Abstract
It has been known since the development of nearest neighbor analysis that the frequency of the dinucleotide CpG is markedly suppressed in vertebrate DNA (i.e. less than %C x %G). This suppression appears to be heterogeneous since it was shown some years ago that three vertebrate tRNA genes did not exhibit CpG suppression. We have analyzed 13 different human tRNA genes and found that they also do not exhibit CpG suppression. Because CpG suppression has been linked, to some extent at least, to the methylation-deamination process by which a methylated CpG is mutated to TpG, we investigated whether the lack of suppression of CpG in tRNAs could originate from an absence of methylation. Three human tRNA genes were selected from Genbank (lysine, Proline, and Phenylalanine) and examined for methylation at HpaII sites by polymerase chain reaction (PCR) and Southern blot analysis. The observed patterns were consistent with the absence of methylation at the seven HpaII sites analyzed in and around the tRNA genes, and we predict that the remaining CpGs in these genes will be unmethylated. Since GC-rich promoter regions also escape CpG suppression and since they are generally unmethylated, avoidance of methylation may be a general explanation for the absence of CpG suppression in selected regions of vertebrate genomes.
Full text
PDF
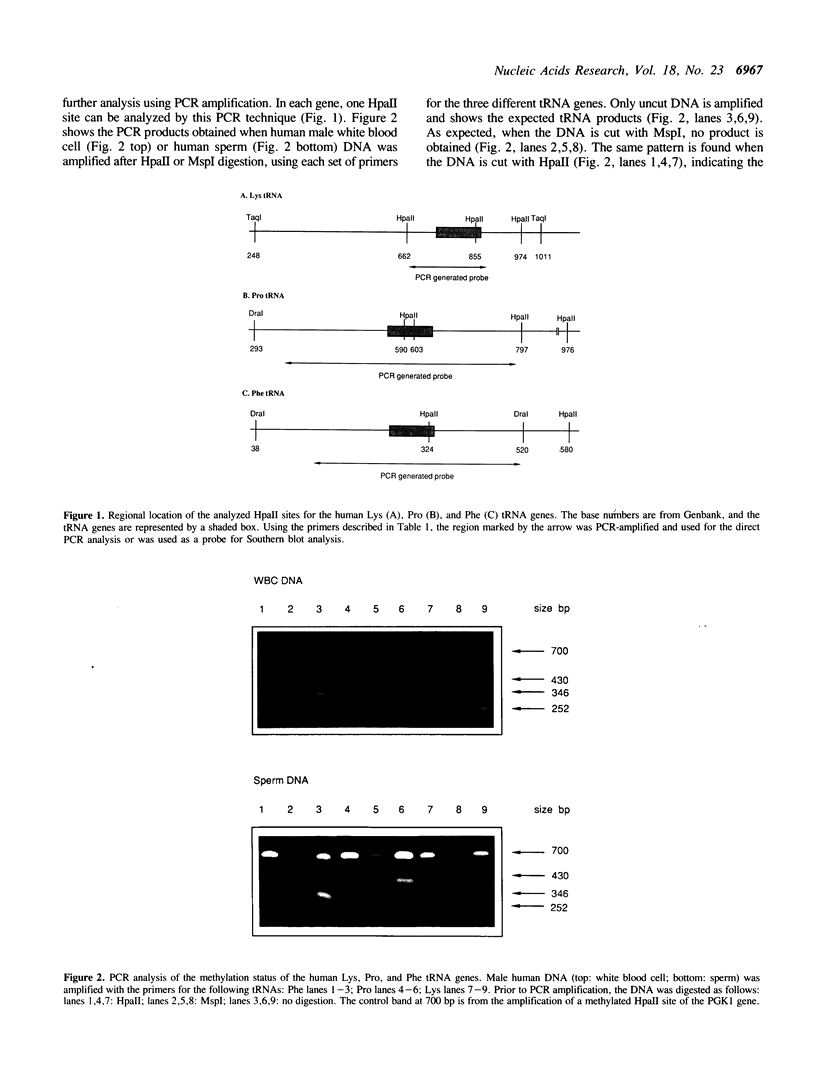


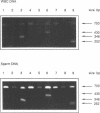
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., Eason R. Increased G + C content of DNA stabilizes methyl CpG dinucleotides. Nucleic Acids Res. 1984 Jul 25;12(14):5869–5877. doi: 10.1093/nar/12.14.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell. 1987 Sep 11;50(6):945–950. doi: 10.1016/0092-8674(87)90521-6. [DOI] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988 Aug 26;54(5):705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- Chang Y. N., Pirtle I. L., Pirtle R. M. Nucleotide sequence and transcription of a human tRNA gene cluster with four genes. Gene. 1986;48(1):165–174. doi: 10.1016/0378-1119(86)90362-8. [DOI] [PubMed] [Google Scholar]
- Cooper D. N., Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988 Feb;78(2):151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Doran J. L., Wei X. P., Roy K. L. Analysis of a human gene cluster coding for tRNA(GAAPhe) and tRNA(UUULys). Gene. 1987;56(2-3):231–243. doi: 10.1016/0378-1119(87)90140-5. [DOI] [PubMed] [Google Scholar]
- JOSSE J., KAISER A. D., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. VIII. Frequencies of nearest neighbor base sequences in deoxyribonucleic acid. J Biol Chem. 1961 Mar;236:864–875. [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Rideout W. M., 3rd, Coetzee G. A., Olumi A. F., Jones P. A. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990 Sep 14;249(4974):1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Russell G. J., Walker P. M., Elton R. A., Subak-Sharpe J. H. Doublet frequency analysis of fractionated vertebrate nuclear DNA. J Mol Biol. 1976 Nov;108(1):1–23. doi: 10.1016/s0022-2836(76)80090-3. [DOI] [PubMed] [Google Scholar]
- SWARTZ M. N., TRAUTNER T. A., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem. 1962 Jun;237:1961–1967. [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Tykocinski M. L., Max E. E. CG dinucleotide clusters in MHC genes and in 5' demethylated genes. Nucleic Acids Res. 1984 May 25;12(10):4385–4396. doi: 10.1093/nar/12.10.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]