Abstract
Perhaps the most complex step of macroautophagy is the formation of the double-membrane autophagosome. The majority of the autophagy-related (Atg) proteins are thought to participate in nucleation and expansion of the phagophore, and/or the completion of this compartment. Monitoring this part of the process is difficult, and typically involves electron microscopy analysis; however, unless three-dimensional tomography is performed, even this method cannot be used to easily determine if the phagophore is completely enclosed. Accordingly, a complementary approach is to examine the accessibility of sequestered cargo to exogenously added protease. This type of protease protection analysis has been used to monitor the formation of cytoplasm-to-vacuole targeting (Cvt) vesicles and autophagosomes by examining the protease sensitivity of precursor aminopeptidase I (prApe1). For determining the status of autophagosomes formed during nonselective autophagy, however, prApe1 is not the best marker protein. Here, we describe an alternative method for examining autophagosome completion using GFP-Atg8 as a marker for protease protection.
Keywords: autophagy, lysosome, stress, vacuole, yeast
1. Introduction
In the last few years, great achievements have been made in understanding the molecular mechanisms of macroautophagy (hereafter autophagy) in Saccharomyces cerevisiae and in various higher eukaryotic systems. This exciting work has also shed light on the role of autophagy in maintaining normal physiological function, and the significance of this process in various diseases. It is therefore critical to establish reliable methods to monitor and quantify autophagy. In S. cerevisiae, as in most other systems, most of these methods rely on an analysis of the autophagy-related ubiquitin-like protein Atg8, and its mammalian homolog microtubule-associated protein 1 light chain 3 (LC3).
Atg8/LC3 is conjugated primarily to the lipid phosphatidylethanolamine (PE), through a cascade involving the action of the cysteine protease Atg4, the ubiquitin-like conjugation system components Atg7 and Atg3, and the putative E3 ligase made up of the Atg12–Atg5-Atg16 complex. In yeast, Atg8–PE conjugation is regulated by the nutritional status of the cell. In nutrient-rich conditions, Atg8 mainly exists in the unconjugated form; however, upon nitrogen starvation most of the Atg8 is conjugated to PE.1,2 Atg8–PE is recruited to the phagophore assembly site (PAS) and localizes to both the inner and outer phagophore membranes, but is generally not detected on the surface of completed autophagosomes.3 Upon autophagosome completion, Atg8–PE that lines the inner autophagosome membrane is delivered to the vacuole where it is degraded by vacuolar proteases as part of the autophagic body.1-4 The population of Atg8 that is present on the outer membrane of the autophagosome is released via the deconjugation of Atg8–PE by a second Atg4-dependent cleavage step.3 GFP-Atg8 shows the same behavior as Atg8, and autophagic flux can be followed by monitoring the vacuolar delivery and subsequent breakdown of GFP-Atg8. When autophagic flux is normal, GFP-Atg8 that is present inside the autophagosome is cleaved after the autophagic body membrane is lysed and the contents are exposed to vacuolar hydrolases. The proteolysis of GFP-Atg8 releases an intact GFP moiety, which accumulates in the vacuole as autophagy proceeds, because it is relatively resistant to degradation. The increase in free GFP levels can be detected and quantified by western blot analysis and correlated with the autophagic rate.5-8 Alternatively, the degradation of the nonselective cargo Pgk1-GFP,9 or the quantitative Pho8Δ60 assay can be used to monitor the magnitude of autophagy.10,11
This itinerary of GFP-Atg8 can be used to determine whether a complete autophagosome has formed, by carrying out a protease protection assay.12,13 Previously, such an analysis was often done by determining the sensitivity of the primary Cvt complex cargo protein, prApe1, to externally added protease.14,15 Briefly, lysates or membrane fractions are prepared from spheroplasts of three different strains: (1) A positive control strain that is wild type for autophagosome biogenesis, but accumulates autophagosomes or autophagic bodies such as the temperature sensitive (ts) vam3 strain (vam3ts) or a pep4Δ strain, respectively; (2) a negative control strain such as atg1Δ that is defective in autophagosome formation; and (3) an experimental strain in which we want to determine the status of autophagosome biogenesis.16 The lysates or membrane fractions from each of these strains are split into three aliquots and subjected to different treatments: (1) no treatment; (2) treatment with protease alone; and (3) treatment with protease and detergent (it is also possible to include a fourth sample, in which only detergent is added). In the positive control strain, prApe1 should be protected from cleavage by protease in the absence, but not the presence, of detergent (Fig. 1A). In the negative control strain, exogenously added proteases have full access to prApe1 even in the absence of detergent, resulting in cleavage of the propeptide and the generation of one or more bands of lower molecular mass (Fig. 1B).
