Abstract
We have previously shown the association of AXIN2 with oral clefts in a US population. Here, we expanded our study to explore the association of 11 AXIN2 markers in 682 cleft families from multiple populations. Alleles for each AXIN2 marker were tested for transmission distortion with clefts by means of the Family-based Association Test. We observed an association with SNP rs7224837 and all clefts in the combined populations (p = 0.001), and with SNP rs3923086 and cleft lip and palate in Asian populations (p = 0.004). We confirmed our association findings in an additional 528 cleft families from the United States (p < 0.009). We tested for gene-gene interaction between AXIN2 and additional cleft susceptibility loci. We assessed and detected Axin2 mRNA and protein expression during murine palatogenesis. In addition, we also observed co-localization of Axin2 with Irf6 proteins, particularly in the epithelium. Our results continue to support a role for AXIN2 in the etiology of human clefting. Additional studies should be performed to improve our understanding of the biological mechanisms linking AXIN2 to oral clefts.
Keywords: cleft lip and palate, genetics, gene expression, gene-gene interactions, wnt pathway, craniofacial anomalies
Introduction
Oral-facial clefts are common birth defects of complex etiology involving the interplay of genetic predisposition and environmental exposures. Most oral-facial clefts are non-syndromic, isolated defects, which can be separated into two different phenotypes: cleft lip with or without cleft palate (CL/P) and cleft palate only (CPO). Genetics and embryology suggest that CL/P and CPO are different entities (Murray, 2002). CL/P is more common, with a prevalence of 1 to 2/1000 births, with Asians and Native Americans having the highest rate and Africans the lowest, whereas CPO affects approximately 1/1500 to 2000 births and does not vary in different racial backgrounds (Mossey and Little, 2002). Numerous lines of evidence now suggest that the phenotypic spectrum of non-syndromic CL/P is more complex than previously realized and should include a variety of subclinical phenotypic features observed in either an individual with a cleft and/or his/her ‘unaffected’ relatives (Weinberg et al., 2006; Letra et al., 2007).
Several genes—including, but not limited to, MSX1, IRF6, CRISPLD2, FOXE1, and members of the WNT and FGF gene families—have been associated with oral clefts (Jezewski et al., 2003; Zucchero et al., 2004; Chiquet et al., 2007; Riley et al., 2007; Menezes et al., 2009, 2010; Moreno et al., 2009). Recently, genome-wide association studies have implicated additional loci on 1p22, 8q24, 10q25.3, 17q22, and 20q12 in the etiology of oral clefts (Birnbaum et al., 2009; Beaty et al., 2010; Mangold et al., 2010). Current efforts now include unveiling the etiologic variants and their biological functions that may contribute to the cleft phenotype (Dixon et al., 2011).
Members of the WNT gene family have been associated with clefts in humans and mice (Lan et al., 2006; Chiquet et al., 2008; Menezes et al., 2009). The AXIN2 (axis inhibition protein 2) gene is a negative regulator of the WNT pathway due to its role in the degradation of β-catenin. Mutations in AXIN2 have been associated with increased susceptibility to cancer (Lammi et al., 2004; Kanzaki et al., 2006; Yook et al., 2006), and in some cases have been detected segregating together with familial tooth agenesis (Lammi et al., 2004). We have previously reported the association of a missense mutation in AXIN2 (rs2240308, P50S) with CL/P in US cleft families (Menezes et al., 2009).
Here, we performed a family-based association study of AXIN2 polymorphisms and oral clefts in multiple populations. Additionally, an independent family-based dataset was used for confirmation of the association findings. We investigated Axin2 expression at critical periods during mouse palatogenesis and looked for statistical evidence of gene-gene interaction with additional cleft susceptibility genes.
