Abstract
The ability to respond and adapt to changes in the physical environment is a universal and essential cellular property. Here we demonstrated that cells respond to mechanical compressive stress by rapidly inducing autophagosome formation. We measured this response in both Dictyostelium and mammalian cells, indicating that this is an evolutionarily conserved, general response to mechanical stress. In Dictyostelium, the number of autophagosomes increased 20-fold within 10 min of 1 kPa pressure being applied and a similar response was seen in mammalian cells after 30 min. We showed in both cell types that autophagy is highly sensitive to changes in mechanical pressure and the response is graduated, with half-maximal responses at ~0.2 kPa, similar to other mechano-sensitive responses. We further showed that the mechanical induction of autophagy is TOR-independent and transient, lasting until the cells adapt to their new environment and recover their shape. The autophagic response is therefore part of an integrated response to mechanical challenge, allowing cells to cope with a continuously changing physical environment.
Keywords: autophagy, homeostasis, mechanical stress, mechanobiology
Introduction
Macroautophagy (hereafter referred to as autophagy) is a catabolic process whereby cytosolic components are first sequestered in autophagosomes, then digested by fusion with the lysosomal system. Autophagic degradation fulfills a number of roles including the removal of excess or damaged organelles, degrading misfolded and aggregated proteins as well as promoting cell survival and adaptation to metabolic and cytotoxic stresses.1-3
Autophagy is induced by a number of different stresses. The best understood of these is during starvation, when autophagy maintains viability by supplying nutrients and amino acids from the digested cytosolic material4 but it is also induced by diverse stresses such as hypoxia, DNA damage, ER stress and pathogen infection.5-8 In these latter cases, the functional role of autophagy is less clear and can contribute to either survival and adaptation, or cell death, depending on the circumstances (for a review see ref. 9).
In addition to these chemical stresses, cells are also frequently exposed to mechanical stresses caused by sudden physical changes. It is therefore essential for cells to continuously detect and respond to mechanical forces, and adapt their physiology to maintain proper cellular function and protect against mechanical injury.10,11
Cells respond to mechanical stress in many ways. A number of groups have shown that mechanical forces rapidly induce a proportional stiffening of the cytoskeletal cortex10,12 and exposure of Dictyostelium cells to shear forces induces motility.13,14 Proper cytoskeletal function is critical for a number of processes including migration, growth and development and therefore dynamic adaptation to mechanical change is crucial.
In other cell types, the response to mechanical stress induces more specialized changes. For example, shear stress elicits a dramatic transcriptional response in endothelial cells,15 and mechanical stimulation is a potent regulator of osteoblast differentiation, regulating the density of bone.16,17 Responses to mechanical stimulation therefore govern a broad range of physiological processes at both the cellular and whole organism levels.
Despite the biological inevitability of mechanical challenge, little is known in detail about the strategies employed by cells to respond to this stress and survive. In this study, we showed that when subjected to compressive stress, both Dictyostelium and mammalian cells rapidly induce autophagy. This response is transient, lasting until the cell has remodeled its cortex to relieve the stress, indicting that autophagy is part of a physiological response that is activated during adaption to mechanical stress.
Results
The induction and quantitation of autophagy in Dictyostelium
Previous work using Dictyostelium has identified and disrupted a number of genes essential for autophagy.18-22 However, in order to use Dictyostelium to study this process in detail, it is important to first define the conditions required for, and the dynamics of, the normal autophagic response.
Autophagy is best understood as a starvation response, where the digestion of cytosolic components supplies nutrients to keep the cell alive. In order to starve Dictyostelium cells in a defined way, we used synthetic SIH medium23 lacking both lysine and arginine. In this amino acid-deficient medium, Dictyostelium cells are unable to grow and require autophagy to maintain viability for more than a few days (Fig. S1).
To monitor autophagy in living cells, we used GFP-Atg8 as a marker for phagophore formation. When autophagy is induced, the Atg8 protein is processed and lipidated, becoming incorporated into the expanding phagophore membrane.24-26 GFP-Atg8 is therefore frequently used as a marker for autophagy; in particular it translocates from a mainly cytosolic to a punctate localization upon autophagosome accumulation.
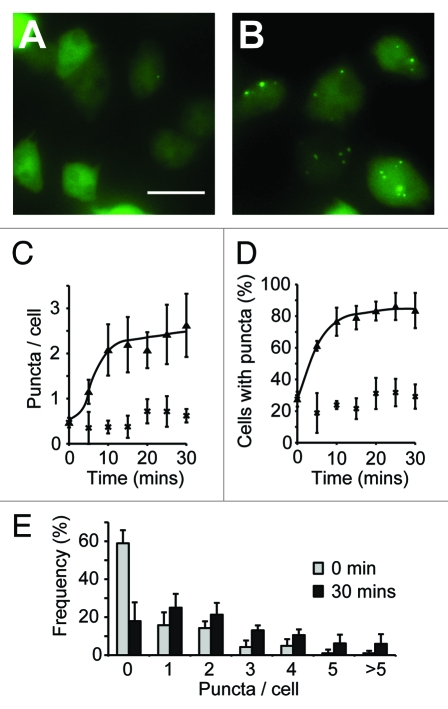
When we expressed GFP-Atg8 in Dictyostelium cells grown in complete SIH medium, the GFP signal was largely diffuse, with only 25% of cells containing any puncta, with an average of 0.5 puncta per cell (Fig. 1). However, 15 min after transfer to medium lacking arginine and lysine, 80% of cells contained between one and ten puncta, with an average of 2.4 per cell (Fig. 1). This number of autophagosomes was constant after 24 h indicating that this is the steady-state level, and that accumulation of autophagosomes had reached equilibrium after just 15 min. Placing the cells in non-nutrient phosphate buffer induced autophagy to the same extent (2.5 puncta per cell after 1 h), demonstrating that this level of induction is the maximum that can be achieved by starvation.
Figure 1.
