Abstract
Immuno-laser capture microdissection (immuno-LCM) enables highly selective retrieval of designated cell populations from their in situ locations in complex tissue like the brain. However, the amount of tissue acquired by immuno-LCM is extremely limited, and the RNA purification, amplification and labeling steps necessary for expression analysis by hybridization microarray are tedious and time consuming. This report therefore describes a protocol in which these RNA steps are eliminated altogether, yet allows for global gene profiling. Specifically, immuno-LCM tissue was solubilized and the extract directly subjected to reverse transcription to generate cDNA. Pre-amplification of cDNA was performed next, and then relative expression of 96 different immune-related genes simultaneously determined by quantitative real-time PCR using a microfluidic card TaqMan® Low Density Array (TLDA). This protocol was highly reproducible and extremely sensitive, demonstrating high correlation of raw Ct values among both technical and biological replicate samples when using only 1/32 of total pre-amplified cDNA obtained from as little as 500 LCM `shots.' As this abridged protocol takes only approximately 7 hr from LCM tissue acquisition to analysis by TLDA, it can prove a very effective tool for both screening and validation purposes when investigating gene regulation in health and disease of the nervous system and other tissues.
Keywords: laser capture microdissection, brain microvascular endothelial cells, gene expression profiling, microarray, TLDA
1. Introduction
The endothelium shows considerable phenotypic and molecular diversity along the CNS vascular tree (Ge et al., 2005). Segmental heterogeneity exists between the endothelium of parenchymal microvessels (e.g., arterioles, capillaries and venules) and surface macrovessels (e.g., arteries and veins), and even between the endothelium of the respective microvascular tributaries. There is also regional heterogeneity between the endothelium of similar type vessel segments within different CNS regions.
Endothelial heterogeneity poses challenges to studying CNS vascular gene expression. Whole brain and/or spinal cord homogenates suffer from pooling different vascular segments from different CNS regions, providing at best a mosaic of endothelial subtypes. This can lead to significant misinterpretation of vascular function. A means to enrich for specific endothelial populations, coupled to a platform to quantitatively analyze gene expression efficiently, is needed to accurately assess the CNS endothelium in health and disease.
Laser capture microdissection (LCM) enables selective isolation of cells from microscopic samples of tissue (Espina et al., 2007) – and presents opportunity to exploit CNS endothelial heterogeneity in depth. This laboratory showed that, when guided by combined immunohistochemistry/immunofluorescence and coupled to SYBR® Green-based, quantitative real-time PCR (qrt-PCR), immuno-LCM allowed analysis of endothelial gene expression in separate populations of CNS arterioles, capillaries and venules (Macdonald et al., 2010).
Coupling gene expression to LCM has nonetheless been encumbered by the rigors of isolating intact RNA from mere fractions of cells and, thus, largely limited to analyzing only a few genes at a time. Moreover, isolating RNA on such a small scale from multiple samples can become unmanageable in time and cost. A method that would avoid time-consuming and inefficient RNA isolation, yet allow for more extensive gene profiling, would thus be advantageous.
Here we describe a novel protocol, whereby brain microvascular endothelial cell (BMEC) tissue retrieved by LCM is lysed and directly reverse-transcribed, then subject to pre-amplification of cDNA, and analyzed by TaqMan Low Density Array® (TLDA) for simultaneous assessment of 96 different genes. Significantly, the high sensitivity of this approach enables analysis of a modest amount of LCM tissue that can be quickly acquired, allowing in situ gene profiling on this global scale to be completed in approximately 7.0 hr from beginning to end.
2. Materials and methods
Specific aspects of immuno-LCM, including examples of immunostained tissue, discussion of tissue preservation and confirmation of endothelial purity, have been elaborated in previous reports from this laboratory (Kinnecom and Pachter, 2005; Macdonald et al., 2008), and not reiterated here. Where appropriate, approximate times are indicated below next to individual steps in the immuno-LCM/TLDA protocol, to allow comparison to other procedures used for global gene profiling.