Materials & Methods
Sample Population
Data and samples from 682 non-syndromic cleft pedigrees (2,916 individuals) were ascertained from study sites in the United States, Argentina, Guatemala, Spain, Hungary, Turkey, Beijing, Shanghai, and India. This study was approved by the institutional review boards at each site and by the University of Pittsburgh. Pedigrees were ascertained through probands presenting overt clefts. Individuals provided written informed consent and saliva samples as source of genomic DNA. Details of the study families are summarized in the Table.
Table.
Details of the Studied Populations and Number of Pedigrees in Each Cleft Subgroup
| Lip Involvement |
No Lip Involvement |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Population | Mean Pedigree Size | Min-Max | Unaffected Types | Affected Types | CLO | CLP | CL/P | CPO | All |
| Latin America | Argentina | 5.24 ± 2.9 | 1-16 | 192 | 81 | 11 | 60 | 71 | 7 | 78 |
| Guatemala | 5.82 ± 3.6 | 1-20 | 377 | 115 | 21 | 99 | 120 | 3 | 123 | |
| Europe | USA | 5.07 ± 2.6 | 1-14 | 303 | 141 | 14 | 89 | 103 | 14 | 117 |
| Madrid | 3.82 ± 0.9 | 3-7 | 88 | 45 | 5 | 28 | 33 | 3 | 36 | |
| Hungary | 4.96 ± 1.1 | 3-8 | 154 | 79 | 20 | 53 | 73 | 16 | 89 | |
| Turkey | 9.68 ± 2.7 | 3-17 | 72 | 44 | 7 | 20 | 27 | 11 | 38 | |
| Asian | Shanghai | 10.61 ± 4.7 | 3-28 | 480 | 171 | 12 | 54 | 66 | 0 | 66 |
| Beijing | 3 | 3-3 | 168 | 84 | 14 | 46 | 60 | 24 | 84 | |
| India | India | 14.00 ± 6.7 | 3-38 | 222 | 100 | 22 | 29 | 51 | 0 | 51 |
| All populations | 6.51 ± 4.5 | 1-38 | 2,056 | 860 | 126 | 478 | 604 | 78 | 682 | |
We formed cleft groups by looking at the type(s) of cleft(s) present in each pedigree.
Cleft subgroups include:
CLO (pedigrees with Cleft Lip only)
CLP (pedigrees with Cleft Lip with Cleft Palate)
CL/P (pedigrees with Cleft Lip with or without Cleft Palate)
CPO (pedigrees with Cleft Palate only)
All (combination of CL/P and CPO groups)
Association Analysis
Details of the investigated genes and polymorphisms are provided in the Appendix. Alleles at each AXIN2 SNP were tested for association with all clefts, cleft lip and palate (CLP), and cleft lip with or without cleft palate (CL/P) phenotypes according to the Family-based Association Test (FBAT) (Horvath et al., 2001). We performed analyses for each population individually, combined, and in the following population groups: Asians (Beijing and Shanghai), Europeans (US, Spain, Hungary and Turkey), Latin Americans (Argentina and Guatemala), and non-Asians (i.e., European plus Latin American). India was included only in the combined population. We performed haplotype analysis to test for linkage disequilibrium between haplotypes in each cleft group. We used Bonferroni correction considering the number of variables and tests performed (19), and a p ≤ 0.003 was considered statistically significant.
Confirmation Dataset
We tested 6 SNPs of interest in AXIN2 for confirmation of association findings in an independent dataset of 528 CL/P nuclear families ascertained in Texas (344 non-Hispanic white and 184 Hispanic). These included SNPs rs2240308 and rs7224837 associated in the original dataset and other suggestive SNPs. The Pedigree Disequilibrium test (PDT), Geno-PDT, and Association in the Presence of Linkage (APL) were used to test for association as previously described (Chiquet et al., 2007). APL was used to test for overtransmission of pairwise haplotypes. A p ≤ 0.006 was considered statistically significant under Bonferroni correction.