Quantification of autophagic induction in Dictyostelium. Images of wild-type (ax3) cells expressing GFP-Atg8 grown in either complete SIH medium (A), or transferred into medium lacking arginine and lysine for 30 min (B). Images shown are the maximum intensity projection of a Z-series of images, bar = 20 μm. The number of GFP-Atg8 puncta was quantified over time and expressed as both the mean number per cell (C) and the proportion of cells containing at least one puncta (D) (cells in full medium, or lacking arginine and lysine are shown as crosses and triangles respectively). The frequency distribution of puncta in each cell is shown in (E). Values plotted are the means ± standard deviation of three independent experiments.
Autophagy is induced by mechanical compression
One problem visualizing vesicular compartments in Dictyostelium is that the cells are relatively round and vesicles move in the Z-plane, frequently going out of focus. A commonly employed technique to reduce this movement and improve imaging is to compress cells under a layer of agarose to promote spreading and reduce the cell depth.27
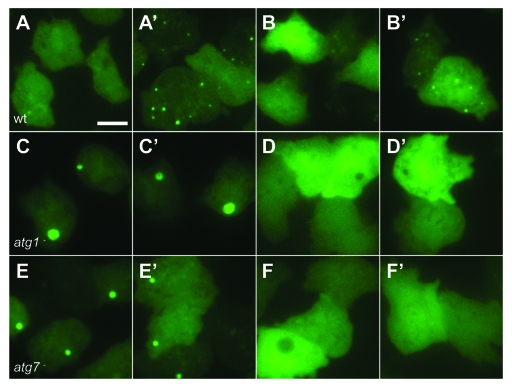
When cells expressing GFP-Atg8 were compressed however, numerous puncta were induced within 5 min (Fig. 2A). When cells lacking either atg1 or atg7 (required for autophagy)21 were compressed, only the large ubiquitinated protein aggregates described previously19 were present, and there was no induction of other puncta (Fig. 2).
Figure 2.
Autophagy is induced by mechanical stress. Images of (A and B) wild-type (DH1) cells, (C and D) atg1-null and (E and F) atg7-null Dictyostelium cells expressing either (A, C and E) GFP-Atg8, or (B,D and F) GFP-Atg18. Cells were compressed under 1.15 kPa and images taken either 2 min (A–F) or 10 min (A’–F’) after compression. Bar indicates 10 μm.
To confirm that the GFP-Atg8 puncta observed were true autophagosomes and discount the possibility that the puncta observed are simply aggregates of GFP-Atg8, we also used an additional marker GFP-Atg18. In both yeast and mammalian cells, Atg18 (known as WIPI in mammalian cells) is recruited to early autophagosomal structures by directly binding PtdIns(3)P generated by the vps34 complex, and is required for autophagosome formation.28,29 Consistent with this, starvation induced the formation of puncta in Dictyostelium cells expressing GFP-Atg18, with similar dynamics to that observed with GFP-Atg8 (see Fig. S2).
Importantly, when these cells were compressed, GFP-Atg18 puncta were also rapidly induced (Fig. 2), and with similar dynamics to GFP-Atg8 (Fig. 4B). This was also dependent upon both atg1 and atg7 (although GFP-Atg18 does not localize to the protein aggregates) (Fig. 2) and compression-induced tagRFP-Atg18 and GFP-Atg8 puncta colocalize (Fig. 3G and Vid. S1), demonstrating that the accumulation of these puncta acts through the canonical autophagic machinery.
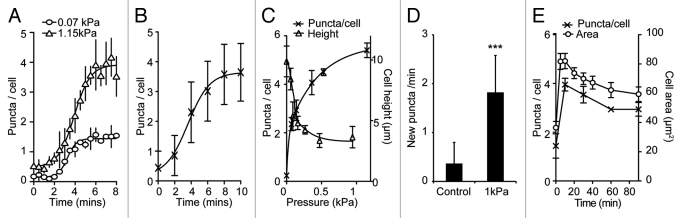
Figure 4.
Dynamics of the mechanical induction of autophagy in Dictyostelium. Timecourse of the accumulation of (A) GFP-Atg8 puncta under both 1.15 kPa (triangles) and 0.07 kPa (circles) pressure and (B) GFP-Atg18 puncta under 1.15 kPa. Both the number of GFP-Atg8 puncta per cell (crosses), and the average cell height (triangles) after 10 min compression under different pressures is plotted is shown in (C). Values plotted are the mean ± standard deviation of three independent experiments. (D) The rate of new puncta formation is increased upon cell compression. Maximum intensity projections of Z-stack movies were used to identify the appearance of new puncta in cells either uncompressed, or compressed under 1.15 kPa for 10 min. Values plotted are the means ± standard deviation of 45 cells for each condition, observed over three independent experiments (***p < 0.001 Student’s t-test). (E) Adaptation of cell morphology and the autophagic response. The number of GFP-Atg8 puncta in cells compressed under 0.2 kPa was quantified at the times indicated (crosses). Also plotted is the cross-sectional area of cells at each time point (circles), an indicator of cytoskeletal reorganization. At least 50 cells were scored for each data point. Values plotted are the mean ± standard deviation of three independent experiments.
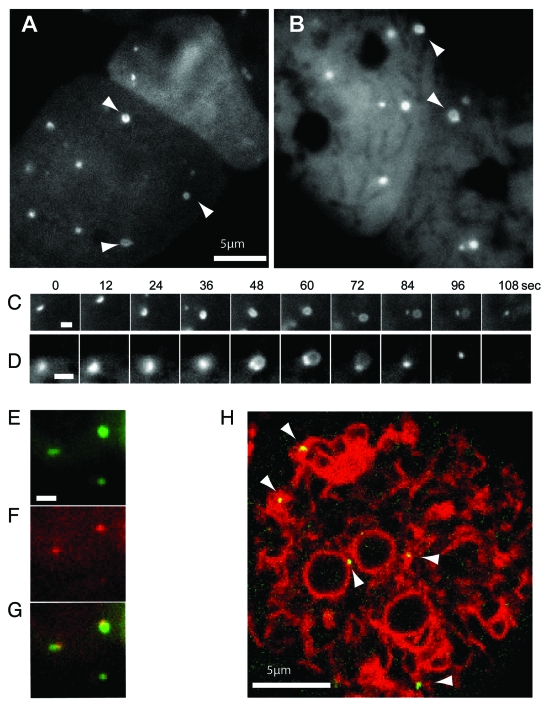
Figure 3.
