are known to be involved.
Fourth, genetic analysis consists of correlations between genetic differences and phenotypic differences. We cannot find a gene encoding a given phenotype, but we can detect alleles linked to individual phenotypic differences. Genetic analysis cannot deal with ‘invariants'; it operates with variations. We can discover a group of genes involved in learning and memory, but together, these genes do not constitute the ‘genetics' of memory in a given species.
The same absence of isomorphism observed for genotype and phenotype is also found for the relationship between the brain and behaviour. The elementary components of a neuron do not predict behavioural complexity (Fig 1). The number of ion channel families in C. elegans is greater than in Drosophila, but Drosophila is capable of more complex behaviour. C. elegans has a greater variety of G proteins than Drosophila, but only the latter is capable of discriminative learning.
It is the higher number of neurons in mammalian species, and particularly in humans, that leads to greater complexity and thus makes a difference in behaviour. The large number of possible connections, and therefore the large number of synapses, means there are more possibilities for neuronal integration, the impact of alternative splicing and more frequent cascades.
For several decades, scientists accepted Henry Dale's principle of ‘one neuron releases one transmitter' as they followed the ‘one gene codes for one protein' rule, but, with time, both were questioned. We now know that one neuron can influence more than one transmitter system, and that one neurotransmitter can modulate the release or reuptake of another neurotransmitter. Modern neurochemistry, by focusing on interactions between neurotransmission systems, offers an analogy with epistasis and cascades, which leads to a similar dilution of causality.
The quest to find a relationship between a given transmitter and a given feeling or behaviour has now been abandoned, because the effect of each neurotransmitter can vary depending on where it is released. In two separate cases, for example, the same interneuron can induce different behavioural responses in the mollusc Tritonia diomedea: rhythmic swimming, which uses the muscles; or non-rhythmic crawling, which uses the ciliar system. The versatility or multifunctionality of the interneuron depends on the neuronal context; whether the interneuron triggers swimming or crawling depends on the activity of its adjacent neurons (Popescu & Frost, 2002). Similarly, the concept of a ‘reflex' supported the hypothesis of ultra-specialized, hard-wired neuronal networks. For example, the stretching reflex was seen as the prototype of an autonomous, specialized function, but it now seems that the sensory-motor units provide the appropriate response and are the elementary components of the stretching response, rather than the motor neurons (Clarac et al, 2000).
The concept of multifunctional genes goes together with the concept of multifunctional neurons or neuronal networks. When it is combined with cascades, or with interactions between neurotransmitters, the possibility of predicting behavioural differences on the basis of brain characteristics is undermined. Immediately after the sequence of the human genome was published, scientists thought that there would be a simple relationship between the genome, the nervous system and behaviour. However, this idea quickly became obsolete (Fig 2A) as it was based on linear relationships between genes, the brain and behaviour. A more realistic view holds that each gene, α, β and χ, has several allelic forms (Fig 2B); each allelic form can undergo alternative splicing; and the genes act in cascades. A gene might act either alone or cumulatively, interacting with other genes on one or several brain phenotypes. The resulting brain modules again interact and each module, either individually or in combination with other brain modules, can produce differences in one or more behaviour patterns. The modification in behaviour then regulates brain functioning and modulates gene expression.
Genes are obviously involved in generating behaviour, but genetic determinism is rare. We can only predict the probability of a given phenotype on the basis of a gene or, more usually, a network of genes. Accordingly, we must change our terminology: no gene codes for a behavioural trait; at the very most, we can say that a gene is ‘involved' in a behavioural trait, or that a phenotype has genetic correlates. In the same vein, the terms ‘genetics of behaviour' or ‘behavioural genetics' seem inappropriate as they infer strict determinism; indeed, they are absurd as behaviour carries no genes, for it is the individual who carries the genes. “Behaviour-genetic analysis” (Hirsch, 1963) seems to be a more appropriate term: instead of focusing on relationships between genes and behaviour, it uses individual differences in behavioural phenotypes to discover allelic forms and their function.
What does natural selection then mean in the context of multifunctional genes when gene–phenotype causality is tenuous? It is much more difficult to understand the efficiency of selection when the gene on which selection is supposed to act has several functions, some contributing to positive adaptation and others to negative adaptation.
The nature of causal relationships between genes and behaviour also raises concerns wherever society uses scientific findings to allay ancestral fears, to encourage us to move into a brave new world, or to use the authority and aura of science to conceal ulterior motives. A report by the European Commission discussed the possibility of detecting susceptibility genes for occupational diseases (EGE, 2000); carriers of a gene that increases the risk of developing such a disease might then be discouraged from continuing in their positions. Job applicants might be rejected for the same reason. The emphasis on being able to detect genetic factors and the disregard for weak causality would mean that employers could ignore the need to find a proper solution, such as abandoning the practices and techniques that cause occupational diseases in the first place.