Statistical Analysis of Gene-Gene Interactions
We tested for AXIN2 SNP (rs3923086 and rs7224837) interactions with loci previously associated with oral clefts: IRF6 (rs2235371 and rs642961), ABCA4 (rs560426) and MAFB (rs13041247), FOXE1 (rs3758249 and rs4460498), and chromosome 8q24 (rs987525 and rs16903544). Allele and haplotype Transmission Disequilibrium Tests (TDT) were used to detect Mendelian transmission of alleles from heterozygous parents to affected offspring. Analyses were performed in FBAT, utilizing the extended family option (i.e.: -e) to adjust for multiple affected individuals in a pedigree. Interaction calculations were performed with the per-pedigree TDT Z-score for each SNP, and the correlation between pairs of individual SNPs in specific genes was assessed.
Expression Analysis
Details regarding sample collection, experimental procedures, and analysis are available in the Appendix. In brief, we analyzed Axin2 mRNA and protein expression during mouse palatogenesis using in situ hybridization, real-time PCR, and immunohistochemistry. We also analyzed mRNA and protein expression of Irf6, an acknowledged cleft susceptibility gene, to see if it correlated with Axin2 expression pattern.
Results
Association Analysis
Association results for the original dataset are presented in Fig. 1. The most significant P value and the greatest degree of overtransmission were for a variant located in intron 10 [SNP rs7224837] and all clefts in non-Asian populations (p = 0.0009) and in the combined populations (p = 0.002). Although not formally significant, a suggestive association with this SNP and all clefts was also observed in Europeans (p = 0.005) (Fig. 1). This same SNP was associated with CL/P (p = 0.001) in the combined populations and in non-Asians (p = 0.004) (Fig. 1), and with CLP in the combined populations (p = 0.006) (Fig. 1). In Asians, the best association results were observed for SNP rs3923086 and CLP (p = 0.004) (Fig. 1).
Figure 1.
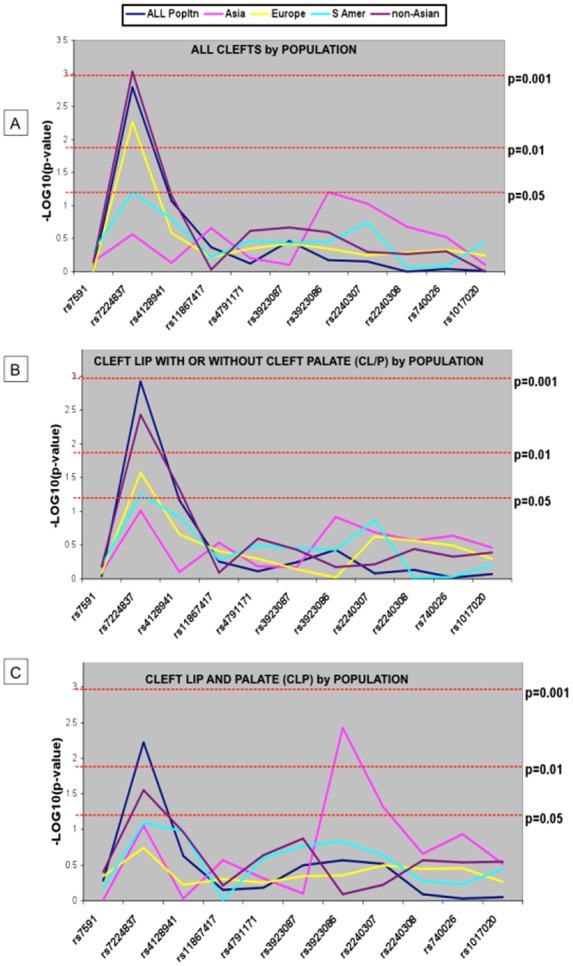
Results of TDT analyses of AXIN2 SNPs and oral clefts by population groups. (A) For all clefts. [Note strongest association for SNP rs7224837 in non-Asians (p = 0.0009) and Europeans (p = 0.005) and in the combined populations (p = 0.002). The lack of association in the other population groups indicates that the association in the combined population is primarily due to contributions from non-Asians.] (B) For CL/P (cleft lip with or without cleft palate). [Note strongest association with rs7224837 in non-Asians (p = 0.004) and in the combined populations (p = 0.001).] (C) For CLP (cleft lip with cleft palate). [Note strongest association for SNP rs3923086 in Asians and rs7224837 in non-Asians (p = 0.004) and in the combined populations (p = 0.006).]