2.1. Animals
Female C57 BL/6 mice, age 8–10 weeks and obtained from Charles River Laboratories, Inc. (Wilmington, MA), were used to minimize microvascular heterogeneity due to genetic variability, sex, and age (Macdonald et al., 2008). Animals were euthanized by CO2 inhalation, following Animal Care and Use Guidelines of the University of Connecticut Health Center (Animal Welfare Assurance # A3471-01).
2.2. Induction of experimental autoimmune encephalomyelitis (EAE)
EAE was induced in mice by active immunization with MOG35–55 peptide (MEVGWYRSPFSRVVHLYRNGK), of murine origin (W. M. Keck Biotechnology Resource Center, Yale University), as described (Juedes et al., 2000). Mice were observed daily and scored on a scale of 0 to 4 with gradations of 0.5 for intermediate scores: 0, no clinical signs; 1, loss of tail tone; 2, wobbly gait; 3, hind limb paralysis; and 4 hind and fore limb paralysis. LCM tissue was acquired at day 16 post-immunization (score ~ 2.0).
2.3. Tissue preparation and sectioning
Following euthanasia, the brain was removed, snap-frozen in dry ice-cooled 2-methylbutane (Acros; Geel, Belgium) and processed for cryosectioning (MacDonald et al., 2008). Coronal sections (7 μm thick) of cerebellum were cut between Bregma −6.56 mm and – 7.32 mm, adhered to glass slides, and stored at −80°C until immuno-LCM.
2.4. Fixation and immunostaining of sections for identification of microvessels
[30 min] To obtain sufficient material for comparing two samples by LCM/qrt-PCR analysis, two slides, each containing two consecutive brain sections, were fixed, stained, and dehydrated simultaneously. Two sections provided sufficient material for each sample. Pairing samples this way enabled both samples to experience the same environmental conditions for the same time (Macdonald et al., 2008). Double immunostaining of BMEC and astrocyte populations, by combined immunohistochemistry and immunofluorescence, respectively, was carried out as detailed (Macdonald et al., 2008).
2.5. Dehydration [8.0 min]
After immunostaining, sections were dehydrated in the following solutions, made by diluting 100% EtOH (Pharmco-AAPER; Brookfield, CT) with DEPC treated water: 75% EtOH for 10s, 95% EtOH for 30s, 100% EtOH for 60 s, 100% EtOH for 90s, xylene (Fisher Scientific; Pittsburgh, PA) for 2 min and a final xylene for 3 min.
2.6. LCM [20 – 30 min]
The Arcturus PixCell IIe microscope (Life Technologies; Bedford, MA) was used to retrieve BMEC according to the LCM conditions described (Kinnecom and Pachter, 2005; Macdonald et al., 2008).). In brief, the LCM parameters utilized were as follows: 7.5 μm spot size; 72 mW power; and 0.950 ms pulse duration. BMEC were captured exclusively from capillaries (~ 5 – 9 μm diameter) within the designated cerebellar boundaries. As LCM can only be performed on one slide at a time, one of the two simultaneously stained slides was left at room temperature until action on the first slide was completed. Tissue samples were captured onto HS® caps (Life Technologies Corp.) by a designated number of `shots' (i.e., laser pulses), with one cap used per sample. An equal number of LCM shots was necessary to standardize the amount of input RNA, and was previously shown to provide a highly reproducible amount of tissue equivalent (Macdonald et al., 2008).
Because LCM acquires material from tissue sections, only “fractions” of cells are obtained with each LCM shot. However, the following sample calculation may be used to estimate the number of endothelial cells captured in an LCM session: Assuming the typical endothelial cell is 10 – 15 μm deep × 25 – 50 μm long (Simionescu and Simionescu, 1977), then in 7 μm-thick sections each capillary endothelial profile cut in longitudinal section would represent 0.47 – 0.7 cells in thickness. Using a 7.5 μm diameter laser spot size, the number of capillary endothelial cells cut lengthwise would be 0.15 – 0.3. Hence, the number of endothelial cells captured/shot = 0.07 – 0.21 cells; 500 shots would thus represent 35 – 105 cells, and 1000 shots represent 70 – 210 cells.