High magnification imaging of the expansion of mechanically-induced autophagosomes. Ax3 cells expressing either (A) GFP-Atg8 or (B) GFP-Atg18 were imaged 20 min after capillary action-compression. Arrows indicate cup or circle structures, the full time-lapse series are shown in Video S2 and S3. Sequential images of individual forming autophagosomes visualized with GFP-Atg8 or GFP-Atg18 are shown in (C and D) respectively. Bars in (A and B) represent 10 μm and 2 μm respectively. Both markers localize to the same structure in compressed cells co-expressing (E) GFP-Atg8 and (F) tagRFP-Atg18; (G) shows the merged image and the complete time-lapse is shown in Video S1. (H) Image from a time-lapse series of compressed cells co-expressing the ER marker vmp1-mRFPmars and GFP-Atg18 observed by confocal microscopy. Arrows indicate forming autophagosomes. The full sequence can be seen in Video S4.
Video S1.
This is the Video S1 legend.
Previously it has been shown that autophagy can be regulated by heterotrimeric G-proteins,30 so to control for the possibility that the induction of autophagy is due to the activation of cell surface receptors by the agarose sheet we used Dictyostelium mutants lacking the single gene encoding the Gβ subunit of heterotrimeric G-proteins. In this mutant, all G-protein coupled receptor signaling is blocked31 but mechanical stress still induced autophagy to a similar extent (see Fig. S4) demonstrating that the autophagic response is not mediated by heterotrimeric G-protein signaling.
The dynamics of Dictyostelium autophagosome formation
Using timelapse imaging of compressed Dictyostelium cells at high magification, we were able to clearly observe GFP-Atg8 structures growing from small puncta into the characteristic cup-shape of an expanding autophagosome (Fig. 3 and Vid. S2). These then expanded into completed, circular structures before the rigid shape of the vesicle relaxed and the GFP signal disappeared, presumably due to fusion with lysosomes as well as delipidation by Atg4.26 This progression is unique to autophagy demonstrating that the GFP-Atg8 structures induced by mechanical stress are bona fide autophagosomes.
Video S2.
This is the Video S2 legend.
Furthermore, the excellent imaging possible under these conditions also allowed us for the first time to analyze the temporal dynamics of this process in Dictyostelium. Expansion of the phagophore from initial punctum to complete circular vesicle took 90 ± 9 (n = 15) seconds after which it took a further 30 ± 7 (n = 30) seconds to mature and remove the GFP-Atg8 signal. Under these conditions, Dictyostelium autophagosomes were remarkably uniform (see Vid. S2), and we were able to estimate their size as 800 ± 40 nm diameter (n = 30), comparable to those in mammalian cells, despite the cells’ smaller size.
Similar structures were also seen with GFP-Atg18; using this marker, a small punctum first enlarges, and expands into a loop structure, with GFP-Atg18 enriched at the collar (Fig. 3B and Vid. S3), strongly resembling the PtdIns(3)P enriched omegasome structures recently described in mammalian cells.32 Omegasomes were shown to form on the surface of the endoplasmic reticulum (ER), providing a platform for phagophore expansion from the ER membrane. In Dictyostelium, GFP-Atg18 also colocalized with the ER marker vmp1-mRFPmars19 indicating that mechanically induced autophagosomes are generated in a similar way (Fig. 3H and Vid. S4).
Video S3.
This is the Video S3 legend.
The mechanical induction of autophagy is graduated within the physiological range
To determine the sensitivity of the autophagic response to mechanical pressure, and see if the response was graduated, we developed an experimental set-up where different, defined pressures could be applied to the cell (Fig. S3). Using this, we observed that the number of puncta was graduated, with larger forces inducing more puncta per cell, although the timing of the response was the same (Fig. 4A). The pressure required to induce autophagy in Dictyostelium was surprisingly low, requiring 0.25 kPa for a half-maximal response (Fig. 4C).
To confirm that the accumulation of puncta is due to the induction of autophagosome formation, and not inhibition of autophagosome degradation, we monitored the appearance of new autophagosomes by time-lapse microscopy. Compression led to a ~5-fold increase in the rate of autophagosome initiation, indicating that autophagy is activated under these conditions (Fig. 4D).
Dictyostelium cells growing in liquid culture feed primarily by macropinocytosis involving the engulfment of nutrients by large actin-generated cups. It is possible that the induction of autophagy could be due to the physical restriction of macropinosome formation, resulting in starvation. However, as the accumulation of GFP-Atg8 puncta is faster under mechanical stress than starvation and the maximal level of induction by mechanical stress is double that of starved cells, this cannot be the case. Therefore the primary inducer of autophagy is a mechanical signal. Accordingly, the level of autophagic induction strongly corresponds to the degree with which the cells are deformed (Fig. 4C), implying that the mechanical deformation itself stimulates autophagy.