When the individual populations were considered, SNP rs2240308 was associated with CL/P in the US (p = 0.003); meanwhile, additional SNPs showed marginal association values (0.01 ≤ p ≤ 0.05) with cleft phenotypes (Appendix Table 2).
Haplotype analysis showed that the allelic combination of SNPs was significant only in those groups where the individual association with SNP rs7224837 was significant, namely, for SNP alleles rs7224837 (A) - rs4128941 (G) with all clefts (p = 0.002), and with CL/P (p = 0.006) in non-Asians. Haplotypes including our previously associated variant, SNP rs2240308, showed nominal association only in Asian families with CLP (p ≤ 0.05) (Appendix Tables 3-5).
Confirmation of Association
Individual SNP analysis showed association of SNP rs11655966 (located at ~1.5Kb from rs7224837) in both Caucasian (p = 0.008) and Hispanic samples (p = 0.009), whereas rs4791171 was also associated in Caucasians (p = 0.008), although these p-values do not meet conservative criteria for multiple testing. In addition, analysis of two-window haplotypes revealed the significant association of SNPs rs7224837 (A) and rs11655966 (T) in the Caucasian group (p = 0.006). Additional haplotypes in both non-Hispanic white and Hispanic populations showed nominal significance values (0.02 ≤ p ≤ 0.05) (Appendix Tables 6 and 7).
Interaction Analysis
We found evidence of statistical interaction between AXIN2 rs3923086 (T) and IRF6 rs2235371 (C) alleles with CLP (p = 0.007) in Asian families, whereas results in non-Asians suggest interaction of AXIN2 rs7224837 (A) and IRF6 rs2235371 (C) alleles with CL/P (p = 0.02). A trend was also observed for AXIN2 rs7224837 (A) and IRF6 rs642961 (A) (p = 0.06) and with IRF6 rs2235371 (C) (p = 0.07) with CLP in non-Asians.
Expression Results
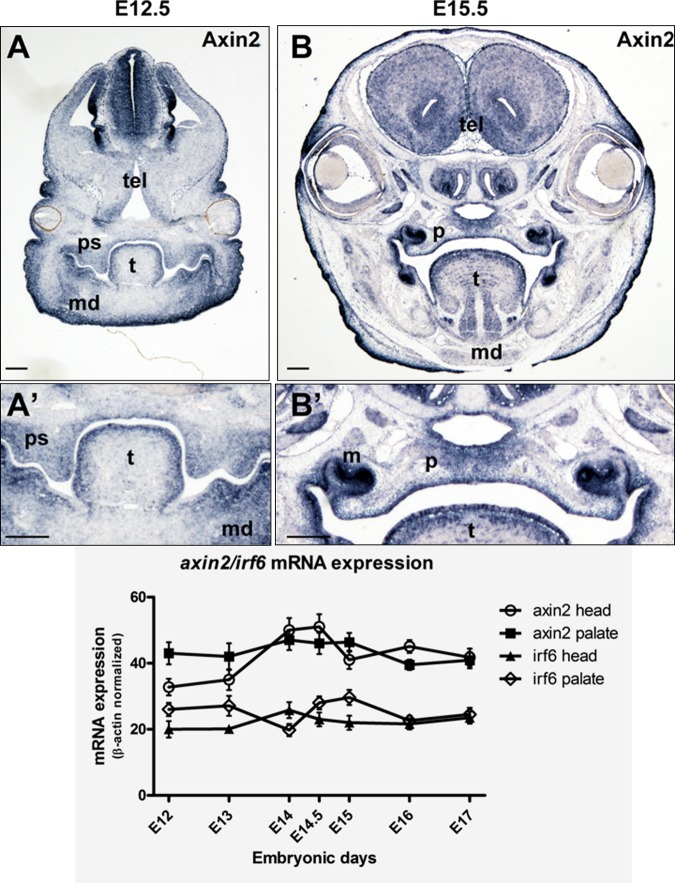
Analysis of Axin2 mRNA expression by in situ hybridization in E12.5 mice showed expression in the mesenchyme of the palatal shelves, tongue, in neural crest cells that surround the myogenic core, as well as the optic nerve, and telencephalon. Near the end of palatogenesis, at E15.5, Axin2 mRNA was markedly expressed in the palatal