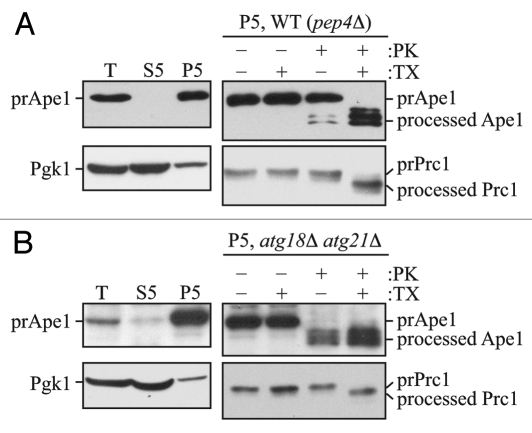
Figure 1.
The prApe1 protease protection assay. (A) Wild-type (atg18Δ atg21Δ pep4Δ) cells expressing Atg18-PA and Atg21 (WT) or (B) atg18Δ atg21Δ pep4Δ cells were grown to exponential phase in YPD medium and converted to spheroplasts.16 The spheroplasts were starved for 1 h in SD-N medium supplemented with 1.2 M sorbitol to induce autophagy, collected by centrifugation, resuspended in osmotically balanced lysis buffer, and then disrupted. In order to remove unbroken cells, the lysate was subjected to centrifugation at 300 × g. The resulting total lysate (T) was further separated into 5,000 × g lysate (S5) and pellet (P5) fractions. The P5 fraction from each strain was split into four parts and subjected to different treatments: No treatment, treatment with 0.4% Triton X-100 (TX), treatment with proteinase K (PK), or treatment with proteinase K in the presence of 0.4% Triton X-100. The samples were precipitated using 10% TCA, acetone washed and analyzed by immunoblot using anti-Ape1 antiserum. Lysis conditions were verified by immunoblot analysis using the anti-Pgk1 and anti-Ape1 antisera. To verify the integrity of organellar membranes in the P5 fractions, maturation of the precursor form of Prc1 was examined. This figure includes data previously published in reference 16, which are reproduced by permission of the American Society for Biochemistry and Molecular Biology and Elsevier, copyright 2010.
With this method, it is also possible to use an internal control (i.e., a control within each sample) to determine the efficiency of spheroplast lysis, and to verify the integrity of intracellular compartments including the vacuole and, by extension, autophagosomes. An ideal marker is Pho8, because this protein contains both a cytosolic tail and a lumenal propeptide. In a pep4Δ strain, the propeptide remains intact. Therefore, the degree of accessibility of the cytosolic tail to exogenous protease indicates the efficiency of spheroplast lysis, whereas resistance of the lumenal propeptide reflects the integrity of the vacuole.17 The vacuole is the organelle most sensitive to osmotic lysis. Accordingly, if the vacuole membrane is intact following spheroplast lysis, it is reasonable to assume that autophagosomes are similarly intact.
As an alternative to monitoring Pho8, it is possible to examine spheroplast lysis and membrane integrity by following other markers. For example, Pgk1 is a soluble, cytosolic protein and therefore should be predominantly present in the total extract and the soluble (S5) fraction following spheroplast lysis (Fig. 1). In contrast, prApe1 is associated with the membrane fraction in a MgCl2-dependent manner18 and therefore should be present to a negligible extent in the soluble fraction (Fig. 1). Carboxypeptidase Y (Prc1) can be monitored to assess the integrity of the vacuole membrane. Prc1 is synthesized as a precursor, and is converted to its mature, active form by the removal of a propeptide after its delivery to the vacuole. In a pep4Δ strain, the precursor form of Prc1 will accumulate within the vacuole lumen; treatment of the pellet fraction (P5) with proteinase K in the absence of detergent does not yield the mature form of Prc1 (Fig. 1). In contrast, the addition of both detergent and proteinase K results in cleavage of the propeptide and the generation of a faster-migrating form. Again, integrity of the vacuole implies that autophagosomes are also intact. In theory, organelle markers such as Kar2 for the ER, or Idh1 for the mitochondria can be used instead of Prc1; however, the membranes of these organelles are more resistant to osmotic lysis, and therefore it is preferable to determine whether or not the vacuolar membrane is intact as a better test of autophagosome integrity. In addition, Prc1 contains a propeptide so that access to exogenous protease can be monitored by a shift in molecular mass rather than the absence or presence of the protein.
