Abstract
INTRODUCTION
Neural tube defects (NTDs) are congenital anomalies caused by a combination of genetic and environmental influences. A defect below the head region resulting in protuberance of meninges and nervous tissue is termed myelomeningocele (MM). MM, the most common NTD compatible with survival, occurs in approximately 1 in 1,000 births worldwide. Maternal pre- and periconceptional folate supplementation reduces the risk of NTDs by up to 70%. A key enzyme in folate metabolism is 5, 10-methylene-tetrahydrofolate reductase (MTHFR).
OBJECTIVES
Sequence the 12 exons of the MTHFR gene among 96 subjects with MM to identify variants potentially contributing to the disease trait.
METHODS
Exons were amplified by polymerase chain reaction and the products were sequenced by Sanger method to reveal sequence variants compared to MTHFR reference sequences. Association of variants was examined by Fisher’s test.
RESULTS
A novel variant c.171+3G>T was identified in intron 1 in one affected subject. The variant was not found in the subject’s unaffected mother’s DNA and the unaffected father’s DNA was unavailable. We found significant differences in allele frequencies for seven SNPs in MM subjects compared to ethnically matched reference populations reported in the single nucleotide polymorphism (SNP) database (dbSNP).
CONCLUSION
We identified a novel variant c.171+3G>T in the MTHFR gene that potentially affects splicing in an affected subject. Also, we observed five SNPs (rs13306561, rs2274976, rs2066462, rs12121543, and rs1476413) in the MTHFR gene not previously shown to associate with MM. The current study provides additional evidence that multiple variations in the MTHFR gene are associated with MM.
INTRODUCTION
Neural tube defects (NTDs) are congenital malformations of the brain and spinal cord caused by failure of the neural tube to close between 21 and 28 days following conception (Blencowe et al., 2010). They are the most common structural malformations of the central nervous system in humans (Copp et al., 2003). The majority of cases can be categorized as either anencephaly (lack of closure in the region of the head) or spina bifida (SB; lack of closure below the head) (Au et al., 2010). These two categories of NTDs occur in approximately equal frequencies at birth (Botto et al., 1999). Anencephaly is lethal, with affected individuals usually dying shortly after birth. SB includes meningocele, lipomeningocele and myelomeningocele (MM) with MM accounting for >90% of SB cases (Au et al., 2010). As a result of major advances in medical care, the majority of babies born with SB survive to adulthood with varying degrees of morbidity (Centers for Disease Control, 1992). Spina bifida represents a significant burden for those affected and their families with an estimated $806,000 in 2007 dollars for lifetime medical care costs when living up to 65 years of age (Grosse et al., 2008). This financial burden based on medical costs alone does not even begin to account for the societal costs or the emotional impact on the affected individuals and their families.
Each year, SB and anencephaly together occur at a rate of 5.37 per 10,000 pregnancies in the United States (Boulet et al. 2008). Worldwide incidence ranges from 1.0 to 10.0 per 1,000 births (Au et al., 2010). The prevalence of anencephaly and SB in the United States has steadily declined since the late 1960s (Yen et al., 1992). In the early 1990s, epidemiologic evidence indicated that maternal folate status affected the occurrence and recurrence of NTDs (MRC 1991; Czeizel and Dudas, 1992). As a result, in January 1998 the United States Food and Drug Administration (US FDA) mandated fortification of food with folate in the United States. Comparison of SB prevalence prior to food fortification (October 1995-December 1996) to SB prevalence after the mandate (October 1998-December 1999), indicates a decrease in SB from 2.62 to 2.02 per 10,000 live births (22.9%) (Honein et al., 2001; Centers for Disease Control, 2004). For all races and ethnicities, the prevalence of SB and anencephaly combined significantly decreased (10%) when comparing rates in 1999–2000 to those in 2003–2004 (Boulet et al., 2008). Additionally, the prevalence of NTDs in the United States has been shown to vary by race/ethnicity with the highest rates among women of Hispanic ethnicity and the lowest rates among black and Asian women (Centers for Disease Control, 2009).
The etiology of NTDs is multifactorial, resulting from genetic and environmental influences occurring during the critical time in embryogenesis when the neural tube is forming (Martinez et al. 2009). Maternal folate status is known to be an important factor in the etiology of NTDs (Au et al., 2010); however, despite an impressive number of folate related investigative studies, the mechanism(s) by which folate deficiency predisposes to NTDs remains unclear (Bassuk and Kibar, 2009). Folate, a water-soluble B-complex vitamin, is an essential nutrient and the only source is from diet. It is critical to the carbon transfer necessary for DNA synthesis, cell division, and tissue growth (Botto and Yang, 2000).
The 5,10-methylene-tetrahydrofolate reductase (MTHFR) gene is the most extensively studied gene as a potential risk factor for NTD susceptibility. This gene is of particular importance as it regulates the levels of 5 methyl tetrahydrofolate available for homocysteine (hcy) remethylation (Bassuk and Kibar, 2009). The MTHFR gene is 2.2 kb in length and has been has been mapped to chromosomal region 1p36.3. It was initially reported by Goyette et al (1998) to have 11 exons; however, subsequent studies reported >11 exons and the existence of multiple transcripts differing in their first exon (Tran et al., 2002; Homberger et al., 2000). The diversity of transcripts is due to alternative transcription initiation and alternative splicing.
