Abstract
Trabectedin, a new antitumor compound originally derived from a marine tunicate, is clinically effective in soft tissue sarcoma. The drug has shown a high selectivity for myxoid liposarcoma, characterized by the translocation t(12;16)(q13; p11) leading to the expression of FUS-CHOP fusion gene. Trabectedin appears to act interfering with mechanisms of transcription regulation. In particular, the transactivating activity of FUS-CHOP was found to be impaired by trabectedin treatment. Even after prolonged response resistance occurs and thus it is important to elucidate the mechanisms of resistance to trabectedin. To this end we developed and characterized a myxoid liposarcoma cell line resistant to trabectedin (402-91/ET), obtained by exposing the parental 402-91 cell line to stepwise increases in drug concentration. The aim of this study was to compare mRNAs, miRNAs and proteins profiles of 402-91 and 402-91/ET cells through a systems biology approach. We identified 3,083 genes, 47 miRNAs and 336 proteins differentially expressed between 402-91 and 402-91/ET cell lines. Interestingly three miRNAs among those differentially expressed, miR-130a, miR-21 and miR-7, harbored CHOP binding sites in their promoter region. We used computational approaches to integrate the three regulatory layers and to generate a molecular map describing the altered circuits in sensitive and resistant cell lines. By combining transcriptomic and proteomic data, we reconstructed two different networks, i.e. apoptosis and cell cycle regulation, that could play a key role in modulating trabectedin resistance. This approach highlights the central role of genes such as CCDN1, RB1, E2F4, TNF, CDKN1C and ABL1 in both pre- and post-transcriptional regulatory network. The validation of these results in in vivo models might be clinically relevant to stratify myxoid liposarcoma patients with different sensitivity to trabectedin treatment.
Introduction
Trabectedin (Yondelis®) is a tetrahydroisoquinoline molecule originally derived from the Caribbean marine tunicate, Ecteinascidia turbinata, with peculiar in vitro and in vivo cytotoxic activity in a wide range of tumor type: it has been approved by EMEA for the second line therapy of soft tissue sarcomas and ovarian cancer [1]–[7].
From a mechanistic point of view, trabectedin binds to the minor groove of DNA, differently from the conventional alkylating agents, forming DNA adducts on N2 position of guanine, and bends DNA towards the major groove; two of the three fused rings of this molecule are involved in the minor groove binding while the third (ring C) plays an important role in the process of interaction with different DNA-binding proteins [8], [9]. It is now widely accepted that trabectedin-DNA interaction affects transcription regulation although the level of selectivity is not completely elucidated [8], [10], [11].
Although trabectedin has shown activity against several types of sarcomas, its higher activity has been observed in patients with myxoid liposarcoma (MLS).
The high sensitivity displayed by MLS tumors is probably correlated to the ability of trabectedin to block the aberrant transcription activity of FUS-CHOP, the chimeric protein hallmark of MLS, generated by the chromosomal translocation t(12;16)(q13;p11) [12], [13].
Despite its activity, it is becoming a clinical experience that patients with long lasting trabectedin treatment develop resistance over time. To our knowledge there are only few studies focused on the mechanism of resistance and even fewer are based on a systems biology approach.
To investigate the molecular pathways involved in trabectedin resistance, the 402-91/ET cell line, resistant to trabectedin, was generated from the parental type I myxoid liposarcoma cell line 402-91 by stepwise increasing drug concentrations [14]. Based on the assumption that trabectedin impairs mechanisms of transcription regulation, we exploited several high-throughput technologies to combine different layers of regulation to elucidate the molecular pathways involved in the mechanism of trabectedin resistance in a well defined in vitro model of myxoid liposarcoma, previously described [14].
Materials and Methods
Cells and experimental design
The MLS type I 402-91 cell line was kindly supplied by Dr. P. Aman [15], while the trabectedin resistant 402-91/ET cell line was obtained in our laboratory as previously described [14]. For the array experiments, different batches of exponentially growing 402-91 and 402-91/ET cells were seeded and then total RNA enriched in microRNA (miRNA) fraction and protein lysates were harvested according to standard protocols. Gene, miRNA and protein expression profiles were generated from three independent biological replicates. Each experiment was performed using four technical replicates.
RNA purification and quantitative Reverse Transcription PCR (qRT-PCR)
Total RNA enriched in miRNA fraction was purified using the miRNeasy Mini Kit according to the manufacturer's instructions (Qiagen, Milan, Italy). RNA was quantified by spectrophotometry (Nanodrop; Thermo Scientific, Wilmington, USA) and quality assessed by on-chip electrophoresis (Bioanalyser RNA 6000 Nano, Agilent Technologies, Palo Alto, USA). Mature miRNA and mRNA expression levels were examined by qRT-PCR using Applied Biosystems 7900HT. Briefly, cDNA was generated from 1 µg of purified total RNA using the miScript® Reverse Transcription Kit, following the manufacturer's instructions (Qiagen). miRNA expression was quantified using QuantiTect SYBR Green PCR Master Mix and the specific miScript® Primer Assay (Qiagen); RU-6b was used as reference gene. mRNA expression was analyzed using QuantiFast SYBR Green PCR Master Mix (Qiagen) and dedicated primers that were designed as described [16]; cyclophilin A was used as reference gene. Experiments were run in triplicate for each case to assess technical variability. Analysis was performed by using the 2−ΔΔCt protocol and the median values were compared using the non parametric Mann-Whitney t-test. Differences were considered statistically significant with a two sided p-value<0.05. All tests and data plots were done using GraphPad Prism Version 5.01 (GraphPad Software, La Jolla, Ca, USA).
Western blot analysis
Total protein extracts were prepared by lysing cell extracts in lysis buffer (50 mM Tris-HCl, pH 8.0, 1% Triton X-100, 150 mM NaCl, 1 mM EGTA, 100 mM NaF, 10% glycerol (100%) and 1 mM MgCl2) with 2 mM phenylmethanesulfonyl fluoride (Sigma Aldrich, St. Louis, MO) as protease inhibitor, in agitation for 30 minutes at 4°C. Insoluble material was pelleted at 13 000 rpm at 4°C and the total protein concentration was determined by Bradford assay. The cellular proteins (40 µg) were separated using 8% and 15% (for detection of HMGA2 protein) SDS-PAGE and transferred electrophoretically to polyvinylidenedifluoride membranes (GE Healthcare, CA, USA). Blots were probed at 4°C overnight with primary antibodies in 5% milk/Tris Buffer Saline Tween20 (TBST). Immunoblotting was carried out with Cyclin D1 (Pharmingen, BD, La Jolla, CA, USA- 554181), E2F5 (abcam, Cambridge, UK - ab59769), SEMA4C (R&D Systems, Minneapolis MN, USA – AF6125), HMGA2 (kindly provided by Prof. Alfredo Fusco, Institute of Experimental Endocrinology and Oncology, CNR Naples, Italy) and PDCD4 (Sigma-Aldrich – SAB1407349) antibodies. Lamin B (Santa Cruz Biotechnology – sc6217) and Actin (Santa Cruz Biotechnology, CA, USA– sc130301) were used for loading control. Antibody binding was revealed by peroxidase labelled secondary antibodies and visualized using enhanced chemiluminescent (ECL) substrate (SuperSignal* West Pico, Thermo Scientific, Rockford IL USA). Experiments were repeated twice.
Gene expression analysis
Gene expression (GE) was analyzed using Agilent slides on which 44,000 oligonucleotides were spotted (Agilent Technologies, Santa Clara, USA). Experiments were performed in four replicates, with dye-swapping duplication-scheme. The arrays were washed and scanned with a laser confocal scanner (G2565BA, Agilent Technologies) according to manufacturer's instructions. Oligo microarrays underwent standard post hybridization processing and the intensities of fluorescence were calculated by Feature Extraction software v.11 (Agilent Technologies). The microarray raw data are deposited in GEO (experiment number GSE20350) according to MIAME (Minimum Information About a Microarray Experiment) guidelines. GE signals were filtered according to the signal-to-background flag; features with more than 60% of signal below the local background were removed from the analysis. Then, lowess normalization was applied [17]. The identification of differentially expressed genes was performed using Empirical Bayes test [18] implemented in Bioconductor package limma in R software (http://www.r-project.org) with Benjamini and Hochberg False Discovery Rate (FDR) [19] to control for multiple testing problem. Given the large number of differentially expressed genes, we decided to adopt a more conservative strategy with respect to that applied for differentially expressed miRNAs and proteins: FDR≤0.01 and log fold change greater/lower than 1/-1 were considered to select differentially expressed features. Enrichment analysis of Gene Ontology (GO) categories and KEGG pathways was performed with DAVID web tool [20]. Cytoscape 2.7 [21] was used to reconstruct miRNA-target network.
Transcription factor binding site enrichment analysis was performed using oPOSSUM web tool [22].
miRNA expression analysis
100 ng of total RNA enriched in miRNA fraction was Cy5-labelled and hybridized with a miRNA labelling and hybridization kit according to the manufacturer's instructions (Agilent Technologies). We used the commercially available G4470B human miRNA microarray kit (Agilent Technologies), which consists of 15 K features printed in an 8-plex format (8 of 15 K features printed in known human miRNAs :723 human and 76 human-viral miRNAs) sourced from the Sanger miRBASE public database, release 10.1. The arrays were washed and scanned (with a laser confocal scanner) according to the manufacturer's instructions. miRNA microarrays underwent standard post hybridization processing and the intensities of fluorescence were calculated by Feature Extraction software version 11 (Agilent Technologies). miRNA expression values were filtered as described in the previous section, and vsn algorithm has been applied to normalize the data [23]. miRNA data have been submitted to Array Express (accession number E-MTAB-965).
