Abstract
Ascidians, primitive chordates that have retained features of the likely progenitors to all vertebrates, are a useful model to study the evolutionary relationship of chordates to other animals. We have selected the well characterized ribosomal RNA (rRNA) genes to investigate this relationship, and we describe here the cloning and characterization of an entire ribosomal DNA (rDNA) tandem repeat unit from a lower chordate, the ascidian Herdmania momus. rDNA copy number and considerable sequence differences were observed between two H. momus populations. Comparison of rDNA primary sequence and rRNA secondary structures from H. momus with those from other well characterized organisms, demonstrated that the ascidians are more closely related to other chordates than invertebrates. The rDNA tandem repeat makes up a larger percentage (7%) of the genome of this animal than in other higher eukaryotes. The total length of the spacer and transcribed region in H. momus rDNA is small compared to most higher eukaryotes, being less than 8 kb, and the intergenic spacer region consists of smaller internal repeats. Comparative analysis of rDNA sequences has allowed the construction of secondary structures for the 18S, 5.8S and 26S rRNAs.
Full text
PDF


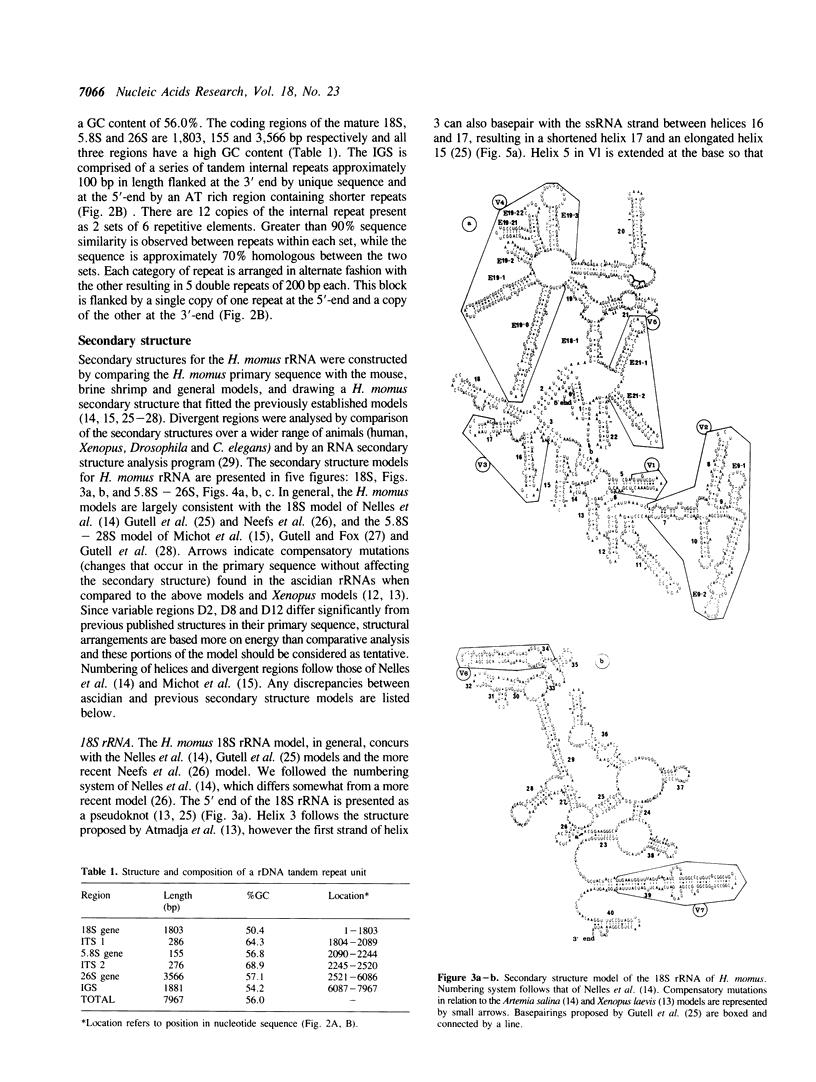


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atmadja J., Brimacombe R., Maden B. E. Xenopus laevis 18S ribosomal RNA: experimental determination of secondary structural elements, and locations of methyl groups in the secondary structure model. Nucleic Acids Res. 1984 Mar 26;12(6):2649–2667. doi: 10.1093/nar/12.6.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Chipchase M., Speirs J. The ribosomal RNA cistrons. Prog Nucleic Acid Res Mol Biol. 1971;11:351–389. doi: 10.1016/s0079-6603(08)60332-3. [DOI] [PubMed] [Google Scholar]
- Cesarone C. F., Bolognesi C., Santi L. Improved microfluorometric DNA determination in biological material using 33258 Hoechst. Anal Biochem. 1979 Nov 15;100(1):188–197. doi: 10.1016/0003-2697(79)90131-3. [DOI] [PubMed] [Google Scholar]
- Chambon P. Eukaryotic nuclear RNA polymerases. Annu Rev Biochem. 1975;44:613–638. doi: 10.1146/annurev.bi.44.070175.003145. [DOI] [PubMed] [Google Scholar]
- Clark C. G., Tague B. W., Ware V. C., Gerbi S. A. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 1984 Aug 10;12(15):6197–6220. doi: 10.1093/nar/12.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. A., Flavell R. B. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984 Oct;38(3):622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Sulston J. E., Coulson A. R. The rDNA of C. elegans: sequence and structure. Nucleic Acids Res. 1986 Mar 11;14(5):2345–2364. doi: 10.1093/nar/14.5.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. On spacers. Cell. 1979 Apr;16(4):697–710. doi: 10.1016/0092-8674(79)90086-2. [DOI] [PubMed] [Google Scholar]
- Field K. G., Olsen G. J., Lane D. J., Giovannoni S. J., Ghiselin M. T., Raff E. C., Pace N. R., Raff R. A. Molecular phylogeny of the animal kingdom. Science. 1988 Feb 12;239(4841 Pt 1):748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- Gans C., Northcutt R. G. Neural crest and the origin of vertebrates: a new head. Science. 1983 Apr 15;220(4594):268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Gorski J. L., Gonzalez I. L., Schmickel R. D. The secondary structure of human 28S rRNA: the structure and evolution of a mosaic rRNA gene. J Mol Evol. 1987;24(3):236–251. doi: 10.1007/BF02111237. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Fox G. E. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res. 1988;16 (Suppl):r175–r269. doi: 10.1093/nar/16.suppl.r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Noller H. F., Woese C. R. Higher order structure in ribosomal RNA. EMBO J. 1986 May;5(5):1111–1113. doi: 10.1002/j.1460-2075.1986.tb04330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. M., Dover G. A. Molecular coevolution among cryptically simple expansion segments of eukaryotic 26S/28S rRNAs. Mol Biol Evol. 1988 Jul;5(4):377–391. doi: 10.1093/oxfordjournals.molbev.a040505. [DOI] [PubMed] [Google Scholar]