The enzyme 5,10-methylenetetrahydrofolate reductase (MTHFR) catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, a co-substrate for homocysteine remethylation to methionine. Normal MTHFR activity helps maintain the pool of circulating folate and methionine and prevents a buildup of homocysteine (Botto and Yang, 2000). The C677T SNP in the MTHFR gene was the first genetic variant to be implicated as possibly important in NTD susceptibility and remains the most studied (Beaudin and Stover, 2007). The C to T substitution results in an amino acid change (alanine to valine). The resulting enzyme is termed “thermolabile” because the activity of the encoded enzyme is reduced at 37°C or higher. Thus, MTHFR activity among C677T homozygotes is 50–60% lower at 37°C and approximately 65% lower at 46°C than in similarly treated controls (Botto and Yang, 2000). It is associated with reduced levels of enzyme activity, elevated levels of plasma Hcy, and an increased NTD risk in some populations (Bassuk and Kibar, 2009). Meta-analysis by Botto and Yang (2000) and other authors (van der Put, Eskes and Blom, 1997) strongly implicated the MTHFR 677TT genotype as a risk factor for NTDs in mothers (50%–70% increase) and fetuses (80%–90% increase).
Another SNP in the MTHFR gene, A1298C, has also been described and studied for its relationship to NTDs. Available data suggest that the A1298C variant alone is probably not a major risk factor for MM. In a study by Trembeth et al (1999), a significant association between the A1298C allele and MM risk was found in a subset of cases and controls (OR = 2.4; 95 percent CI: 1.4, 4.1); however, this finding could not be replicated in other subsets within the same study. Data also suggest that compound heterozygosity for the C677T and A1298C alleles might be associated with an increased risk for MM in comparison with the presence of two homozygous wild-type alleles (Trembath et al., 1999; Lievers et al., 2001).
In a recent report by Martinez et al (2009), a third SNP (rs3737965), this one in the promoter region of the MTHFR gene, was found to be associated with MM risk. A genome-wide SNP association study of genes important in folate metabolism found that rs3737965 was associated with lower plasma folate levels but not elevated plasma homocysteine (Hcy) levels (Hazra et al., 2009).
All of the evidence together points to the importance of variation in the MTHFR gene in the etiology of MM indicating that a comprehensive study of the MTHFR gene in an MM population is important. In a recent review paper by Au et al. (2010), it was noted that of the 32 published studies of the MTHFR gene within different NTD-affected populations, 29 of the 32 tested only (Mulinare et al., 1988) two specific nonsynonymous SNPs (i.e., c.677C>T/p.Ala222Val and/or 1298A>C/p.Glu429Ala) located within the gene.
The objective of the current study was to deep sequence the 12 exons of the MTHFR gene in DNAs from 96 MM subjects to obtain a more comprehensive picture of variants in the MTHFR gene potentially contributing to MM risk.
MATERIALS AND METHODS
Study Population
The MM subjects tested in the current study are a subset of a larger cohort of MM subjects participating in genetic studies in our laboratory. The study was approved by the Institutional Review Board of the University of Texas Health Science Center at Houston (UTHSC). The study population characteristics are further described in Au et al., 2008. The 96 MM subjects tested in this study included 49 Caucasians of European descent and 47 Hispanics of Mexican descent living in the United States or Canada who were randomly selected from our study cohort based on ethnicity and age relative to the date of the US FDA mandate on food fortification with folic acid in 1998. We only included MM affected subjects born between 1968 and 1993, prior to mandatory folic acid fortification.
DNA Sequencing
Blood and/or saliva samples were obtained from the patients and their parents. Genomic DNA was extracted from whole blood using the Puregene DNA Purification Kit (Gentra Systems, Minneapolis, MN). DNA from saliva was extracted using the Oragene DNA Preparation Kit (DNA Genotek, Ontario, Canada).
Polymerase chain reaction (PCR) and nested-sequencing primers were designed based on the reference genomic sequences of MTHFR (NM_005957) extracted from the University of Santa Cruz Genome Browser (UCSC Genome Browser ; http://genome.ucsc.edu/cgi-bin/hgGateway). We included approximately 50–100 bases flanking the boundaries of exons to also examine the splice donors and acceptors. A total of 11 PCR primer pairs were designed (available upon request) and synthesized by Integrated DNA Technologies USA. PCR amplification of exons was done using the MJ Research PTC-100 ™ Programmable Thermal Cycler (MJ Research Inc., Waltham, MA, USA). We confirmed the PCR product sizes by 1.4 % agarose gel electrophoresis. The amplified exon DNA was then treated with T7 exonuclease 1 and Shrimp Alkaline Phosphatase (United States Biochemicals, Affymetrix Inc., Cleveland, OH, USA). We sequenced the exons using the BigDye®Terminator (Applied Biosystems Inc., ABI, Foster City, CA, USA) reagents using a nested-sequencing primer and resolved the sequenced products on the ABI3100 Genetic Analyzer (ABI).
Data Analysis
Sequence results were analyzed using the DNA Sequencing Analysis Software v5.1 from ABI. To identify variants in sequences, we first manually compared results to the reference sequences (NM 005957) obtained from UCSC Genome Browser. We then used Biolign 4.0.6 software (http://en.bio-soft.net/dna/BioLign.html) to cross check. The Caucasian reference population (CEU) includes Caucasians from Utah included in the Centre d’Étude du Polymorphisme Humain Collection for mapping genetic markers. The reference Mexican American (MEX) population includes Mexican Americans from Los Angeles used in the HapMap project, data from Martin et al. (2006) and data from 1000 Genomes Project in dbSNP. Allele frequencies of variants were tallied, and any variant not reported in dbSNP was considered a novel variant. The allele frequencies of known SNPs of MM subjects were compared to the ethnically matched population frequencies of the same SNPs reported in dbSNP using Fisher’s test. A two-tailed p value of 0.05 and below was considered significant.