The mechanical induction of autophagy is conserved in mammalian cells
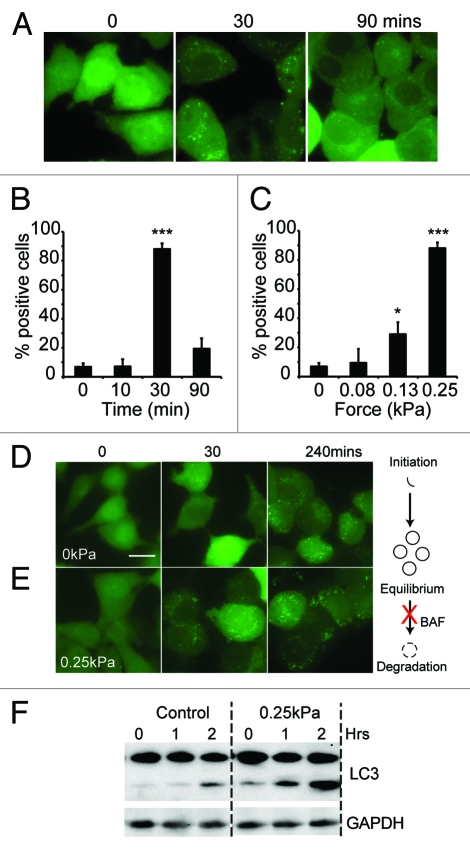
In order to determine if the mechanical induction of autophagy is a general property of cells or is specific to free-living organisms such as Dictyostelium, we repeated the compression experiments above using human cells. When we examined MDA-MB-231 cells, expressing EGFP-LC3 (a mammalian ortholog of atg8), only 6.8 ± 2% of cells contained > 50 autophagosomes prior to treatment (Fig. 5A and B). After 30 min compression under 0.25 kPa, there was a dramatic accumulation of autophagosomes and 88.1 ± 4% of cells contained a large number of puncta. Once more, this response was titratable, with a half-maximal response induced at 0.2 kPa, almost identical to the value obtained with Dictyostelium (Fig. 5C).
Figure 5.
The induction of autophagy by mechanical stress in mammalian cells. (A) MDA-231 cells expressing EGFP-LC3 were compressed under 0.25 kPa, and images taken at the times indicated; bar indicates 20 μm. Quantification of the same experiment is shown in (B). The proportion of cells responding after 30 min under different compressive loads is plotted in (C). Positive cells are defined as those with > 50 EGFP-LC3 puncta. Values plotted are the means ± standard deviation of three independent experiments. (***p < 0.0001, *p < 0.05 compared with control using Student’s t-test). MDA-231 cells expressing EGFP-LC3 were also treated with 200 nM bafilomycin A1. Images were taken at the times indicated for cells either uncompressed (D) or subjected to 0.25 kPa compression (E); Bar represents 20 μm. (F) LC3 I/II levels of cells treated with 200 nM bafilomycin alone, or also compressed under 0.25 kPa. Cells were lysed after 0, 1 and 2 h treatment and LC3 levels measured by western blot.
The accumulation of autophagosomes could be caused by either increased autophagosome initiation, or by decreased maturation and fusion with the lysosomal system. To differentiate between these two possibilities, we treated MDA-MB-231 cells with bafilomycin A, an inhibitor of the vacuolar V+-ATPase, which blocks lysosomal function.33 As expected, bafilomycin A treatment alone led to the accumulation of autophagosomes although at basal levels of autophagy this took several hours (Fig. 5D). When bafilomycin A-treated cells were subjected to mechanical stress however, the accumulation of autophagosomes was much faster, with large numbers visible after just 30 min (Fig. 5E) indicating that mechanical stress causes the formation of new autophagosomes.
Correspondingly, when bafilomycin-treated cells were analyzed for LC3 lipidation, uncompressed cells only had a slow accumulation of the lipidated (LC3 II) form. This was significantly increased upon compression, biochemically confirming the results obtained with GFP-LC3 (Fig. 5F).
Autophagy is transiently induced during adaptation to stress
Although MDA-MB-231 cells accumulated large numbers of autophagosomes over the first 30 min of compression, at longer time points these gradually reduced, and after 90 min had reverted back to basal levels (Fig. 5A and B). Interestingly, when we looked at cell morphology, there was a clear correlation between the adaptation of the cytoskeleton and the level of autophagy.
When initially compressed, numerous bleb-like protrusions formed over the plasma membrane, indicating that the cortex was unable to resist the elevated pressure, violently delaminating from the membrane (See Fig. 6F). Subsequently there was clear evidence of cytoskeletal adaption and over 90 min, the blebbing gradually subsided (Fig. 5A). This adaptation, mitigating the structural disruption and presumably the stress caused by the compression, correlates well with the return of autophagy to basal levels.
Figure 6.
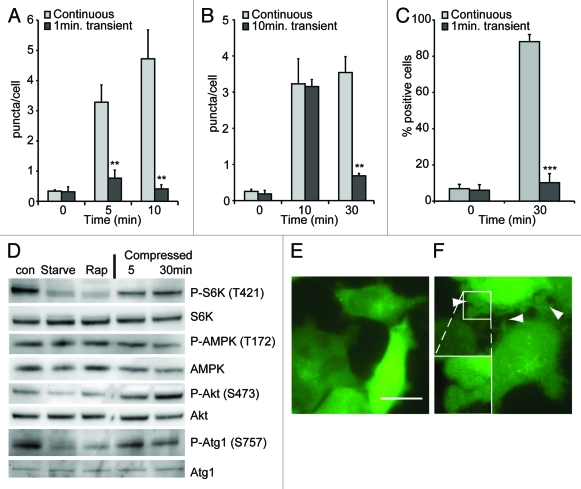
The induction of autophagy requires continuous pressure. Dictyostelium cells expressing GFP-Atg8 were compressed continuously or for either 1 (A) or 10 (B) min before the weight was removed. (C) MDA-231 cells expressing EGFP-LC3 were transiently compressed for 1 min. Images were taken at the timepoints indicated and the number of fluorescent puncta quantified, values are the mean ± standard deviation of three independent experiments. *p < 0.005, ***p < 0.0001 compared with constitutively compressed cells (Student t-test). (D) Western blot analysis of S6 kinase, AMPKα and Akt (PKB) and ULK1/atg1 phosphorylation in MDA-MB-231 cells after compression under 0.25 kPa for the times indicated. Control cells were either untreated, starved in EBSS or treated with 500 nM rapamycin for 1 h. Images of MDA-MB-231 cell morphology when uncompressed (E), or after 1 min compression (F). Dictyostelium and MDA-231 cells were compressed by 1.15 and 0.25 kPa respectively.
