Abstract
Glucocorticoids may play a significant role in the etiology of neuropsychiatric illnesses. Abnormalities in plasma cortisol levels, glucocorticoid sensitivity, and HPA-axis function often accompany clinical symptoms of stress-related illnesses such as PTSD and depression. Of particular interest are genetic association studies that link single nucleotide polymorphisms of HPA-axis genes with illnesses only in the context of an early-life trauma exposure such as child abuse. These studies suggest that dysregulation of HPA-axis function can have lasting repercussions in shaping mood and anxiety, long after termination of the traumatic experience. As persistent glucocorticoid-induced loss of DNA methylation in Fkbp5 was previously observed in the hippocampus and blood and in the neuronal cell line HT-22, we asked whether these epigenetic alterations occur in non-neuronal, HPA-axis relevant cells. We used the pituitary adenoma cell line AtT-20 to demonstrate that the intronic enhancer region of Fkbp5 undergoes loss of DNA methylation in response to dexamethasone treatment in a dose-dependent manner. We also focused on the mouse hippocampal dentate gyrus to test whether these changes would be enriched in a region implicated in the HPA-axis stress response, neurogenesis, and synaptic plasticity. We observed an increase in enrichment of DNA methylation loss in the dentate gyrus, as compared to whole hippocampal tissues that were similarly treated with glucocorticoids. We then asked whether Dnmt1, a methyltransferase enzyme involved in maintaining DNA methylation following cell division, is involved in the observed epigenetic alterations. We found a dose-dependent decrease of Dnmt1 expression in the AtT-20 cells following dexamethasone treatment, and a similar decrease in corticosterone-treated mouse hippocampus. Taken together, we provide evidence that these glucocorticoid-induced epigenetic alterations have a broader validity in non-neuronal cells and that they may involve the DNA methylation machinery.
Keywords: Fkbp5, Dnmt1, epigenetics, DNA methylation, AtT-20, pituitary, hippocampus, HPA-axis
1. Introduction
Glucocorticoids (GCs) may play a significant role in the etiology of several neuropsychiatric illnesses. A model for evaluating the role of GCs in mood disorders is Cushing’s syndrome (CS), due to an endogenous increase in cortisol secretion, or secondary to iatrogenic GC treatment. Remarkably, up to 60~90% of patients with endogenous CS develop depression [1,2,3], and the depressive symptoms often disappear with resolution of hypercortisolemia [2,4]. In addition, non-CS patients suffering from major depressive disorder (MDD) often present with elevated levels of plasma cortisol and glucocorticoid resistance [5], further implicating glucocorticoids in mood disorder biology.
Effects of glucocorticoids are mediated in part by the glucocorticoid receptor (GR) and its associated chaperone protein complex consisting of FKBP5, HSP70, and HSP90. Upon activation by GC binding, GR dissociates from the chaperone complex, homodimerizes with other GR molecules, and translocates into the nucleus by interactions with FKBP4 and members of the dynein family of motor proteins [6]. In the nucleus, the GR dimer acts as a potent activator of transcription by binding glucocorticoid response elements (GREs) and recruiting transcription factors to gene promoters [7]. Importantly, FKBP5 is one of the immediate-early target genes of GC action, and GC induction of FKBP5 provides a short intracellular negative feedback loop, where FKBP5 reduces its own transcription by impeding further translocation of the GR complex. As a result, FKBP5 has been shown to be a strong modulator of GC-signaling in vitro, as levels of FKBP5 reduce GR’s affinity for GCs in a dose-dependent manner [6]. This event, along with GC-induced changes in levels of GR [8,9], forms the molecular basis for glucocorticoid resistance.