RESULTS
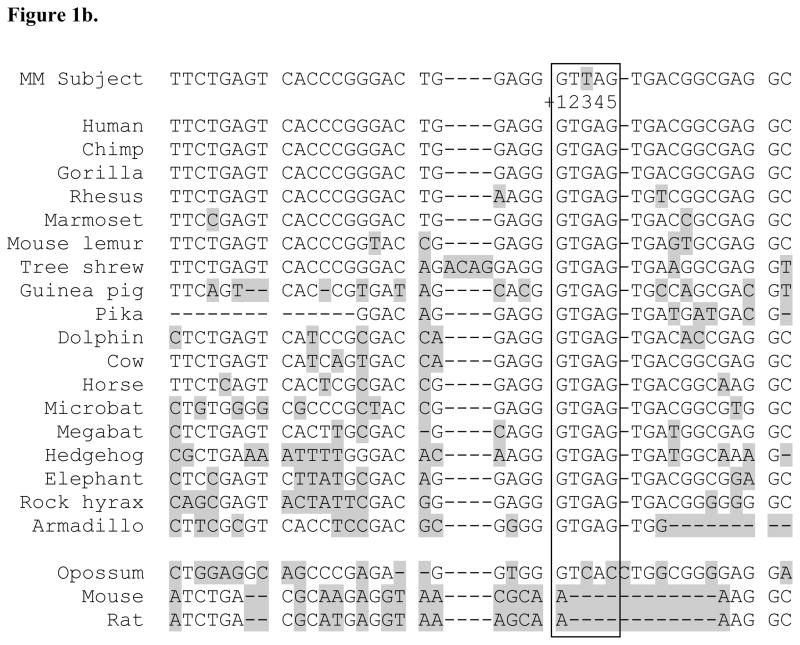
In sequencing the 12 exons of the MTHFR gene among 96 MM affected subjects, we found a novel variant at c.171+3G>T three bases downstream from exon 1 within the splice donor consensus sequence (Buratti et al,, 2007) not previously reported in public SNP databases (Figure 1a). Aligning sequences of the MTHFR gene exon 1 splice donor region of 44 different vertebrate species using the UCSC Genome Browser showed the +3G sequence is highly conserved except in mouse, rat and opossum (Figure 1b). DNA of the affected individual’s mother was available for sequencing and this variant was not present. The father’s DNA was not available for testing.
Figure 1.
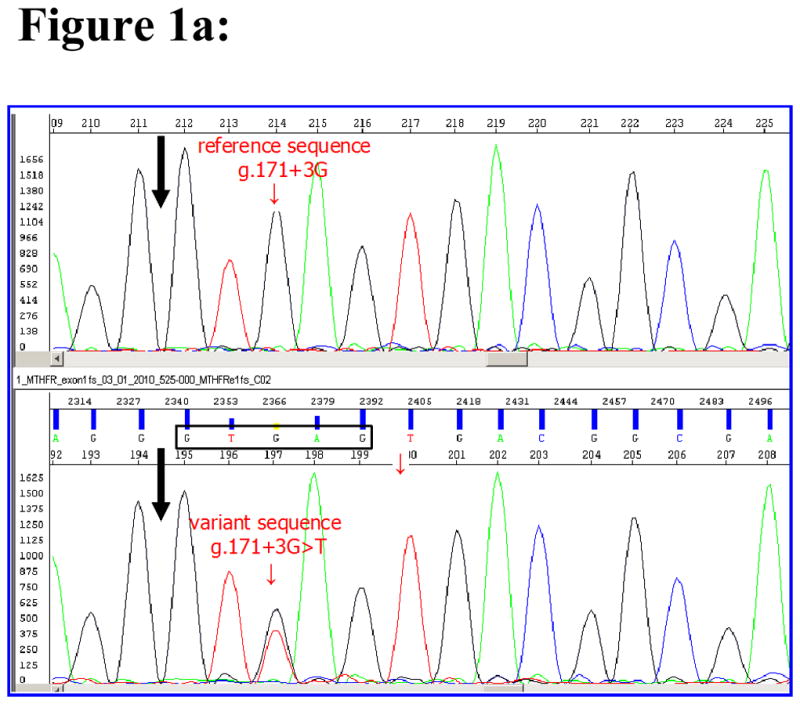
Properties of the novel MTHFR gene intron 1 splice donor variant found in MM patient SB525-000. 1a. sequencing result of the novel variant within intron 1 splice donor sequence of MM subject. Black arrow indicates end of exon 1 sequences of MTHFR gene of a reference sample (top panel) and an MM sample (bottom panel) carrying the novel c.171+3G>T mutation. Splice donor consensus sequences from +1 to +5 (GTGAG) of MM subject are boxed. Small arrow points to the position of the third nucleotide G of splice donor site in the reference sample and shows G and T in MM subject sample. 1b. The +3G nucleotide in the MTHFR intron 1 splice donor site is highly conserved. Sequences of the MTHFR gene intron 1 splice donor from +1 to +5 (GTGAG in box) of 21 different vertebrate species were aligned by the UCSC Genome Browser conservation track using the latest version of genome sequences of each species (http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=216042193&c=chr1&g=cons44way). The human MTHFR reference sequence NM_005957 (chr1:11768374-11788747, GRCH36/hg18) was used to extract conservation information. The sequence including the c.171+3T variant from an MM subject is added and shown as MM subject at top. Sequences differing from the human sequences are indicated by shading.
Currently, there are 122 SNPs reported within the exon regions we sequenced, among them, 84 are coding SNPs (cSNPs) and 38 are intronic non-coding SNPs (ncSNPs). The cSNPs include 45 nonsynonymous SNPs (nsSNPs), 36 synonymous SNPs (sSNPs) and three nonsense SNPs. We observed variations in allele frequencies for 18 SNPs (4 nsSNPs, 4 sSNPs and 10 ncSNPs) for the Caucasian and Mexican American MM subject groups we examined (Table 1). Four of the 18 SNPs do not have population frequencies reported for the reference Caucasian (CEU) population and one of the 18 SNPs does not have a frequency reported for the reference Mexican American (MEX) populations.
Table 1.
