Abstract
Neocentromeres (NCs) are fully functional centromeres that arise ectopically in noncentromeric regions lacking α-satellite DNA. Using telomere-associated chromosome truncation, we have produced a series of minichromosomes (MiCs) from a mardel(10) marker chromosome containing a previously characterized human NC. These MiCs range in size from ≈0.7 to 1.8 Mb and contain single-copy intact genomic DNA from the 10q25 region. Two of these NC-based Mi-Cs (NC-MiCs) appear circular whereas one is linear. All demonstrate stability in both structure and mitotic transmission in the absence of drug selection. Presence of a functional NC is shown by binding a host of key centromere-associated proteins. These NC-MiCs provide direct evidence for mitotic segregation function of the NC DNA and represent examples of stable mammalian MiCs lacking centromeric repeats.
Mammalian artificial chromosomes have several potential biotechnological and therapeutic applications arising from their ability to exist episomally, carry large DNA inserts, and allow expression of genes independently of the host genome. By analogy with their yeast counterparts, it has been assumed that mammalian artificial chromosomes require a functional mammalian centromere, telomeres, and DNA replication origins for proper segregation. At present, the least understood and most complex of these three components is the centromere.
The identification of many protein components necessary for correct centromere activity, and the characterization of centromere DNA sequences in a variety of species, have greatly increased our knowledge of the mechanisms underlying centromere formation and function (1–3). This knowledge has facilitated the development of a number of strategies for mammalian artificial chromosome construction. One strategy involves the de novo formation of artificial chromosomes by transfection of large arrays of human α-satellite into human cells (4–7). Although some of the generated artificial chromosomes were linear in structure (4), others were consistently circular (5, 7, 8) and all were typically 1 or more orders of magnitude larger than the input DNA.
A second strategy involves the use of telomere-associated chromosome truncation to remove nonessential chromosome arms to produce minichromosomes (MiCs) in situ. Sequential truncation of a human X and Y chromosome has yielded a number of α-satellite-containing MiCs of varying stability and sizes ranging from ≈0.7 Mb to more than 4 Mb (9–15). A final strategy for production of mammalian artificial chromosomes involves the amplification of pericentric DNA followed by controlled breakage of chromosomes to produce satellite DNA-based artificial chromosomes of between 60 and 400 Mb (16, 17). These artificial chromosomes express exogenous genes and can be stably introduced into different mammalian cell lines and transgenic mice (18–20).
Neocentromeres (NCs) lacking the repeat sequences traditionally associated with centromere function recently have been described (21, 22). Characterization of NCs in humans suggests epigenetic mechanism of formation independent of primary DNA sequence composition (23–26). The discovery of NCs provides an alternative approach to the construction of artificial chromosomes. We describe here the production of mitotically stable NC-based human MiCs containing a fully functional human NC derived from the 10q25 region of the mardel(10) marker chromosome (27, 28).
Experimental Protocols
Cell Culture and Transfection.
BE2Cl-18–5f (abbreviated 5f) was cultured as described (28). HT1080 and derivatives were cultured in DMEM (GIBCO/BRL) with 10% FCS. Hygromycin (Roche Molecular Biochemicals), puromycin (Sigma), or zeocin (Invitrogen) were added to medium at 250 μg/ml, 1 μg/ml, or 200 μg/ml, respectively. Microtubule-depolymerizing agents colcemid (GIBCO/BRL) or nocadazole (Sigma) were added at 10 μM or 0.1 μg/ml for 1 or 6–12 h, respectively, before cell harvesting.
Transfection of 5f and ZB30 cells was performed by electroporation (1.2 kV, 25 uF; Bio-Rad Gene Pulser Electroporator). Transfection of HT1080 or derivatives was performed by electroporation (29) or lipofection. For lipofection, cells were plated 1 day before transfection to give 60–70% confluency at time of transfection. Two milliliters of diluted Fugene 6 transfection reagent (100 μl in a total of 2 ml containing 20 μg DNA) (Roche Molecular Biochemicals) was added onto cells drop-wise, and selection was applied 24–48 h posttransfection.
Microcell-Mediated Chromosome Transfer.
Microcell fusion was carried out as described (30). Log-phase donor ZB30 cells arrested in colcemid for 48 h were resuspended in percoll/serum-free DMEM (1:1) supplemented with 20 μg/ml of cytochalasin B (Sigma). The cell suspension was subjected to centrifugation at 18,000 rpm, 90 min, 32°C. Both bands of cell-mix were pelleted, washed with serum-free DMEM, and filtered through isopore membranes of 30, 8, and 5 μM (Millipore). Microcells were resuspended in serum-free DMEM containing 10 μg/ml phytohemagglutinin-P (Difco), agglutinated with recipient HT1080 cells for 45 min at 37°C, and fused by addition of 50% polyethylene glycol (Roche Molecular Biochemicals) for 2 min at room temperature. Cells then were rinsed and cultured overnight in DMEM containing 10% FCS followed by addition of selection.
Fluorescence in Situ Hybridization (FISH), Immunofluorescence, and Pulsed-Field Gel Electrophoresis (PFGE).