An animal model of hypercortisolemia and GC resistance provides additional in vivo support for FKBP5 as a key mediator of GC sensitivity. GC resistance in squirrel monkeys, New World primates of the genus Saimiri, has been attributed to elevated levels of FKBP5, and transfection assays implicate species- and sequence-specific FKBP5 as the key modulator of GC resistance in this primate [10,11]. In humans, several single nucleotide polymorphisms (SNPs) in FKBP5 have been associated with depression, altered HPA-axis function, and rapid response to antidepressants. MDD patients that carry these SNPs exhibit symptoms of GC resistance in that they often fail to properly suppress their endogenous cortisol levels following dexamethasone suppression test [12,13,14]. Specifically, SNP rs1360780 confers decreased sensitivity to GCs by causing elevated FKBP5 protein levels [12], consistent with symptoms of GC resistance often comorbid with MDD.
Recently, results from several candidate gene association studies have linked FKBP5 to MDD, suicide, or PTSD, only in the context of previous stressors such as early-life trauma or child abuse events, and have begun to highlight gene-environment interactions as crucial factors for disease development [15,16,17,18,19]. These studies suggest that the HPA-axis may become dysregulated during early-life traumatic events and that this alteration persists through many years, even decades, to help precipitate mental illnesses later in life. These findings also suggest that long-term consequences of these early-life traumatic events may be potentiated by DNA sequence-independent, non-mutational events that chronically alter function of HPA-axis genes. To comprehend the long-term consequences of GC exposure on the HPA-axis and mood disorder biology, an epigenetic approach may be useful.
During our efforts to identify GC-induced epigenetic alterations on candidate HPA-axis genes, we made several discoveries in Fkbp5: (i) GC administration decreases brain and blood DNA methylation (DNAm) in the Fkbp5 gene; (ii) DNAm alterations in Fkbp5 are associated with behavioral deficits in an animal model of Cushing’s syndrome; and (iii) there is a persistence of GC-induced DNAm change in both blood and brain for up to 4 weeks following GC withdrawal [20,21].
To address whether the observed loss of DNAm is confined to cells of lymphocytic and neuronal origins, we chose the murine AtT-20 anterior pituitary corticotroph cells [22], as these cells synthesize adrenocorticotropic hormone (ACTH), and its transcription is suppressed by GCs [23]. We also tested this cell line for GC-induced changes in mRNA levels of DNA methyltransferase Dnmt1, hypothesizing that GCs would decrease Dnmt1 expression.
2. Materials and methods
2.1 Animals
Four-week-old male C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) were housed in a temperature- and humidity-controlled room. At five weeks of age, animals were given ad libitum access to solutions containing corticosterone (Sigma-Aldrich, St. Louis, MO; 100 μg/ml with 1% ethanol; “CORT” group) or 1% ethanol (“VEHICLE” group) in place of their normal drinking water, and this treatment continued for four weeks. Solutions were made fresh daily. On the morning of the last day of treatment, tail blood was collected (~20 μl) and centrifuged to separate plasma. Plasma was frozen and stored at −80°C until assayed for corticosterone by radioimmunoassay (RIA). Following four weeks of treatment, the animals were euthanized, and their brains frozen on powdered dry ice and subsequently stored at −80°C. All procedures were approved by the Institutional Animal Care and Use Committee at Johns Hopkins University School of Medicine and were performed in accordance with guidelines established in the National Research Council’s Guide for the Care and Use of Laboratory Animals.
2.2 Brain dissections
Frozen brains were sectioned using a cryostat, and ~500 μm sections were mounted on plain glass slides. 19-gauge needles (0.686 mm inner diameter and 1.086 mm outer diameter) were used to punch out the dentate gyrus from sections containing the hippocampus (bregma −0.98 mm through −1.5 mm and bregma −1.5 mm through −2.0 mm) [24]. Dissected samples were stored at −80°C until processed for genomic DNA and mRNA.
2.3 Radioimmunoassay (RIA)
Plasma hormone levels were determined by commercially available RIA kits for corticosterone (MP Biomedicals, Costa Mesa, CA), according to manufacturer’s instructions as previously described [25]. All samples were run in duplicate and comparisons were made within assay with a coefficient of variance of 3.9%.