2.7. Tissue extraction [15 minutes]
Once LCM was completed, tissue was solubilized in Cell Lysate Buffer® (Signosis; Sunnyvale, CA) for direct reverse transcription. Cell Lysate Buffer®, pre-heated to 75°C, was added and the resulting lysate heated at 75°C for an additional 15 min. Samples of both extracts were immediately frozen at −80°C.
2.8. DNase treatment and cDNA synthesis [2 hours]
Cell Lysate Buffer® extracts were treated with Turbo DNase (Ambion; Austin, TX) according to the manufacturer's instructions. Specifically, Turbo DNase buffer and DNase were added and samples incubated at 37°C for 30 min. Next, DNAse inactivation reagent was added for 2 min at room temperature. Samples were then reverse transcribed using the SuperScript III (Invitrogen) standard protocol with random hexamers (Roche; Indianapolis, IN), and employing an extension temperature of 42°C – optimal for random hexamers – for 60 min. cDNA was stored at −20°C until used for analysis.
2.9 cDNA Pre-Amplification [1 hr 40 min]
Pre-amplification was carried out using TaqMan® PreAmp Master Mix and a custom PreAmp Pool containing all the primers for detection by TaqMan® Gene Expression Assays. Relative amplification was validated using single gene (single-plex) probes for CCL2, VCAM1 and GAPDH prior to analysis by TaqMan® Low Density Array (TLDA) Microfluidic Cards. Both 10 and 14 cycles of pre-amplification were evaluated initially, with an initial hold at 95°C for 10 min, followed by 10/14 cycles at 95°C for 15 sec and 60°C for 4 min. For TLDA analysis, 14 cycles of pre-amplification were used.
2.10. qrt-PCR [2 hours]
Relative cDNA levels were quantified by qrt-PCR using an ABI PRISM 7500 Sequence Detection System Version 2.3, and custom TaqMan® MGB probes. For single-plex assays, separate controls included no template or no reverse transcriptase, and standard curves were constructed for all primers used. Custom TaqMan® primers/probes were used for both single-plex qrt-PCR and the Mouse Immune Panel TLDA (Life Technologies Corp.). TLDA analysis was conducted as per the manufacturer's protocol, with 100 μl sample volumes containing a 1/32 or 1/4 dilution of pre-amplified cDNA added to each port of the microfluidic card.
2.11. Statistical analysis
Data from single-plex reactions were analyzed in SDS 2.3 and Microsoft Excel 2007, while that from TLDA plates were imported to the SDS 2.3 companion software RQ Manager and exported to Microsoft Excel 2007 for calculations. Linear regression analysis was conducted and a Pearson product-moment correlation (Pearson r) determined using GraphPad Prism 5.0. Pearson r was calculated from paired Ct values reflecting expression of each of the 96 genes represented in the Immune Panel TLDA. A Spearman-Rho rank order correlation (Spearman r) was calculated to determine if these 96 gene expression values were comparable in rank order between samples.
3. Results
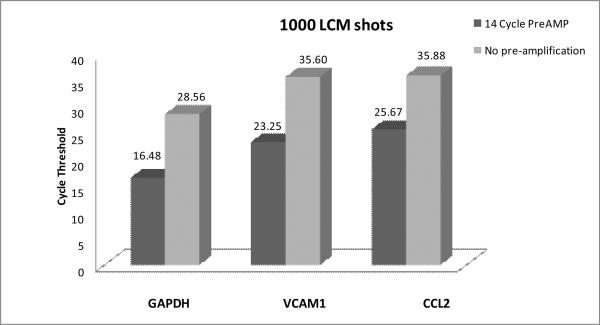
Because of the extremely low level of RNA from LCM samples, the first step was to determine if cDNA generated directly from LCM tissue extracted in Cell Lysate Buffer® – without RNA purification – could be amplified using a commercially available pre-amplification kit prior to quantification by qrt-PCR. A sampling of three genes represented on the Mouse Immune-TLDA (GAPDH, VCAM-1 and CCL2) was individually analyzed in single-plex format. These genes were selected as they exhibit high (GAPDH), moderate (VCAM-1), and relatively low expression (CCL2) by BMEC during acute EAE, the animal model for the neuroinflammatory disease multiple sclerosis (MS), (Furlan et al., 2009) – and thus provide a wide spectrum by which to judge efficacy of the pre-amplification. Fig. 1 demonstrates a 14-cycle pre-amplification significantly lowered Ct values for all three genes. Both GAPDH and VCAM-1 had their Ct values similarly lowered by 12 cycles, while Ct value of CCL2 decreased by 10.2 cycles, thus indicating the relative abundance of amplicons was generally preserved through preamplification. The apparent lesser amplification of CCL2 signal may have resulted from expression of this gene being lower than that which was reliably detectable in the non-amplified sample.