The identification of differentially expressed miRNAs has been performed using Empirical Bayes test [18] implemented in Bioconductor package limma of R software with Benjamini and Hochberg FDR to control for multiple testing problem. FDR≤0.1 were considered to select significant miRNAs.
mirandaSVR [24] was used to select putative miRNA target genes, while integration of mRNA and miRNA data was performed using MAGIA [25] web tool and dedicated in-house software.
Enrichment analysis of GO categories and KEGG pathways [26] of differentially expressed miRNA targets was performed with DAVID web tool [20] and MetaCore software [27].
CHOP transcription factor binding site identification on putative miRNA promoter regions was performed using miRGen2.0 [28] and CircuiDB database [29]. Cytoscape 2.7 [21] was used to reconstruct miRNA-target regulatory networks. KEGG database has been used to retrieve gene-gene interactions.
Antibody microarray analysis
The Panorama Ab Microarray–XPRESS kit (Sigma-Aldrich) used in this study contains 725 different antibodies, spotted in duplicate on nitrocellulose-coated glass slides. These antibodies represent families of proteins known to be involved in a variety of different biological pathways (www.sigma.com/arrays). All reagents and solutions for protein purification were supplied within the kit and experiment was performed on four independent biological replicates, according to manufacturer's instructions. Briefly, total cell lysates were obtained from almost 10 million of exponentially growing 402-91 and 402-91/ET cells. 10 mg of total protein lysate, measured using Bradford protocol, were labeled with Cy3 and Cy5 fluorescent dyes (Amersham Biosciences, Buckinghamshire, UK) and their specific activity and concentration measured at the Nanodrop (Nanodrop, Thermo Scientific) after affinity column purification, provided within the kit. 150 µg of Cy3 and Cy5 labeled samples were co-hybridized on the same slides for 45′ at room temperature, accordingly to manufacturer's instructions. Washing and scanning (GENEPIX, version B) was performed according to the manufacturer's instructions. To increase the robustness of data analysis we performed technical replicates using dye-swap.
Scanned data files were first background subtracted using the normexp method [30]. Positive and negative control spots were removed, and the data were normalized using a modified rank-invariant selection algorithm (in-vMod) [31].
Determination of differential proteins between the two cell lines was carried out with linear models for microarray analysis [30], correcting the resulting p-value for multiple testing with FDR. To balance statistical significance and biological relevance, given the relatively low number of spots on the arrays, proteins were selected as differential expressed when their adjusted p-value was less than or equal to 0.1, and they were subsequently annotated with the EntrezGene and RefSeq identifiers of the corresponding genes to facilitate data integration.
Network analysis andGO enrichment of differential proteins was performed using the MetaCore software [27], using both public data and internal MetaCore curated maps, and with DAVID web tool [20]. Networks and GO terms were deemed significant if their p-value was less than or equal to 0.1.
Results
In order to deeply understand the molecular differences between 402-91 and 402-91/ET cells, we performed multistep omics comparative analyses evaluating gene, miRNA and protein expressions.
Gene expression data
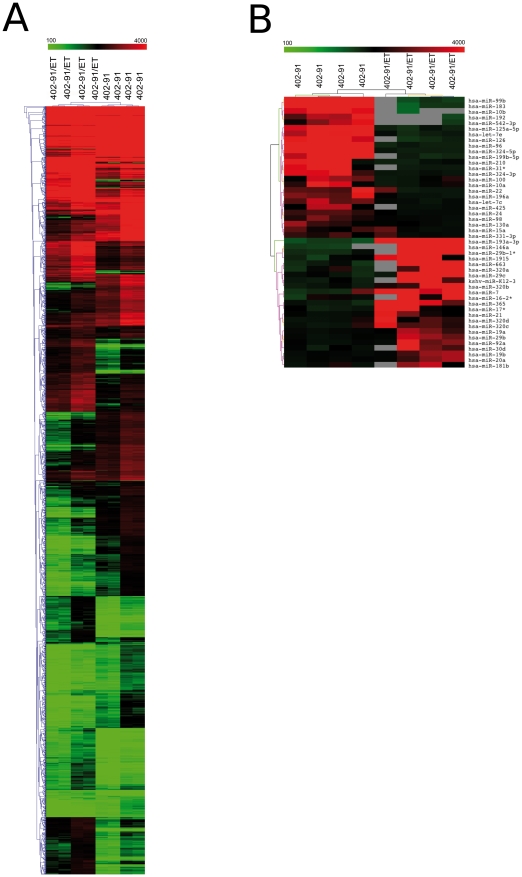
A list of 2676 significant genes (corresponding to 3,083 non-unique probes) has been identified as differentially expressed between 402-91/ET and 402-91 cell lines ( Figure 1A ): 1473 (55%) underexpressed and 1203 (45%) overexpressed. Multiple probes in the microarray platform can be representative of a single gene; in our analysis more than 80% of probes belonging to the same gene behave concordantly. Only 505 genes represented by more than one probe in the platform have been identified as significant by a single probe. The complete list of differentially expressed probes is reported in Table S1. Remarkable is that a significant portion of the underexpressed (6%) and of the overexpressed (2%) genes are zinc finger proteins, i.e. proteins able to bind DNA (p = 0.0003 with hypergeometric distribution).
Figure 1. Heat maps of genes and miRNAs differentially expressed between 402-91/ET and 402-91 cells.
Panel A. Heat map and cluster analysis of the 3,083 differentially expressed probes between resistant and sensitive cell lines. Red and green represent respectively differentially upregulated and downregulated genes in 402-91/ET cell lines. Panel B. Heat map and cluster analysis of the 48 miRNA differentially expressed between resistant and sensitive cell lines. Red and green represent respectively differentially upregulated and downregulated genes in 402-91/ET cell lines. Gray is for missing values.
Functional enrichment has been performed separately for up and downregulated genes, both on GO categories ( Table 1 ) and KEGG and MetaCore pathways ( Table 2 ).
Table 1. Biological processes enrichment analysis within up and downregulated genes using Gene Ontology categories.
| Category | Name | Adj p-value | Expression 402-91/ET vs 402-91 |
| GO:0048522 | positive regulation of cellular process | 0,02 | up |
| GO:0006915 | apoptosis | 0,03 | up |
| GO:0008284 | positive regulation of cell proliferation | 0,04 | up |
| GO:0031325 | positive regulation of cellular metabolic process | 0,04 | up |
| GO:0051093 | negative regulation of developmental process | 0,05 | up |
| GO:0001525 | angiogenesis | 0,05 | up |
| GO:0001944 | vasculature development | 0,05 | up |
| GO:0048523 | negative regulation of cellular process | 0,05 | up |
| GO:0009893 | positive regulation of metabolic process | 0,05 | up |
| GO:0001558 | regulation of cell growth | 0,05 | up |
| GO:0010941 | regulation of cell death | 0,05 | up |
| GO:0001501 | skeletal system development | 0,05 | up |
| GO:0002684 | positive regulation of immune system process | 0,05 | up |
| GO:0051249 | regulation of lymphocyte activation | 0,05 | up |
| GO:0006954 | inflammatory response | 0,05 | up |
| GO:0048513 | organ development | 0,05 | up |
| GO:0043067 | regulation of programmed cell death | 0,06 | up |
| GO:0050865 | regulation of cell activation | 0,06 | up |
| GO:0051252 | regulation of RNA metabolic process | 0,0002 | down |
| GO:0006350 | transcription | 0,0002 | down |
| GO:0010468 | regulation of gene expression | 0,0006 | down |
| GO:0080090 | regulation of primary metabolic process | 0,0007 | down |
| GO:0060255 | regulation of macromolecule metabolic process | 0,0007 | down |
| GO:0034645 | cellular macromolecule biosynthetic process | 0,0008 | down |
| GO:0010556 | regulation of macromolecule biosynthetic process | 0,002 | down |
| GO:0031326 | regulation of cellular biosynthetic process | 0,003 | down |
| GO:0009889 | regulation of biosynthetic process | 0,003 | down |
| GO:0051171 | regulation of nitrogen compound metabolic process | 0,003 | down |
| GO:0031323 | regulation of cellular metabolic process | 0,003 | down |
| GO:0019219 | regulation of nucleobase. nucleoside. nucleotide and nucleic acid metabolic process | 0,003 | down |
| GO:0009887 | organ morphogenesis | 0,03 | down |
| GO:0010563 | negative regulation of phosphorus metabolic process | 0,03 | down |
| GO:0000082 | G1/S transition of mitotic cell cycle | 0,03 | down |
| GO:0051329 | interphase of mitotic cell cycle | 0,05 | down |
Table 2. Pathways enrichment analysis within up and downregulated genes using KEGG and Metacore databases.