Similar results were obtained with Dictyostelium, where the autophagic response to compression also reduced over time. Although the level of autophagy did not return completely to basal levels, but stabilized at the same level as amino acid-starved cells (presumably due to a continued inhibition of macropinocytosis), this again corresponded closely with cytoskeletal adapatation as indicated both by the restoration of cell shape (Fig. 4E), as well as the suppression of small blebs. Therefore autophagy is activated during adaptation, induced by changes in localized mechanical pressure and not simply pressure per se.
Induction of autophagy requires continual compression
A major role of autophagy is to remove damaged or superfluous organelles. Therefore one hypothesis would be that the autophagic response to compression is due to mechanical damage to organelles such as mitochondria. If this were true, a short transient compression would cause similar levels of damage to a sustained pressure and induce a similar autophagic response.
However, when a load of 1.15 kPa was applied to Dictyostelium cells for 1 min and then removed, there was no stimulation of autophagy (Fig. 6A). Additionally, if the stress was removed after autophagy was already induced (by compression for 10 min) the number of autophagosomes rapidly reverted to basal levels (Fig. 6B).
Similar results were obtained with MDA-MB-231 cells, where 1 min transient compression also did not elicit an autophagic response (Fig. 6C), despite clear signs of stress—illustrated by large numbers of blebs (Fig. 6E and F). This is consistent with the cessation of the autophagic response after the stress is relieved by adaptation. As the autophagic response requires a continuous force, it cannot be simply clearing mechanically damaged organelles, implying a directly regulation by mechano-sensitive signaling.
Mechanical induction of autophagy is mTOR-independent
The best understood pathway for the regulation of autophagy is via the target of rapamycin complex 1 (TORC1), which directly phosphorylates and negatively regulates the activity of the Atg1 complex.34 TORC1 therefore acts as a central component of a number of stress pathways, linking them to the autophagic machinery and its activity can be inferred by the phosphorylation levels of its target protein, ribosomal S6 kinase (S6K). Interestingly, mechanical stress of MDA-MB-231 cells did not change the phosphorylation of S6K, indicating that TOR activity was not inhibited (Fig. 6D). Recently it has been shown that TORC1 directly phosphorylates Atg1 on serine 757;35 again we saw no change in phosphorylation at this site following compression (Fig. 6D).
Upstream, TOR activity and a number of stress responses can be regulated via Akt (PKB), however Akt phosphorylation (and therefore activity) was also unaffected by mechanical stress (Fig. 6D). Consistent with this, Dictyostelium PKB-null mutants remained able to induce autophagy upon compression (Fig. S3) indicating that the induction of autophagy is both TOR and Akt(PKB)-independent.
Atg1 can also be regulated by phosphorylation by the key energy-sensor AMP-activated protein kinase (AMPK).35,36 In MDA-MB-231 cells however, P-AMPK levels were not changed by either starvation, or compression (Fig. 6D) consistent with the fact that these cells lack the major AMPK regulator LKB1.37 We were unable to detect any phosphorylation of the proposed AMPK regulatory site of Atg1 (S555) in these cells using commercially available antibodies.
Although the TOR-independent regulation of autophagy has been observed in other circumstances, the signaling pathways involved are unclear.38 Recent work has shown that the Atg1 complex can be regulated independently of TOR by cAMP-dependent protein kinase A (PKA);39 however Dictyostelium mutants in which the catalytic subunit of PKA was disrupted40 also remained able to mechanically induce autophagy (Fig. S3) indicating that other, alternative pathways must exist.
Discussion
In order to work efficiently and survive, cells have to continuously detect and adapt to their environment. Autophagy is induced under a number of stresses, including starvation, organelle/DNA damage, hypoxia, ER stress, and pathogen infection.5-8 Here we demonstrate that autophagy also plays a conserved role in the response to mechanical stress.
The pressure required to induce autophagy lies precisely within the expected physiological range; both Dictyostelium and mammalian cells respond strongly to pressures of ~0.2 kPa. Importantly, this is comparable to that required to induce transcriptional changes in osteoblasts,17 and significantly less than estimates of the pressure within muscle during massage (> 1.2 kPa).41 Therefore the autophagic response occurs within the range of pressure that cells will encounter in vivo.
What is the role of autophagy in the response to mechanical stress? The catabolism of cytosolic components by autophagy could potentially serve a number of functions. Autophagy constitutes a major cytoprotective mechanism, defending the cell from damaged organelles, pathogens and toxic, misfolded proteins1,42 and elevated autophagy has been proposed to extend life span by generally improving cellular health.43 By definition, all stresses disrupt cellular function to some degree, potentially leading to the accumulation of damaging agents; therefore inducing an autophagic response would be a wise precaution to aid cellular survival.
Additionally, autophagy may assist adaptation. It is unlikely that the role of autophagy plays a direct role in the rapid cytoskeletal adaptation seen upon compression; indeed, we detected no significant defects in the recovery of cell shape in Dicytostelium atg1-null mutants (Fig. S5). However, when cells are challenged they must change a number of aspects of their physiology to fit new environmental demands and preserve function.
In yeast, autophagy is required for the upregulation of several starvation-induced genes and therefore it has been proposed that an important role of autophagy is to facilitate stress adaptation.4 Autophagy also appears to serve as a mechanism to enhance cellular re-programming, and is important for the developmental remodeling of yeast, Dictyostelium, Caenorhabditis elegans and flies20,44-46 and required for the early development and erythrocyte differentiation of mice.47,48 Therefore autophagy may play a role in supporting the adaptation to mechanical changes.
Mechanical forces are able to regulate a number of cellular functions; probably the best understood is in osteoblast physiology, where differentiation is triggered by mechanical stress.16 Interestingly, autophagy has recently been shown to be important for osteoblast differentiation49 and therefore the link between mechanical loading and autophagy may strongly impact skeletal remodeling. There is also evidence indicating that autophagy is important for the cortical remodeling of Drosophila hemocytes and mouse macrophages50 and it is possible that autophagy plays a more specific role in cytoskeletal adaptation.