2.4 Cell line
AtT-20 cell line (Atcc.org, Manassas, VA) derived from a mouse pituitary tumor was cultured using DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) under standard conditions (5% CO2, 37°C). Cells were trypsinized and replated in six-well plates before treatment with dexamethasone (DEX; Sigma). Control samples were treated with an equal volume of EtOH as those treated with 1 μM DEX. Respective media were changed daily. Cells were split on the third day of treatment to maintain them in log phase of growth. After 5 days of treatment, cells were harvested for genomic DNA, and mRNA.
2.5 Gene expression
Messenger RNA from hippocampus and AtT-20 cell line was obtained using the RNeasy Lipid Tissue Mini (Qiagen, Valencia, CA). QuantiTect Reverse Transcription Kit (Qiagen) was used to generate cDNA for quantitative real-time PCR. Negative reverse transcriptase samples were used to ensure the absence of contaminating DNA. All reactions were carried out in triplicate using 1X Taqman master mix (Applied Biosystems, Foster City, CA), 1X Taqman probes for each gene [Fkbp5, Dnmt1, and Actb (β-actin)], and 30 ng of cDNA in a total volume of 20 μl. Real-time reactions were performed on an Applied Biosystems 7900HT fast real-time PCR system with standard PCR conditions (50°C for 2 min; 95°C for 10 min; and 60°C for 1 min for 40 cycles). To determine relative expression values, the −ΔΔCt method (Applied Biosystems) was used, where triplicate threshold cycle (Ct) values for each sample were averaged and subtracted from those derived from the housekeeping gene Actb. The Ct difference for a calibrator sample was subtracted from those of the test samples, and the resulting −ΔΔCt values were raised to a power of two to determine normalized relative expression.
2.6 DNA extraction and bisulfite conversion
Genomic DNA (gDNA) from mouse brain tissues and cell line was isolated with the Masterpure DNA Purification Kit, according to manufacturer’s instructions (Epicentre Biotechnologies, Madison, WI). Concentration of the gDNA was determined using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Rockford, IL), and 500 ng of the DNA was used for bisulfite conversion according to manufacturer’s protocol (EZ DNA Methylation Gold Kit; Zymo Research, Irvine, CA).
2.7 Bisulfite PCR and pyrosequencing
We measured DNAm by pyrosequencing of the PCR products, which measures methylation variation at >90% precision [26]. Two sets of primers (outside: Forward 5′-GAAAAGTTTTTGAGAATTAAGTTTAT-3′ and Reverse 5′-ATAACAAAACACCAAAAACCTCTA-3′; and nested: Forward 5′-TTGTTGTGGGTATGTATTGATGTT-3′ and Reverse 5′-CTCTCTCAACAATATAACTATAAA-3′) were designed to ultimately amplify a 206 bp intron 1 region of mouse Fkbp5. Another set of primers (outside: Forward 5′-GATGATTAGTTTTTTTTAGTAGTGATGT-3′ and Reverse 5′-CTTATTATTCTCTTACTACCCTAA -3′; and nested: Forward 5′-TAGTTTTTGGGGAAGAGTGTAGAGTTAT-3′ and Reverse 5′-ATTTTAAAAAACACAAAACACCCTATT-3′) were designed to amplify a 307 bp intron 5 region of mouse Fkbp5. 25 ng of bisulfite treated DNA was used for each PCR reaction. An additional nested PCR was performed with 2 μL of the outside PCR reaction and one biotinylated primer (other primer being unmodified). Amplification for both PCR steps consisted of 40 cycles (94°C for 1 min, 53°C for 30 sec, 72°C for 1 min). PCR products were confirmed on agarose gels. Pyro Gold reagents were used to prepare samples for pyrosequencing according to manufacturer’s instructions (Qiagen). Percentage of methylation at each CpG as determined by pyrosequencing was compared between DNA from CORT- and VEHICLE-treated brain tissues, or DEX- and VEHICLE-treated cell line samples.