Combined FISH/immunofluorescence was performed as described (25). FISH using pan-α-satellite probe pTRA7 and PNA-FISH of telomeric sequences (PerSeptive Biosystems, Framingham, MA) were performed as described (31, 32). Chromosome painting was performed by using a WCP Chromosome Paint Kit (Vysis). Subchromosome-10 DNA paints were derived from somatic cell radiation hybrid obtained from M. Rocchi (University of Bari, Bari, Italy). InterAlu amplification of somatic cell hybrid DNA was carried out by using primers 5′-GGATTACAGGYRTGAGCCA and 5′-RCCAYTGCACTCCAGCCTG as described (33).
Polyclonal anti-CENP-A, monoclonal anti-CENP-B, polyclonal anti-CENP-C, and CREST6 antisera were as described (28, 34, 35, 36). Polyclonal anti-CENP-E (37), anti-CENP-F (38), and anti-hBUB1 (39) were kindly provided by T. Yen (Fox Chase Cancer Center, Philadelphia), polyclonal anti-hZW10 (40) by B. Williams and M. Goldberg (Cornell University, Ithaca, NY), polyclonal p55CDC (41) by J. Weinstein (Amgen Biologicals), and polyclonal anti-TRF1 (42) by T. deLange (The Rockefeller University, New York).
High molecular weight genomic DNA was prepared (43) and run on pulsed-field gels (Bio-Rad CHEF Mapper System) in 0.5 × Tris-acetate/EDTA with buffer changes every 2 days.
Size Determination by 4′,6-Diamidino-2-phenylindole (DAPI) Staining.
Metaphase spreads were stained in 600 ng/ml DAPI for 10 min. NC-MiCs were identified by FISH using E8 probe. Intensity of DAPI signal was determined by using ip lab software (Signal Analytics) as the total area of DAPI staining multiplied by the average intensity. Twenty cells were scored for each NC-MiC. The sizes of NC-MiCs 4 and 5 were calculated as average DAPI intensity divided by the average intensity for NC-MiC-3, multiplied by the size of NC-MiC3 as determined by pulsed-field gel (1.7 Mb).
Truncation Constructs.
Truncation constructs contained either pGK:hygromycin, pGK:puromycin, or pGK:neomycin resistance gene cassettes. A 2-kb array of human telomeric repeats was obtained from pBS Sal-tel(5) plasmid (44, 45). Targeting DNA fragments (5–10 kb) lacking high-copy repeats were subcloned into the truncation vectors. All truncation constructs were made in pAlter (Promega) vector backbone.
Concatamerization of Zeocin Resistance Marker.
A plasmid containing zeocin resistance cassette [pZeoSV2(+); Invitrogen] was digested with NotI, purified, and concatamerized by using 10 μl ligase buffer, 40 units T4 ligase, and 5 μl 100 mM ATP in a total volume of 100 μl. Ligation was performed overnight. One microliter of ligated product was tested on pulsed-field gel, and only DNA over 50 kb was used in transfections.
p′ Arm Truncation Screening Using Dot Blot Hybridization.
Cells in 24-well plates were harvested and transferred to HybondN+ (Amersham Pharmacia) in a 96-well Minifold dot blotting apparatus (Schleicher & Schuell). After transfer, membrane was denatured for 10 min (1 M NaOH, 1 M NaCl), neutralized twice for 5 min (1 M Tris⋅HCl, 1.5 M NaCl) and washed in 2× SSC. Membranes were baked for 1.5 h at 80°C and probed with 32P-labeled pAlter vector DNA.
Results
In Vitro Approaches for Artificial Chromosome Construction.
A previously described 80-kb bacterial artificial chromosome (BAC) clone (E8) containing the centromere-binding domain of the 10q25 NC (23, 43) was transfected into HT1080 cells either alone, or in combination with cloned human telomeric repeats, human genomic DNA, and/or cloned DNA flanking the E8 region. Except for the test DNA, procedures were essentially those used reproducibly to generate α-satellite-based human artificial chromosomes in HT1080 cells (5–7). Although we routinely observed the uptake and insertion of the introduced DNA into the host genome by FISH, screening of more than 450 drug-resistant cell lines failed to identify any containing an activated NC either in the form of a stable MiC or as the result of insertion of the introduced DNA into HT1080 genome (data not shown). In an alternative approach, a 640-kb yeast artificial chromosome (YAC) clone (YAC-3) (28) containing the core NC region was retrofitted with human telomeric DNA and selectable markers by using the plasmid vectors pRANT 11 and pLGTEL 1, following published procedures (46). Repeated transfection of correctly retrofitted YACs into human and Chinese hamster ovary cells using spheroplast fusion resulted in YAC DNA uptake, insertion into the Chinese hamster ovary genome, YAC DNA amplification, and double minute formation, but no activation of NC or artificial chromosome formation (data not shown). Despite extensive efforts, our inability to recover functional artificial chromosomes by using in vitro assembly approaches prompted us to focus on the use of a different strategy involving in situ chromosome truncation to generate MiCs.
Mardel(10) Tagging and Transfer into HT1080 Cells.