SNPs with rare alleles in the MTHFR gene of MM subjects categorized by ethnicity
| SNP | A1/A2 | significance | Caucasian (A1/A2) | Mexican (A1/A2) | ||
|---|---|---|---|---|---|---|
| MM | controls | MM | controls | |||
| rs3834043 | G/- | Intron 1 | 94/4 | No data | 94/0 | 120/0 c |
| rs45594036 | G/T | Intron 1 | 91/5 | 114/6 d | 92/2 | 114/6 c |
| rs13306561 | T/C | Intron 1 | 89/7a | 82/24 | 92/2 | 109/7 |
| rs2066470 | C/T | Pro39Pro | 94/4 | 104/12 | 93/1 | 109/5 |
| rs2066466 | G/A | Thr139Thr | 98/0 | No data | 92/2 | 119/1 c |
| rs1801133 | C/T | Ala222Val | 58/40a | 91/29 | 43/51 | 69/47 |
| rs2066462 | C/T | Ser352Ser | 94/4a | 100/14 | 93/1 | 113/7 c |
| rs1994798 | T/C | Intron 7 | 61/37 | 55/49 | 74/20 | 95/25 c |
| rs12121543 | G/T | Intron 7 | 73/21 | 87/33 | 89/5b | 96/20 |
| rs1801131 | A/C | Glu429Ala | 68/26 | 77/43 | 87/7b | 92/24 |
| rs4846051 | T/C | Phe435Phe | 92/2 | 120/0 | 92/2 | 113/3 |
| rs17375901 | G/A | Intron 9 | 93/5 | 113/7 | 94/0 | 118/2 c |
| rs1476413 | G/A | Intron 9 | 77/21 | 84/36 | 89/5b | 94/22 |
| rs55686944 | G/A | Intron 11 | 90/8 | 111/9 d | 88/4 | No data |
| rs3818762 | C/G | Intron 11 | 78/20 | 80/36 | 89/5 | 113/7 c |
| rs45622739 | G/A | Intron 11 | 98/0 | No data | 93/1 | 120/0 c |
| rs2274976 | G/A | Arg594Gln | 98/0a | 113/7 | 93/1 | 111/5 |
| rs35737219 | C/T | Thr653Met | 96/2 | No data | 92/2 | 120/0 c |
Notes: A1/A2 represents alleles 1 and 2 of the SNPs. SNPs and allele counts with significant difference compared to reference population are bolded.
p<0.05 comparing Caucasian MM subject to dbSNP Caucasian control allele frequencies.
p <0.05 comparing Mexican American MM subject to Mexican American control allele frequencies.
Controls = reference population from dbSNP or other sources include
control population data from Martin et al. 2006, and
control population data from the 1000 Genome Project.
We compared the allele frequencies for the SNPs between MM subjects to the ethnically-matched reference population and observed a statistically significant higher minor allele frequency for rs1801133 (c.677C>T) among Caucasian subjects (Table 2). A higher 677T allele frequency was also observed in Mexican American MM subject but the difference did not reach significance (p=0.0525). In addition, statistically significant lower minor allele frequencies among Caucasian subjects were observed for three additional SNPs (rs13306561, rs2066462, and rs2274976) (Table 2). We also observed significantly lower minor allele frequencies among the Mexican American subjects for three SNPs, (rs12121543, rs1801131 and rs1476413) (Table 2).
Table 2.
SNPs in the MTHFR gene with significant association with MM
| MTHFR | SNP | Allele A1/A2 | Allele frequency A1/A2 | ||
|---|---|---|---|---|---|
| MM | control | p value | |||
| Caucasian American | |||||
| Intron 1 | rs13306561 | T/C | 0.927/0.073 | 0.774/0.226 | p=0.003 |
| Exon 5 | rs1801133 (Ala222Val)) | C/T | 0.592/0.408 | 0.758/0.242 | p=0.013 |
| Exon 7 | rs2066462 (Ser352Ser) | C/T | 0.958/0.042 | 0.877/0.123 | p=0.047 |
| Exon 12 | rs2274976 (Arg594Gln) | G/A | 1.000/0.000 | 0.942/0.058 | p=0.018 |
| Mexican American | |||||
| Intron 7 | rs12121543 | G/T | 0.974/0.053 | 0.828/0.172 | p=0.009 |
| Exon 8 | rs1801131 (Glu429Ala) | A/C | 0.926/0.074 | 0.793/0.207 | p=0.010 |
| Intron 10 | rs1476413 | G/A | 0.947/0.053 | 0.810/0.190 | p=0.003 |
Note: p values – Fisher Exact two-tailed test results.
When we compared allele frequencies by ethnicity within our MM subjects, there was a significant difference between Caucasians and Mexican Americans for five SNPs (rs12121543, rs1801131, rs1476413, rs3818762 and rs1994798).
The MM subjects in the study showed only the reference allele for the other 104 known SNPs in the 12 sequenced exon regions.
DISCUSSION
Despite the fact that many studies have demonstrated variants such as c.677C>T and c.1298A>C of the MTHFR gene are associated with NTDs, the presence of these variants in the general population without NTDs suggests these variants are not directly causative for NTDs. It is important to identify previously unreported MTHFR variants present in the genome of individuals with MM and evaluate whether these variants contribute to the disease phenotype. In the study reported here, we identified a novel variant (c.171+3G>T) within the splice donor consensus −3 to +7 sequence MAG|GUGAGU (Buratti et al., 2007) of intron 1 of the MTHFR gene in one MM subject. Exon 1 of MTHFR codes for a major part of the 5′ untranslated region (5′-UTR) of MTHFR mRNA. Splicing defects of the 5′-UTR potentially affects transcription of functional mRNA and influencing stability and/or translation efficiency of MTHFR. The +3G of the splice donor site sequence is highly conserved in 18 placental mammals with the exceptions being in mouse, rat and opossum suggesting functional importance of maintaining the nucleotide G in position +3. Using the online Human Splicing Finder program (http://www.umd.be/HSF/; Desmet et al., 2009), we submitted the wildtype exon 1 intron 1 boundary sequences ( with +3G) of MTHFR (NM_005957) to identify potential splicing related sites. Then we introduced a mutation to substitute +3G with +3T and found the novel +3T variant is predicted to affect several splice donor activities of exon 1. Pathological mutations occurring within the extended consensus sequences of exon–intron splice junctions account for 10% of all inherited lesions included in The Human Gene Mutation Database (http://www.hgmd.org) and are frequently encountered in mutations screening studies (Krawczak et al., 2007; Stenson et al., 2009). Introns probably represent a substantially larger mutational target than has hitherto been appreciated because it is now known that they contain a multiplicity of functional elements, including intron splice enhancers and silencers that regulate alternative splicing, trans-splicing elements and other regulatory elements ((Tress et al., 2007; Gingeras, 2009; Wang et al., 2009). Some of these elements may be deeply embedded within very large introns (Solis et al., 2008). Studying the functional implication of non-coding SNPs (ncSNPs) that are located in the introns is expensive and challenging. The potential of an ncSNP influencing splicing cannot be validated without functional assays. Obtaining lesion tissue from the MM subject to test for the presence of a splice variant is not possible because the lesion was surgically repaired before enrollment in the study. Further study utilizing in vitro methods such as minigene construct expression in disease relevant cell lines may reveal the functional significance of the novel variant c.171+3T.