mesenchyme, oral epithelia, and in the molar tooth buds (Fig. 2). Similarly, real-time PCR showed Axin2 mRNA expression in the mouse head and palate throughout the experimental periods, although inversely correlated. At the initial stages, while Axin2 expression increased from E12 to E13 in the mouse head, it decreased in the palate. At the later stages, Axin2 expression in both the head and palate appeared to reach similar levels gradually. This was also observed for Irf6. Although not significant, an increase in Axin2 mRNA expression was observed at later stages of palatogenesis (E14-15), decreasing thereafter. Axin2 mRNA expression was significantly higher when compared with that of Irf6 (p = 0.0001) (Fig. 2).
Figure 2.
Axin2 is expressed in the mouse palate. Top, mouse embryos were sectioned coronally. (A) At E12.5, Axin2 expression (blue) is found in the palatal shelf (ps) and tissues of the developing head, including the mandible (md), tongue (t), and telencephalon (tel). (A′) Magnified view of the palatal shelves. (B) After palatal fusion at E15.5, Axin2 expression is maintained in the palate (p). (B′) Magnified view of the palate. Axin2 expression is also found in the molar tooth buds (m). Scale bars = 125 microns (bottom). Graphical representation of Axin2 and Irf6 mRNA expression levels by real-time PCR. Both gene transcripts are expressed in the murine head and palate at embryonic days (E) 12-17.
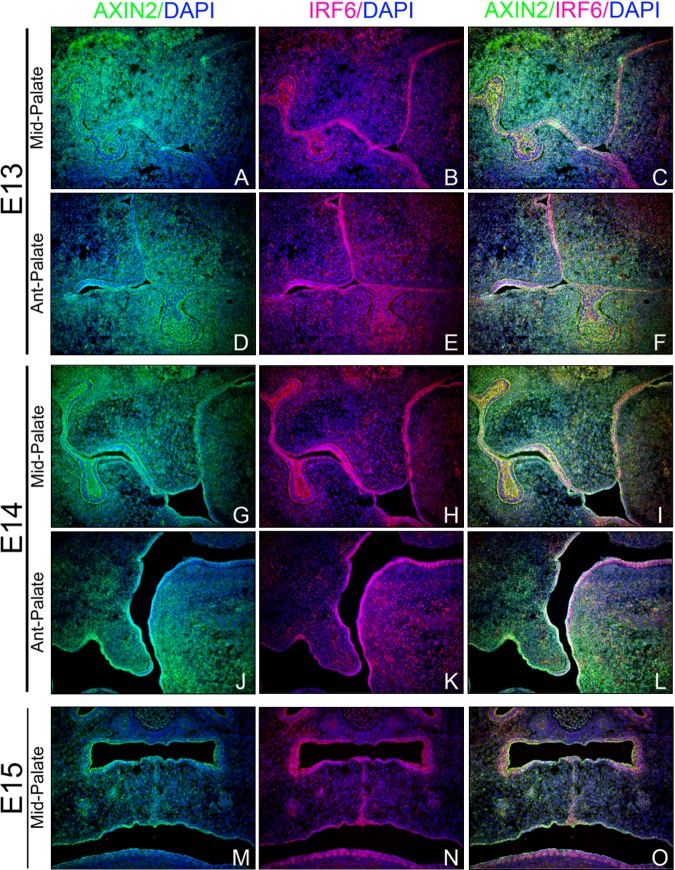
Analysis of Axin2 protein expression showed a pattern consistent with its levels of mRNA expression, including epithelial expression in the palate shelves, teeth, tongue, and also palate and facial mesenchyme, and tongue musculature (Fig. 3). Expression in the brain, eye, olfactory epithelium, nasal cartilage, ocular muscles, and optic nerve was also evident (Appendix Fig. 2). Irf6 protein expression was restricted to epithelial cell layers of the oral cavity, teeth, and palate shelves. Although Axin2 appears to be expressed in a more dispersed pattern than Irf6, both proteins appear to co-localize in relevant orofacial epithelial cell layers critical to normal palate development (Fig. 3).