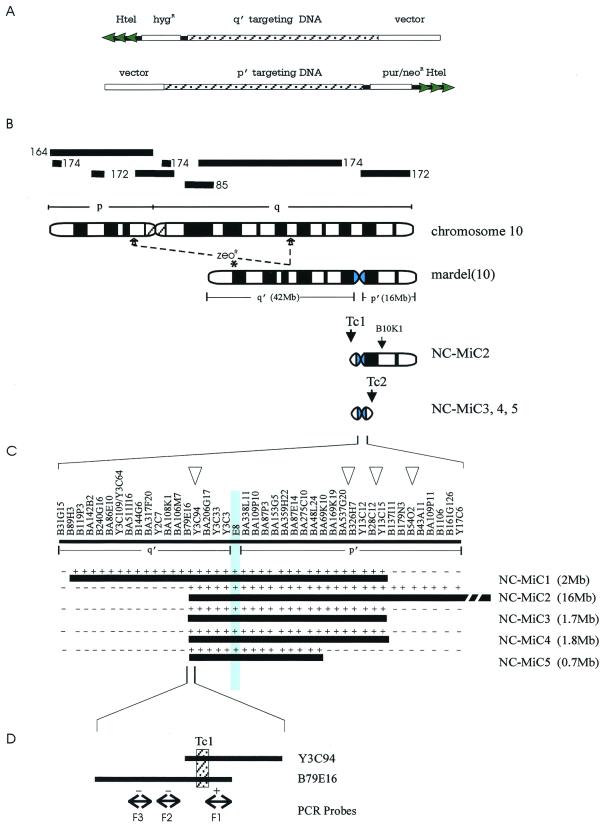
We previously have created a somatic cell hybrid line (designated 5f) containing mardel(10) (28) in a Chinese hamster ovary background. Telomere-associated chromosome truncation (TACT) (9, 12, 44) was performed on this line by using the truncation vectors shown in Fig. 1A. Exhaustive screening of >25,000 drug-resistant colonies transfected with truncation vectors containing different targeting sequences yielded no truncation events, suggesting that 5f cells were not a suitable host for TACT. We decided to transfer mardel(10) into human HT1080 cells because these cells are known to be homologous recombination proficient (29, 47), show telomerase activity (42, 48, 49), and are good recipients for microcell-mediated chromosome transfer (MMCT) (50, 51). A random insertion approach was first used to tag mardel(10) in 5f cells with a zeocin resistance gene. Screening 63 individual zeocin-resistant cell lines identified one (designated ZB30) in which mardel(10) was tagged at the distal q′ region (Figs. 1B and 6, which is published as supplemental data on the PNAS web site, www.pnas.org). This cell line was used as a donor for MMCT into HT1080 cells. FISH and immunofluorescence analyses indicated that 15 of 60 resulting cell lines contained an intact copy of the zeocin-tagged mardel(10) (data not shown). One cell line, ZBHT-14, was used in a subsequent truncation experiment. In addition, a number of lines containing randomly truncated derivatives of mardel(10) were isolated. One of these (NC-MiC1) was shown by FISH to carry an ≈2-Mb MiC containing the 10q25 NC region (28) (Fig. 1C; data not shown). This cell line was retained for further studies.
Figure 1.
NC-MiC formation from mardel(10) chromosome. (A) Telomere-associated chromosome truncation targeting constructs containing targeting DNA from p′ and q′ arms of mardel(10) and small arrays of human telomeric DNA (Htel) adjacent to a mammalian selectable marker. Homologous recombination with host chromosome would result in the loss of vector arm, which provided a screening assay for possible recombination events. (B) Schematic formation of NC-MiCs. The long and short arms of mardel(10) are denoted as q′ and p′, respectively. Tc1 and Tc2 indicate truncation of q′ and p′ arms, respectively. Site of insertion of the zeocin resistance gene in mardel(10) in the ZB30 cell line is indicated by *. Position of BAC B10K1 used to screen for loss of p′ arm is shown. Bold lines above chromosome 10 denote the locations and designations of subchromosomal DNA paints. (C) FISH mapping of NC-MiCs. Ordered cosmid and BAC clones covering ≈3 Mb of the 10q25 NC region are shown (52). Vertical shaded area represents the E8 BAC containing the previously defined CREST-binding NC domain (23, 28). Open arrowheads indicate intended positions of targeted truncation. + denotes a positive FISH result for a probe, while − indicates a negative result. Approximate sizes of the NC-MiCs are shown in parentheses. (D) Characterization of the targeted q′ truncation site. The targeting DNA (hatched box) for q′ truncation is a 6-kb XbaI fragment subcloned from cosmid Y3C94 (also present in BAC B79E16). Double-headed arrows indicate locations of PCR probes (denoted F1, F2, F3) from B79E16. Hybridization status of these probes is denoted by + or −, confirming that the q′ truncation was a targeted event.
Truncation of q′ Arm.
We prepared a complete physical map containing more than 50 BAC and cosmid clones covering ≈3 Mb (Fig. 1C) (52). Based on this map, truncation constructs containing different targeting DNA, mammalian selectable markers, and human telomeric DNA were prepared (Fig. 1A). Initial telomere-associated chromosome truncation experiments were performed on the q′ arm in ZBHT-14 and NC-MiC1 cells by using a hygromycin-resistant truncation construct containing a 6-kb targeting DNA derived from the Y3C94 cosmid (Fig. 1C).
For ZBHT-14, initial screening involved the identification of cell lines that had lost the zeocin-containing portion of mardel(10). Of 7,300 hygromycin-resistant cell lines, 210 were zeocin sensitive. FISH analysis revealed that most of these lines contained random truncations or other unknown rearrangements. One cell line (NC-MiC2) appeared to have undergone targeted truncation and was characterized extensively by FISH. Cosmids and BACs proximal to the targeting site were found to be present, whereas all clones distal to this site were absent (summarized in Fig. 1C). When three PCR fragments (F1–F3) immediately adjacent to either side of the targeting site (Fig. 1D) were used in FISH, only the fragment (F1) located proximal to the targeting site gave a positive signal. No α-satellite or CENP-B was detected on the truncated chromosome (Fig. 6). These data therefore support a targeted truncation event that removed most of the q′ arm of mardel(10) in NC-MiC2.