2.8 Statistical Analysis
DNAm and relative gene expression measurements were analyzed by t-tests to compare CORT-treated and VEHICLE-treated groups (Microsoft Excel). A p-value < 0.05 was considered statistically significant.
3. Results
3.1 Glucocorticoid-induced changes in Fkbp5 gene expression and DNA methylation in cell lines
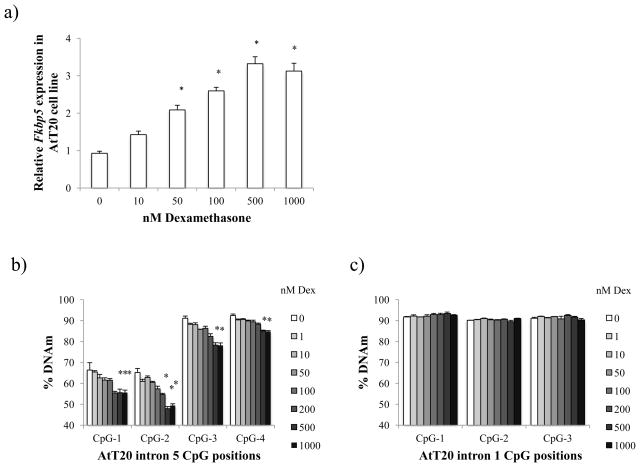
We first attempted to establish whether the previously observed glucocorticoid-induced changes in gene expression and DNAm in brain and lymphocyte could also be observed in the AtT-20 cell line. Several concentrations of DEX (0, 10, 50, 100, 500, and 1000 nM) were used to treat the AtT-20 cells for five days. We observed a steady dose-dependent increase in relative Fkbp5 mRNA expression (Fig. 1A). The highest expression occurred at 500 nM concentration of DEX, with a 257% (p = 0.004) increase over controls, suggesting that 500 nM may have saturated the glucocorticoid-signaling system. We then assessed the gDNA for any changes in DNAm in the two Fkbp5 intronic regions previously characterized as glucocorticoid response elements (GREs, Suppl. Fig. 1) [27]. Using bisulfite pyrosequencing, we found subtle but significant dose-dependent decreases in DNAm at all four CpGs flanking the intron 5 GRE (Fig. 1B). The largest reduction was found at the second CpG position at 500 nM DEX, where we observed a 17.3% decrease in DNAm (p = 0.002). No further reduction was observed at 1000 nM DEX concentration (15.8%, p = 0.01), further supporting our speculation that GR-signaling has become saturated at 500 nM. In contrast, we failed to observe any significant differences between control and treated cells at any of the doses of DEX when we assessed for DNAm in the intron 1 GRE (Fig. 1C).
Fig. 1.
Glucocorticoid-induced increase in Fkbp5 gene expression and decrease in DNAm in the AtT-20 cell line. (A) Fkbp5 gene expression increases with dexamethasone (DEX) treatment in a dose-dependent manner in the AtT-20 cell line. (B) Increase in gene expression was accompanied by statistically significant dose-dependent loss of DNAm of four CpG dinucleotides flanking the intron 5 GRE (glucocorticoid response element). (C) No dose-dependent change in DNAm was observed in the intron 1 GRE. Data are shown as mean ± SEM. * p ≤ 0.05: compared to 0 nM DEX-treated samples.