We showed that the mechanical induction of autophagy requires a continuous mechanical signal, but what is its nature? Mechano-sensation is complex and incompletely understood, leading to the activation of a wide range of signaling pathways.51-54 We have demonstrated that the mechanical induction of autophagy is independent of classical TOR/Akt or AMPK signaling. Others have shown that TOR-independent autophagy can be induced by a number of drugs,38 but little is known about the signaling pathways involved or the physiological relevance of this pathway. Our observation that the mechanical activation of autophagy is TOR-independent therefore describes the first physiological role for this pathway and indicates that we still do not know the full range of signals that regulate the autophagic machinery.
We have demonstrated that autophagy is part of the cellular response to mechanical stress. The ability to respond and adapt to mechanical challenge is crucial for cell survival and plays an important role in the differentiation of a number of specialized cells. Therefore the mechanical induction of autophagy may have widespread implications.
Materials and Methods
Cell-culture conditions
Unless otherwise stated, the Dictyostelium wild-type strain used was ax3 and were routinely subcultured in synthetic SIH medium (Formedium, SIH0102). The atg1, atg5 and atg7-null cells have been previously described20,21 their parental strain DH1 (a uracil auxotroph) was used as control for these mutants, and these strains were maintained in 10 μg/ml exogenous uracil (Sigma, U1128). For amino acid starvation, cells were washed by three changes of medium into SIH medium lacking arginine and lysine (custom order from Formedium). MDA-MB-231 cells were grown in DMEM (Invitrogen, 10313-039) and starved in Earles Balanced Salt Solution (EBSS) (Invitrogen, 24010-043). EGFP-LC3 expressing cells were generated by transduced with the pBabe-puro EGFP-ratLC3B retroviral construct that has been described previously55 and stable transfectants selected with G418.
Dictyostelium viability assay
This was performed as previously described.20 Dictyostelium cells grown in full medium were washed three times in SIH medium lacking arginine and lysine, resuspended at 5 × 105 per ml and shaken in flasks. Every 2 d, samples were removed and cells plated onto SM agar (Formedium, SMA0102) plates in conjunction with a lawn of klebsiella aerogenes. Viability was assessed by scoring the number of colonies formed.
Plasmid construction and transformation
Dictyostelium tagRFP expression vectors were generated by amplifying the tagRFP gene from pTagRFP-N (Evrogen, FP142) using the primers: GGA TCC AAA ATG GTT TCT TTA AAA GGT GAA GAA CTA ATT AAG GAG / ACT AGT TTT AGA TCT ATT AAG TTT GTG CCC CAG TTT GC and subsequent cloning as a BamHI/SpeI fragment into pDM358.56 The plasmid was further optimised by inserting the oligo linker GAT CCG ATG ATG ACG ATA GAT CTA GTA / CTA GTA CTA GAT CTA TCG TCA TCA TCG into the BglII/SpeI sites between the tagRFP and fusion gene, giving the plasmid pJSK464. The Dictyostelium Atg8-1 (DDB_G0286191) and atg18 coding sequences were amplified from cDNA using the primers GGA TCC ATG GTT CAT GTA TCA AGC / TCT AGA TTA TAA ATC ACT ACC AAA AG and GGA TCC ATG AAT GTT GGA GGT AAA TTC / TCT AGA TTA TAA TAT TTT TGC TGT AAC ATC ATT GTC respectively. Atg8 was cloned as a BamHI/XbaI fragment into the BglII/SpeI sites of pDM351 and pDM41056 to give pDM430 (GFP-Atg8 expression) and pJSK421 (GFP-Atg8 shuttle) respectively, and Atg18 was cloned as a BamHI/XbaI fragment into the BglII/SpeI sites of pDM448, pDM41056 and pJSK468 to give pJSK489 (GFP-expression), pJSK490 (GFP-shuttle) and pJSK468 (tagRFP-expression) respectively. The GFP-Atg8 and tagRFP-Atg18 co-expression plasmid was constructed by ligating the NgoMIV expression fragment from pJSK421 into the NgoMIV site of pJSK489, giving plasmid pJSK498. The vmp1 coding sequence was amplified from cDNA using the primers GGA TCC AAA ATG GGT AAA AGT AAT ACG ATA G / TCT AGA TTT TGT TTT TTT AGT TTC TTT TGG TTG TTG and cloned as a BamHI/XbaI fragment into the BglII/SpeI sites of pDM451.56 The NgoMIV expression fragment from pJSK490 was then ligated into this to give the vmp1-mRFPmars / GFP-Atg18 co-expression plasmid pJSK542.
Cells were transformed by electroporation using standard methods and transformants selected in 10 μg/ml G418 or 20 μg/ml hygromycin as appropriate, in HL5 medium (Formedium, HL50102).
Fluorescence microscopy and quantitation
Unless otherwise stated, cells were imaged using an Olympus IX81 wide-field fluorescence microscope equipped with 60× 1.42NA Plan ApoN objective. Images were captured using a QuantEM:512SC EMCCD camera (Photometrics). For high-magnification imaging, cells were imaged using a Nikon Eclipse TE 2000-U microscope equipped with a 100×/1.45 Nikon TIRF oil immersion objective, 473 nm diode and 561 laser illumination (Omicron) and a Cascade 512F EMCCD camera (Photometrics). Dual-color images were obtained simultaneously using a Multi-Spec dual emission splitter (Optical Insights, NM) with a 595 nm dichroic and two bandpass filters (510–565 for green and 605–655 nm for red) to separate both emissions.
For quantification of fluorescent puncta, a Z-series of images in 0.25 μm steps was captured. A maximum intensity projection of each series was used to score autophagosome number. More than 100 cells were scored for each experiment, which were repeated in triplicate. To quantitate the rate of new puncta formation, a fast series of Z-sections (in 0.5 μm steps) were obtained at 5 sec intervals. Maximum intensity projections of these images were then used to follow the puncta over time and detect the appearance of new ones.