Although the prApe1 protease protection assay is extremely reliable, it does have certain drawbacks. For example, only mutants that have a clear defect in import of prApe1 through the Cvt pathway can be analyzed using this method; otherwise, the background accumulation of mature Ape1 makes the analysis problematic. This situation can be bypassed by knocking out a gene such as VAC8, which is not essential for the Cvt pathway; in a vac8Δ strain prApe1 accumulates in vegetative conditions, and is only sequestered upon autophagy induction.19 Another drawback in analyzing the protease sensitivity of prApe1 is seen in the case of ts strains in which prApe1 processing is only blocked at the nonpermissive temperature (NPT). In such strains, in order to ensure that only the population of prApe1 that accumulates after the shift to NPT is examined, it is necessary to radiolabel the cells at the NPT, followed by analysis of the lysates or isolated membrane fractions. Because these additional steps may be time consuming, less desirable, or difficult in some strains, we established a protease protection assay using GFP-Atg8 as a marker protein.
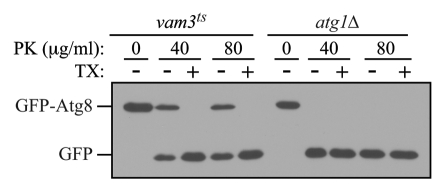
As an internal control for this assay, it is possible to monitor the accessibility of Pho8 as described above; however, this necessitates the use of a pep4Δ background. As an alternative, cells that accumulate autophagosomes in their cytosol such as ypt7Δ or vam3ts cells should be included as a positive (but external) control.20 Fusion of completed autophagosomes with the vacuolar membrane requires the small Rab GTPase Ypt7 and the vacuolar Q-SNARE Vam3. In the ypt7Δ mutant, or in the vam3ts strain (at the NPT), autophagosomes accumulate in the cytosol and their content is protected against externally added proteases such as trypsin or proteinase K (Fig. 2). Thus, in the case of the positive control strains, GFP-Atg8 that is enwrapped within completed autophagosomes is insensitive to exogenously added protease in spheroplast lysates prepared from hypotonically lysed. In these strains, only under conditions that disrupt membrane integrity, such as the addition of detergent, will GFP-Atg8 be protease sensitive. In contrast, in an autophagy-deficient strain such as atg1Δ, autophagosome biogenesis is blocked, and exogenously added proteases will have complete access to GFP-Atg8, resulting in its cleavage to free GFP. Therefore, a protease protection assay analyzing the sensitivity of GFP-Atg8 to proteases in a mutant that is defective for autophagy can be used to distinguish whether the block is in autophagosome biogenesis or in a later step such as fusion with the vacuole. One drawback of the method as described here is that it does not separate the cell lysate into membrane and soluble fractions. Accordingly, there is no internal control to verify the integrity of organelles after the spheroplast lysis. Rather, it is necessary to rely on the positive control strain for this purpose.
Figure 2.
The GFP-Atg8 protease protection assay. Cultures of vam3ts and atg1Δ cells expressing GFP-Atg8 grown to mid-log phase at 25°C were preincubated at 37°C for 30 min, and then shifted to starvation temperature at 37°C, for 1 h. Osmotically lysed cell extracts were analyzed for sensitivity to proteinase K (PK) in the presence or absence of 0.2% Triton X-100 (TX).
2. Materials
2.1. Cells and culture
1. Strains: GFP-Atg8 can be integrated into the genome of the strains of interest, or the strains may be transformed with a plasmid expressing GFP-Atg8 such as pRS316 GFP-AUT721 [note that “AUT7” and “APG8” were previous names for “ATG8”; this plasmid is now referred to as GFP-Atg8(316)] or an equivalent plasmid. Always include atg1Δ, and ypt7Δ or vam3ts strains as negative and positive controls, respectively (see Note 1).