| Database | Name | Adj p-value | Expression 402-91/ET vs 402-91 |
| KEGG | NOD-like receptor signaling pathway | 0,01 | up |
| KEGG | Small cell lung cancer | 0,03 | up |
| KEGG | Vitamin B6 metabolism | 0,05 | up |
| KEGG | p53 signaling pathway | 0,05 | up |
| KEGG | PPAR signaling pathway | 0,05 | up |
| Metacore | Immune response_IL-17 signaling pathways | 0 | up |
| Metacore | Immune response_Alternative complement pathway | 0 | up |
| Metacore | Cytokine production by Th17 cells in CF | 0,00021 | up |
| Metacore | Cell cycle_Regulation of G1/S transition (part 1) | 0,00023 | up |
| Metacore | Cell cycle_Regulation of G1/S transition (part 2) | 0,0006 | up |
| Metacore | Development_TGF-beta-dependent induction of EMT via MAPK | 0,0007 | up |
| Metacore | Cytokine production by Th17 cells in CF (Mouse model) | 0,00085 | up |
| Metacore | Cell cycle_Nucleocytoplasmic transport of CDK/Cyclins | 0,00122 | up |
| Metacore | Development_TGF-beta-dependent induction of EMT via SMADs | 0,00256 | up |
| Metacore | Immune response_Th17-derived cytokines | 0,00262 | up |
| Metacore | Development_PEDF signaling | 0,00361 | up |
| Metacore | Cytoskeleton_Regulation of cytoskeleton rearrangement | 0,00381 | up |
| KEGG | Focal adhesion | 0,001 | down |
| KEGG | Notch signaling pathway | 0,002 | down |
| KEGG | Prion diseases | 0,003 | down |
| KEGG | ECM-receptor interaction | 0,007 | down |
| KEGG | Lysosome | 0,01 | down |
| KEGG | Gap junction | 0,011 | down |
| KEGG | Axon guidance | 0,012 | down |
| KEGG | Inositol phosphate metabolism | 0,015 | down |
| KEGG | Bladder cancer | 0,03 | down |
| KEGG | Tight junction | 0,033 | down |
| KEGG | Neurotrophin signaling pathway | 0,034 | down |
| KEGG | Insulin signaling pathway | 0,035 | down |
| KEGG | Valine, leucine and isoleucine degradation | 0,038 | down |
| KEGG | Chronic myeloid leukemia | 0,044 | down |
| KEGG | Endocytosis | 0,044 | down |
| KEGG | N-Glycan biosynthesis | 0,047 | down |
| KEGG | Adherens junction | 0,051 | down |
| Metacore | Cell adhesion_Amyloid proteins | 0 | down |
| Metacore | Immune response_Histamine signaling in dendritic cells | 0,00001 | down |
| Metacore | Signal transduction_cAMP signaling | 0,00001 | down |
| Metacore | Development_S1P2 and S1P3 receptors in cell proliferation and differentiation | 0,00001 | down |
| Metacore | Development_Blood vessel morphogenesis | 0,00001 | down |
| Metacore | Development_Notch Signaling Pathway | 0,00002 | down |
| Metacore | Development_A2A receptor signaling | 0,00002 | down |
| Metacore | Blood coagulation_GPCRs in platelet aggregation | 0,00002 | down |
| Metacore | Cell adhesion_Platelet aggregation | 0,00002 | down |
| Metacore | Cytoskeleton_Actin filaments | 0,00002 | down |
| Metacore | Development_Neurogenesis_Axonal guidance | 0,00006 | down |
| Metacore | Development_A2B receptor: action via G-protein alpha s | 0,00008 | down |
| Metacore | Signal transduction_Activation of PKC via G-Protein coupled receptor | 0,00012 | down |
| Metacore | Transcription_CREB pathway | 0,00013 | down |
| Metacore | PGE2 pathways in cancer | 0,00018 | down |
| Metacore | Signal Transduction_Cholecystokinin signaling | 0,00052 | down |
| Metacore | Cytoskeleton_Regulation of cytoskeleton rearrangement | 0,00105 | down |
| Metacore | Muscle contraction_Nitric oxide signaling in the cardiovascular system | 0,00197 | down |
| Metacore | Development_Hedgehog signaling | 0,00287 | down |
| Metacore | Cell adhesion_Leucocytechemotaxis | 0,00747 | down |
Most of the genes overexpressed in 402-91/ET cell line are implicated in apoptosis, regulation of cell proliferation, angiogenesis, vascular development and negative regulation of developmental processes (enclosed the adipocyte differentiation category). On the other hand, pathways related to metabolism, gene expression and cell cycle progression are mainly populated by downregulated genes. Pathways associated to cancer (p53 signaling pathway and small cell lung cancer) are involved in trabectedin resistance as well as immunoregulatory mechanisms like PPARs and NOD-like receptors signaling. Some key genes involved in cell cycle and regulation of apoptosis have been validated with qRT-PCR (see Table S2 and section below).
miRNA expression data
Inferential analysis identified a list of 47 miRNAs that showed an altered regulation between 402-91/ET and 402-91 cell lines ( Figure 1B ): 23 overexpressed and 24 underexpressed (see Table S3). Among these, we found two miRNA clusters (defined as miRNAs whose chromosomal position is less than 1000 kb) on chromosome 13 (miR-17*, miR-19a, miR-19b, miR-20a, miR-92a) and 19 (miR-99b, let-7e, miR-125a-5p) and the miRNA families miR-320, miR-19 and miR-29.
Moreover, we performed a functional enrichment analysis on putative targets of significantly altered miRNAs in 402-91/ET cell line (see Table 3 ). The in silico predicted targets were cleaned out of those genes without differential expression. Gene targets of miRNAs regulation belong to categories involved in immune response, regulation of cell death, apoptosis and anti-apoptosis and G1/S transition of mitotic cell cycle. Among these genes, there were several known oncogenes playing a key role in tumor progression in melanoma, bladder and lung cancer. Some key miRNAs involved in cell cycle and regulation of apoptosis have been validated with qRT-PCR (see Table S2 and section below).
Table 3. GO biological processes and KEGG and Metacore pathway enrichment analysis within miRNA targets.
| Category | Name | Adj p-value |
| Gene Ontology | ||
| GO:0001525 | angiogenesis | 0,01 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 0,01 |
| GO:0043067 | regulation of programmed cell death | 0,02 |
| GO:0051252 | regulation of RNA metabolic process | 0,02 |
| GO:0042981 | regulation of apoptosis | 0,02 |
| GO:0006355 | regulation of transcription. DNA-dependent | 0,02 |
| GO:0009887 | organ morphogenesis | 0,02 |
| GO:0001944 | vasculature development | 0,03 |
| GO:0006916 | anti-apoptosis | 0,03 |
| GO:0048705 | skeletal system morphogenesis | 0,03 |
| GO:0001568 | blood vessel development | 0,04 |
| GO:0046395 | carboxylic acid catabolic process | 0,07 |
| GO:0048514 | blood vessel morphogenesis | 0,07 |
| GO:0016477 | cell migration | 0,09 |
| Pathway | ||
| Metacore | PGE2 pathways in cancer | 0 |
| Metacore | Immune response_Histamine signaling in dendritic cells | 0,00013 |
| Metacore | Signal transduction_cAMP signaling | 0,00015 |
| Metacore | Development_S1P1 receptor signaling via beta-arrestin | 0,00019 |
| Metacore | Muscle contraction_Regulation of eNOS activity in endothelial cells | 0,00021 |
| Metacore | Immune response_Histamine H1 receptor signaling in immune response | 0,00022 |
| Metacore | Cell adhesion_Chemokines and adhesion | 0,00022 |
| Metacore | Development_Mu-type opioid receptor signaling via Beta-arrestin | 0,00032 |
| Metacore | Immune response_PGE2 signaling in immune response | 0,00038 |
| Metacore | Development_S1P2 and S1P3 receptors in cell proliferation and differentiation | 0,00055 |
| KEGG | ECM-receptor interaction | 0,009 |
Signature validation
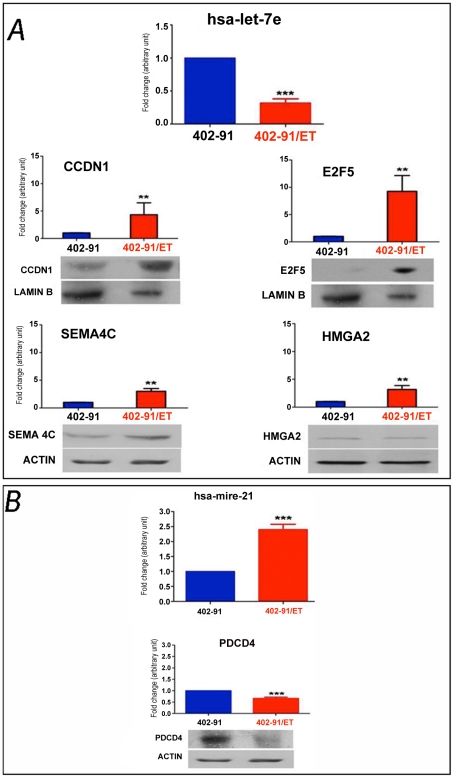
Apoptosis and cell cycle seem to be the most involved pathways in trabectedin resistance in 402-91/ET cell line either on transcriptional or on translational levels. Thus, to support the robustness of our analysis, differences in expression of genes and miRNA, belonging to the apoptosis and cell cycle pathways were validate in an independent batch of 402-91 and 402-91/ET cells. Representative results for let-7e and miR-21 are reported in Figure 2 . As expected, let-7e resulted three folds downregulated (p<0.001) and miR-21 two folds upregulated (p<0.0001) in 402-91/ET compared to 402-91cells. qRT-PCR analysis of let-7e downstream targets confirmed four folds upregulation of CCDN1 (p<0.01), three folds upregulation for SEMA4C (p<0.01) and nine folds upregulation for E2F5 (p<0.001) in the resistant compared to the sensitive cell line ( Figure 2A ). Western blot analysis confirmed at protein level such differences. Analysis of PDCD4, a downstream target of miR-21, was confirmed to be downregulated in the resistant cells (1.5 times, p<0.0001) at both mRNA and protein levels ( Figure 2B ). These results confirmed our omics analysis even if the effect of let-7e on HMGA2 needs to be discussed separately. The three folds upregulation of HMGA2 at the mRNA level in 402-91/ET cells (p<0.01) was not further confirmed by western blot analysis ( Figure 2A ), suggesting that other mechanisms, not directly related to control of mRNA stability by miR-21, are involved in the HMGA2 protein stability.