The decline in gene–phenotype causality and the emergence of multifunctionality must also raise questions in medical applications, including medically assisted reproduction, gene therapy, therapeutic cloning and cell cloning. These techniques have not yet taken into account all the possible ramifications, such as epistatic effects, between the corrected gene and the host genome. This is particularly true for therapeutic cloning, for which the role of mitochondrial DNA and its modulating effects on nuclear genes have been underestimated. Mitochondrial DNA—which, in most cases, is of unknown origin—has been shown to have unexpected effects (Roubertoux et al, 2003). Cloning might therefore generate phenotypes that differ from those expected from the nuclear genotype alone. There is a clear need for basic research on the epistatic events that are generated by mitochondrial DNA before we can safely move on to generate tissue for transplantation by therapeutic cloning.
Other medical issues, such as genetic counselling, prenatal diagnosis and embryo selection, are also affected if the strict relationship between gene and phenotype dissolves. Direct-to-consumer genetic tests can screen for monogenic disorders, susceptibility genes and polygenic disorders. However, the causality between susceptibility genes, the components of polygenes and phenotype is weak. Most disorders are caused by several genes, and these genes vary from one population to another. How is it possible to make any predictions? Similarly, rejecting an embryo with undesirable characteristics on the basis of a prenatal diagnostic test is not a simple decision because it does not depend solely on negative parameters, as was previously thought with the ‘one gene, one protein, one function' perspective. An individual is not determined by one single deleterious gene but through personal development, and living with and reacting to the genetic disease. If direct-to-consumer genetic testing and prenatal diagnostic tests had been available centuries ago, and if people carrying genes associated with epilepsy had been eliminated, the world would never have known Leonardo da Vinci, Leo Tolstoy, Gustave Flaubert, Graham Greene or Vincent van Gogh.
Acknowledgments
This article is dedicated to Professor Jean-Marc Monteil with esteem and friendship.
References
- Carlier M, Roubertoux PL, Wahlsten D (1999) Maternal effects in behavior genetic analysis. In Jones BC, Mormède P (eds) Neurobehavioral Genetics: Methods and Applications, pp 187–197. Boca Raton, FL, USA: CRC [Google Scholar]
- Changeux JP, Danchin A (1976) Selective stabilisation of developing synapses as a mechanism for the specification of neuronal networks. Nature 264: 705–712 [DOI] [PubMed] [Google Scholar]
- Clarac F, Cattaert D, Le Ray D (2000) Central control components of a ‘simple' stretch reflex. Trends Neurosci 23: 199–208 [DOI] [PubMed] [Google Scholar]
- EGE (2000) Genetic testing in the workplace. Proceedings of the round table debate held at the Brochette Center, Brussels. Brussels, Belgium: European Group on Ethics in Science and New Technologies, European Commission [Google Scholar]
- Ganesh S et al. (2002) Genotype–phenotype correlations for EPM2A mutations in Lafora's progressive myoclonus epilepsy: exon 1 mutations associate with an early-onset cognitive deficit subphenotype. Hum Mol Genet 11: 1263–1271 [DOI] [PubMed] [Google Scholar]
- Hacia JG (2001) Genome of the apes. Trends Genet 17: 637–645 [DOI] [PubMed] [Google Scholar]
- Hirsch J (1963) Behavior genetics and individuality understood. Science 142: 1436–1442 [DOI] [PubMed] [Google Scholar]
- Maxson SC (1996) Searching for candidate genes with effects on an agonistic behavior, offense, in mice. Behav Genet 26: 471–476 [DOI] [PubMed] [Google Scholar]
- Popescu IR, Frost WN (2002) Highly dissimilar behaviors mediated by a multifunctional network in the marine mollusk Tritonia diomedea. J Neurosci 22: 1985–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubertoux PL (2004) Existe-t-il des Gènes du Comportement? Paris, France: Odile Jacob [Google Scholar]
- Roubertoux PL et al. (2003) Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nat Genet 35: 65–69 [DOI] [PubMed] [Google Scholar]
- Roubertoux PL, Guillot PV, Mortaud S, Pratte M, Jamon M, Cohen-Salmon C, Tordjman S (2005) Attack behaviors in mice: from factorial structure to quantitative trait loci mapping. Eur J Pharmacol 526: 172–185 [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL (2000) Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671–684 [DOI] [PubMed] [Google Scholar]
- Szczypka MS, Kwok K, Brot MD, Marck BT, Matsumoto AM, Donahue BA, Palmiter RD (2001) Dopamine production in the caudate putamen restores feeding in dopamine-deficient mice. Neuron 30: 819–828 [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB (2006) An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]



 Pierre L. Roubertoux is in the Functional Genomics Group, CNRS Aix-Marseille University in Marseille, France.
Pierre L. Roubertoux is in the Functional Genomics Group, CNRS Aix-Marseille University in Marseille, France.