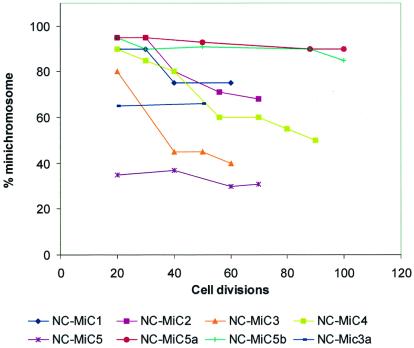
Mitotic stability of NC-MiC2 was 85% over 20–30 cell divisions, and 70% over 70 cell divisions with or without selection, suggesting that NC-MiC2 was largely mitotically stable (Fig. 2). Immunofluorescence using CREST-6 autoimmune serum (28) and specific antibodies to CENP-A, CENP-C, and CENP-E confirmed NC activity on NC-MiC2 (see Fig. 6).
Figure 2.
Stability of NC-MiCs. Cell lines were cultured for >20 divisions in the absence of selection before they were harvested at different intervals for determination of retention rates. Vertical axis denotes percentage of cells containing a MiC as shown by FISH.
Transfection of NC-MiC1 cells with the Y3C94 truncation-construct resulted in more than 1,000 hygromycin-resistant colonies that were screened for loss of vector DNA contained in the construct, indicative of a targeted truncation event (see Fig. 1A and Experimental Protocols). Detailed FISH analysis of one resulting cell line, NC-MiC3, with cosmid, BAC, and PCR probes demonstrated a correctly targeted truncation event as in NC-MiC2 (data not shown). Further p′ mapping and analysis of NC-MiC3 are described below.
Truncation of p′ Arm.
NC-MiC2 was subjected to further truncation using targeting DNA derived from three different p′ regions (Fig. 1C). Extensive screening of drug-resistant cell lines for possible targeting yielded none with the correct event. However, two cell lines (NC-MiC4 and NC-MiC5) were identified that showed apparently random truncation at p′ sites relatively close to the NC region (28).
Structure of NC-MiCs 3, 4, and 5.
Fig. 1C summarizes the detailed FISH mapping results for NC-MiCs 3, 4, and 5 (some examples are shown in Figs. 3, 4, and 7, which is published as supplemental data). All three NC-MiCs showed the intended q′ truncation within Y3C94. On the p′ arm, chromosome truncation was seen between probes Y13C12 (present; Fig. 3A)/BA179N3 (absent; Fig. 3B), Y13C12 (present)/BA179N3 (absent; Fig. 7), and BA48L24 (present; Fig. 4A)/BA69K10 (absent; Fig. 4B), for NC-MiC3 (and its predecessor NC-MiC1), NC-MiC4, and NC-MiC5, respectively. Signal intensity on the NC-MiCs was indistinguishable from that seen on the normal chromosomes 10 in HT1080 cells for all probes tested, suggesting that no major duplication of DNA has occurred. Of 24 different human whole-chromosome paints tested, only the chromosome 10-paint produced positive signals on the NC-MiCs. Similarly, a number of subchromosome 10-paints (see Fig. 1B for locations) showed no signal on the NC-MiCs, demonstrating the absence of non-10q25 genomic regions on the NC-MiCs (see Fig. 8, which is published as supplemental data). We conclude that, within the FISH detection sensitivity, NC-MiCs 3, 4, and 5 contain single-copy DNA derived solely from the 10q25 NC region.
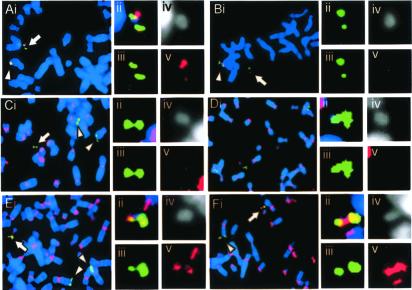
Figure 3.
FISH and/or immunofluorescence analysis on NC-MiC3. Green and red denote E8 FISH signal and test probe signal, respectively. (A) FISH using Y13C12, positive on NC-MiC3. (B) FISH using B179N3, negative on NC-MiC3. (C) FISH using pTRA7, showing absence of α-satellite sequences on NC-MiC3. (D) Immunofluorescence using anti-CENP-B antibody, negative on NC-MiC3. (E) Immunofluorescence using anti-CENP-C antibody, positive on NC-MiC3. (F) Immunofluorescence using anti-CENP-E antibody, positive on NC-MiC3. (ii–v) Combined image, and split images for green, DAPI, and red, for the NC-MiCs.
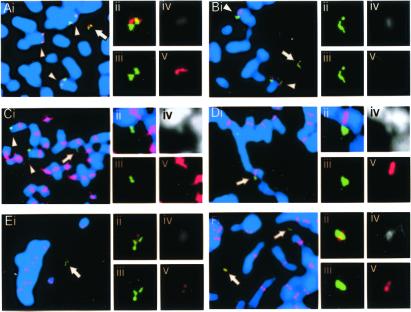
Figure 4.