Eighteen of the 122 known SNPs within the 12 exons and the flanking intron sequences of the MTHFR gene have observed rare alleles in the MM subjects while the remaining 104 SNPs revealed only the reference allele as reported in the reference populations. Interestingly, the rare alleles of all SNPs except for rs1801133 are underrepresented in the MM subjects. Seven of the 18 SNPs demonstrated significant differences between the ethnically matched reference populations and MM subjects. Two of the significant cSNPs (rs1801133 and rs1801131) were shown to associate with MM in the current study as well as in previous studies with NTDs. Five of the seven significant SNPs (rs13306561, rs2066462, and rs2274976 for Caucasian MM subjects; rs12121543 and rs1476413 for Mexican American MM subjects) have not previously been reported to associate with MM. The study results suggest that further investigation of the five SNPs in future studies of MM subjects who have different ethnic backgrounds is warranted.
Only four nsSNPs (rs1801133, rs1801131, rs2274976, and rs35737219, coding for rare variants p.Ala222Val, p.Glu429Ala, p.Arg594Gln and p.Thr653Met, respectively) and four sSNPs (rs2066466, rs2066462, rs2066432 and rs4846051) showed the rare variants in the MM subjects thus allowing for further statistical analysis. Three nsSNPs (rs1801133, rs1801131 and rs2274976) and one sSNP rs2066462 demonstrated statistical significance in either Caucasian or Mexican MM subjects. Among the intronic SNPs, we identified 11 ncSNPs with a rare allele in the MM subjects. Three ncSNPs (rs13306561 for Caucasian MM subjects; rs12121543 and rs1476413 for Mexican American subjects) showed significant association with MM subjects. It is possible that the five newly identified SNPs with demonstrated significance in the current study were in linkage disequilibrium (LD) with the two known NTD associated nsSNPs (rs1801131 and rs1801133). All seven SNPs with demonstrated significance here are in LD with a D′ values over 0.8 (HapMap, http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap3r2_B36). However, their correlation coefficients R^2 are much less than 0.5 implying the LD relationships suggested by the D′ values are not well correlated.
The MTHFR enzyme activity resulting from the four rare missense variants (p.Ala222Val, p.Glu429Ala, p.Arg594Gln, and p.Thr653Met) found in MM subjects have been examined, potentially shedding some light on the biological significance of the presence of the rare variants in MM subjects (van der Put et al., 1995; Martin et al., 2006; Marini et al., 2008). In one study, Marini et al (2008) identified 14 nonsynonymous changes including 11 having minor allele frequencies <1% and three with more frequent rare alleles (p.Ala222Val, p.Glu429Ala, and p.Arg594Gln). Six nsSNP variants (p.Met110Ile, p.Arg134Cys, p.His213Arg, p.Ala222Val, p.Asp223Asn and p.Asp291Asn) fell within the catalytic domain of MTHFR and the authors found only the p.His213Arg variant was benign when expressed in yeast. The variants p.Met110Ile, p.Ala222Val, p.Asp223Asn and p.Asp291Asn when expressed in yeast displayed folate remedial behavior, i.e., these enzyme variants expressed similar to the major allele at higher concentrations of folate supplementation (50–200ug/ml folinic acid) but were considerably weakened as folate levels decreased.
In 2006, an MTHFR exon resequencing study by Martin et al. (2006) examined 240 DNA samples from four reference ethnic groups including 60 Los Angeles Mexican Americans for MTHFR gene variants in the HapMap project. Martin et al. also performed functional studies of the MTHFR variants they identified using the monkey kidney cell line COS-1. The MTHFR variants with p.Glu429Ala or p.The653Met exhibited comparable enzyme activity to the wild type MTHFR while the variants with p.Ala222Val or p.Arg594Gln showed reduced activity (Martin et al., 2006). In contrast to Martin et al., two other studies showed a decrease in enzyme activity for the 429Ala variant (Weisberg et al., 2001; Frosst et al., 2006). The MM subjects in our study have a higher than reported frequency of the p.Ala222Val variant in Caucasian MM subjects (40.8%) and Mexican American subjects (54.3%), consistent with previous findings associating the variant with MM risk (van der Put et al., 1995; Ou et al., 1996; Shields et al., 1999).
Interestingly, Martin et al. (2006) also tested the effect of the presence of two MTHFR variants in cis and found that the p.Glu429Ala variant when paired with p.Thr653Met resulted in reduced MTHFR activity. Additionally, the authors found reduction in the amount of double mutants (p.Glu429Val and p.Thr653Met) containing MTHFR in the COS cell to be another reason for loss of enzyme activity suggesting the double mutant may be poorly translated or unstable. There are four MM subjects heterozygous for both p.Glu429Ala in exon 8 and p.Thr653Met in exon 12 of MTHFR. Unfortunately, we can not determine whether these variants are in cis or trans because mRNAs from these subjects are not available for further analysis.