Figure 2. Signature validation of miRNA-mRNA and proteins found differentially expressed between 402-91/ET and 402-91 cell line.
Panel A. qRT-PCR and Western blot analysis showing differences in the expression levels of let-7e and its downstream targets (CCDN1, E2F5, SEMA4C, HMGA1 and HMGA2). Panel B. qRT-PCR and Western blot for miR-21 and its downstream target, PDCD4. qRT-PCR data are the mean of three independent experiments performed in triplicate and calculated with the 2−ΔΔCt method as described in the material and method section. The control 402-91 cells values are arbitrarily set as 1. Bars are +/− SD. * is significant with p<0.05, ** p<0.01, ***p<0.001, as assessed with Student T-test. Western blot is representative of at least two independent experiments.
Antibody array data
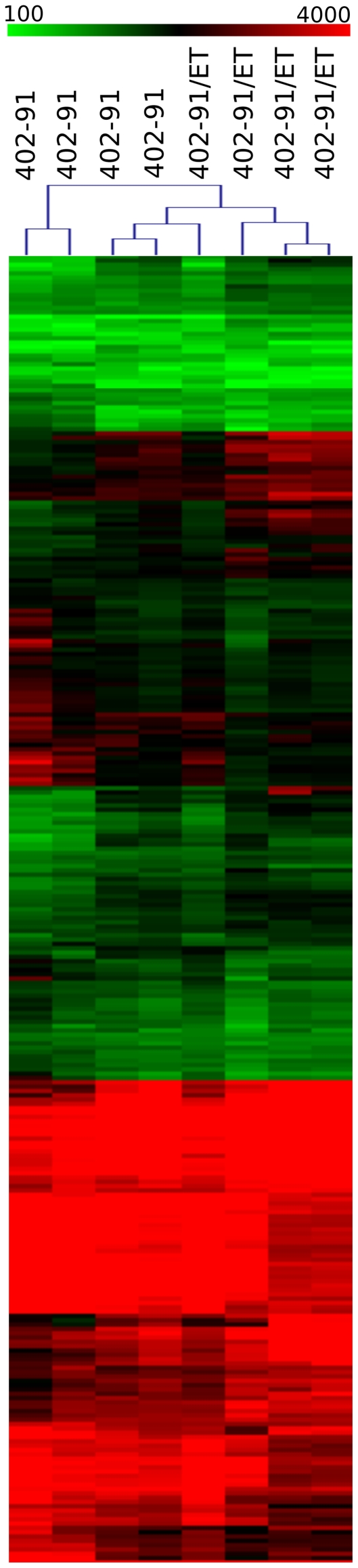
Protein profiling identified 336 proteins that were significantly (FDR<0.1) different between sensitive and resistant cell lines (Table S4): 148 upregulated and 188 downregulated (see Figure 3 ). Cluster dendrograms of Figure 1A, 1B and 3 were performed on different data sources (genes, miRNAs and proteins expression levels) characterised by different sizes, means and range of values. Since hierarchical clusteringis highly sensitive to data structure, some differences in dendrograms are expected, given the intrinsic characteristics of the data. According to the experimental design, gene, miRNA and protein expression profiles were quantified using three different biological replicates, while within each batch technical replicates were performed. Thus, the difference in the dendrogram structures is due to the combination of these effects: biological and intrinsic data differences.
Figure 3. Heat map and cluster analysis of the 336 differentially expressed proteins between resistant and sensitive cell lines.
Red and green represent respectively differentially upregulated and downregulated proteins in 402-91/ET cell lines.
In order to assess the biological meaning of the differences in the protein expression, we performed enrichment analysis on GO terms, cellular networks and pathway maps using DAVID web tool and MetaCore software.
MetaCore analysis showed a prevalence of apoptosis-survival related functions for the upregulated proteins: the top three categories from enrichment analysis on GO terms showed prevalence of terms associated to cell death (p<0.0001). Of the top three pathways, two were associated with apoptosis (FAS signaling cascades and TNF family pathways), and “death domain receptors and caspases in apoptosis” was identified as significantly enriched in process networks. On the other hand, GO terms over-represented in downregulated proteins mainly involved cellular component reorganization and affected pathways and networks including AKT signaling, cytoskeleton remodeling and G1/S cell cycle regulation ( Table 4 ). Other involved functions were related to cell cycle, cytoskeletal remodeling and growth factor regulation.
Table 4. Gene Ontology categories and KEGG, PANTHER and Metacore pathway enrichment analysis within up and down regulated proteins.
| Category | Name | p-value | Expression 402-91/ET vs 402-91 |
| GeneOntology | |||
| GO | apoptosis | 0,00000 | up |
| GO | programmedcelldeath | 0,00000 | up |
| GO | celldeath | 0,00000 | up |
| GO | cellular component organization | 0,00000 | down |
| GO | cellular component organization or biogenesis | 0,00000 | down |
| GO | cellular component organization at cellular level | 0,00000 | down |
| Pathway | |||
| KEGG | hsa04115:p53 signalingpathway | 0,00027 | up |
| KEGG | hsa04210:Apoptosis | 0,00140 | up |
| KEGG | hsa05219:Bladder cancer | 0,00397 | up |
| KEGG | hsa05218:Melanoma | 0,04945 | up |
| KEGG | hsa05212:Pancreatic cancer | 0,05277 | up |
| KEGG | hsa04010:MAPK signalingpathway | 0,07292 | up |
| KEGG | hsa05200:Pathways in cancer | 0,07840 | up |
| KEGG | hsa04650:Natural killer cell mediated cytotoxicity | 0,14232 | up |
| PANTHER | P00006:Apoptosis signalingpathway | 0,00052 | up |
| PANTHER | P00059:p53 pathway | 0,01579 | up |
| Metacore | Apoptosis and survival_p53-dependent apoptosis | 0 | up |
| Metacore | Apoptosis_Death Domain receptors &caspases in apoptosis | 0 | up |
| Metacore | Cell adhesion_Amyloid proteins | 0 | up |
| Metacore | Cell cycle_G1-S Growth factor regulation | 0,00001 | up |
| Metacore | Apoptosis and survival_Role of IAP-proteins in apoptosis | 0,00026 | up |
| Metacore | Translation _Regulation of EIF2 activity | 0,00084 | up |
| KEGG | hsa04110:Cell cycle | 0,00000 | down |
| KEGG | hsa05200:Pathways in cancer | 0,00235 | down |
| KEGG | hsa04510:Focal adhesion | 0,00681 | down |
| KEGG | hsa04210:Apoptosis | 0,01537 | down |
| KEGG | hsa05212:Pancreatic cancer | 0,03870 | down |
| PANTHER | P00006:Apoptosis signalingpathway | 0,00022 | down |
| PANTHER | P00020:FAS signalingpathway | 0,00130 | down |
| Metacore | Cytoskeleton remodeling_TGF, WNT and cytoskeletal remodeling | 0,00000 | down |
| Metacore | Signaltransduction_AKTsignaling | 0,00000 | down |
| Metacore | Cytoskeletonremodeling_Cytoskeletonremodeling | 0,00000 | down |
| Metacore | Cell cycle_G1-S Growth factor regulation | 0,00000 | down |
| Metacore | Cell cycle_G1-S Interleukin regulation | 0,00001 | down |
| Metacore | Cell cycle_Core | 0,00015 | down |
Likewise, the analysis performed with DAVID identified among the highest significant results pathways related to apoptosis, stress induction of HSP and the p53 pathway for upregulated proteins, and cell cycle, AKT signaling, and FAS signaling for downregulated proteins ( Table 4 ).
CHOP transcription factor binding site analysis
It has been reported that FUS-CHOP is the main molecular target of trabectedin in myxoid liposarcoma cell line [12], [13]. Under the assumption that the FUS-CHOP binding site is the same of CHOP (then of the heterodimer CHOP-C/EBPá), we tried to identify putative miRNAs transcriptionally regulated by the chimera and therefore having an enriched CHOP motif in their promoter regions.