3.2 Corticosterone-induced changes in hippocampal Fkbp5 intronic DNA methylation
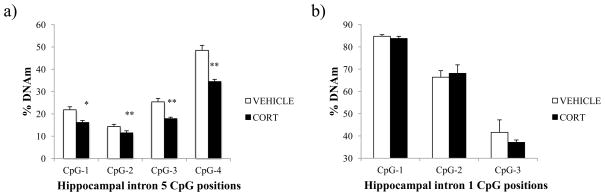
We have previously observed corticosterone-induced loss of Fkbp5 DNA methylation in the whole hippocampus and hypothalamus. This time, we asked whether the epigenetic activity that we have also observed in AtT-20 cells would be enriched in the dentate gyrus, a region of the hippocampus that has been implicated in stress-response, neurogenesis, and antidepressant response [28,29,30]. Further, we were interested in whether focusing on a specific sub-region would allow us to observe glucocorticoid-induced DNAm differences in the intron 1 GRE. Mice were either treated with 1% EtOH vehicle-solution (“VEHICLE”; N=12) or 100 μg/mL of corticosterone in their drinking water (“CORT”; N=12) for four weeks. At the end of the treatment period, plasma corticosterone levels were 454.7 ± 78.3 ng/mL in corticosterone-treated animals and 22.9 ± 6.0 ng/mL in the vehicle-treated animals. We performed bisulfite pyrosequencing following gDNA extraction from the hippocampal punches. At intron 5, we observed a significant 14.0% decrease in DNAm (p = 4.5×10−6) at CpG position 4, and more subtle but still significant DNAm differences at CpG-1 (5.7%, p = 0.03), CpG-2 (2.9%, p = 0.001), and CpG-3 (7.6%, p = 9.3×10−5) (Fig. 2A). As in the AtT-20 cell line, we did not observe any significant differences in DNAm between VEHICLE-treated and CORT-treated mice at the intron 1 GRE, which suggested site-specific methylation changes both in vitro and in vivo (Fig. 2B).
Fig. 2.
Corticosterone-induced changes in hippocampal Fkbp5 intronic DNAm (A) Loss of DNAm was found in the intron 5 GRE in the hippocampal tissue of mice treated with corticosterone (“CORT”; N=12), compared to vehicle-solution treated controls (“VEHICLE”; N=12). (B) No differences were observed in the intron 1 GRE. Data are shown as mean ± SEM. ** p ≤ 0.001, * p ≤ 0.05
3.3 Decrease in Dnmt1 expression in the AtT-20 cell line and mouse hippocampus
Given the crucial role played by DNA methyltransferase enzymes in maintaining status quo of DNAm in the cells, we asked whether the observed loss of DNAm could be explained by glucocorticoid-induced suppression of Dnmt1 expression. We observed a dose-dependent decrease in Dnmt1 expression with a maximum of 36.7% decrease (p = 0.0002) at 500 nM DEX concentration in AtT-20 cells. Interestingly, we observed no further decrease at the 1000 nM concentration, further supporting our previous observation with Fkbp5 expression that GR-signaling may have become saturated at 500 nM DEX concentration (Fig. 3A). We also assessed for Dnmt1 expression changes in the hippocampal tissue of mice treated with corticosterone. We found a 14.1% decrease in Dnmt1 expression (p = 0.02), suggesting a similar effect of glucocorticoids in the brain as in the cell line (Fig. 3B).
Fig. 3.
Glucocorticoid-induced decrease in Dnmt1 gene expression in the AtT-20 cell line and mouse hippocampus. (A) Dnmt1 gene expression decreased with DEX treatment in a dose-dependent manner in the AtT-20 cell line. * p ≤ 0.05: compared to 0 nM DEX-treated samples. (B) Statistically significant decrease was also observed in the hippocampal tissue of mice treated with corticosterone (“CORT”; N=12). Data are shown as mean ± SEM. * p < 0.05.
4. Discussion
In this study, we explored two aspects of FKBP5’s role in mediating the stress-response, namely its glucocorticoid-induced DNAm loss in non-neuronal cells that participate in the HPA-axis system and a potential epigenetic mechanism that may underlie the observed loss in DNAm. We found GC-dependent transactivation and loss of DNAm of Fkbp5 in the AtT-20 pituitary cell line that mirrored what were previously observed in other cells [20,21], suggesting a common mechanism of action. Our findings demonstrate that GC-induced loss of DNAm can be generalized to include such tissues and cells involved in the HPA-axis system.