Our study and others have demonstrated that two common polymorphisms, rs1801133 (c.677C>T; p.Ala222Val) and rs1801131 (c.1298A>C; p.Glu429Ala), of the MTHFR gene associate independently with NTDs (Gonzalez-Herrera et al., 2007). The MTHFR SNP rs1801133 resulting in the p.Ala222Val thermolabile protein variant is the most frequently investigated polymorphism in NTDs with a positive association observed in some populations (van der Put et al., 1995; Ou et al., 1996; Shields et al., 1999; O’Leary et al., 2005). One large study (Shields et al., 1999) of 271 NTD cases in 218 families in Ireland concluded that their results favored a biological model of MTHFR-related NTD pathogenesis in which suboptimal maternal folate status imposes biochemical stress on the developing embryos possessing the TT genotype. The “TT” genotype in that study was present in 18.8% of cases but only in 8.3% of controls. Similarly, our finding is that the Caucasian MM subjects have the TT genotype 16.7% compared to 6.7% in the Caucasian reference population in dbSNP. Compared to the ethnically matched reference population in public databases (dbSNP and HapMap), our study found a significantly higher incidence of the 677T allele in MM Caucasians (p=0.013), an observation consistent with findings from other studies that presence of the 677T allele contribute to risk of NTDs.
On the other hand, other studies have shown no evidence that the 677T allele associates with NTDs. One study examined 65 subjects with SB, 60 of their mothers and 110 control subjects in the State of Yucatan, Mexico. This study did not observe statistical significance in the genotype or allele frequencies between cases and controls suggesting that the thermolabile variant C677T is not an associated risk factor for the development of NTDs in that population (Gonzalez-Herrera et al., 2002). We found in the current study that the 677T allele did not associate with MM development in Mexican American subjects. However, we observed the 677T allele frequency for the MM subjects is higher than the HapMap MEX reference population (54.3% and 40.5% respectively) with a two-tailed Fisher Exact p=0.0525 approaching significance level. Similar to the Gonzalez-Herrera et al study, the power of our study is limited by the small sample size. We need to examine a larger MM population to validate the finding.
A few studies have evaluated the impact of NTD association of 429Ala (c.A1298C), a SNP localized within the C-terminal regulatory domain of the MTHFR protein ( De Marco et al., 2001; De Marco et al., 2002; Gonzalez-Herrera et al., 2007), with conflicting results. In studies by De Marco et al., the A1298C polymorphism is concluded to be a contributing risk factor for NTDs in the Italian population (De Marco et al., 2001; De Marco et al., 2002). In contrast, we find significantly lower incidence of 429Ala among Mexican American MM subjects in this study. The role that 429Ala plays in NTD development remains to be elucidated.
In conclusion, we identified a novel variant c.171+3G>T in exon 1 of the MTHFR gene of an MM affected subject that can potentially affect splicing. More studies need to be done to determine whether splicing or translation is affected. We found significant differences in the allele frequencies in seven SNPs when we compared our MM subjects with the reference population in dbSNP. Our results provide further evidence to support previous findings of the association of rs1801133 with SB. Five of these SNPs have not previously been identified by other studies as associated with MM or NTDs. Additionally, we showed differences in allele frequencies between the two studied subject populations arguing the importance for matched-controls to be used in similar studies.
The strength of this study is that we resequenced all 12 exons of the MTHFR gene including 50–100 flanking bases by Sanger sequencing in 96 MM subjects from the two ethnic American groups with the highest prevalence of NTDs. All selected patients were born before the FDA mandated folic acid fortification of food crops.
There are several limitations to the study. One limitation is the small sample size that will exclude finding of rare variants with frequency < 2% for each ethnic MM subject group. Examining a larger sample size by deep sequencing is currently cost prohibitive. Another limitation of our small sample size is the limited statistical power of the study to identify significant association of SNPs with low rare allele frequency. We are currently studying additional SNPs within the MTHFR gene by genotyping with SNPlex to test for the presence of the other five SNPs (rs13306561, rs2066462, and rs2274976 rs12121543 and rs1476413) in our large MM population and their parents.
Acknowledgments
This work was supported by grants from the National Institutes of Health (P01 HD35946-06A2) and the Shriners Hospital for Children (project 8580). Additional support was provided by Dr. Kathleen Kennedy, Richard W. Mithoff Professor, Division of Neonatology, Department of Pediatrics, The University of Texas Medical School at Houston.
We thank the patients and their families for their participation in this study. We are also grateful to Phong X Tran and Michelle O’Byrne for technical assistance.