- Hancock J. M., Tautz D., Dover G. A. Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster. Mol Biol Evol. 1988 Jul;5(4):393–414. doi: 10.1093/oxfordjournals.molbev.a040501. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hindenach B. R., Stafford D. W. Nucleotide sequence of the 18S-26S rRNA intergene region of the sea urchin. Nucleic Acids Res. 1984 Feb 10;12(3):1737–1747. doi: 10.1093/nar/12.3.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery W. R. Specification of cell fate by cytoplasmic determinants in ascidian embryos. Cell. 1985 May;41(1):11–12. doi: 10.1016/0092-8674(85)90052-2. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P. Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochimie. 1987 Jan;69(1):11–23. doi: 10.1016/0300-9084(87)90267-7. [DOI] [PubMed] [Google Scholar]
- Michot B., Hassouna N., Bachellerie J. P. Secondary structure of mouse 28S rRNA and general model for the folding of the large rRNA in eukaryotes. Nucleic Acids Res. 1984 May 25;12(10):4259–4279. doi: 10.1093/nar/12.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Qu L. H., Bachellerie J. P. Evolution of large-subunit rRNA structure. The diversification of divergent D3 domain among major phylogenetic groups. Eur J Biochem. 1990 Mar 10;188(2):219–229. doi: 10.1111/j.1432-1033.1990.tb15393.x. [DOI] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., Hendriks L., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles L., Fang B. L., Volckaert G., Vandenberghe A., De Wachter R. Nucleotide sequence of a crustacean 18S ribosomal RNA gene and secondary structure of eukaryotic small subunit ribosomal RNAs. Nucleic Acids Res. 1984 Dec 11;12(23):8749–8768. doi: 10.1093/nar/12.23.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisson P. E., Hickey R. J., Boshar M. F., Crain W. R., Jr Identification of a repeated sequence in the genome of the sea urchin which is transcribed by RNA polymerase III and contains the features of a retroposon. Nucleic Acids Res. 1988 Feb 25;16(4):1431–1452. doi: 10.1093/nar/16.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Enhancers and ribosomal gene spacers. Cell. 1984 Sep;38(2):349–351. doi: 10.1016/0092-8674(84)90489-6. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Enhancers and ribosomal gene spacers. Cell. 1984 Sep;38(2):349–351. doi: 10.1016/0092-8674(84)90489-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Stumph W. E., Wu J. R., Bonner J. Determination of the size of rat ribosomal deoxyribonucleic acid repeating units by electron microscopy. Biochemistry. 1979 Jun 26;18(13):2864–2871. doi: 10.1021/bi00580a030. [DOI] [PubMed] [Google Scholar]
- Tautz D., Dover G. A. Transcription of the tandem array of ribosomal DNA in Drosophila melanogaster does not terminate at any fixed point. EMBO J. 1986 Jun;5(6):1267–1273. doi: 10.1002/j.1460-2075.1986.tb04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Tautz C., Webb D., Dover G. A. Evolutionary divergence of promoters and spacers in the rDNA family of four Drosophila species. Implications for molecular coevolution in multigene families. J Mol Biol. 1987 Jun 5;195(3):525–542. doi: 10.1016/0022-2836(87)90181-1. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks D. P., Beerman N., Griffith O. M. A small-scale five-hour procedure for isolating multiple samples of CsCl-purified DNA: application to isolations from mammalian, insect, higher plant, algal, yeast, and bacterial sources. Anal Biochem. 1986 Feb 1;152(2):376–385. doi: 10.1016/0003-2697(86)90423-9. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H., Dawid I. B., Brown D. D. Arrangement of length heterogeneity in repeating units of amplified and chromosomal ribosomal DNA from Xenopus laevis. J Mol Biol. 1976 Aug 25;105(4):487–505. doi: 10.1016/0022-2836(76)90230-8. [DOI] [PubMed] [Google Scholar]