FISH and/or immunofluorescence analysis on NC-MiC5. Green and red denote E8 FISH signal and test probe signal, respectively. (A) FISH using B48L24, positive on NC-MiC5. (B) FISH using B69K10, negative on NC-MiC5. (C) FISH using pTRA7, showing absence of α-satellite sequences on NC-MiC5. (D) Immunofluorescence using anti-CENP-B antibody, negative on NC-MiC5. (E) Immunofluorescence using anti-CENP-C antibody, positive on NC-MiC5. (F) Immunofluorescence using anti-CENP-E antibody, positive on NC-MiC5. (ii–v) Combined image, and split images for green, DAPI, and red, for the NC-MiCs.
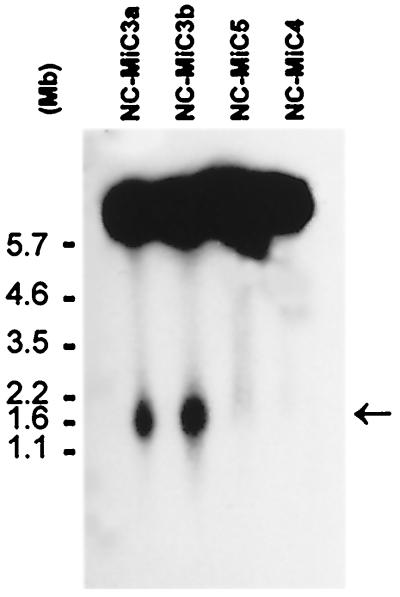
PFGE under conditions that resolved DNA of up to 6 Mb failed to resolve NC-MiCs 4 and 5, suggesting a circular structure of these MiCs. In contrast, linearity of NC-MiC3 was demonstrated by its migration into the gel. Furthermore, the resulting 1.7-Mb NC-MiC3 band (Fig. 5) closely matched the size predicted by mapping and sequencing (Fig. 1C). FISH using a pan-telomere probe or immunofluorescence using an antibody to the telomere repeat-binding factor TRF1 failed to detect telomeres on any of the NC-MiCs (see Discussion).
Figure 5.
PFGE and Southern hybridization of NC-MiCs. Uncut genomic DNA of NC-MiCs 3a, 3b, 4, and 5 was run on 0.6% gel, 0.5× Tris-acetate/EDTA buffer, at 1 V/cm, 192 h, transferred to HybondN+ and probed with 32P-labeled E8 DNA. NC-MiC3a and b migrated at a size of ≈1.7 Mb (arrowed), whereas NC-MiCs 4 and 5 failed to enter the gel.
Based on our FISH and pulsed-field gel analyses, and recently available genome sequence data, we estimated the sizes of NC-MiCs 4 and 5 at 1.8 and 0.7 Mb, respectively (Fig. 1C). These sizes were supported by a direct comparison of the DAPI staining intensities of NC-MiCs 4 and 5 with NC-MiC3, which gave size estimates of 1.8 ± 0.4 Mb and 0.64 ± 0.2 Mb, respectively. No α-satellite DNA or CENP-B protein was detected on any of the NC-MiCs (some examples are shown in Figs. 3, 4, and 7).
NC-MiC Stability and NC Activity.
The mitotic stability of NC-MiCs 3, 4, and 5 and their subclones was assayed with and without selection for up to 4 months in culture. For NC-MiC3 and NC-MiC4, retention rates of >80% were observed after 20 cell divisions, with 40% at 60 divisions for NC-MiC3 and 50% after 90 divisions for NC-MiC4 in the absence of drug selection (Fig. 2). This finding implies a small, but definite, loss over time. However, a low loss rate of <0.3% per division between divisions 40 and 70 for NC-MiC3 and between divisions 55 and 90 for NC-MiC 4 suggested the subsequent attainment of stability in the absence of selection. For the NC-MiC5 cell line, a constant 35–36% retention rate was observed between 20 and 70 divisions in the absence of selection, again suggesting a reasonable level of mitotic stability over time. The low initial retention rates observed for these cell lines were most likely related to the inherent genomic instability seen in our HT1080 parental cell line (see below), although in all three cases, relatively stable retention rates for each of the MiCs was achieved after prolonged culture.
Subcloning of NC-MiC5 and NC-MiC3 produced cell lines showing increased stability of the NC-MiCs (Fig. 2). After 50 cell divisions in the absence of selection, NC-MiC5a and NC-MiC5b subclones demonstrated retention rates of 93% and 90%, respectively, comparable to that seen with selection (90% and 91%, respectively; data not shown), and at 100 cell divisions retention rates of 90% and 85% were observed without selection. Similarly, preliminary data for a single subclone derived from the linear NC-MiC3 (NC-MiC3a) showed a constant retention rate of ≈65% after 20 and 50 cell divisions in the absence of selection. In control experiments using the different chromosome-specific paints, we have established that the NC-MiC-containing HT1080 cell lines exhibited relatively high chromosomal loss rates for all human chromosomes. For example, examination of 100 cells from NC-MiC4 and NC-MiC5a cell lines revealed that 38% and 26%, respectively, showed random gain or loss of one or more host chromosomes. These relatively high levels of inherent genome instability further highlighted the relative stability of our NC-MiCs.