Transcription factor binding sites enrichment analysis performed on separately up or downregulated genes (oPOSSUM web tool with Z-score>5 and p-value<0.5) identified respectively 10 and 11 binding sites enriched (see Table S5). CHOP binding site was not identified as enriched.
The identification of miRNA promoter regions is still a challenging task, since putative miRNA regulatory regions could be several thousands of bp upstream of the pri-miR. There are only a limited number of experimentally validated miRNA promoters. In particular only few of the differentially expressed miRNAs have an experimentally validated promoter region. In this context, TFBS enrichment analysis on the promoter regions of the differentially expressed miRNAs would be misleading. However, we could identify miRNAs among those differentially expressed with an experimentally validated promoter with a putative CHOP binding site. Using mirGen2.0 web tool, we found that miR-130a (downregulated in 402-91/ET cell line) and miR-21 (upregulated in 402-91/ET cell line) have a CHOP-C/EBPá binding site in their experimentally validated promoter regions, while using CircuiDB database [29] we found that miR-7 (upregulated in 402-91/ET cell line) has a CHOP-C/EBPá binding site in its computationally identified promoter region.
Integrative analysis and network reconstruction
Integration of miRNA and mRNA expression data
miRNA effect on mRNA degradation can be detected by the anti-correlation relationship between miRNA and gene expression. To reconstruct miRNA-mRNA network in this section we focused only on miRNAs having an inverse relationship with their respective mRNA targets.
Using only the list of differentially expressed miRNAs and genes we reconstructed the network in which nodes represent both genes and miRNAs (colored according to their expression levels) while edges represent computationally identified miRNA-target interactions. The entire resulting network is available in Figure S1 and Table S6.
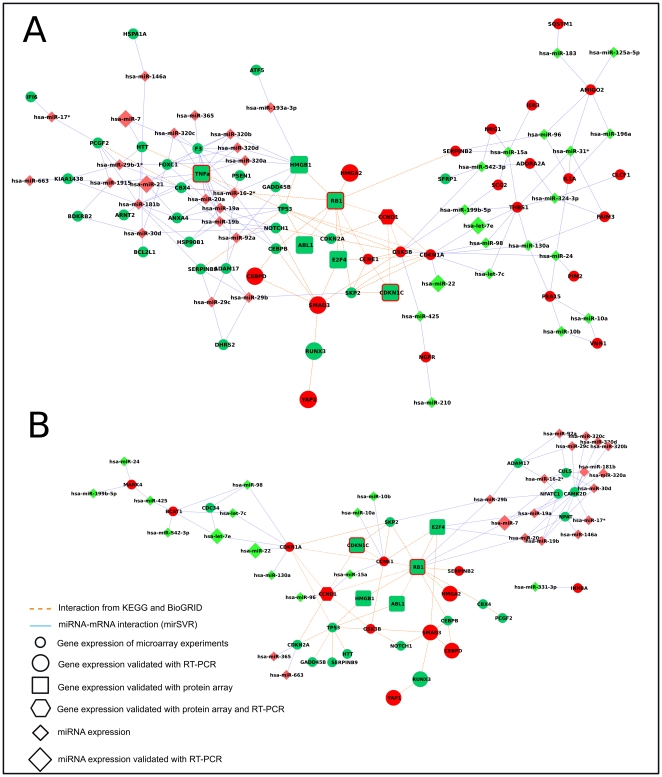
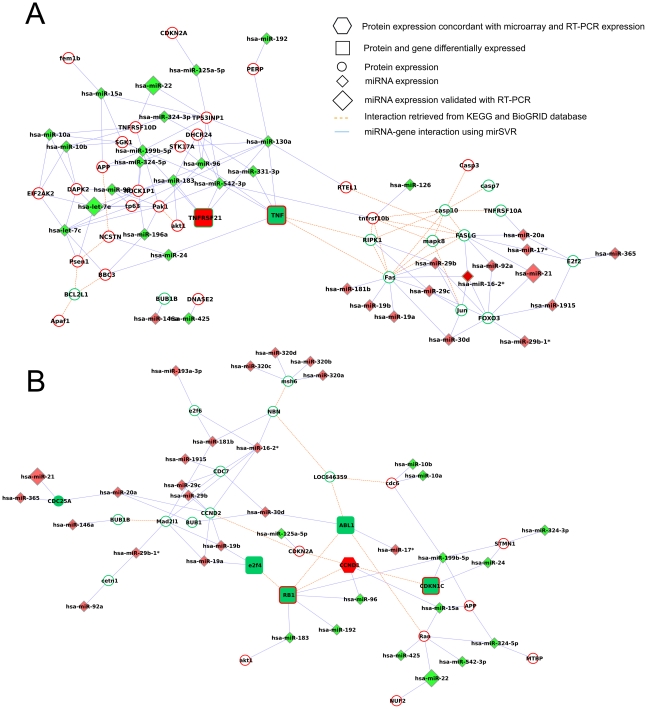
We then focused on specific network modules (sub-networks), corresponding to pathways or specific miRNAs found to be interesting according to the biological problem under study ( Figure 4 ). In particular we selected genes involved in negative regulation of apoptosis ( Figure 4A ) and cell cycle ( Figure 4B ). Then we selected the putative targets of those miRNAs (miR-7, miR-21 and miR-130a) with experimentally validated or computationally identified CHOP- C/EBPá binding sites in their promoter regions ( Figure 5 ). Several miRNAs and genes identified as differentially expressed have been validated in qRT-PCR as previously reported ( Figure 2 ; Table S2).
Figure 4. Post-transcriptional network.
miRNA and mRNA subnetworks representing negative regulation of apoptosis (Panel A) and cell cycle (Panel B). Small circle: differentially expressed genes found by microarray analysis. Big circle: differentially expressed genes validated by qRT-PCR. Square: differentially expressed genes encoding a protein found differentially expressed using protein array. Exagon: differentially expressed genes validated with qRT-PCR encoding a protein found differentially expressed using protein array. Small diamonds: differentially expressed miRNAs found by array analysis. Big diamonds: differentially expressed miRNAs validated by RT-PCR. Filled colors inside represent gene expression, border colors outside represent protein level.
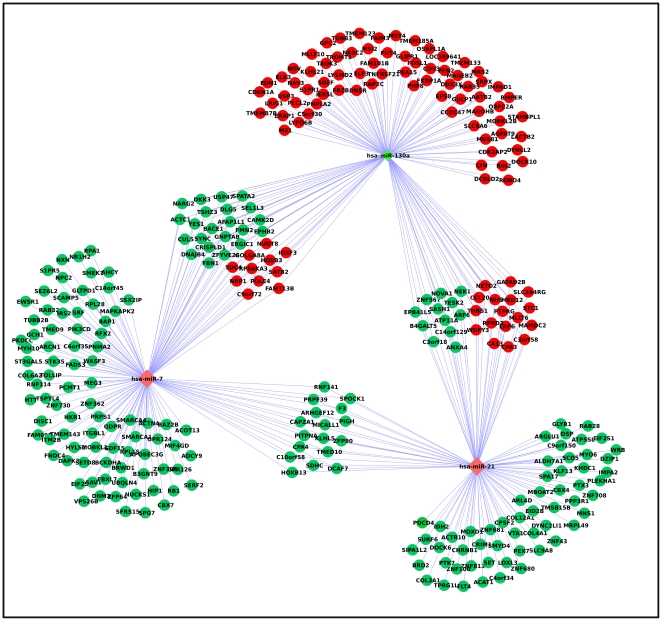
Figure 5. Target genes shared by miR-7, miR-21 and miR-130a.
Nodes represent miRNAs and mRNA gene expression. Red represents gene/miRNA upregulation, green downregulation.
Integration of miRNA-protein expression levels
As the effect of miRNAs does not limit to mRNA degradation but also translational repression, we integrated miRNA and protein expression using the approach described previously (see above). As apoptosis and cell cycle were mainly involved in trabectedin resistance as reported in the previous enrichment results (see Table 4 ), we generated a miRNA-protein network for these two pathways ( Figure 6 , panels A and B respectively).
Figure 6. Pre-translational network. miRNA and protein subnetworks representing apoptosis (Panel A) and cell cycle (Panel B).
Small circle: differentially expressed genes found by microarray analysis. Square: differentially expressed genes encoding a protein found differentially expressed using protein array. Exagon: differentially expressed genes validated with qRT-PCRencoding a protein found differentially expressed using protein array. Small diamonds: differentially expressed miRNAs found by array analysis. Big diamonds: differentially expressed miRNAs validated by qRT-PCR. Filled colors inside represent gene expression, border colors outside represent protein level.
Integration of mRNA and protein levels
The simultaneous analysis of differentially expressed genes and proteins showed a core of 22 genes for which not only the mRNA expression but also the protein levels were significantly altered between 402-91/ET and 402-91 cell lines. Within this core it is possible to clearly identify two sets of genes: 54% of genes with mRNA positively correlated with protein levels (r = 0.96) and 45% of genes with mRNA negatively correlated with protein levels (r = −0.6).
This result can be easily explained considering two points: i. mRNA and protein levels are not quantified from exactly the same biological sample but rather from two different biological replicates ii. mRNAs and proteins have different turnover.
Integration of miRNA-mRNA-protein expression levels
miRNA, mRNA and protein can be combined in modules in which a miRNA acts on messenger targets and the resultant effect is measured on the protein.