Figure 3.
Axin2 co-localizes with Irf6 in mouse palate epithelium. Coronal sections through mid- and anterior-palate regions of wild-type embryos pre- (E13, A-F; E14, G-L) and post-palate fusion (E15, M-O) were subject to immunofluorescent labeling with Axin2- and Irf6-specific polyclonal antibodies. Axin2 expression (green) is evident in palate and tooth epithelium and mesenchyme, in addition to tongue and oral epithelium and tongue musculature throughout palatogenesis (A, D, G, J, and M). Irf6 (pink) is expressed primarily in palate, tooth, oral, and tongue epithelium (B, E, H, K, and N). Merged images (yellow; C, F, I, L, and O) show co-localized expression of Axin2 and Irf6 in these epithelial cell layers critical to normal palate and tooth development.
Discussion
In this study, we confirmed our previously reported association of AXIN2 with oral clefts (Menezes et al., 2009) with additional variants in multiple populations. While the associated variant in our previous study in a US population was comprised of a missense mutation (rs2240308) in exon 2 of the AXIN2 gene, in the present study, a variant located in intron 10 (SNP rs7224837) yielded the most significant association values in families presenting CL/P in non-Asian and European populations, and in the overall combined populations as well. Meanwhile, SNP rs3923086, located at the 3′ UTR, was most significant in Asian families with CLP. When compared with the individual populations, our previously associated SNP (rs2240308) continued to show association with CL/P in an expanded sample set from the same US population. An independent dataset of CL/P families and additional SNPs were used for confirmation of the association findings with AXIN2. At ~1.5-kb bp from rs7224837, another intronic SNP rs1165596 showed association in both Caucasian and Hispanic populations, whereas SNP rs4791171 was also associated in Caucasians. Intriguingly, in both original and confirmation datasets, the haplotype analysis was most interesting in views of 2- and 3-SNP sliding windows across the AXIN2 gene in the area of SNP rs7224837 [for non-Asians with all clefts and CL/P in the original dataset, and for Caucasians in the confirmation dataset], and rs3923086 [for Asian families with CLP]. Of note, the associated haplotypes are always those composed of alleles showing positive association in the individual SNP analysis, regardless of whether the TDT result was significant. Haplotypes including our previously associated variant rs2240308 also showed nominal association with Asian families with CLP.