Fig. 1. Pre-amplification of cDNA from Cell Lysate Buffer®proportionally enhances LCM/qrt-PCR of BMEC.
At day 16 following MOG35–55 peptide immunization to induce EAE (disease score ~ 2.0), mouse cerebellar tissue was processed for immuno-LCM, and BMEC captured by 1000 LCM shots then extracted with Cell Lysate Buffer®. BMEC extract then underwent reverse transcription reaction without prior RNA isolation, and equal aliquots of the resulting cDNA were either not pre-amplified or subject to 14 cycles of pre-amplification prior to single-plex qrt-PCR detection with TaqMan® probes/primers for the genes GAPDH, VCAM1 and CCL2. Each qrt-PCR reaction was run in duplicate (technical replicates), and the mean Ct values for each gene are depicted. Pre-amplification of 14 cycles with extract from 1000 shots of LCM-derived BMEC yielded the following mean ΔCt [cDNA – pre-AMP] values: GAPDH (12.087), VCAM1 (12.34), and CCL2 (10.21). Inherent lower expression of CCL2 may explain the lesser-detected amplification of this gene.
It was next determined if pre-amplified BMEC cDNA, generated from Cell Lysate Buffer extracts® without RNA isolation, could be coupled to the Immune Panel TLDA for simultaneous analysis of 96 genes related to inflammation/autoimmunity. This specific TLDA was selected as it contains a cadre of genes thought to be involved in MS/EAE.
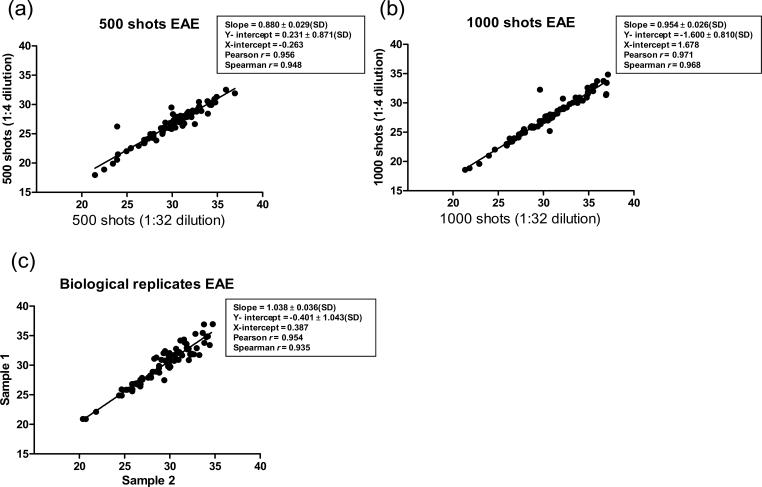
Two different dilutions of the pre-amplified cDNA were assayed to cover a broad range of gene expression and test the limits of sensitivity. Fig. 2a,b depicts linear regression analyses of raw Ct values for the genes represented in the TLDA, contrasting values obtained from 1/4 and 1/32 dilutions of pre-amplified cDNA samples generated from 500 or 1000 LCM shots. Notably, raw Ct values – as apposed to values normalized to a reference gene – reflect the absolute values of input RNA for qrt-PCR and, thus, reveal an accurate picture of the reproducibility of the immuno-LCM/TLDA approach (Chen et al., 2009).