Immunofluorescence detection was used to investigate the functional status of the NCs on the NC-MiCs. Antisera to a host of centromere-associated proteins, including CENP-A, CENP-B (Figs. 3D and 4D), CENP-C (Figs. 3E and 4E), CENP-E (Figs. 3F and 4F), CENP-F, hZW10, p55CDC, and BUB1, were tested. All of the NC-MiCs gave identical protein-binding profiles to those previously established for the parental mardel(10) chromosome (25, 28), thereby confirming the presence of full NC activity on these NC-MiC derivatives.
Discussion
Existing human artificial chromosomes or MiCs have been created by using α-satellite DNA to provide centromere function (4–8, 9–15). We have investigated the use of two different in vitro assembly approaches to construct NC-based human MiCs. These approaches involved either cotransfection of an 80-kb NC DNA-containing BAC with cloned telomeric sequences, or direct transfection of a 640-kb NC DNA-containing YAC that had been retrofitted with human telomere sequences. Neither strategy led to an activated NC or a functional artificial chromosome, suggesting that NC activation is highly inefficient or not possible under the conditions used. This finding contrasts with the relatively high frequency (>50%) of centromere/artificial chromosome formation seen in studies using α-satellite DNA (7, 8).
Better success was obtained when we combined targeted telomere-associated truncation of the q′ arm and apparently random truncation of the p′ arm of the mardel(10) chromosome in human HT1080 cells. Detailed mapping using known probes allowed the truncation sites to be defined. Intensity of FISH probe signals on the NC-MiCs suggests that no duplication of 10q25 material has occurred. Extensive FISH analysis using pan-α-satellite DNA, whole-chromosome paints for all human chromosomes, and different subchromosome-10 paints suggest that none of the NC-MiCs have acquired detectable amounts of α-satellite DNA or other human genomic sequences. Based on PFGE analysis, DAPI image comparison, physical mapping, and sequencing data, the sizes of NC-MiCs 3, 4, and 5 are estimated at ≈1.7, 1.8, and 0.7 Mb, respectively.
PFGE analysis demonstrates that NC-MiC3 is a linear MiC whereas NC-MiCs 4 and 5 are likely to be circular. The significance of our inability to detect telomeric DNA and protein on these NC-MiCs is unclear, given that NC-MiC3 is linear and therefore presumably contains telomeric ends, although it is possible that telomere length may be below our detection threshold. Previous studies also have described circular mammalian artificial chromosomes (5, 7, 8). At present, the mechanisms for the formation of these circular chromosomes are unclear. However, despite their apparent circular nature, NC-MiCs 4 and 5 are largely stable in the absence of selection. Furthermore, we demonstrate that through subcloning, cell lines with significantly greater stability can be obtained, presumably due to selection of a subpopulation of cells in a more stable genomic background. Analysis of NC-MiC5a and NC-MiC5b subclones after more than 100 cell divisions without selection reveals mitotic retention rates of over 85%, equivalent to a loss rate per division of less than 0.15% per generation. Similarly, analysis of NC-MiC3a subclone revealed no loss over 30 cell divisions without selection. These loss rates are comparable with that seen in other human chromosomes in the same genetic background or in nonessential chromosomes of other human cells (53). In addition, this level of stability is comparable with or higher than that reported for α-satellite-based artificial chromosomes created in HT1080 cells [0.48–1.6% (5, 8); 0.1–0.5% (4)].
Because the NC-MiCs have not acquired detectable α-satellite DNA, mitotic segregation activity most likely is conferred by the 10q25 NC that resides in the mardel(10) chromosome. NC activity was confirmed by positive immunofluorescence staining for a host of functionally important centromere proteins: CENP-A, CENP-C, CENP-E, CENP-F, hZW10, p55CDC, and BUB1. The only protein showing absence of binding is CENP-B, agreeing with the analphoid nature of the NC (27, 28). Thus, the NC-MiCs appear to have fully functional centromere activity, which is unlikely to be the cause of the small loss rate observed over time in some cell lines.
The most plausible reason for this small loss rate is the inherent genome instability of the cell lines. The HT1080 line containing mardel(10) chromosome used in the targeting experiments has a highly variable chromosome complement. Chromosome painting studies have demonstrated that this and other NC-MiC-containing cell lines are unstable in their genomic content, randomly losing and gaining genomic materials from a variety of chromosomal origins. Further support comes from subcloning experiments, which yielded derivative cell lines that show highly varying MiC retention rates despite originating from the same genomic background.
The construction of NC-based human MiC is significant for a number of reasons. First, the NC-MiCs provide direct functional evidence for the mitotic activity of the 10q25 NC DNA. Our mapping data suggests that a maximum of ≈0.7 Mb of DNA within this region is sufficient to confer full NC activity. These NC-MiCs therefore provide an excellent model system for further detailed investigation of the functional elements of the 10q25 NC, such as through direct deletion and mutagenesis studies.
A major impetus for the production of the NC-based MiCs is their potential utility as gene delivery and expression vectors for human gene therapy. These MiCs could offer a number of unique advantages over similar entities constructed from normal repetitive centromere DNA. Because the NC-MiCs are made up of single-copy genomic DNA, they are amenable to full sequence characterization to provide a well-defined tool. This characterization step should be greatly facilitated by the completion of the human genome sequence. The availability of full sequence information also should allow the identification of genes that may be contained within the NC-MiCs, thereby allowing direct investigation of any effects neocentromerization may have on gene expression. Finally, our smallest MiC appears as small or smaller than any previously reported stable mammalian artificial chromosomes (4–7, 16), perhaps reflecting a lower DNA size requirement for NC-based MiCs. Future studies should determine whether these features will make NC-MiCs suitable vectors for therapeutic gene delivery.