We also performed statistical interaction analysis between AXIN2 and other cleft susceptibility loci. In complex diseases, gene-gene interactions should be considered as additional thresholds for genetic predisposition. The results suggested possible allelic interactions between AXIN2 and IRF6. IRF6 is one of the largest contributors to the underlying genetics of human non-syndromic CL/P, contributing to as much as 12% of cleft cases in the background of other genes (Zucchero et al., 2004). AXIN2 SNP rs7224837 appears to interact with 2 IRF6 SNPs, rs2235371, and rs642961. The associated A and C alleles in AXIN2 rs7224837 and IRF6 rs2235371, respectively, are both the reference alleles for these loci. For rs2235371, which facilitates a Valine to Isoleucine substitution at position 274 (V274I) of IRF6, the common C allele is also overtransmitted in cleft families (Zucchero et al., 2004). In contrast, the trend for interaction with IRF6 rs642961 is with the rare A allele, which was significantly overtransmitted in cleft families, particularly those with cleft lip (Rahimov et al., 2008).
AXIN2 belongs to the WNT pathway, which plays a critical and evolutionary conserved role in directing cell fates during craniofacial morphogenesis (Parr et al., 1993). AXIN2 mutations have been linked to cancer and developmental defects as well (Lammi et al., 2004; Yu et al., 2005; Yook et al., 2006). Intriguingly, Axin2-null mice present skeletal defects resembling craniosynostosis but do not present a cleft (Yu et al., 2005; Liu et al., 2007)—hence our question if Axin2 was expressed in the palatal tissues during murine palatogenesis. We observed Axin2 mRNA expression throughout palatogenesis stages, and particularly around E14.5, when the palatal shelves fuse in the midline. These results corroborate previous findings where Axin2 expression was observed most intensely in the mesenchyme underlying the oral and dental epithelium of mouse embryos at E11-13 (Lammi et al., 2004), at which time-points the lip and primary palate have formed. At more advanced stages of tooth morphogenesis (i.e., E17), Axin2 expression was also detected in the odontoblasts and in the enamel knot (Lammi et al., 2004). Unlike Axin mRNA, which is expressed ubiquitously throughout embryogenesis, Axin2 protein expression has been described in a more restricted pattern during mouse embryogenesis, such as in the dorsal neural tube and branchial arches from embryonic days 8.5 to 11.5, and at later stages in the epithelial components of several developing organs (Jho et al., 2002; Yu et al., 2007). In our study, Axin2 protein expression was markedly evident at E14.5 in the epithelial cell layers of the palate shelves, teeth, tongue, facial mesenchyme, and tongue musculature.
We also investigated the expression of Axin2 protein and its possible co-localization with Irf6 in the developing palatal tissue, to verify if the genetic interaction data in humans would correlate with a biological scenario. We found Axin2 and Irf6 proteins co-localizing particularly in the developing secondary palate, nasal, oral, and eye epithelia. Of note, these observations of simultaneous expression of Axin2 and Irf6 in these relevant tissues do not necessarily implicate these molecules as interacting during their programmed functions. Previous evidence has shown that interaction between Axin2 and Axin1 genes, through targeted deletion of Axin2 in an Axin1 heterozygous background in mice (Axin2−/−; Axin1+/−), results in severe craniofacial deformities with truncation of the neural-crest-derived skeletal structures (Yu et al., 2007). Hence, Axin2 may play a role in craniofacial development, individually or interactively with other genes. Nevertheless, the role of Axin2 as well as Axin2/Irf6 interactions in lip fusion still needs to be defined in animal models.
Overall, our results support a likely role for AXIN2 in the etiology of human clefting. The different variants associated with distinct population groups, and the observed differences in minor allele frequencies of the investigated polymorphisms, justify the individual associations in the particular populations, and also highlight the importance of allelic heterogeneity when results of genetic studies are interpreted. It is possible that more than one variant might contribute to a particular condition, and that these variants may be different across populations. Additionally, a specific combination of variants on a single chromosome may be required for a person to exhibit a particular phenotype. While the reported genetic interactions between AXIN2 and IRF6 reflect statistical calculations, additional research should be performed to verify if correlate biological interactions exist in animal models. Nonetheless, the contribution of variants in single genes to oral clefts is an important consideration in genetic counseling.
Acknowledgments
The authors thank the study families and Children of the Americas.