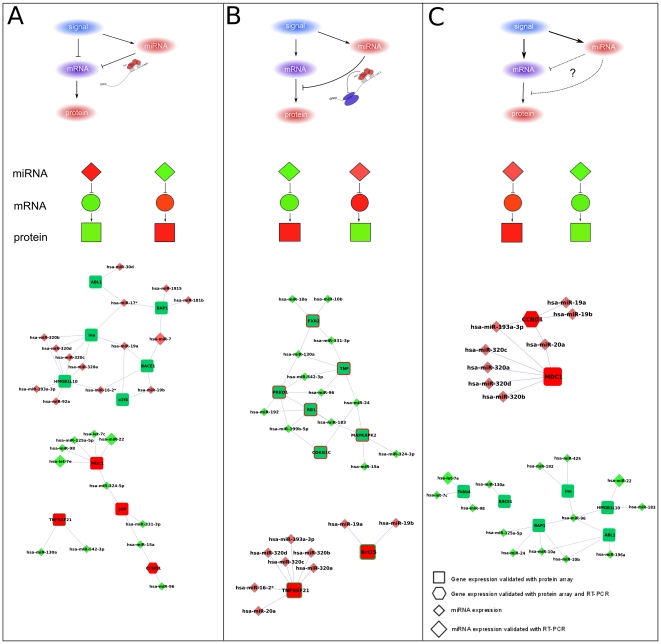
In our study the combination of post-transcriptional and pre-translational regulatory networks allows the identification of triplets (miRNA-mRNA-protein) representing different miRNA action scenarios. Mapping the levels of expression on these modules allows us to identify putative different types of regulatory loops [32]. The possible loops to be considered regard i) an up/down miRNA regulating the target mRNA through degradation (type A, Figure 7A ), ii) an up/down miRNA regulating the protein level through translational repression (type B, Figure 7B ), iii) an up/down miRNA whose expression does not support an action on the predicted targets or an up/down miRNA whose effect is not sufficient to overcome the effect of another external signal (and thus is impossible to hypothesize if miRNA acts as mRNA degradation or translational repressor) (type C, Figure 7C ).
Figure 7. Regulatory loops identified through the combination of miRNA, mRNA and protein expression levels.
Panel A. Coherent regulatory loops: up/down miRNA regulating the target mRNA through degradation. Panel B. Incoherent regulatory loops: an up/down miRNA regulating the protein level through translational repression. Panel C. Incoherent regulatory loops: an up/down miRNA whose effect is not sufficient to overcome the effect of another external signal and thus it is impossible to hypothesize if miRNA acts as mRNA degradation or translational repressor.
Depending on the signal, loops can be divided into two classes: coherent (loop of type A; characterized by mutually exclusive expression between miRNA and its targets) and incoherent (loops of type B and C; characterized by coexpression of miRNA and its target) [33]. In the coherent loops both pathways from the signal to the target have the same effect (both repressing or activating target expression), while in the incoherent ones the two pathways have opposite effects [33]. From a biological point of view, incoherent loops of type B have an easy interpretation while incoherent loops of type C have a less clear explanation and then usually require further investigations. Figure 7 shows in details these possibilities jointly with the loops found in our data. In particular we found 38 putative loops of type A, 34 of type B and 28 of type C. Looking at the coherent loops, we identified 3 loops involving differentially expressed miRNAs with roles in drug resistance and/or potentially regulated by FUS-CHOP chimera (let-7e, miR-130a, miR-98 and miR-7). These loops have as potential targets MDC1, TNFRSF21 (overexpressed in 402-91/ET), BACE1 and BAP1 (underexpressed in 402-91/ET). MDC1 activates the intra-S phase and G2/M phase cell cycle checkpoints, in response to DNA damage. It has been demonstrated that MDC1 is implicated in cellular apoptotic response [33]. TNFRSF21 encodes a protein that is a member of the TNF-receptor superfamily. This receptor has been shown to activate NF-kappaB and MAPK8/JNK and induce cell apoptosis. The protein encoded by BAP1 localizes to the nucleus and it interacts with the RING finger domain of the breast cancer 1, early onset protein (BRCA1). This gene is thought to be a tumor suppressor gene that functions in the DNA repair. As far as the incoherent circuits of type B concerns we identified two loops with miR-130a that have PRKD1 and TNF as potential targets and one loop regarding a cluster of miRNAs (miR-320) with TNFRSF21. On the other hand, as far as circuits of type C concerns we identified 3 loops with let-7e and miR-98, FXR2, TUBB4 genes and maintenance of genomic stability pathway.
Discussion
Despite its complex molecular heterogeneity, cancer results from the integration of at least ten common pathways that drive tumor progression and therapy response [34]. It is now becoming clear that pathway integration rather than single gene manipulation could represent the basis to understand malignant transformation and therapy response.
Based on this hypothesis, in the current study we exploited different transcriptomic (miRNA and GE array) and proteomic (protein array) platforms to generate, in a well defined in vitro model of tumor cells, an integration map of the key pathways possibly involved in the mechanism of resistance to trabectedin.
In this perspective, our study is one of the first attempts towards the combination of the three layers of gene regulation (transcriptional, post-transcriptional and translational) for the identification of those pathways and networks playing a critical role in trabectedin action.
In particular, in this work, we looked for changes in molecular behavior due to an intensive and prolonged treatment with trabectedin that was performed to induce drug resistance. Then, using a systems biology approach we systematically screened the possible sources of variability in mRNAs, miRNAs and protein expressions. Our integration strategy started by extracting individual results from the three different layers and then combining them to increase their power.
Gene expression data
Regarding gene expression data, a significant portion of the underexpressed (6%) and overexpressed (2%) genes are zinc finger (ZF) proteins. ZF proteins are multiple and play important role in the metabolism of normal cells. There are many reports on the inhibition of ZF transcription factors in early as well as advanced stages of oncogenesis, including the impairment of signal transduction [35]. These results are also in agreement with the recent findings of Duan et al. [36] who performed gene expression analysis on chondrosarcoma cell lines resistant to trabectedin. Remarkably, the authors found some ZF proteins to be differentially expressed and suggest that ZF proteins are involved in the mechanism of trabectedin resistance. However, differently from Duan et al. [36] we did not find overexpression of ZNF93 and ZNF43 in the resistant 402-91/ET cell line.
Interestingly, we found BIRC2 (which encodes for c-IAP1 protein) overexpressed in 402-91/ET cells. c-IAP1 inhibits apoptosis by binding to TNF receptor-associated factors TRAF1 (that we found overexpressed in resistant cells) and TRAF2. It has recently been reported that BIRC2 is associated with resistance of esophageal squamous cell carcinomas to drug-induced apoptosis [37] and that c-IAP1 could be a novel predictive marker for resistance to radiotherapy in cervical squamous cell carcinomas [37].
The 402-91/ET cell line seems to be characterized by a reduced apoptosis and an increased cell proliferation. It is noteworthy that pathways involved in cell adhesion are enriched in downregulated genes, suggesting a loss of 402-91/ET intracellular communications and altered cellular processes such as proliferation, migration and differentiation.
miRNA expression data
miRNAs are involved in processes such as development, carcinogenesis, cell survival, and apoptosis, as well as cellular sensitivity to anticancer drugs [38]. However, opposite effects can be seen towards the same compound in different tumor types, suggesting a complex relationship between miRNAs and drug resistance [39].
Among differentially expressed miRNAs in 402-91/ET we found miR-21, let-7e, miR-192, miR-130a and miR-98, whose ability to affect the potencies of a number of anticancer agents have been recently reported. Several studies recently reported a relation between the overexpression of miR-21 and the resistance to a variety of anticancer drugs. In particular, Shi and colleagues [40] demonstrated a role of miR-21 in temozolomide resistance in glioblastoma cells. Moreover, the overexpression of miR-21 has also been found to induce resistance against cytosine arabinoside (Ara-C) and arsenic trioxide. Targeting of PDCD4 (underexpressed in 402-91/ET cells) by miR-21 prevented Ara-C-induced apoptosis in the leukemic HL60 cell line [41], whereas in several myelogenous cell lines reduced expression of PDCD4 by miR-21 targeting prevented arsenic trioxide-induced apoptosis [42].
Let-7e, and miR-130a were found underexpressed in a panel of paclitaxel- and cisplatin-resistant cells lines [43]. Furthermore, Herbert et al. [44] predicted HGMA2 (overexpressed in microarray and validated with qRT-PCR) as a target for miR-98, and showed their involvement in promotion of resistance to doxorubicin and cisplatin, while Boni and colleagues suggested a role of miR-192 in 5-fluorouracil resistance in colorectal cancer [45].
In general, pathway analysis on microRNA target genes highlights a subset of the pathways previously found in gene expression analysis. This means that in some specific pathways both levels of transcription are co-involved in the development of resistance to trabectedin. Cell cycle, angiogenesis, regulation of transcription and apoptosis-survival are the mostly over-represented pathways.
CHOP transcription factor binding site
We investigated the presence of differentially expressed miRNAs, potentially transcriptionally regulated by FUS-CHOP chimera with an enriched CHOP motif in their promoter regions. We found three differentially expressed miRNAs (miR-21, miR-130a, miR-7) having either computationally or experimentally validated CHOP binding sites. Remarkably two of them, miR-21 and miR-130a, were previously discussed because of their documented involvement in drug resistance.