Next, we examined whether focusing on a stress-relevant hippocampal region such as the dentate gyrus would allow us to observe a larger magnitude of epigenetic changes compared to bulk hippocampal tissues. In three of the four intron 5 CpGs tested, we observed a greater decrease in DNAm in this region compared to bulk hippocampal tissue. This phenomenon may be due to a larger ratio of granule neurons vs. glial cells that populate this region of the hippocampus, as we also observed greater corticosterone-induced DNAm changes in a homogenous neuronal cell line [20]. Recently, fluorescence activated cell sorting (FACS) was employed by Iwamoto et al. [31] to demonstrate that DNAm patterns from neurons were distinct from those of non-neurons and bulk tissues. In our experiments, we found that selective dissection of hippocampal tissue that enriched for granule neurons of the dentate gyrus preferentially represented the DNAm change that we initially observed in the AtT-20 cells. Since this DNAm change is shared by both granule neurons of the hippocampus and the pituitary cell line, additional experiments are needed to identify the common mechanism that governs this activity.
In an effort to identify one of the potential modulators of GC-dependent loss of DNAm, we assayed both the cell line and the dentate gyrus for expression levels of Dnmt1. We found that GCs caused a dose-dependent decrease in Dnmt1, providing a potential mechanism for the observed loss of DNAm. There is emerging evidence that active demethylation of 5-methylcytosines (5-mC) involves the formation of an intermediate 5-hydroxymethylcytosine (5-hmC), followed by a base-excision repair process that exchanges the 5-hmC with cytosine [32,33]. On the other hand, there’s also evidence that nuclear hormone receptors employ a different mechanism of demethylation, particularly one involving methyl-CpG binding domain 4 protein MBD4, and DNA methyltransferases DNMT1 and DNMT3B [34]. Since Dnmt3b is a de novo methyltransferase, and therefore unlikely to play a role in regions that are already methylated, we focused on the maintenance methyltransferase Dnmt1 that methylates newly synthesized daughter strands following DNA replication. Given that cell lines and the dentate gyrus are capable of active cell proliferation [28,29], we propose a passive demethylation process that is mediated by reduction in Dnmt1 expression coupled with cell replication that results in a failure to methylate the newly synthesized strand. It remains to be determined whether reduction in Dnmt1 levels translates to global reduction of DNAm, and whether restoring its levels can rescue the loss of DNAm. Further, site-specific loss of DNAm in the intron 5 GRE, but not in the intron 1 GRE, suggests an interaction between the GR and DNMT1 that may include mutually exclusive occupancy of the intron 5 GRE. Carefully designed chromatin immunoprecipitation (ChIP) experiments are necessary to confirm this relationship.
In this study, we have broadened the validity of GC-induced DNAm loss to include pituitary cells and have identified a potential epigenetic mediator of the stress response. A clearer understanding of GC target tissues and mechanism of gene function alterations are essential for the development of better therapeutic strategies for treatment of mood disorders and stress exposure.
Supplementary Material
Genomic organization of mouse Fkbp5 gene including its two upstream CpG islands, according to the UCSC Genome Browser NCBI37/mm9 assembly. Glucocorticoid response elements (GREs) in two intronic regions, Intron 1 and Intron 5, and the Promoter region are represented by dark gray horizontal bars. Exons (Ex) are represented by vertical black boxes and alternate upstream exons (AltEx) are represented by light gray bars. Three CpG islands, two upstream and one within intron 1, are represented by boldfaced boxes.
Highlights.