References
- Au KS, Ashley-Koch A, Northrup H. Epidemiologic and genetic aspects of spina bifida and other neural tube defects. Dev Disabil Res Rev. 2010;16:6–15. doi: 10.1002/ddrr.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KS, Tran PX, Tsai CC, O’Byrne MR, Lin JI, Morrison AC, Hampson AW, Cirino P, Fletcher JM, Ostermaier KK, Tyerman GH, Doebel S, Northrup H. Characteristics of a spina bifida population including North American Caucasian and Hispanic individuals. Birth Defects Res A Clin Mol Teratol. 2008;82:692–700. doi: 10.1002/bdra.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, Kibar Z. Genetic basis of neural tube defects. Semin Pediatr Neurol. 2009;16:101–110. doi: 10.1016/j.spen.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Beaudin AE, Stover PJ. Folate-mediated one-carbon metabolism and neural tube defects: balancing genome synthesis and gene expression. Birth Defects Res C Embryo today. 2007;81:183–203. doi: 10.1002/bdrc.20100. [DOI] [PubMed] [Google Scholar]
- Blencowe H, Cousens S, Modell B, Lawn J. Folic acid to reduce neonatal mortality from neural tube disorders. Internat J of Epidemiol. 2010;39(Suppl 1):i110–21. doi: 10.1093/ije/dyq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, Moore CA, Khoury MJ, Erickson JD. Neural-tube defects. New Engl J of Med. 1999;341:1509–1519. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- Botto LD, Yang Q. 5,10-Methylenetetrahydrofolate reductase gene variants and congenital anomalies: a HuGE review. Am J of Epidemiol. 2000;151:862–877. doi: 10.1093/oxfordjournals.aje.a010290. [DOI] [PubMed] [Google Scholar]
- Boulet SL, Yang Q, Mai C, Kirby RS, Collins JS, Robbins JM, Meyer R, Canfield MA, Mulinare J National Birth Defects Prevention Network. Trends in the post fortification prevalence of spina bifida and anencephaly in the United States. Birth Defects Res A Clin Mol Teratol. 2008;82:527–532. doi: 10.1002/bdra.20468. [DOI] [PubMed] [Google Scholar]
- Buratti E, Chivers M, Kralovicova J, Romano M, Baralle M, Krainer AR, Vorechovsky I. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nuclei Acid Res. 2007;35(13):4250–4263. doi: 10.1093/nar/gkm402. PMID. PMID: 17576681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Recommendations for the use of folic acid to reduce the number of cases of spina bifida and other neural tube defects. Morbidity and Mortality Weekly Report (MMWR) 1992;41(RR-14):1–7. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Spina bifida and anencephaly before and after folic acid mandate--United States, 1995–1996 and 1999–2000. MMWR. 2004;53:362–365. [PubMed] [Google Scholar]
- Center for Disease Control and Prevention. Racial/Ethnic Differences in the Birth Prevalence of spina bifida - United States, 1995--2005. MMWR. 2009;57:1409–1413. [PubMed] [Google Scholar]
- Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4:784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- Czeizel AE, Dudas I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. New Engl J of Med. 1992;327:1832–1835. doi: 10.1056/NEJM199212243272602. [DOI] [PubMed] [Google Scholar]
- De Marco P, Calevo MG, Moroni A, Arata L, Merell E, Finnell RH, Zhu H, Andreussi L, Cama A, Capra V. Study of MTHFR and MS polymorphisms as risk factors for NTD in the Italian population. J of Hum Genet. 2002;47:319–324. doi: 10.1007/s100380200043. [DOI] [PubMed] [Google Scholar]
- De Marco P, Calevo MG, Moroni A, Arata L, Merello E, Cama A, Finnell RH, Andreussi L, Capra V. Polymorphisms in genes involved in folate metabolism as risk factors for NTDs. Eur J of Pediat Surgery : Suppl. 2001;1:S14–17. doi: 10.1055/s-2001-19739. PMID:118131127. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acid Res. 2009 May;37(9):e67. doi: 10.1093/nar/gkp215. Epub 2009 Apr 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Mathews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP, Rozen R. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206– 211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Herrera L, Garcia-Escalante G, Castillo-Zapata I, Canto-Herrera J, Ceballos-Quintal J, Pinto-Escalante D, Diaz-Rubio F, Del Angel RM, Orozco-Orozco L. Frequency of the thermolabile variant C677T in the MTHFR gene and lack of association with neural tube defects in the State of Yucatan, Mexico. Clin Genet. 2002;62:394–398. doi: 10.1034/j.1399-0004.2002.620507.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Herrera L, Castillo-Zapata I, Garcia-Escalante G, Pinto-Escalante D. A1298C polymorphism of the MTHFR gene and neural tube defects in the state of Yucatan, Mexico. Birth Defects Res A Clin Mol Teratol. 2007;79:622–626. doi: 10.1002/bdra.20381. [DOI] [PubMed] [Google Scholar]
- Goyette P, Pai A, Milos R, Frosst P, Tran P, Chen Z, Chan M, Rozen R. Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR) Mamm Genome. 1998;9:652–656. doi: 10.1007/s003359900838. [DOI] [PubMed] [Google Scholar]
- Grosse SD, Ouyang L, Collins JS, Green D, Dean JH, Stevenson RE. Economic evaluation of a neural tube defect recurrence-prevention program. Am J of Preventive Med. 2008;35:572–577. doi: 10.1016/j.amepre.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Hazra A, Kraft P, Lazarus R, Chen C, Chanock SJ, Jacques P, Selhub J, Hunter DJ. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet. 2009;18:4677–4687. doi: 10.1093/hmg/ddp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homberger A, Linnebank M, Winter C, Willenbring H, Marquardt T, Harms E, Koch HG. Genomic structure and transcript variants of the human methylenetetrahydrofolate reductase gene. Eur J of Hum Genet. 2000;8:725–729. doi: 10.1038/sj.ejhg.5200522. [DOI] [PubMed] [Google Scholar]