Supplementary Material
Acknowledgments
We thank T. deLange, M. Goldberg, J. Weinstein, B. Williams, and T. Yen for generous gifts of antibodies, H. Cooke for cloned human telomere array, M. Rocchi for chromosome-10 radiation hybrid DNA, J. Craig and E. Earle for helpful discussion, and E. Earle, K. Tainton, and L. Hii for assistance in the preliminary investigation of de novo artificial chromosome construction strategies. This work was funded by the National Health and Medical Research Council, Australian Medical Research and Development Corp., and AusIndustry.
Abbreviations
- NC
neocentromere
- MiC
minichromosome
- FISH
fluorescence in situ hybridization
- PFGE
pulse-field gel electrophoresis
- DAPI
4′,6-diamidino-2-phenylindole
- BAC
bacterial artificial chromosome
- YAC
yeast artificial chromosome
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 5374.
References
- 1.Dobie K W, Hari K L, Maggert K A, Karpen G H. Curr Opin Genet Dev. 1999;9:206–217. doi: 10.1016/S0959-437X(99)80031-8. [DOI] [PubMed] [Google Scholar]
- 2.Pidoux A L, Allshire R C. Curr Opin Cell Biol. 2000;12:308–319. doi: 10.1016/s0955-0674(00)00094-6. [DOI] [PubMed] [Google Scholar]
- 3.Tyler-Smith C, Floridia G. Cell. 2000;102:5–8. doi: 10.1016/s0092-8674(00)00004-0. [DOI] [PubMed] [Google Scholar]
- 4.Harrington J J, Van Bokkelen G, Mays R W, Gustashaw K, Willard H F. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 5.Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill N I, Cooke H, Masumoto H. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 6.Henning K A, Novotny E A, Compton S T, Guan X Y, Liu P P, Ashlock M A. Proc Natl Acad Sci USA. 1999;96:592–597. doi: 10.1073/pnas.96.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebersole T A, Ross A, Clark E, McGill N, Schindelhauer D, Cooke H, Grimes B. Hum Mol Genet. 2000;9:1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- 8.Masumoto H, Ikeno M, Nakano M, Okazaki T, Grimes B, Cooke H, Suzuki N. Chromosoma. 1998;107:406–416. doi: 10.1007/s004120050324. [DOI] [PubMed] [Google Scholar]
- 9.Farr C J, Stevanovic M, Thomson E J, Goodfellow P N, Cooke H J. Nat Genet. 1992;2:275–282. doi: 10.1038/ng1292-275. [DOI] [PubMed] [Google Scholar]
- 10.Farr C J, Bayne R A, Kipling D, Mills W, Critcher R, Cooke H J. EMBO J. 1995;14:5444–5454. doi: 10.1002/j.1460-2075.1995.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills W, Critcher R, Lee C, Farr C J. Hum Mol Genet. 1999;8:751–761. doi: 10.1093/hmg/8.5.751. [DOI] [PubMed] [Google Scholar]
- 12.Heller R, Brown K E, Burgtorf C, Brown W R. Proc Natl Acad Sci USA. 1996;93:7125–7130. doi: 10.1073/pnas.93.14.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J W, Pendon C, Yang J, Haywood N, Chand A, Brown W R. Hum Mol Genet. 2000;9:1891–1902. doi: 10.1093/hmg/9.12.1891. [DOI] [PubMed] [Google Scholar]
- 14.Shen M H, Yang J, Loupart M L, Smith A, Brown W. Hum Mol Genet. 1997;6:1375–1382. doi: 10.1093/hmg/6.8.1375. [DOI] [PubMed] [Google Scholar]
- 15.Loupart M L, Shen M H, Smith A. Chromosoma. 1998;107:255–259. doi: 10.1007/s004120050305. [DOI] [PubMed] [Google Scholar]
- 16.Kereso J, Praznovszky T, Cserpan I, Fodor K, Katona R, Csonka E, Fatyol K, Szeles A, Szalay A A, Madlaczky G, et al. Chromosome Res. 1996;4:226–239. doi: 10.1007/BF02254964. [DOI] [PubMed] [Google Scholar]
- 17.Csonka E, Cserpan I, Fodor K, Hollo G, Katona R, Kereso J, Praznovszky T, Szakal B, Telenius A, deJong G, et al. J Cell Sci. 2000;113:3207–3216. doi: 10.1242/jcs.113.18.3207. [DOI] [PubMed] [Google Scholar]
- 18.Telenius H, Szeles A, Kereso J, Csonka E, Praznovszky T, Imreh S, Maxwell A, Perez C F, Drayer J I, Hadlaczky G. Chromosome Res. 1999;7:3–7. doi: 10.1023/a:1009215026001. [DOI] [PubMed] [Google Scholar]
- 19.DeJong G, Telenius A H, Telenius H, Perez C F, Drayer J I, Hadlaczky G. Cytometry. 1999;35:129–133. [PubMed] [Google Scholar]
- 20.Co D O, Borowski A H, Leung J D, van der Kaa J, Hengst S, Platenburg G J, Pieper F R, Perez C F, Jirik F R, Drayer J I. Chromosome Res. 2000;8:183–191. doi: 10.1023/a:1009206926548. [DOI] [PubMed] [Google Scholar]
- 21.Choo K H A. Am J Hum Genet. 1997;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams B C, Murphy T D, Goldberg M L, Karpen G H. Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 23.Barry A E, Bateman M, Howman E V, Cancilla M R, Tainton K M, Irvine D V, Saffery R, Choo K H A. Genome Res. 2000;10:832–838. doi: 10.1101/gr.10.6.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choo K H A. Trends Cell Biol. 2000;10:182–188. doi: 10.1016/s0962-8924(00)01739-6. [DOI] [PubMed] [Google Scholar]