Dose dependent increase in Fkbp5 and decrease in Dnmt1 expression by dexamethasone
Site-specific loss of methylation of Fkbp5 in pituitary cell line and hippocampus
Glucocorticoid-induced epigenetic activity in hippocampus enriched in dentate gyrus
Acknowledgments
Funding and grants
This study was funded by NIH grants AA10158 (GSW), HD055030 (KLKT), and T32MH015330 (RSL), the Kenneth Lattman Foundation (GSW), a NARSAD Young Investigator Award (RSL), and the Dalio Family for Mood Disorders Research (RSL).
Abbreviations and acronyms
- Fkbp5
FK506 binding protein 5
- DNAm
DNA methylation
- Dnmt1
DNA methyltransferase 1
- HPA-axis
hypothalamic-pituitary-adrenal axis
- CS
Cushing’s syndrome
- GC
glucocorticoid
- DEX
dexamethasone
Footnotes
Conflict of interest: None
Financial disclosure: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cohen SI. Cushing’s syndrome: a psychiatric study of 29 patients. Br J Psychiatry. 1980;136:120–124. doi: 10.1192/bjp.136.2.120. [DOI] [PubMed] [Google Scholar]
- 2.Dorn LD, Burgess ES, Friedman TC, et al. The longitudinal course of psychopathology in Cushing’s syndrome after correction of hypercortisolism. J Clin Endocrinol Metab. 1997;82:912–919. doi: 10.1210/jcem.82.3.3834. [DOI] [PubMed] [Google Scholar]
- 3.Flitsch J, Spitzner S, Ludecke DK. Emotional disorders in patients with different types of pituitary adenomas and factors affecting the diagnostic process. Exp Clin Endocrinol Diabetes. 2000;108:480–485. doi: 10.1055/s-2000-8144. [DOI] [PubMed] [Google Scholar]
- 4.Starkman MN, Schteingart DE, Schork MA. Cushing’s syndrome after treatment: changes in cortisol and ACTH levels, and amelioration of the depressive syndrome. Psychiatry Res. 1986;19:177–188. doi: 10.1016/0165-1781(86)90096-x. [DOI] [PubMed] [Google Scholar]
- 5.Rubin RT, Phillips JJ, McCracken JT, et al. Adrenal gland volume in major depression: relationship to basal and stimulated pituitary-adrenal cortical axis function. Biol Psychiatry. 1996;40:89–97. doi: 10.1016/0006-3223(95)00358-4. [DOI] [PubMed] [Google Scholar]
- 6.Wochnik GM, Ruegg J, Abel GA, et al. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J Biol Chem. 2005;280:4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- 7.Paakinaho V, Makkonen H, Jaaskelainen T, et al. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol Endocrinol. 2010;24:511–525. doi: 10.1210/me.2009-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao HM, Ma LY, McEwen BS, et al. Regulation of glucocorticoid receptor and mineralocorticoid receptor messenger ribonucleic acids by selective agonists in the rat hippocampus. Endocrinology. 1998;139:1810–1814. doi: 10.1210/endo.139.4.5896. [DOI] [PubMed] [Google Scholar]
- 9.Makino S, Smith MA, Gold PW. Increased expression of corticotropin-releasing hormone and vasopressin messenger ribonucleic acid (mRNA) in the hypothalamic paraventricular nucleus during repeated stress: association with reduction in glucocorticoid receptor mRNA levels. Endocrinology. 1995;136:3299–3309. doi: 10.1210/endo.136.8.7628364. [DOI] [PubMed] [Google Scholar]
- 10.Scammell JG, Denny WB, Valentine DL, et al. Overexpression of the FK506-binding immunophilin FKBP51 is the common cause of glucocorticoid resistance in three New World primates. Gen Comp Endocrinol. 2001;124:152–165. doi: 10.1006/gcen.2001.7696. [DOI] [PubMed] [Google Scholar]
- 11.Denny WB, Valentine DL, Reynolds PD, et al. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- 12.Binder EB, Salyakina D, Lichtner P, et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 13.Horstmann S, Lucae S, Menke A, et al. Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology. 2010;35:727–740. doi: 10.1038/npp.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zobel A, Schuhmacher A, Jessen F, et al. DNA sequence variants of the FKBP5 gene are associated with unipolar depression. Int J Neuropsychopharmacol. 2010;13:649–660. doi: 10.1017/S1461145709991155. [DOI] [PubMed] [Google Scholar]