Fig. 2. Expression profiling of BMEC by immuno-LCM/TLDA with no RNA purification shows high reproducibility.
Immuno-LCM of BMEC was carried out on separate EAE cerebellar sections, and captured tissue extracted with Cell Lysate Buffer® then processed through reverse transcription and cDNA pre-amplification, as in Fig. 1. To gauge sensitivity and exclusively highlight technical reproducibility, different dilutions (1/4 or 1/32) of pre-amplified cDNA derived from 500 (a) or 1000 (b) LCM shots were analyzed by qrt-PCR using the Immune Panel TLDA, and linear regression performed on the two corresponding raw Ct values obtained for each gene at the respective dilutions. To highlight overall reproducibility, while accounting for all potential variation due to both biological and technical components, the results of 500 LCM shots from two different tissue sections were compared (c), and linear regression performed on the two corresponding raw Ct values obtained for each gene from the respective sections.
Raw Ct values considered “undetermined” by the software, or at a level ≥ 40 cycles, were excluded from analysis. In the case of 1000 LCM shots, 14 genes were so excluded, while only 6 genes were omitted from the 500 LCM shot sample. Both 1000 and 500 LCM shot samples yielded Pearson r values of >0.95, signifying extremely high linear correlation between technical replicates. These technical replicates originated from the same tissue sections and resultant cDNA pools, their only difference being they represented either 1/4 or 1/32 dilutions of pre-amplified cDNA applied to the TLDA. The high Pearson r thus highlights that diluting cDNA samples over an eight-fold range did not distort the relationship between amplicon measurements of the different genes. Such technical replicates showed a high Spearman r value as well, denoting that raw Ct scores for each gene maintained their rank position relative to all other genes in the TLDA when 1/4 and 1/32 dilutions of pre-amplified cDNA were compared.
Immuno-LCM/TLDA further demonstrated both a high Pearson r (0.95) and Spearman r (0.93) among raw Ct values obtained from “biological” replicates (Fig. 2c). To obtain biological replicates, 500 LCM shots were performed on separate brain sections of the same animal, and a 1/32 dilution of pre-amplified cDNA derived from these respective sections was used in each case. As such, these biological replicates were subject to potential technical variability as well. Brain tissue from different animals was intentionally not compared here, as the immunization protocol itself could produce varying disease states in individual mice, and the objective was to isolate any variability due solely to analysis of random BMEC populations. Despite undergoing a multitude of separate acquisition and processing steps, however, biologic replicates also demonstrated high fidelity. Specifically, linear regression yielded a line with a slope of 1.03 ± 0.038(SD) and Y and X intercepts of near zero – reflecting that raw scores of any specific gene were virtually identical in the biological replicates. These results collectively demonstrate – for the first time – high efficiency and reproducibility of linking immuno-LCM to TLDA format without need of tedious RNA isolation and amplification steps.
4. Discussion
Coupling LCM to genomic profiling platforms has greatly expanded the capacity to probe gene expression within the most discrete CNS domains in situ. Linking LCM to hybridization-based microarrays, in particular, has enabled transcriptional profiling of tens of thousands of genes from select cell groups in CNS tissue samples (Rossner et al., 2006). Nevertheless, this particular combinatorial approach is labor intensive, expensive, and presents difficulties with quantification and statistical evaluation.
Much of this labor stems from RNA isolation and downstream amplification steps needed to generate sufficient labeled complementary RNA (cRNA) for detection (usually > 1μg). These steps can become unmanageable when dealing with multiple samples. Thus, proceeding directly from solubilization of LCM tissue to reverse transcription and then cDNA pre-amplification, would save considerable time and effort when comparing a number of variables (e.g., stages of disease progression or different drug dosages). Concerns have also been raised regarding bias in the RNA amplification process (Li et al., 2005). Furthermore, while the wide breadth of genes that can be analyzed with hybridization-based microarrays may be advantageous in certain applications (e.g. whole-cell transcriptome analysis), it may not be warranted in others. A limited analysis may be more desirable when investigating a specific biological process or tissue property. As qrt-PCR is often used to confirm results of hybridization arrays – which frequently illuminate no more than a few hundred significantly variable genes – coupling LCM to a qrt-PCR-based array format without RNA purification would also prove an efficient validation or screening tool.