- Honein MA, Paulozzi LJ, Mathews TJ, Erickson JD, Wong LY. Impact of folic acid fortification of the US food supply on the occurrence of neural tube defects. J of Am Med Assoc. 2001;285:2981–2986. doi: 10.1001/jama.285.23.2981. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- Lievers KJ, Boers GH, Verhoef P, den Heijer M, Kluijtmans LA, van der Put NM, Trijbels FJ, Blom HJ. A second common variant in the methylenetetrahydrofolate reductase (MTHFR) gene and its relationship to MTHFR enzyme activity, homocysteine, and cardiovascular disease risk. J Mol Med. 2001;79:522–528. doi: 10.1007/s001090100253. [DOI] [PubMed] [Google Scholar]
- Marini NJ, Gin J, Ziegle J, Keho KH, Ginzinger D, Gilbert DA, Rine J. The prevalence of folate-remedial MTHFR enzyme variants in humans. Proc Natl Acad Sci USA. 2008;105:8055–8060. doi: 10.1073/pnas.0802813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin YN, Salavaggione OE, Eckloff BW, Wieben ED, Schaid DJ, Weinshilboum RM. Human methylenetetrahydrofolate reductase pharmacogenomics: gene resequencing and functional genomics. Pharmacogenet and Genom. 2006;16:265–277. doi: 10.1097/01.fpc.0000194423.20393.08. [DOI] [PubMed] [Google Scholar]
- Martinez CA, Northrup H, Lin JI, Morrison AC, Fletcher JM, Tyerman GH, Au KS. Genetic association study of putative functional single nucleotide polymorphisms of genes in folate metabolism and spina bifida. Am J Obstet Gyn. 2009;201:394, e1–11. doi: 10.1016/j.ajog.2009.06.042. PMID:19683694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MRC Vitamin Study Research Group. Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. Lancet. 1991;338:131–137. [PubMed] [Google Scholar]
- Mulinare J, Cordero JF, Erickson JD, Berry RJ. Periconceptional use of multivitamins and the occurrence of neural tube defects. J Am Med Assoc. 1988;260:3141–3145. [PubMed] [Google Scholar]
- O’Leary VB, Mills JL, Parle-McDermott A, Pangilinan F, Molloy AM, Cox C, Weiler A, Conley M, Kirke PN, Scott JM, Brody LC. Birth Defects Research Group. Screening for new MTHFR polymorphisms and NTD risk. Am J Med Genet A. 2005 Oct 1;138A(2):99–106. doi: 10.1002/ajmg.a.30846. [DOI] [PubMed] [Google Scholar]
- Ou CY, Stevenson RE, Brown VK, Schwartz CE, Allen WP, Khoury MJ, Rozen R, Oakley GP, Jr, Adams MJ., Jr 5,10 Methylenetetrahydrofolate reductase genetic polymorphism as a risk factor for neural tube defects. Am J Med Genet. 1996;63:610–614. doi: 10.1002/(SICI)1096-8628(19960628)63:4<610::AID-AJMG15>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Shields DC, Kirke PN, Mills JL, Ramsbottom D, Molloy AM, Burke H, Weir DG, Scott JM, Whitehead AS. The “thermolabile” variant of methylenetetrahydrofolate reductase and neural tube defects: An evaluation of genetic risk and the relative importance of the genotypes of the embryo and the mother. Am J Hum Genet. 1999;64:1045–1055. doi: 10.1086/302310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis AS, Shariat N, Patton JG. Splicing fidelity, enhancers, and disease. Front in Biosci. 2008;13:1926–1942. doi: 10.2741/2812. [DOI] [PubMed] [Google Scholar]
- Solis AS, Shariat N Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P, Leclerc D, Chan M, Pai A, Hiou-Tim F, Wu Q, Goyette P, Artigas C, Milos R, Rozen R. Multiple transcription start sites and alternative splicing in the methylenetetrahydrofolate reductase gene result in two enzyme isoforms. Mamm Genome. 2002;13:483–492. doi: 10.1007/s00335-002-2167-6. [DOI] [PubMed] [Google Scholar]
- Trembath D, Sherbondy AL, Vandyke DC, Shaw GM, Todoroff K, Lammer EJ, Finnell RH, Marker S, Lerner G, Murray JC. Analysis of select folate pathway genes, PAX3, and human T in a Midwestern neural tube defect population. Teratology. 1999;59:331–341. doi: 10.1002/(SICI)1096-9926(199905)59:5<331::AID-TERA4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PI, Albrecht M, Hegyi H, Giorgetti A, Raimondo D, Lagarde J, Laskowski RA, Lopez G, Sadowski MI, Watson JD, Fariselli P, Rossi I, Nagy A, Kai W, Storling Z, Orsini M, Assenov Y, Blankenburg H, Huthmacher C, Ramirez F, Schlicker A, Denoeud F, Jones P, Kerrien S, Orchard S, Antonarakis SE, Reymond A, Birney E, Brunak S, Casadio R, Guigo R, Harrow J, Hermjakob H, Jones DT, Lengauer T, Orengo CA, Patthy L, Thornton JM, Tramontano A, Valencia A. The implications of alternative splicing in the ENCODE protein complement. Proc Natl Acad Sci USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Put NM, Steegers-Theunissen RP, Frosst P, Trijbels FJ, Eskes TK, van den Heuvel LP, Mariman EC, den Heyer M, Rozen R, Blom HJ. Mutated methylenetetrahydrofolate reductase as a risk factor for spina bifida. Lancet. 1995;346:1070–1071. doi: 10.1016/s0140-6736(95)91743-8. [DOI] [PubMed] [Google Scholar]
- van der Put NM, Eskes TK, Blom HJ. Is the common 677C>T mutation in the methylenetetrahydrofolate reductase gene a risk factor for neural tube defects? A meta-analysis. QJM. 1997;90:111–115. doi: 10.1093/qjmed/90.2.111. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang K, Radovich M, Wang Y, Wang G, Feng W, Sanford JR, Liu Y. Genome-wide prediction of cis-acting RNA elements regulating tissue-specific pre-mRNA alternative splicing. BMC Genomics. 2009;10(Suppl 1):S4. doi: 10.1186/1471-2164-10-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg IS, Jacques PF, Selhub J, Bostom AG, Chen Z, Curtis Ellison R, Eckfeldt JH, Rozen R. The 1298A>C polymorphism in methylenetetrahydrofolate reductase (MTHFR): in vitro expression and association with homocysteine. Atherosclerosis. 2001;156:409–415. doi: 10.1016/s0021-9150(00)00671-7. [DOI] [PubMed] [Google Scholar]
- Yen IH, Khoury MJ, Erickson JD, James LM, Waters GD, Berry RJ. The changing epidemiology of neural tube defects. United States, 1968–1989. Am J of Dis Child. 1992;146:857–861. doi: 10.1001/archpedi.1992.02160190089028. [DOI] [PubMed] [Google Scholar]