- 15.Binder EB, Bradley RG, Liu W, et al. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299:1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koenen KC, Uddin M. FKBP5 polymorphisms modify the effects of childhood trauma. Neuropsychopharmacology. 2010;35:1623–1624. doi: 10.1038/npp.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy A, Gorodetsky E, Yuan Q, et al. Interaction of FKBP5, a stress-related gene, with childhood trauma increases the risk for attempting suicide. Neuropsychopharmacology. 2010;35:1674–1683. doi: 10.1038/npp.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie P, Kranzler HR, Poling J, et al. Interaction of FKBP5 with childhood adversity on risk for post-traumatic stress disorder. Neuropsychopharmacology. 2010;35:1684–1692. doi: 10.1038/npp.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appel K, Schwahn C, Mahler J, et al. Moderation of adult depression by a polymorphism in the FKBP5 gene and childhood physical abuse in the general population. Neuropsychopharmacology. 2011;36:1982–1991. doi: 10.1038/npp.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee RS, Tamashiro KL, Yang X, et al. Chronic corticosterone exposure increases expression and decreases deoxyribonucleic acid methylation of Fkbp5 in mice. Endocrinology. 2010;151:4332–4343. doi: 10.1210/en.2010-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee RS, Tamashiro KL, Yang X, et al. A measure of glucocorticoid load provided by DNA methylation of Fkbp5 in mice. Psychopharmacology (Berl) 2011;218:303–312. doi: 10.1007/s00213-011-2307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison RW, Yeakley J. Corticosterone binding in AtT-20 pituitary tumor cell cytosol: evidence for one class of binding site for both natural and synthetic glucocorticoids. Biochim Biophys Acta. 1979;583:360–369. doi: 10.1016/0304-4165(79)90460-4. [DOI] [PubMed] [Google Scholar]
- 23.Drouin J, Trifiro MA, Plante RK, et al. Glucocorticoid receptor binding to a specific DNA sequence is required for hormone-dependent repression of pro-opiomelanocortin gene transcription. Mol Cell Biol. 1989;9:5305–5314. doi: 10.1128/mcb.9.12.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2. San Diego: Academic Press; 2001. [Google Scholar]
- 25.Yang X, Wang S, Rice KC, et al. Restraint stress and ethanol consumption in two mouse strains. Alcohol Clin Exp Res. 2008;32:840–852. doi: 10.1111/j.1530-0277.2008.00632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colella S, Shen L, Baggerly KA, et al. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 27.Magee JA, Chang LW, Stormo GD, et al. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- 28.van Praag H, Schinder AF, Christie BR, et al. Functional neurogenesis in the adult hippocampus. Nature. 2002;415:1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santarelli L, Saxe M, Gross C, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- 30.Snyder JS, Soumier A, Brewer M, et al. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature. 2011;476:458–461. doi: 10.1038/nature10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwamoto K, Bundo M, Ueda J, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 2011;21:688–696. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams K, Christensen J, Pedersen MT, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JU, Su Y, Zhong C, et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim MS, Kondo T, Takada I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic organization of mouse Fkbp5 gene including its two upstream CpG islands, according to the UCSC Genome Browser NCBI37/mm9 assembly. Glucocorticoid response elements (GREs) in two intronic regions, Intron 1 and Intron 5, and the Promoter region are represented by dark gray horizontal bars. Exons (Ex) are represented by vertical black boxes and alternate upstream exons (AltEx) are represented by light gray bars. Three CpG islands, two upstream and one within intron 1, are represented by boldfaced boxes.