Given these considerations, this report described a simplified and expeditious approach whereby immuno-LCM of BMEC was followed by tissue solubilization and direct reverse transcription (in the absence of RNA purification), pre-amplification of the resulting cDNA, and then simultaneous qrt-PCR analysis of 96 immune-related genes by TLDA. Because RNA was not isolated in this study, equal number of LCM shots served as basis for sample comparisons. Previous work from this laboratory showed utilizing equal numbers of LCM shots was an effective means to standardize RNA input (Macdonald et al., 2008), this approach yielding no statistically significant variance in relative gene expression values between technical or biological replicates as detected by single-plex qrt-PCR. The number of shots in the present study was capped at 1000 to minimize the time between immunostaining and processing of RNA for analysis, as restricting this period to < 30 min has been shown to be critical for reliable qrt-PCR detection (Macdonald et al., 2008). Notably, 500 LCM shots was able to be performed in < 20 min, and yielded a higher number of detectable genes than did 1000 LCM shots (90genes vs. 82 genes, respectively) – perhaps reflecting loss of RNA with increased LCM time. Both 500 and 1000 LCM shots nevertheless showed high reproducibility in technical and biological replicates among 96 different genes assayed simultaneously by TLDA – even in the absence of RNA purification. These results support the findings of Keays et al. (2005), who coupled LCM to single-plex qrt-PCR without purifying RNA. But, to the best of our knowledge, this is the first report to describe linkage of LCM to microarray while bypassing RNA purification and employing cDNA pre-amplification.
While there have been more than 100 published reports of LCM/microarray analysis, the array format used has near exclusively been genomic DNA chip hybridization – necessitating RNA purification and amplification. Most recently, Balogh et al. (2007) described coupling LCM of hemotoxylin/eosin-stained tissue to the Immune Panel TLDA used in this study. However, they utilized a four-step RNA amplification process: first purifying RNA, then amplifying it based on dT-T7 and switch T7 primers followed by in vitro transcription, and finally purifying the synthetic RNA population. The present study averted these steps entirely. It is further significant that high reproducibility of immuno-LCM/TLDA was demonstrated with raw Ct scores – not gene expression values normalized to a housekeeping gene.
The validity of the immuno-LCM/TLDA approach was underscored by the distribution of raw Ct scores obtained – from the low 20's to upper 30's – reflecting the variable expression patterns of genes during evolving EAE. Moreover, preliminary comparison between naïve and EAE mice revealed several significant differences. Among these were elevated expression of STAT6 and CCR4 (data not shown), both of which have been shown to be up-regulated in EAE (Zaheer et al., 2007; Forde et al., 2011).
In summary, the immuno-LCM/TLDA approach described offers a relatively quick, reliable and inexpensive means to globally profile gene expression patterns of select cell groups in situ. Given the significant cellular heterogeneity within the nervous system, immuno-LCM/TLDA holds high promise for elaborating gene regulatory mechanisms in the neurobiology of health and disease, and could provide a highly efficient clinical diagnostic tool for neuropathologists (McShea et al., 2006).