Footnotes
This work was supported by NIH grants R00DE018413 (to RM), R00DE018954 (to AL), R01DE016148 (to MLM, ARV), R01DE009886 (to MLM, LLF), R01DE012472 (to MLM).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. (2010). A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet 42:525-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, et al. (2009). Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet 41:473-477 [DOI] [PubMed] [Google Scholar]
- Chiquet BT, Lidral AC, Stal S, Mulliken JB, Moreno LM, Arcos-Burgos M, et al. (2007). CRISPLD2: a novel NSCLP candidate gene. Hum Mol Genet 16:2241-2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Blanton SH, Burt A, Ma D, Stal S, Mulliken JB, et al. (2008). Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Mol Genet 17:2212-2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. (2011). Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet 12:167-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. (2001). The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet 9:301-306 [DOI] [PubMed] [Google Scholar]
- Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O’Brien SE, et al. (2003). Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet 40:399-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. (2002). Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22:1172-1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki H, Ouchida M, Hanafusa H, Yano M, Suzuki H, Aoe M, et al. (2006). Single nucleotide polymorphism of the AXIN2 gene is preferentially associated with human lung cancer risk in a Japanese population. Int J Mol Med 18:279-284 [PubMed] [Google Scholar]
- Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, et al. (2004). Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet 74:1043-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, et al. (2006). Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn 235:1448-1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, Menezes R, Granjeiro JM, Vieira AR. (2007). Defining subphenotypes for oral clefts based on dental development. J Dent Res 86:986-991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yu HM, Hsu W. (2007). Craniosynostosis caused by Axin2 deficiency is mediated through distinct functions of beta-catenin in proliferation and differentiation. Dev Biol 301:298-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, et al. (2010). Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet 42:24-26 [DOI] [PubMed] [Google Scholar]
- Menezes R, Marazita ML, Goldstein McHenry T, Cooper ME, Bardi K, Brandon C, et al. (2009). AXIS inhibition protein 2 orofacial clefts and a family history of cancer. J Am Dent Assoc 140:80-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes R, Letra A, Kim AH, Küchler EC, Day A, Tannure PN, et al. (2010). Studies with Wnt genes and nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol 88:995-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno LM, Mansilla MA, Bullard SA, Cooper ME, Busch TD, Machida J, et al. (2009). FOXE1 association with both isolated cleft lip with or without cleft palate and isolated cleft palate. Hum Mol Genet 18:4879-4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey PA, Little J. (2002). Epidemiology of oral clefts: an international perspective. In: Cleft lip and palate: from origin to treatment. Wyszynski DF, editor. New York, NY: Oxford University Press, pp. 127-158 [Google Scholar]
- Murray JC. (2002). Gene/environment causes of cleft lip and/or palate. Clin Genet 61:248-256 [DOI] [PubMed] [Google Scholar]
- Parr BA, Shea MJ, Vassileva G, McMahon AP. (1993). Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development 119:247-261 [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, et al. (2008). Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet 40:1341-1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, et al. (2007). Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci USA 104:4512-4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SM, Neiswanger K, Martin RA, Mooney MP, Kane AA, Wenger SL, et al. (2006). The Pittsburgh Oral-Facial Cleft study: expanding the cleft phenotype. Background and justification. Cleft Palate Craniofac J 43:7-20 [DOI] [PubMed] [Google Scholar]
- Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, et al. (2006). A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 8:1398-1406 [DOI] [PubMed] [Google Scholar]
- Yu HM, Jerchow B, Sheu TJ, Liu B, Costantini F, Puzas JE, et al. (2005). The role of Axin2 in calvarial morphogenesis and craniosynostosis. Development 132:1995-2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HM, Liu B, Costantini F, Hsu W. (2007). Impaired neural development caused by inducible expression of Axin in transgenic mice. Mech Dev 124:146-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, et al. (2004). Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med 351:769-780 [DOI] [PubMed] [Google Scholar]