Measurement of autophagosome size and dynamics were done from multiple movies obtained on at least three independent experiments, and the numbers stated are the combined results from at least six individual cells. In each case, only autophagosomes that we were clearly able to follow over the complete course of expansion and maturation were included in the analysis.
To measure cell height, a confocal Z-series in 0.25 μm steps was taken at different positions on the dish using a Nikon A1R microscope with a 60× PlanApo 1.40NA objective. This was used to measure the cell height at each position, and at least six positions were observed for each data point. The data plotted are the means ± standard deviations of at least 20 images, obtained over three independent experiments.
Compression of cells
Two different methods were used to compress the cells. Unless otherwise stated, cells were compressed as follows. One day prior to the experiments, cells were seeded into 3 cm glass-bottomed dishes (Iwaki, 3930-035). Thin agarose sheets were prepared by pouring 10 ml of 1% agarose in normal growth medium into a 10 cm Petri dish. For use with MDA-MB-231 cells this was placed in a 37°C / 5% CO2 incubator overnight to equilibriate. For compression, 3 cm discs were cut out of the agarose sheet and placed upon the cells. A custom plastic insert was then placed on top, followed by appropriate weight metal discs (see Fig. S3). For high-magnification imaging of Dictyostelium the capilliary action compression method previously described was used.27 Agarose was prepared as above using LoFlo medium (Formedium, LF05) and cells were incubated in LoFlo medium for 2 h prior to imaging. A small (1 cm × 1 cm) square of agarose was then cut out, placed on the cells and most of the medium removed. Remaining medium was then removed through the surface of the agarose using blotting paper, compressing the cells underneath.
Western blotting
Analysis of protein phosphorylation was done by western blotting. 80% confluent 3 cm dishes of MDA-MD-231 cells were compressed as described above, under 0.25 kPa for the times indicated. The weights and well insert were then removed, and with the agarose sheet in place, the dish was washed twice in ice-cold PBS. The PBS and agarose sheet were then removed and the cells lysed in 200 μl RIPA buffer (10 mM Tris pH 7.4, 1 mM EDTA, 0.15 M NaCl, 1% NP40, 0.1% sodium deoxycholate, 0.1% SDS) supplemented with Halt protease cocktail (Pierce, 78430) and phosphatase inhibitor cocktail set 2 (Calbiochem, 524625), and analyzed by SDS-PAGE. All samples were separated on 4–12% NuPage Bis-Tris minigels (Invitrogen, NP0321), except LC3 lipidation assays, which were analyzed on large 15% gels. Blots were probed with anti-phospho p70 S6 kinase (Thr421/ser424) (Cell Signaling, 9204), p70-S6 kinase (2708), phospho-Atk (Ser473) (9271), Atk (9272), phospho-AMPKα (Thr172) (2535), AMPKα (2532), phospho-ULK1 (Ser757) (6888) antibodies from Cell Signaling, and anti-ULK1 (Sigma Aldrich, A7481) and anti-LC3 (Nanotools, 2G6).
Video S4.
This is the Video S4 legend.
Supplementary Material
Supplementary PDF file supplied by authors.
Acknowledgments
We thank Drs. Kevin Ryan and Simon Wilkinson (Beatson Institute) for supplying the MDA-MB-231 cell lines, and critical reading of the manuscript, as well as the Dictyostelium stock center (www.dictybase.org) for supplying mutant strains. This work was funded by Cancer Research UK.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/17924
References
- 1.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 3.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 4.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–6. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 5.Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61:439–44. [PubMed] [Google Scholar]
- 6.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15:1572–81. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 7.Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–68. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 8.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 10.Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88:39–48. doi: 10.1016/S0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 11.Franke RP, Grafe M, Schnittler H, Seiffge D, Mittermayer C, Drenckhahn D. Induction of human vascular endothelial stress fibres by fluid shear stress. Nature. 1984;307:648–9. doi: 10.1038/307648a0. [DOI] [PubMed] [Google Scholar]
- 12.Matthews BD, Overby DR, Mannix R, Ingber DE. Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J Cell Sci. 2006;119:508–18. doi: 10.1242/jcs.02760. [DOI] [PubMed] [Google Scholar]
- 13.Décave E, Rieu D, Dalous J, Fache S, Brechet Y, Fourcade B, et al. Shear flow-induced motility of Dictyostelium discoideum cells on solid substrate. J Cell Sci. 2003;116:4331–43. doi: 10.1242/jcs.00726. [DOI] [PubMed] [Google Scholar]
- 14.Lombardi ML, Knecht DA, Lee J. Mechano-chemical signaling maintains the rapid movement of Dictyostelium cells. Exp Cell Res. 2008;314:1850–9. doi: 10.1016/j.yexcr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Chappell DC, Varner SE, Nerem RM, Medford RM, Alexander RW. Oscillatory shear stress stimulates adhesion molecule expression in cultured human endothelium. Circ Res. 1998;82:532–9. doi: 10.1161/01.res.82.5.532. [DOI] [PubMed] [Google Scholar]
- 16.Huang CY, Hagar KL, Frost LE, Sun Y, Cheung HS. Effects of cyclic compressive loading on chondrogenesis of rabbit bone-marrow derived mesenchymal stem cells. Stem Cells. 2004;22:313–23. doi: 10.1634/stemcells.22-3-313. [DOI] [PubMed] [Google Scholar]
- 17.Kanzaki H, Chiba M, Shimizu Y, Mitani H. Periodontal ligament cells under mechanical stress induce osteoclastogenesis by receptor activator of nuclear factor kappaB ligand up-regulation via prostaglandin E2 synthesis. J Bone Miner Res. 2002;17:210–20. doi: 10.1359/jbmr.2002.17.2.210. [DOI] [PubMed] [Google Scholar]
- 18.Calvo-Garrido J, Carilla-Latorre S, Kubohara Y, Santos-Rodrigo N, Mesquita A, Soldati T, et al. Autophagy in Dictyostelium: Genes and pathways, cell death and infection. Autophagy. 2010;6:686–701. doi: 10.4161/auto.6.6.12513. [DOI] [PubMed] [Google Scholar]
