Abstract
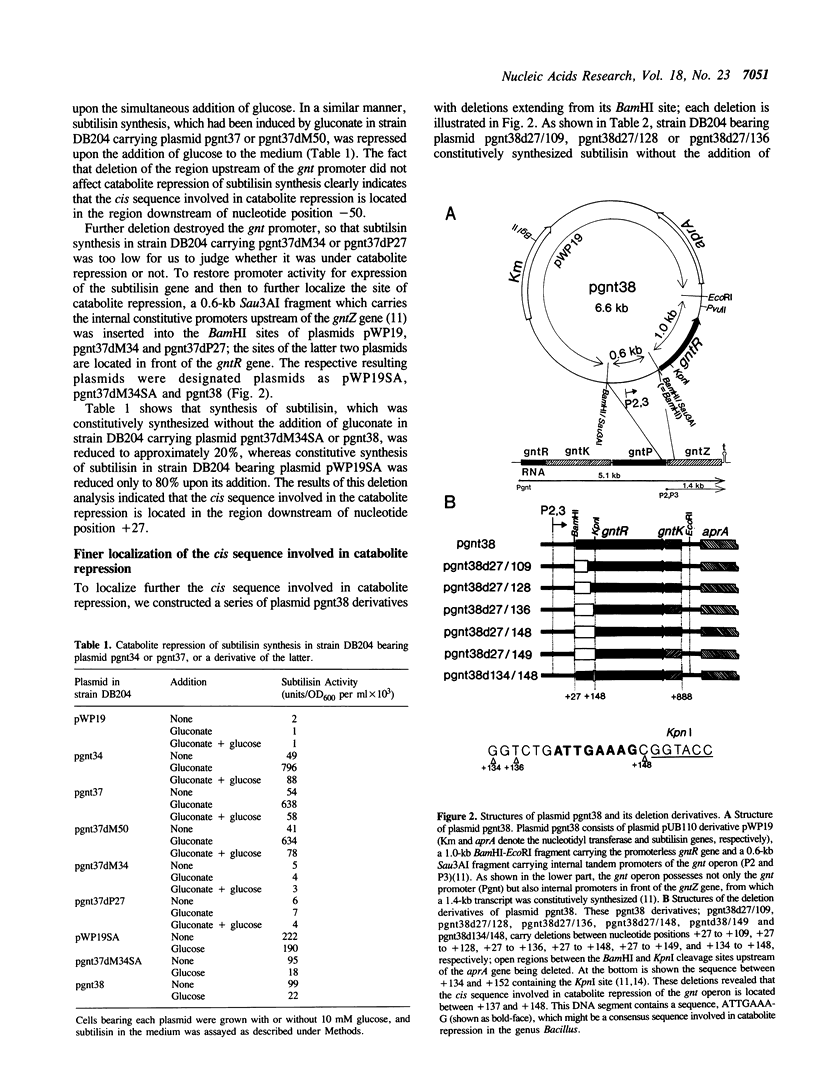
The mechanism underlying catabolite repression in Bacillus species remains unsolved. The gluconate (gnt) operon of Bacillus subtilis is one of the catabolic operons which is under catabolite repression. To identify the cis sequence involved in catabolite repression of the gnt operon, we performed deletion analysis of a DNA fragment carrying the gnt promoter and the gntR gene, which had been cloned into the promoter probe vector, pWP19. Deletion of the region upstream of the gnt promoter did not affect catabolite repression. Further deletion analysis of the gnt promoter and gntR coding region was carried out after restoration of promoter activity through the insertion of internal constitutive promoters of the gnt operon before the gntR gene (P2 and P3). These deletions revealed that the cis sequence involved in catabolite repression of the gnt operon is located between nucleotide positions +137 and +148. This DNA segment contains a sequence, ATTGAAAG, which may be implicated as a consensus sequence involved in catabolite repression in the genus Bacillus.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biville F., Guiso N. Evidence for the presence of cAMP, cAMP receptor and transcription termination factor rho in different gram-negative bacteria. J Gen Microbiol. 1985 Nov;131(11):2953–2960. doi: 10.1099/00221287-131-11-2953. [DOI] [PubMed] [Google Scholar]
- Dingman D. W., Sonenshein A. L. Purification of aconitase from Bacillus subtilis and correlation of its N-terminal amino acid sequence with the sequence of the citB gene. J Bacteriol. 1987 Jul;169(7):3062–3067. doi: 10.1128/jb.169.7.3062-3067.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita T. Identification and nucleotide sequence of the promoter region of the Bacillus subtilis gluconate operon. Nucleic Acids Res. 1986 Feb 11;14(3):1237–1252. doi: 10.1093/nar/14.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita T., Miwa Y. Evidence for posttranscriptional regulation of synthesis of the Bacillus subtilis Gnt repressor. FEBS Lett. 1990 Jul 2;267(1):71–74. doi: 10.1016/0014-5793(90)80290-y. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita T., Miwa Y., Nihashi J., Aratani Y. Organization and transcription of the gluconate operon, gnt, of Bacillus subtilis. J Biol Chem. 1986 Oct 15;261(29):13744–13753. [PubMed] [Google Scholar]
- Fujita Y., Fujita T. The gluconate operon gnt of Bacillus subtilis encodes its own transcriptional negative regulator. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4524–4528. doi: 10.1073/pnas.84.13.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Miwa Y. Identification of an operator sequence for the Bacillus subtilis gnt operon. J Biol Chem. 1989 Mar 5;264(7):4201–4206. [PubMed] [Google Scholar]