Among our differentially expressed genes we found 149 putative targets of miR-7 (with an anti-correlated expression profile), which are mainly involved in cell cycle, chromatin and cytoskeleton organization. Among them we found PIK3CD, RB1, CAMK2D, CUL5. There are emerging evidences about the involvement of PI3K signaling cascade in myxoid liposarcoma. The overexpression of receptor tyrosine kinases MET, IGF1R and IGF2 promote cell survival through both the PI3K/Akt and the Ras-Raf-ERK/MAPK pathways [46].
Mir-21 had 99 putative anti-correlated targets mostly involved in the regulation of transcription (CBX4, KLF13, PDCD4 and several ZF proteins).
Among the 118 anti-correlated putative targets of miR-130a, we identified genes involved in vasculature development (ROBO4, SIPR1, NRP1, ELK3, EDN1, THBS1) and cell motion, in particular FOSL1, PLCL2 and LYN. FOSL1 dimerizes with proteins of the JUN family (JUNB is overexpressed in resistant cell line) forming the transcription factor complex AP-1; FOS proteins have been implicated as regulators of cell proliferation, differentiation, and transformation. Furthermore, LYN has been recently shown to act as a mediator of tumor invasion.
Antibody array data
Gene and miRNA expression profiles are able to separate 402-91/ET from 402-91 cell lines ( Figure 1A and 1B ), while protein expression dendrogram ( Figure 3 ) has a more heterogeneous behaviour. 402-91 cell line is divided into two groups; one 402-91/ET sample clusters with 402-91 ones. It is much more difficult to quantify protein expression than gene expression in a multiplex manner than for gene expression, due to the larger variability in the physico-chemical properties of proteins [31]. Therefore, this discrepancy could be due to the higher variability of protein arrays than that of mRNA and miRNA assessment.
GO enrichment analysis on differentially expressed proteins seems to be much more focused than the same analysis on genes. In particular, apoptosis-survival and cell cycle are categories capable of collecting the greatest number of differences between sensitive and resistant cells. Among the over-represented pathways we found p53, apoptosis and MAPK.
In particular we found CCND1, CCND2, BBC3, CDKN2A, PERP involved in p53 signaling, RB1, E2F2, E2F3, E2F4, SMAD4, CDC14, CDC25A, CDC6, CDC7 involved in cell cycle. Some of these proteins are involved in the regulation of transcription in G1/S phase of the mitotic cell cycle. G1/S transition is a rate-limiting step in cell cycle progression [47].
The cyclin D1 proto-oncogene is a key regulator of G1 to S phase progression in several cell types. Together with CDK4 and CDK6, cyclin D1 forms active complexes that promote cell cycle progression by phosphorylating and inactivating the retinoblastoma protein (RB1). RB1 in turn leads to the release of the E2F family of transcription factors driving the expression of several genes associated with S phase progression [48]. A number of therapeutic agents have been observed to induce cyclin D1 degradation in vitro, indicating that such an induction may offer a useful avenue for therapeutic intervention.
Integrative analysis and network reconstruction
After the analysis and the discussion of each regulatory aspect (mRNAs, miRNAs and proteins) separately, we combined all the biological evidences within a systems biology approach in order to have a wider perspective of the regulatory phenomena acting during trabectedin resistance.
miRNAs act as post-transcriptional regulators of gene expression. The reduction of protein levels is the final result of the regulation that can be achieved through two strategies: (i) mRNA degradation or (ii) translational repression. The effects of the two modes of action can be evaluated at different points of the transcription-translation pathway. Focusing on the two main highly correlated pathways that seem to be involved in trabectedin resistance (negative regulation of apoptosis and cell cycle), we reconstructed two different networks: i) the network composed of differentially expressed mRNAs targeted by differentially expressed miRNAs that allow us to study the regulation at the transcriptional level, hereafter the “post-transcriptional regulatory network” and ii) the network composed by differentially expressed miRNAs leading to differentially expressed proteins, hereafter “pre-translational regulatory network”.
It is clear the central role that CCND1, RB1, E2F4, TNF, CDKN1C, ABL1, TNFRSF21 play in both post-transcriptional and pre-translational regulatory networks. In particular, they act as a bridge between the two parts of the network with opposite regulations: upregulated genes/protein and downregulated miRNA and downregulated genes/protein and upregulated miRNA.
In some cases the involvement of miRNAs can explain the discordant relation between the transcriptome and the proteome. In a seminal review Inui et al. [32] describe the role of miRNAs in pathways as decision maker elements to discriminate real versus too weak or too transient signals. In this study we identified 3 loops involving differentially expressed miRNAs with roles in drug resistance; these loops have as potential targets MDC1 (overexpressed in 402-91/ET), BACE1 and BAP1 (underexpressed in 402-91/ET).
Concluding remarks
The present study is one of the first works that try to screen the behavior of genes, microRNAs (miRNAs) and proteins in order to reconstruct in silico the regulatory networks leading to resistance to anticancer agents such as trabectedin. Diverse sources of omic data have been used to describe altered regulatory mechanisms within a transcriptional, post-transcriptional and translational perspective. The combination of the three layers of regulation gave us the possibility to reconstruct putative gene-miRNA-protein circuits altered in trabectedin resistance. We found that transcriptome and proteome data agreed in recognizing anti-apoptosis and cell cycle proliferation as the two main biological processes characterizing the difference between 402-91 and 402-91/ET cell lines. All the hallmarks of cancer seem to be altered in the resistant cell line, raising the hypothesis that the mechanism of resistance is acting with more strength on the same molecular markers triggered by cancer development.
On the other hand, post-transcriptional analysis revealed two miRNAs (miR-21 and miR-130a) already identified as playing a role in drug resistance and putatively regulated by the FUS-CHOP chimera, which could be good markers for future functional studies. Most of miR-21 and miR-130a targets are known oncogenes or oncosuppressors highlighting their involvement in malignancies.
Once validated on in vivo models this approach might allow the identification of druggable targets for trabectedin-resistant malignancies.
Supporting Information
miRNA and mRNA network reconstruction using mirSVR as target prediction tool. Colors represent expression (green and red for under and overexpressed genes, light green and light red for under and overexpressed miRNA) and edges between miRNA and gene represent mirSVR predictions, while gene-gene edges represent validated gene-gene interactions from KEGG database.
(PDF)
Complete list of probes identifying differentially expressed genes.
(XLSX)
Complete list of validated mRNA and miRNA expression levels using qRT-PCR.
(PDF)
Complete list of differentially expressed miRNAs.
(PDF)
Complete list of differentially expressed proteins.
(XLSX)
Complete list of predicted transcription factor binding sites enriched in the promoter region of up and downregulated gene in 402-91/ET cell lines.
(PDF)
Simple interactions format (sif) file for gene-gene and miRNA-gene interactions of the whole network reported in Figure S1.