- 19.Calvo-Garrido J, Escalante R. Autophagy dysfunction and ubiquitin-positive protein aggregates in Dictyostelium cells lacking Vmp1. Autophagy. 2010;6:100–9. doi: 10.4161/auto.6.1.10697. [DOI] [PubMed] [Google Scholar]
- 20.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem. 2003;278:17636–45. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 21.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J Biol Chem. 2004;279:15621–9. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- 22.Tung SM, Unal C, Ley A, Pena C, Tunggal B, Noegel AA, et al. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell Microbiol. 2010;12:765–80. doi: 10.1111/j.1462-5822.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- 23.Han SI, Friehs K, Flaschel E. Cultivation of Dictyostelium discoideum on an improved synthetic medium in a conventional bioreactor. Process Biochem. 2004;39:925–30. doi: 10.1016/S0032-9592(03)00215-2. [DOI] [Google Scholar]
- 24.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 25.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yumura S, Mori H, Fukui Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J Cell Biol. 1984;99:894–9. doi: 10.1083/jcb.99.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krick R, Henke S, Tolstrup J, Thumm M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy. 2008;4:896–910. doi: 10.4161/auto.6801. [DOI] [PubMed] [Google Scholar]
- 29.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–22. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 30.Ogier-Denis E, Couvineau A, Maoret JJ, Houri JJ, Bauvy C, De Stefanis D, et al. A heterotrimeric Gi3-protein controls autophagic sequestration in the human colon cancer cell line HT-29. J Biol Chem. 1995;270:13–6. doi: 10.1074/jbc.270.1.13. [DOI] [PubMed] [Google Scholar]
- 31.Lilly P, Wu L, Welker DL, Devreotes PN. A G-protein beta-subunit is essential for Dictyostelium development. Genes Dev. 1993;7:986–95. doi: 10.1101/gad.7.6.986. [DOI] [PubMed] [Google Scholar]
- 32.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowman EJ, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA. 1988;85:7972–6. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganley IG. Lam du, H, Wang, J, Ding, X, Chen, S, Jiang, X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen Z, Wen XF, Lan F, Shen ZZ, Shao ZM. The tumor suppressor gene LKB1 is associated with prognosis in human breast carcinoma. Clin Cancer Res. 2002;8:2085–90. [PubMed] [Google Scholar]
- 38.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–11. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA. 2009;106:17049–54. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mann SK, Yonemoto WM, Taylor SS, Firtel RA. DdPK3, which plays essential roles during Dictyostelium development, encodes the catalytic subunit of cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:10701–5. doi: 10.1073/pnas.89.22.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldberg J, Sullivan SJ, Seaborne DE. The effect of two intensities of massage on H-reflex amplitude. Phys Ther. 1992;72:449–57. doi: 10.1093/ptj/72.6.449. [DOI] [PubMed] [Google Scholar]
- 42.Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16:1313–20. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 43.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12:842–6. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 44.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 45.Meléndez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 46.Juhász G, Csikos G, Sinka R, Erdelyi M, Sass M. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543:154–8. doi: 10.1016/S0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 47.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–20. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 48.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitehouse CA, Waters S, Marchbank K, Horner A, McGowan NW, Jovanovic JV, et al. Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc Natl Acad Sci USA. 2010;107:12913–8. doi: 10.1073/pnas.0913058107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadandale P, Stender JD, Glass CK, Kiger AA. Conserved role for autophagy in Rho1-mediated cortical remodeling and blood cell recruitment. Proc Natl Acad Sci USA. 2010;107:10502–7. doi: 10.1073/pnas.0914168107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dassouli A, Sulpice JC, Roux S, Crozatier B. Stretch-induced inositol trisphosphate and tetrakisphosphate production in rat cardiomyocytes. J Mol Cell Cardiol. 1993;25:973–82. doi: 10.1006/jmcc.1993.1109. [DOI] [PubMed] [Google Scholar]
- 52.Gudi S, Huvar I, White CR, McKnight NL, Dusserre N, Boss GR, et al. Rapid activation of Ras by fluid flow is mediated by Galpha(q) and Gbetagamma subunits of heterotrimeric G proteins in human endothelial cells. Arterioscler Thromb Vasc Biol. 2003;23:994–1000. doi: 10.1161/01.ATV.0000073314.51987.84. [DOI] [PubMed] [Google Scholar]
- 53.Lavandero S, Cartagena G, Guarda E, Corbalan R, Godoy I, Sapag-Hagar M, et al. Changes in cyclic AMP dependent protein kinase and active stiffness in the rat volume overload model of heart hypertrophy. Cardiovasc Res. 1993;27:1634–8. doi: 10.1093/cvr/27.9.1634. [DOI] [PubMed] [Google Scholar]
- 54.Lee HS, Millward-Sadler SJ, Wright MO, Nuki G, Salter DM. Integrin and mechanosensitive ion channel-dependent tyrosine phosphorylation of focal adhesion proteins and beta-catenin in human articular chondrocytes after mechanical stimulation. J Bone Miner Res. 2000;15:1501–9. doi: 10.1359/jbmr.2000.15.8.1501. [DOI] [PubMed] [Google Scholar]
- 55.Wilkinson S, O'Prey J, Fricker M, Ryan KM. Hypoxia-selective macroautophagy and cell survival signaled by autocrine PDGFR activity. Genes Dev. 2009;23:1283–8. doi: 10.1101/gad.521709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veltman DM, Akar G, Bosgraaf L, Van Haastert PJ. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009;61:110–8. doi: 10.1016/j.plasmid.2008.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary PDF file supplied by authors.