2. Growth medium: If the strains used contain integrated GFP-Atg8 [generated with GFP-Atg8(306)], the cells may be grown in YPD (Bacto-yeast extract 10 g; Bacto-peptone 20 g; deionized or double distilled H2O to 900 ml; 2% dextrose; total volume 1 L). If the strains are transformed with GFP-Atg8(316), the cells should be grown in SMD-URA medium (0.67% yeast nitrogen base without amino acids; 2% D-glucose; and appropriate amino acids and nucleic acid bases to satisfy any auxotrophies). Instead of these minimal requirements, a complete amino acid mix can be used to improve cell growth (0.017% of the following L-amino acids: alanine, arginine, asparagine, aspartic acid, cysteine, glutamine, glutamic acid, glycine, isoleucine, methionine, phenylalanine, proline, serine, threonine, tyrosine, and valine; 0.3 mM l-histidine; 1.7 mM l-leucine; 1 mM l-lysine; and 0.4 mM l-tryptophan) along with 0.3 mM adenine; 2.22 mM myoinositol; and 0.00117% p-aminobenzoic acid. If a plasmid is used that has a different auxotrophic marker, uracil should be added to 0.02% and the appropriate base amino acid left out of the medium.
3. Culture conditions: Cells are grown in YPD or SMD at 30°C to mid-log stage, then starved in SD(-N) medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose).
2.2. Chemicals
Zymolyase 20T (Seikagaku, NC9934469), trypsin (sequencing grade, (11047841001) and proteinase K (03115879001) are from Roche, and other chemicals are from Sigma or Merck. The ECL detection kit (Amersham, RPN2135) and a Fuji LAS-3000 imaging system (Tokyo, Japan) or the equivalent are used for immunoblots.
2.3. Solutions
2x Laemmli buffer: 116 mM TRIS-HCl, pH 6.8, 12% (w/v) glycerol, 3.42% SDS (w/v), 1% β-mercaptoethanol, 0.01% bromophenol blue.
DTT buffer: 10 mM Tris-Sulfate, pH 9.4, 10 mM DTT.
PS200: 20 mM K-PIPES, pH 6.8, 200 mM Sorbitol, 5 mM MgCl2.
SP buffer: 1 M Sorbitol, 20 mM PIPES, pH 6.8.
2.4. Plasmid
2.5. Antibody
GFP (green fluorescent protein) antibody from Roche (11814460001) or YFP (yellow fluorescent protein) antibody from Clontech (632381) can be used to detect the GFP-Atg8 chimera and free GFP.
3. Methods
1. Yeast strains transformed with the plasmid GFP-Atg8(316)21 are cultured in 5 ml SMD-URA medium and grown in a 30°C shaker at 250 rpm (for ts strains choose an appropriate permissive temperature; for the vam3ts strain the permissive temperature is 24°C). Yeast strains containing an integrated version of GFP-Atg8 are grown in YPD. Overnight cultures are diluted to an OD600 = 0.1–0.2 and cultured until the cells reach exponential phase (OD600 = 0.8–1.0).
2. Harvest 60 OD600 units of cells from each strain by centrifugation at 2,000 rpm for 5 min, wash the cells once with 10 ml SD(-N) medium, resuspend them in 60 ml fresh SD(-N) medium and incubate the samples for 2 to 6 h at 30°C while shaking (see Note 2). For ts strains, choose an appropriate temperature to inactivate the temperature sensitive protein (for the vam3ts strain the NPT is 37°C).
3. Harvest cells from each strain by centrifugation at 2,000 rpm for 5 min and wash once with 4 ml of DTT buffer.
4. Discard the supernatant fraction, resuspend the cells in 30 ml DTT buffer (to 2 OD/ml) and incubate them at 30°C (for ts strains choose an appropriate NPT), shaking at 180 rpm for 15 min.
5. Harvest the cells by centrifugation at 600xg for 6 min and remove the supernatant fraction thoroughly (residual DTT may affect later reactions).
6. Resuspend the cells in 6 ml of SP buffer containing 1.2 mg of Zymolyase 20T (pre-incubated at 30°C) for 30 min while shaking (see Note 3).