- Fujita Y., Nihashi J., Fujita T. The characterization and cloning of a gluconate (gnt) operon of Bacillus subtilis. J Gen Microbiol. 1986 Jan;132(1):161–169. doi: 10.1099/00221287-132-1-161. [DOI] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoide B. M., Chambliss G. H., McConnell D. J. Bacillus licheniformis alpha-amylase gene, amyL, is subject to promoter-independent catabolite repression in Bacillus subtilis. J Bacteriol. 1989 May;171(5):2435–2442. doi: 10.1128/jb.171.5.2435-2442.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoide B. M., McConnell D. J. cis sequences involved in modulating expression of Bacillus licheniformis amyL in Bacillus subtilis: effect of sporulation mutations and catabolite repression resistance mutations on expression. J Bacteriol. 1989 May;171(5):2443–2450. doi: 10.1128/jb.171.5.2443-2450.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovaara P., Ulmanen I., Palva I. In vivo transcription initiation and termination sites of an alpha-amylase gene from Bacillus amyloliquefaciens cloned in Bacillus subtilis. Gene. 1984 Oct;30(1-3):11–16. doi: 10.1016/0378-1119(84)90099-4. [DOI] [PubMed] [Google Scholar]
- Melin L., Magnusson K., Rutberg L. Identification of the promoter of the Bacillus subtilis sdh operon. J Bacteriol. 1987 Jul;169(7):3232–3236. doi: 10.1128/jb.169.7.3232-3236.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J. Characterization of proteinases excreted by Bacillus subtilis Marburg strain during sporulation. J Appl Bacteriol. 1970 Mar;33(1):207–219. doi: 10.1111/j.1365-2672.1970.tb05245.x. [DOI] [PubMed] [Google Scholar]
- Miwa Y., Fujita Y. Purification and characterization of a repressor for the Bacillus subtilis gnt operon. J Biol Chem. 1988 Sep 15;263(26):13252–13257. [PubMed] [Google Scholar]
- Nicholson W. L., Park Y. K., Henkin T. M., Won M., Weickert M. J., Gaskell J. A., Chambliss G. H. Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence. J Mol Biol. 1987 Dec 20;198(4):609–618. doi: 10.1016/0022-2836(87)90204-x. [DOI] [PubMed] [Google Scholar]
- Nihashi J., Fujita Y. Catabolite repression of inositol dehydrogenase and gluconate kinase syntheses in Bacillus subtilis. Biochim Biophys Acta. 1984 Mar 22;798(1):88–95. doi: 10.1016/0304-4165(84)90014-x. [DOI] [PubMed] [Google Scholar]
- Oda M., Sugishita A., Furukawa K. Cloning and nucleotide sequences of histidase and regulatory genes in the Bacillus subtilis hut operon and positive regulation of the operon. J Bacteriol. 1988 Jul;170(7):3199–3205. doi: 10.1128/jb.170.7.3199-3205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastan I., Adhya S. Cyclic adenosine 5'-monophosphate in Escherichia coli. Bacteriol Rev. 1976 Sep;40(3):527–551. doi: 10.1128/br.40.3.527-551.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Inability of detect cyclic AMP in vegetative or sporulating cells or dormant spores of Bacillus megaterium. Biochem Biophys Res Commun. 1973 May 15;52(2):365–372. doi: 10.1016/0006-291x(73)90720-1. [DOI] [PubMed] [Google Scholar]
- Shibata T., Saito H. Repair of ultraviolet-induced DNA damage in the subcellular systems of Bacillus subtilis. Mutat Res. 1973 Nov;20(2):159–173. doi: 10.1016/0027-5107(73)90186-3. [DOI] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. F., Doi R. H. Promoter switching during development and the termination site of the sigma 43 operon of Bacillus subtilis. Mol Gen Genet. 1987 Apr;207(1):114–119. doi: 10.1007/BF00331498. [DOI] [PubMed] [Google Scholar]
- Weickert M. J., Chambliss G. H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]