- 25.Saffery R, Irvine D V, Griffiths B, Kalitsis P, Wordeman L, Choo K H A. Hum Mol Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- 26.Warburton P E, Dolled M, Mahmood R, Alonso A, Li S, Naritomi K, Tohma T, Nagai T, Masegawa T, Ohashi H, et al. Am J Hum Genet. 2000;66:1794–1806. doi: 10.1086/302924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voullaire L E, Slater H R, Petrovic V, Choo K H A. Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 28.du Sart D, Cancilla M R, Earle E, Mao J I, Saffery R, Tainton K M, Kalitsis P, Martyn J, Barry A E, Choo K H A. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 29.Itzhaki J E, Porter A C. Nucleic Acids Res. 1991;19:3835–3842. doi: 10.1093/nar/19.14.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Killary A M, Lott S T. Methods Companion Methods Enzymol. 1996;9:3–11. [Google Scholar]
- 31.Lo A W I, Choo K H A. BioTechniques. 1999;26:408–412. doi: 10.2144/99263bm08. [DOI] [PubMed] [Google Scholar]
- 32.Lansdorp P M, Verwoerd N P, van de Rijke F M, Dragowska V, Little M T, Dirks R W, Raap A K, Tanke H J. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 33.Liu P, Siciliano J, Seong D, Craig J, Zhao Y, de Jong P J, Siciliano M J. Cancer Genet Cytogenet. 1993;65:93–99. doi: 10.1016/0165-4608(93)90213-6. [DOI] [PubMed] [Google Scholar]
- 34.Saffery R, Earle E, Irvine D V, Kalitsis P, Choo K H A. Chromosome Res. 1999;7:261–265. doi: 10.1023/a:1009222729850. [DOI] [PubMed] [Google Scholar]
- 35.Hudson D F, Fowler K J, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford N G, de Kretser D M, Cancilla M R, et al. J Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalitsis P, Fowler K J, Earle E, Hill J, Choo K H A. Proc Natl Acad Sci USA. 1998;95:1136–1141. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thrower D A, Jordan M A, Schaar B T, Yen T J, Wilson L. EMBO J. 1995;14:918–926. doi: 10.1002/j.1460-2075.1995.tb07073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao H, Winkfein R J, Mack G, Rattner J B, Yen T J. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jablonski S A, Chan G K, Cooke C A, Earnshaw W C, Yen T J. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- 40.Starr D A, Williams B C, Li Z, Etemad-Moghadam B, Dawe R K, Goldberg M L. J Cell Biol. 1997;138:1289–1301. doi: 10.1083/jcb.138.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstein J, Jacobsen F W, Hsu-Chen J, Wu T, Baum L G. Mol Cell Biol. 1994;14:3350–3363. doi: 10.1128/mcb.14.5.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Steensel B, de Lange T. Nature (London) 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 43.Cancilla M R, Tainton K M, Barry A E, Larinov V, Kouprina N, Resnick M A, Du Sart D, Choo K H A. Genomics. 1998;47:399–404. doi: 10.1006/geno.1997.5129. [DOI] [PubMed] [Google Scholar]
- 44.Farr C, Fantes J, Goodfellow P, Cooke H. Proc Natl Acad Sci USA. 1991;88:7006–7010. doi: 10.1073/pnas.88.16.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cross S, Lindsey J, Fantes J, McKay S, McGill N, Cooke H. Nucleic Acids Res. 1990;8:6649–6657. doi: 10.1093/nar/18.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor S S, Larin Z, Smith C T. Hum Mol Genet. 1994;3:1383–1386. doi: 10.1093/hmg/3.8.1383. [DOI] [PubMed] [Google Scholar]
- 47.Porter A C, Itzhaki J E. Eur J Biochem. 1993;218:273–281. doi: 10.1111/j.1432-1033.1993.tb18375.x. [DOI] [PubMed] [Google Scholar]
- 48.Parris C N, Jezzard S, Silver A, MacKie R, McGregor J M, Newbold R F. Br J Cancer. 1999;79:47–53. doi: 10.1038/sj.bjc.6690010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holt S E, Wright W E, Shay J W. Mol Cell Biol. 1996;16:2932–2939. doi: 10.1128/mcb.16.6.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein K G, Bouck N P. Cancer Genet Cytogenet. 1994;73:109–112. doi: 10.1016/0165-4608(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 51.Kugoh H M, Hashiba H, Shimizu M, Oshimura M. Oncogene. 1990;5:1637–1644. [PubMed] [Google Scholar]
- 52.Lo, A. W. I., Craig, J. M., Saffery, R., Kalitsis, P., Irvine, D. V., Earle, E., Magliano, D. J. & Choo, K. H. A. (2001) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 53.Burns E M, Christopoulou L, Corish P, Tyler-Smith C. J Cell Sci. 1999;112:2705–2714. doi: 10.1242/jcs.112.16.2705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.