7. Incubate the samples for 25 min at 30°C (or at the NPT for ts strains) while shaking gently at 180 rpm; occasionally mix by inverting the tube during this incubation time (see Note4). Check O.D. to determine spheroplasting efficiency (see Note 5).
8. Sediment the spheroplasts at 2000xg for 10 min.
9. Resuspend the spheroplasts carefully in 6 ml SP buffer with a pipette (adding one ml at a time) to rinse out the Zymolyase.
10. Sediment the spheroplasts again at 2000xg for 10 min.
11. Hypotonically lyse the spheroplasts by resuspending them in 2 ml PS200 using a cropped tip. Incubate the samples 5 min on ice, and invert the sample 3 times during this incubation.
Alternatively, spheroplasts can be osmotically lysed by resuspending in 6 ml PS200 buffer and disrupting them using a 25-mm Swin-Lok holder assembly (Thomas Scientific, 420200) fitted with a 3.0-μm Nucleopore Track-Etch membrane (Whatman, 110612).
12. Perform two preclearing steps by centrifugation at 300–500xg for 10 min at 4°C to remove unbroken cells and cell debris.
13. Divide the supernatant fraction into aliquots of 300 µl (3 OD600 units/treatment). Add 300 µl of PS200 as a control reaction (total) or 5–10 µg/ml trypsin with and without 0.4% Triton X-100. Add PS200 up to a total volume of 600 µl. Incubate the samples 25 min at 30°C. If you use proteinase K also add 300 µl of PS200 as control reaction (total) or 40–80 µg/ml proteinase K with and without 0.2–0.4% Triton X-100. Incubate samples on ice for 30 min (see Note 6).
14. Add 60 µl of 100% TCA to the samples to stop the reaction, mix thoroughly and incubate at least 10 min on ice. Centrifuge the reactions 10 min at 13,000 rpm at 4°C.
15. Discard the supernatant fraction, and wash the pellet fractions twice with acetone. Dry the pellet, and resuspend it in 2x Laemmli buffer. Incubate the samples for 30 min at 30°C with occasional mixing by vortex.
16. Load 10 μl of the samples for SDS-PAGE.
17. Follow a standard protocol for western blotting. Use anti-GFP or anti-YFP antibody to detect GFP-Atg8 and free GFP, and develop the western blots with appropriate exposure.
4. Notes
1. The thermosensitive vam3ts strain is preferred over the vam3Δ strain for the positive control because the deletion of VAM3 results in indirect effects due to the chronic accumulation of vesicles from multiple pathways that normally deliver cargo to the vacuole in a Vam3-dependent manner. For the negative control strain, any mutant that is defective in autophagosome formation can be used.
2. The appropriate starvation period should be optimized.
3. Pilot experiments may be required to establish the optimal incubation time for the Zymolyase 20T reaction depending on the strain background.
4. Shake at a maximum of 180 rpm to maintain spheroplast integrity.
5. With efficient spheroplasting, the O.D. of a 1:10 dilution in H2O should decrease to 10% of the starting O.D.
6. Pilot experiments should be performed to determine the required trypsin or proteinase K concentration and incubation time.
Acknowledgments
This work was supported by Public Health Service grant GM53396 to D.J.K. from the National Institutes of Health, and by the “Deutsche Forschungsgemeinschaft” to M.T.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/18424
References
- 1.Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–51. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 2.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- 6.He C, Klionsky DJ. Analyzing autophagy in zebrafish. Autophagy. 2010;6:642–4. doi: 10.4161/auto.6.5.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meiling-Wesse K, Barth H, Thumm M. Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett. 2002;526:71–6. doi: 10.1016/S0014-5793(02)03119-8. [DOI] [PubMed] [Google Scholar]
- 8.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–94. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 2010;6:794–7. doi: 10.4161/auto.6.6.12348. [DOI] [PubMed] [Google Scholar]
- 10.Noda T, Klionsky DJ. The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 11.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–32. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 12.Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen E-L, et al. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol. 2010;190:965–73. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen W-L, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–99. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair U, Cao Y, Xie Z, Klionsky DJ. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem. 2010;285:11476–88. doi: 10.1074/jbc.M109.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–50. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–73. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–9. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Klionsky DJ. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem. 2000;69:303–42. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]