(XLSX)
Acknowledgments
The authors acknowledge Prof. Luca Tamagnone (Associate Professor, University of Torino, IRCC) for generously supporting set up SEMA4C western blot analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by University of Padova (CPDR075919 to CR). Furthermore, the authors want to acknowledge the CINECA Award N. HP10BDJ9X8, 2010 for the availability of high performance computing resources and support. The authors acknowledge the generous contribution of the Italian Association for Cancer Research (AIRC). IFN is recipient of a fellowship from ACTO foundation. PO and GC are supported by Compagnia di San Paolo, Torino, Italy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Krasner CN, McMeekin DS, Chan S, Braly PS, Renshaw FG, et al. A Phase II study of trabectedin single agent in patients with recurrent ovarian cancer previously treated with platinum-based regimens. Br. J. Cancer. 2007;97:1618–1624. doi: 10.1038/sj.bjc.6604088. doi: 10.1038/sj.bjc.6604088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monk BJ, Sill MW, Hanjani P, Edwards R, Rotmensch J, et al. Docetaxel plus trabectedin appears active in recurrent or persistent ovarian and primary peritoneal cancer after up to three prior regimens: a phase II study of the Gynecologic Oncology Group. Gynecol. Oncol. 2011;120:459–463. doi: 10.1016/j.ygyno.2010.11.012. doi: 10.1016/j.ygyno.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Grosso F, Jones RL, Demetri GD, Judson IR, Blay J-Y, et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: a retrospective study. Lancet Oncol. 2007;8:595–602. doi: 10.1016/S1470-2045(07)70175-4. doi: 10.1016/S1470-2045(07)70175-4. [DOI] [PubMed] [Google Scholar]
- 4.Del Campo JM, Roszak A, Bidzinski M, Ciuleanu TE, Hogberg T, et al. Phase II randomized study of trabectedin given as two different every 3 weeks dose schedules (1.5 mg/m2 24 h or 1.3 mg/m2 3 h) to patients with relapsed, platinum-sensitive, advanced ovarian cancer. Ann. Oncol. 2009;20:1794–1802. doi: 10.1093/annonc/mdp198. doi: 10.1093/annonc/mdp198. [DOI] [PubMed] [Google Scholar]
- 5.Sessa C, De Braud F, Perotti A, Bauer J, Curigliano G, et al. Trabectedin for women with ovarian carcinoma after treatment with platinum and taxanes fails. J. Clin. Oncol. 2005;23:1867–1874. doi: 10.1200/JCO.2005.09.032. doi: 10.1200/JCO.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 6.Grosso F, Sanfilippo R, Virdis E, Piovesan C, Collini P, et al. Trabectedin in myxoid liposarcomas (MLS): a long-term analysis of a single-institution series. Ann. Oncol. 2009;20:1439–1444. doi: 10.1093/annonc/mdp004. doi: 10.1093/annonc/mdp004. [DOI] [PubMed] [Google Scholar]
- 7.Carter NJ, Keam SJ. Trabectedin: a review of its use in soft tissue sarcoma and ovarian cancer. Drugs. 2010;70:355–376. doi: 10.2165/11202860-000000000-00000. doi: 10.2165/11202860-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 8.D'Incalci M, Galmarini CM. A review of trabectedin (ET-743): a unique mechanism of action. Mol. Cancer Ther. 2010;9:2157–2163. doi: 10.1158/1535-7163.MCT-10-0263. doi: 10.1158/1535-7163.MCT-10-0263. [DOI] [PubMed] [Google Scholar]
- 9.Hurley LH, Zewail-Foote M. The antitumor agent ecteinascidin 743: characterization of its covalent DNA adducts and chemical stability. Adv. Exp. Med. Biol. 2001;500:289–299. doi: 10.1007/978-1-4615-0667-6_46. [DOI] [PubMed] [Google Scholar]
- 10.Minuzzo M, Marchini S, Broggini M, Faircloth G, D'Incalci M, et al. Interference of transcriptional activation by the antineoplastic drug ecteinascidin-743. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6780–6784. doi: 10.1073/pnas.97.12.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minuzzo M, Ceribelli M, Pitarque-Martì M, Borrelli S, Erba E, et al. Selective effects of the anticancer drug Yondelis (ET-743) on cell-cycle promoters. Mol. Pharmacol. 2005;68:1496–1503. doi: 10.1124/mol.105.013615. doi: 10.1124/mol.105.013615. [DOI] [PubMed] [Google Scholar]
- 12.Grosso F, Jones RL, Demetri GD, Judson IR, Blay J-Y, et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: a retrospective study. Lancet Oncol. 2007;8:595–602. doi: 10.1016/S1470-2045(07)70175-4. doi: 10.1016/S1470-2045(07)70175-4. [DOI] [PubMed] [Google Scholar]
- 13.Forni C, Minuzzo M, Virdis E, Tamborini E, Simone M, et al. Trabectedin (ET-743) promotes differentiation in myxoid liposarcoma tumors. Mol. Cancer Ther. 2009;8:449–457. doi: 10.1158/1535-7163.MCT-08-0848. doi: 10.1158/1535-7163.MCT-08-0848. [DOI] [PubMed] [Google Scholar]
- 14.Uboldi S, Bernasconi S, Romano M, Marchini S, Fuso Nerini I, et al. 2011. Characterization of a new trabectedin resistant myxoid liposarcoma cell line that shows collateral sensitivity to methylating agents. International Journal of Cancer. Journal International Du Cancer. Available: http://www.ncbi.nlm.nih.gov/pubmed/21805478.
- 15.Aman P, Ron D, Mandahl N, Fioretos T, Heim S, et al. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11). Genes Chromosomes Cancer. 1992;5:278–285. doi: 10.1002/gcc.2870050403. [DOI] [PubMed] [Google Scholar]
- 16.Marchini S, Marabese M, Marrazzo E, Mariani P, Cattaneo D, et al. DeltaNp63 expression is associated with poor survival in ovarian cancer. Ann. Oncol. 2008;19:501–507. doi: 10.1093/annonc/mdm519. doi: 10.1093/annonc/mdm519. [DOI] [PubMed] [Google Scholar]
- 17.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 18.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. doi: 10.2202/1544–6115.1027. [DOI] [PubMed] [Google Scholar]
- 19.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 20.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 21.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, et al. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Parameter estimation for the calibration and variance stabilization of microarray data. Stat Appl Genet Mol Biol. 2003;2:Article3. doi: 10.2202/1544-6115.1008. doi: 10.2202/1544-6115.1008. [DOI] [PubMed] [Google Scholar]
- 24.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sales G, Coppe A, Bisognin A, Biasiolo M, Bortoluzzi S, et al. MAGIA, a web-based tool for miRNA and Genes Integrated Analysis. Nucleic Acids Res. 2010;38:W352–359. doi: 10.1093/nar/gkq423. doi: 10.1093/nar/gkq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolsky Y, Ekins S, Nikolskaya T, Bugrim A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol. Lett. 2005;158:20–29. doi: 10.1016/j.toxlet.2005.02.004. doi: 10.1016/j.toxlet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Alexiou P, Vergoulis T, Gleditzsch M, Prekas G, Dalamagas T, et al. miRGen 2.0: a database of microRNA genomic information and regulation. Nucleic Acids Res. 2010;38:D137–141. doi: 10.1093/nar/gkp888. doi: 10.1093/nar/gkp888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friard O, Re A, Taverna D, De Bortoli M, Corá D. CircuitsDB: a database of mixed microRNA/transcription factor feed-forward regulatory circuits in human and mouse. BMC Bioinformatics. 2010;11:435. doi: 10.1186/1471-2105-11-435. doi: 10.1186/1471-2105-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 31.Sill M, Schröder C, Hoheisel JD, Benner A, Zucknick M. Assessment and optimisation of normalisation methods for dual-colour antibody microarrays. BMC Bioinformatics. 2010;11:556. doi: 10.1186/1471-2105-11-556. doi: 10.1186/1471-2105-11-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 33.Osella M, Bosia C, Corá D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput. Biol. 2011;7:e1001101. doi: 10.1371/journal.pcbi.1001101. doi: 10.1371/journal.pcbi.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 35.Witkiewicz-Kucharczyk A, Bal W. Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicol. Lett. 2006;162:29–42. doi: 10.1016/j.toxlet.2005.10.018. doi: 10.1016/j.toxlet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 36.Duan Z, Choy E, Harmon D, Yang C, Ryu K, et al. ZNF93 increases resistance to ET-743 (Trabectedin; Yondelis) and PM00104 (Zalypsis) in human cancer cell lines. PLoS ONE. 2009;4:e6967. doi: 10.1371/journal.pone.0006967. doi: 10.1371/journal.pone.0006967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imoto I, Yang ZQ, Pimkhaokham A, Tsuda H, Shimada Y, et al. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- 38.Blower PE, Verducci JS, Lin S, Zhou J, Chung J-H, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol. Cancer Ther. 2007;6:1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 39.Giovannetti E, Erozenci A, Smit J, Danesi R, Peters GJ. 2011. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Critical Reviews in Oncology/Hematology. Available: http://www.ncbi.nlm.nih.gov/pubmed/21546262. Consultato 8 Set 2011.
- 40.Shi L, Chen J, Yang J, Pan T, Zhang S, et al. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010;1352:255–264. doi: 10.1016/j.brainres.2010.07.009. doi: 10.1016/j.brainres.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Zhu X, Gu J, Hu H, Dong D, et al. Anti-miR-21 oligonucleotide enhances chemosensitivity of leukemic HL60 cells to arabinosylcytosine by inducing apoptosis. Hematology. 2010;15:215–221. doi: 10.1179/102453310X12647083620840. doi: 10.1179/102453310X12647083620840. [DOI] [PubMed] [Google Scholar]
- 42.Gu J, Zhu X, Li Y, Dong D, Yao J, et al. miRNA-21 regulates arsenic-induced anti-leukemia activity in myelogenous cell lines. Med. Oncol. 2011;28:211–218. doi: 10.1007/s12032-009-9413-7. doi: 10.1007/s12032-009-9413-7. [DOI] [PubMed] [Google Scholar]
- 43.Sorrentino A, Liu C-G, Addario A, Peschle C, Scambia G, et al. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol. Oncol. 2008;111:478–486. doi: 10.1016/j.ygyno.2008.08.017. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 44.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, et al. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol. Cancer Ther. 2010;9:2265–2275. doi: 10.1158/1535-7163.MCT-10-0061. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- 46.Conyers R, Young S, Thomas DM. Sarcoma 2011: 483154; 2011. Liposarcoma: molecular genetics and therapeutics.483154. doi: 10.1155/2011/483154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 48.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol. Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miRNA and mRNA network reconstruction using mirSVR as target prediction tool. Colors represent expression (green and red for under and overexpressed genes, light green and light red for under and overexpressed miRNA) and edges between miRNA and gene represent mirSVR predictions, while gene-gene edges represent validated gene-gene interactions from KEGG database.
(PDF)
Complete list of probes identifying differentially expressed genes.
(XLSX)
Complete list of validated mRNA and miRNA expression levels using qRT-PCR.
(PDF)
Complete list of differentially expressed miRNAs.
(PDF)
Complete list of differentially expressed proteins.
(XLSX)
Complete list of predicted transcription factor binding sites enriched in the promoter region of up and downregulated gene in 402-91/ET cell lines.
(PDF)
Simple interactions format (sif) file for gene-gene and miRNA-gene interactions of the whole network reported in Figure S1.
(XLSX)