Acknowledgements
This work was supported by grants R0-1-MH061525 from the National Institutes of Health, RG-4503A4/1 from the National Multiple Sclerosis Society, and 2010-0913 from the Connecticut Department of Public Health to J.S.P., and American Heart Association predoctoral fellowship award 0815733D to N.M. Special thanks are owed to Alan Carpino, Rachele Kardon and Paula Miccinesi of Life Technologies Corp. for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balogh GH, Russo IH, Spittle C, Heulings R, Russo J. Immune–surveillance and programmed cell death-related genes are significantly overexpressed in the normal breast epithelium of postmenopausal parous women. Int. J. Oncol. 2007;31:303–312. doi: 10.3892/ijo.31.2.303. [DOI] [PubMed] [Google Scholar]
- Chen Y, Gelfond JAL, McManus LM, Shireman PK. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics. 2009;10:407–416. doi: 10.1186/1471-2164-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espina V, Heiby M, Pierobon M, Liotta L. Laser capture microdissection technology. Expert Rev. Mol. Diagn. 2007;7:647–657. doi: 10.1586/14737159.7.5.647. [DOI] [PubMed] [Google Scholar]
- Forde EA, Dogan R-N, Karpus WJ. CCR4 contributes to the pathogenesis of experimental autoimmune encephalomyelitis by regulating inflammatory macrophage function. J. Neuroimmunol. 2011;236:17–26. doi: 10.1016/j.jneuroim.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan R, Cuomo C, Martino G. Animal models of multiple sclerosis. Methods Mol. Biol. 2009;549:157–173. doi: 10.1007/978-1-60327-931-4_11. [DOI] [PubMed] [Google Scholar]
- Ge S, Song Li, Pachter JS. Where is the blood-brain barrier…. really? J. Neurosci. Res. 2005;74:421–427. doi: 10.1002/jnr.20313. [DOI] [PubMed] [Google Scholar]
- Juedes AE, Hjelstrom P, Bergman CM, Neild AL, Ruddle NH. Kinetics and cellular origin of cytokines in the central nervous system: insight into mechanisms of myelin oligodendrocyte glycoprotein-induced experimental encephalomyelitis. J. Immunol. 2000;164:419–426. doi: 10.4049/jimmunol.164.1.419. [DOI] [PubMed] [Google Scholar]
- Keays KM, Owens GP, Ritchie AM, Gilden DH, Burgoon MP. Laser capture microdissection and single-cell RT-PCR without RNA purification. J. Immunolog. Methods. 2005;302:90–98. doi: 10.1016/j.jim.2005.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnecom K, Pachter JS. Selective capture of endothelial cells and perivascular cells from brain microvessels by laser capture microdissection. Brain Res. Protoc. 2005;16:1–9. doi: 10.1016/j.brainresprot.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Li L, Roden J, Shapiro BE, Wold BJ, Bhatia S, Forman SJ, Bhatia R. Reproducibility, fidelity, and discriminant validity of mRNA amplification for microarray analysis from primary hematopoietic cells. J. Mol. Diagn. 2005;7:48–56. doi: 10.1016/S1525-1578(10)60008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald JA, Murugesan N, Pachter JS. Validation of immunolaser capture microdissection coupled with quantitative RT-PCR to probe blood-brain barrier gene expression in situ. J Neurosci. Meth. 2008;174:219–226. doi: 10.1016/j.jneumeth.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Macdonald J, Murugesan N, Pachter JS. Endothelial cell heterogeneity of blood-brain barrier gene expression along the cerebral microvasculature. J. Neurosci. Res. 2010;88:1457–1474. doi: 10.1002/jnr.22316. [DOI] [PubMed] [Google Scholar]
- McShea A, Marlatt MW, Lee HG, Tarkowsky SM, Smit M, Smith MA. The application of microarray technology to neuropathology: cutting edge tool with clinical diagnostics potential or too much information? J. Neuropathol. Exp. Neurol. 2006;65:1031–1039. doi: 10.1097/01.jnen.0000240471.04920.3c. [DOI] [PubMed] [Google Scholar]
- Rossner MJ, Hirrlinger J, Wichert SP, Boehm C, Newrzella D, Hiemsch H, Eisenhardt G, Stuenkel C, von Ahsen O, Nave K-A. Global Transcriptome Analysis of Genetically Identified Neurons in the Adult Cortex. J. Neuroscience. 2006;26:9956–9966. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionescu N, Simnionescu M. The cardiovascular system. In: Weiss Leon., editor. Histology Cell And Tissue Biology. Fifth Edition Elsevier Biomedical; New York: 1977. pp. 371–433. [Google Scholar]
- Zaheer S, Wu Y, Bassett J, Yang B, Zaheer A. Glia maturation factor regulation of STAT expression: a novel mechanism in experimental autoimmune encephalomyelitis. Neurochem. Res. 2007;32:2123–2131. doi: 10.1007/s11064-007-9383-0. [DOI] [PubMed] [Google Scholar]